Abstract
With the drive toward high throughput molecular dynamics (MD) simulations involving ever-greater numbers of simulation replicates run for longer, biologically relevant timescales (microseconds), the need for improved computational methods that facilitate fully automated MD workflows gains more importance. Here we report the development of an automated workflow tool to perform AMBER GPU MD simulations. Our workflow tool capitalizes on the capabilities of the Kepler platform to deliver a flexible, intuitive, and user-friendly environment and the AMBER GPU code for a robust and high-performance simulation engine. Additionally, the workflow tool reduces user input time by automating repetitive processes and facilitates access to GPU clusters, whose high-performance processing power makes simulations of large numerical scale possible. The presented workflow tool facilitates the management and deployment of large sets of MD simulations on heterogeneous computing resources. The workflow tool also performs systematic analysis on the simulation outputs and enhances simulation reproducibility, execution scalability, and MD method development including benchmarking and validation.
Introduction
Continued advances in computing power and the development of the graphics processing unit (GPU), make the study of proteins, receptors, and other biophysical systems with molecular dynamics simulations increasing accessible (1, 2). Molecular dynamics (MD) simulations are a powerful computational tool that predicts protein dynamics in physiological conditions and allows researchers to study atomic interactions (3), aiding in improved models of molecular recognition and drug design (4).
Several well-known MD simulation packages available for biomolecular simulations, including GROMACS (5), NAMD (6), Desmond (7), OpenMM (8), and AMBER (9), have made significant contributions to scientific discoveries (3, 10, 11, 12). Each can run calculations on a single central processing unit (CPU) or a single GPU, and on clusters using multiple CPUs or GPUs. In contrast to a CPU, a GPU is constructed with significantly reduced cache memory. It is built with thousands of smaller computing cores, designed to handle multiple identical mathematical operations simultaneously. Many GPU MD packages utilize the GPU for the nonbonded force calculation. The AMBER GPU software can perform complex calculations, including semiisotropic pressure scaling, adaptively biased MD, and thermodynamic integration. The AMBER package utilized in this work can support MD simulations on a single GPU and multiple GPUs (1, 2) with performances that outstrip even the most powerful conventional CPU-based supercomputers (13, 14). It takes advantage of the GPU to accelerate classical MD on realistic-sized systems, typically up to a maximum of ∼3,000,000 atoms per simulation, bound by a size limit due to memory restrictions unique to specific GPU models. Nevertheless, systems of this size include many drug targets, including membrane-embedded G-protein coupled receptors, or a protein trimer with bound antibodies (3).
Broadly speaking, a major criticism of MD simulations relates to their reproducibility. Reproducing published MD findings (i.e., an independent group reproducing the same set of results for the same system of interest) can be challenging for many reasons, including complicated run procedures that rely on user-developed scripts, technical numerical reproducibility (which is dependent on a number of factors including the compiler, executable, and dependent libraries, as well as whether the FFTW algorithm is utilized), statistical reproducibility (which can be achieved through the use of a high number of simulation replicates), and incomplete reporting of methods in the published literature. For example, it is common for an end-to-end MD experiment to require multiple dozens of steps; conceptually we can define the set of required steps as a “workflow”. Typically researchers employ various scripts to piece together each of the different steps. The formal encoding of these many scripts into an automated workflow framework is one way to address many challenges related to reproducibility, as well as method development and researcher training.
Multiple workflow platforms are available for scientific research. Kepler is a completely open source option with an extensive set of basic and advanced modules that support various workflow configurations. Kepler is a Java-based platform that delivers an easy-to-navigate graphical user interface (GUI) and user-friendly text file interface. From the GUI platform, the user is given a palette of modules, actors, and directors, with which to visually program their workflow (15). A director controls the execution of the workflow, and actors carry out specific functions in the workflow. Contributors around the world develop modules and other workflows that perform complementary functions. Within the last decade, Kepler has implemented multiple modules that support the applications of statistical analysis, biological research, adaptations of external software, and options to select execution on different platforms and with different schedulers (15, 16). Consequently, Kepler has become a flexible and broad-purpose platform on which to build a workflow that can perform both MD simulations and subsequent statistical analysis. The user-friendly GUI interface also provides a suitable learning apparatus for beginners to understand the components of MD and for experienced users to customize and extend the workflow to their project needs.
Combining these two computational tools, the AMBER GPU simulation code and the Kepler platform, we have developed a workflow tool that allows scientists to navigate, develop, deploy, analyze, and share MD simulation protocols. The workflow tool merges multiple MD steps that are otherwise scattered across disparate software tools or scripts into one central command that controls and executes all steps, starting from the initial energy minimization to multicopy production dynamics and routine analysis. The workflow tool submits jobs to various hardware architectures and copies back the output data to the user’s local machine, helps to systematically run MD simulations, and plots thermodynamic properties to check the integrity of the simulations. The integrated simulation execution and analysis features of the workflow reduces the time needed to monitor the various simulation steps and facilitates handling of the simulation data. Moreover, Kepler is equipped with an automatic execution check function, which will stop and highlight the step if the simulations fail to execute until the end.
The AMBER GPU workflow tool leverages Kepler’s advanced provenance capabilities to assist in achieving reproducibility (17). Each time the workflow tool is executed, a detailed provenance report containing key signatures required to replicate a simulation is generated. The report, which can be used as a basis for method section reporting, contains not only MD configuration parameter information, but also details execution parameters not normally reported, including hardware configuration and software and library version information (AMBER, CUDA, etc.). The latter set of information is not typically reported, although it has the potential to impact reproducibility as much as MD configuration file information.
Our AMBER GPU workflow tool provides some capabilities similar to QwikMD, another software that executes MD simulations, but unlike QwikMD (18), which focuses on VMD and NAMD, our workflow tool utilizes AMBER and its well-known GPU computing power. QwikMD also focuses on assisting the user in building the system itself, whereas our workflow tool focuses on MD job execution and analysis, relying on the user to create the system files on their own. Our workflow tool is also able to execute jobs on various clusters including the XSEDE architectures, and the users can easily control the execution options via the GUI or command-line interface. Furthermore, through its integration with Kepler, the AMBER GPU workflow tool can easily utilize Kepler’s extensive data analytics capabilities, which are continually being developed. CHARMM-GUI (19) is another complimentary tool that provides a web interface for system construction. It is capable of building and generating MD input files for a magnitude of biological systems, including solvated proteins, solvated proteins with ligands attached, membrane proteins, etc. The inputs produced by CHARMM-GUI could be used in the AMBER GPU workflow tool to actually execute the MD simulations. To our knowledge, CHARMM-GUI does not yet provide job execution capabilities.
Finally we note that the workflow tool presented here is freely accessible, open source, and easy to modify and reuse. Each of the modules (a.k.a., Kepler actors) can be reconfigured with different components (e.g., including automated preparation and simulation of receptor-ligand complexes), and additional analysis steps can be developed and integrated into customized experiments. Workflows developed with KNIME can be called from within Kepler and functionality of different components can be extended via interoperability with packages such as HTMD (20), MSMBuilder (21), and PyEMMA (22). The AMBER GPU workflow tool can be found at http://nbcr.ucsd.edu/products/workflow-distribution along with the user guide.
Materials and Methods
The workflow tool requires an input directory containing two subdirectories. One subdirectory, labeled “confDir”, contains all the template simulation input files. The template AMBER input scripts provided in the default workflow tool were converted from the same parameters used in the NAMD MD simulations of Wassman et al. (4), which consisted of four stages of minimization, one step of heating, three stages of equilibrations, and one step of production. Users can also configure their own input files if desired (see Supporting Material for details), e.g., if they would like to run another type of MD simulation such as accelerated or replica exchange MD. The second subdirectory, labeled with the system’s stem name, contains the system’s AMBER topology and coordinate files. For example, the system’s topology and coordinate files are named under “r174h_stictic.top” and “r174h_stictic.crd”, respectively; then, the subdirectory folder name will be “r174h_stictic”.
There are three submission options implemented to run the AMBER GPU MD simulation workflow: LocalExecution on user’s local machine, CometGPUCluster on the San Diego Super Computer (SDSC) Comet cluster, and PrivateGPUCluster on a remote private cluster. The workflow will always start from a user’s local machine, but it can connect to remote clusters, submit MD simulation jobs, and transfer simulation results. Moreover, users can connect the workflow to a GPU cluster of their choice. Regardless of which submission option a user chooses, all outputs at the end of a simulation can be found on the local machine, where users started the workflow.
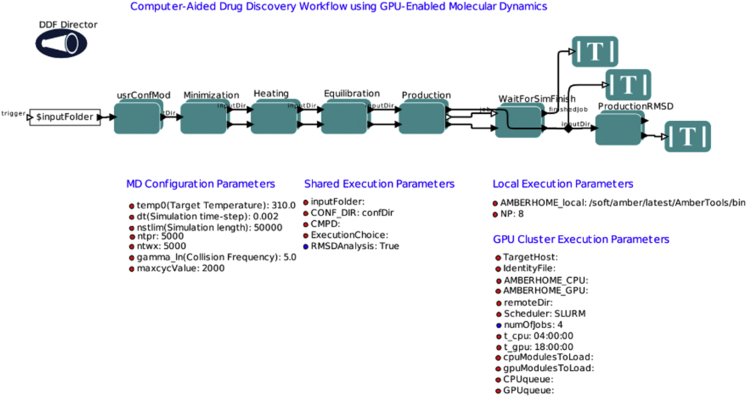
The workflow tool automates MD simulation processes and generates analytical plots that help analyze the system of interest and validate simulation quality. There are two parts to this workflow: simulation and analysis. Simulations can be further broken down into four components executed by four actors: minimization, heating, equilibration, and production. The analysis components are small actors working collectively to call locally implemented software, including cpptraj from the AmberTools suite and R, the programming language (23, 24), to generate time-evolution energy, temperature, pressure, and root-mean-square-deviation (RMSD) plots. The resulting plots are stored in the system subdirectory for analysis and to verify simulations quality.
The first simulation component is the minimization (Fig. 1). In this step, a system is energetically minimized to a local minimum. After the minimizations, the workflow plots the energy for each step and an overall diagram combining all the plots. Because energy is the largest changing parameter during minimization, the energy plots are good indicators of whether minimizations were performed successfully.
Figure 1.
AMBER GPU MD simulation workflow, showing the uppermost level of the workflow construct. To see this figure in color, go online.
Minimizations are useful for finding local minima but typically one would want to simulate a system in its physiological environment or at room temperature. The next heating step (Fig. 1), thus, adds kinetic energy or heats the system from 0 K to a target temperature defined by the user. For the heating step, the temperature plot over time will indicate whether the system did indeed heat from 0 K to a user-defined value. After heating, the equilibration steps (Fig. 1) shift the system in gradual steps toward thermal equilibrium under NPT or NVT conditions.
Both the heating and equilibration steps output several thermodynamic property plots including kinetic energy, potential energy, total energy, pressure, and temperature. The workflow also calculates and plots the time-dependent RMSD of backbone α-carbons of the system. RMSD analyzes the time evolution of a selected set of atoms against some reference frame; in this case, the default is the initial frame. The time-series backbone α-carbon RMSD analysis provides information about the overall global change of the protein backbone structure over time. A collection of these property plots would indicate whether stable equilibration was attained.
The last component, production (Fig. 1), runs nonconstrained NPT or NVT MD simulations (based on the parameters set on users’ input scripts) and derives a dynamic atomic trajectory of the system in a simulated physiological environment. In this step, users can choose to run the workflow on a local machine with one GPU card or on a remote cluster with multiple GPUs; in the latter case, it is possible to deploy multiple copies of the production simulations concurrently. As with heating and equilibration, time-series plots for temperature, pressure, energy, and RMSD plots are also generated in the production stage. Regardless of how many copies of production simulation users would like to run, they will all start with the same starting structure from the last equilibration frame. The best solution to check whether the simulations ran successfully is to visualize the trajectories and examine if there are any abnormalities. In addition, analysis techniques presented here, such as the RMSD plots, can also serve as an investigational tool to identify and quantify any outstanding abnormal structural deviations from the starting structure. At the end of each execution, the workflow prints out a detailed report, which lists the input parameters, hardware specification, the AMBER version used, the random seeds for the equilibrations and productions, and file locations. This report can be appended to a publication as supporting information, and would contain all the requisite details to ensure technical (and statistical, if achieved) reproducibility. Or, it can be used as a guide to assist authors in writing more comprehensive “Materials and Methods” sections.
Application
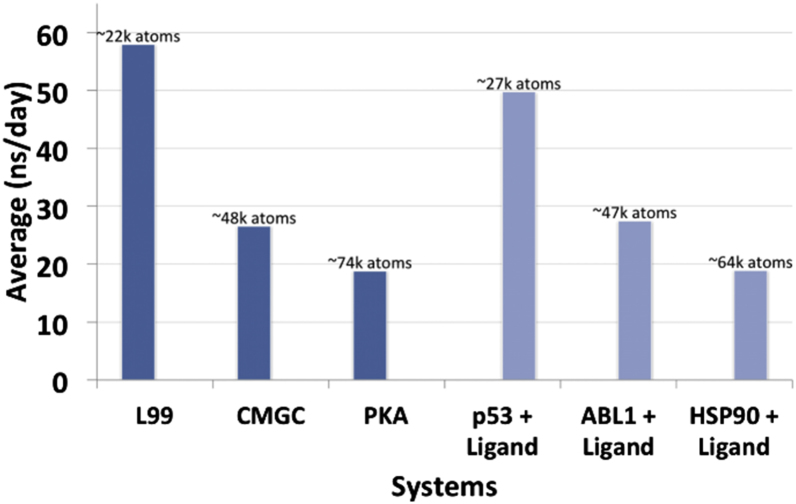
We show the utility of the workflow tool on multiple protein systems of variable size, and with and without bound ligands (Fig. 2). Here we present in detail one of the systems, the p53 protein bound to a small molecule named “stictic acid” (4). P53 is considered to be a potential drug target for anticancer therapy (25, 26, 27), making its simulation with our workflow a relevant test case to perform and scrutinize. Nonactivated and mutant p53 is seen in more than half of human cancer cases (28), and stictic acid is a known compound that activates mutant p53 (4). We are therefore interested in understanding its dynamics when stictic acid is bound.
Figure 2.
Performance of the AMBER GPU workflow on various systems on a local NVIDIA Tesla K20 cluster. To see this figure in color, go online.
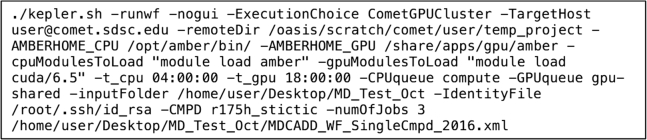
Courtesy of Wassman et al. (4), we obtained the AMBER topology and coordinate files of the p53-stictic acid complex to run MD simulations using the workflow. The simulation consisted of five minimization steps, one heating step, three equilibration steps, and one production step that branched out to perform three independent production simulations. We set up the simulations through the command prompt with Kepler on our local machine (Fig. 3), and Kepler submitted the simulation jobs to the Comet cluster at SDSC. This single command communicates with Kepler to run all the simulation steps consecutively without interruption. For more information on each parameter, please refer to the Kepler Amber GPU Molecular Dynamics Workflow User Manual.
Figure 3.
Command line for initiating MD simulations.
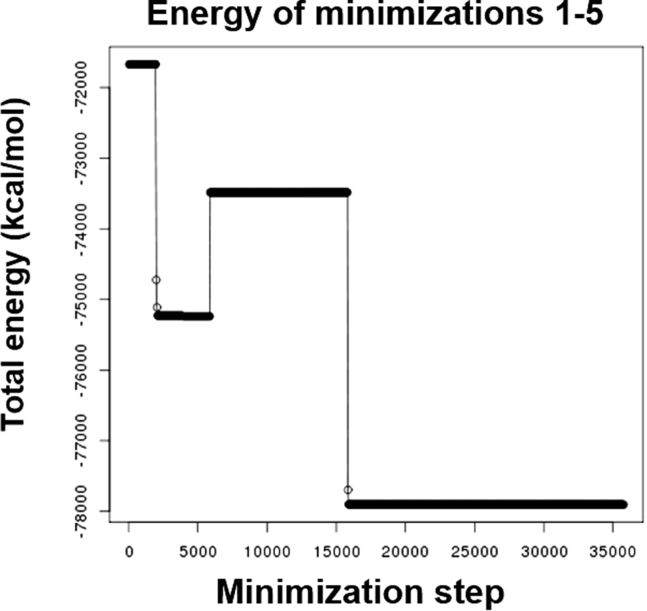
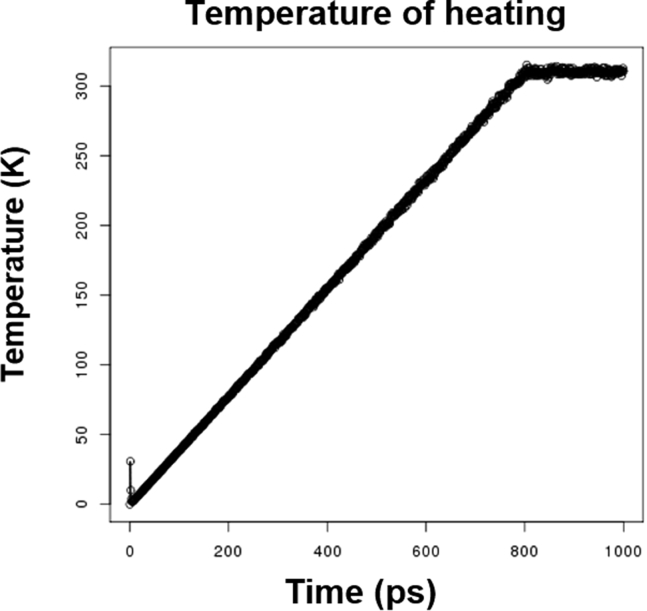
The resulting output data was then copied back to the local machine. We first checked the energy plot generated from the minimization steps (Fig. 4). As shown in Fig. 4, the system did move toward an energy minimum after five rounds of minimization, which suggested that the system was adequately minimized to a local/global minima. Next, we studied the temperature plot from the heating step. We assigned the desired temperature to 310 K to mimic physiological conditions. As shown in Fig. 5, the temperature gradually ramped from 0 to 310 K and plateaued at 310 K. This indicated that the system was successfully heated from 0 to 310 K and held steady at 310 K.
Figure 4.
Energy minimization plot.
Figure 5.
Heating (nonequilibrium) plot.
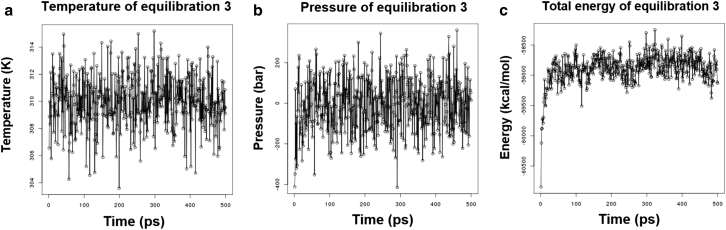
As per the default workflow parameters, for our p53-stictic acid system, the protein backbone was constrained during equilibration, so it was not necessary to study the RMSD during these steps. We looked into the total energy, pressure, and temperature from the last step of equilibration. During production runs, the temperature and pressure oscillated around the defined system values of 310 K and 1.0 bar, respectively, as one would expect. Total energy increased but plateaued at ∼−59,000 kcal/mol, which suggested the system achieved at least a metastable equilibrium for the duration of the run (Fig. 6).
Figure 6.
Time-series analyses (left to right) of the temperature, pressure, and total energy from the third and last stages of equilibration.
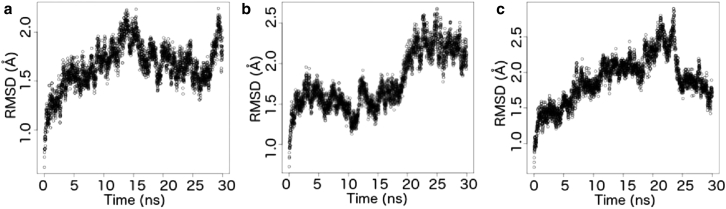
We generated three copies of 30-ns production dynamics for our system. Because the system had already gone through constant pressure simulations in stepwise equilibration steps, we did not expect any drastic global change during production. We were able to verify that the behavior of our system was as expected from the RMSD plot produced by the workflow (Fig. 7). Shown in Fig. 7, we see small deviations (<3 Å) from the starting structure in all three production runs, indicating that the system sampled local conformational space without outstanding abnormal structural deviation.
Figure 7.
Time evolution of the RMSD of (a) copy 1, (b) copy 2, and (c) copy 3 of production MD.
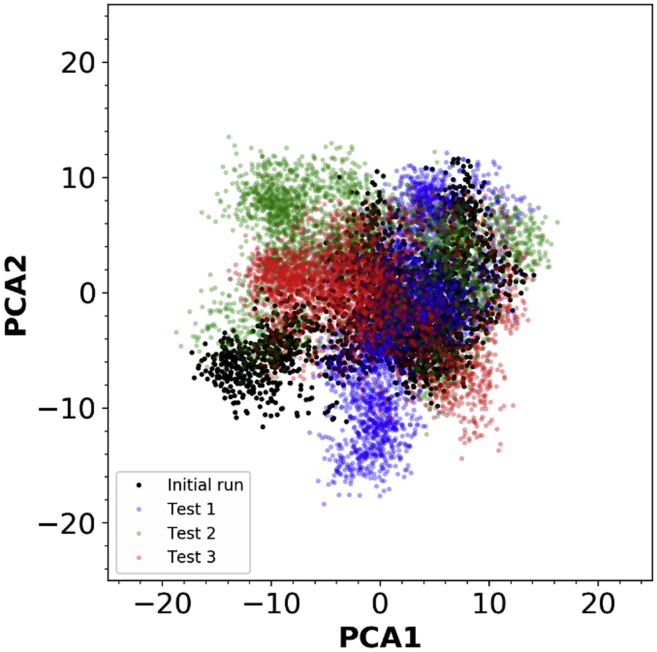
At the end of the execution, the workflow tool outputs a provenance report file consisting of input parameters, hardware specifications, the AMBER version, file locations, and the random seeds used for the simulations. Using the report, we were able to configure the workflow with the same exact random seed to test for reproducibility. For the next execution, we input the random seeds from the report, and the workflow used the values to generate a new set of input files. We tested the workflow on a local machine, on the SDSC Comet supercomputer, and on a private GPU cluster. The three runs resulted in trajectories that were highly similar, but not exactly the same because they were executed on different GPU clusters. Even with the same random seeds, all three runs sample slightly different yet similar conformational spaces (Fig. 8). This result is as expected, because for the runs to be exactly reproducible, we must use the exact same hardware (29). For the local execution, AMBER was compiled with the FFTW algorithm, which introduces uncontrollable variability (29). Nevertheless, we were able to recreate identical input files with the workflow and successfully run additional MD simulations of different trajectories, enhancing statistical and methodological reproducibility.
Figure 8.
Principal component analysis plot of the p53 simulation result. All the trajectories from the initial run (computer-generated random seed) and the three simulation tests (used the same random seed from the initial run) with 1) Comet (Test 1), 2) private GPU cluster (Test 2), and 3) local machine (Test 3) were first aligned. The first two principal components were then calculated and the trajectories from each test were projected onto the PC space. To see this figure in color, go online.
Software availability
The AMBER GPU MD Simulation workflow is available under the GNU Lesser General Public License, version 1.0 or later. Full documentation and examples are available through the National Biomedical Computation Resource download page, http://nbcr.ucsd.edu/products/workflow-distribution, and development is hosted on GitHub at https://github.com/nbcrrolls/workflows/tree/master/Production/AmberGPUMDSimulation. This workflow is also available for download in Zenodo (http://dx.doi.org/10.5281/zenodo.192490) and SciCrunch (RRID SCR_014389).
An open online training course on the Kepler-MDCADD workflow will be available on the Biomedical Big Data Training Collaborative (https://biobigdata.ucsd.edu) in 2017. The course will include lecture contents, videos, and hands-on training utilizing VM toolboxes. The Biomedical Big Data Training Collaborative is a collaborative platform to encourage and facilitate training and education of the Biomedical Big Data community. The platform provides an intuitive portal to upload, distribute, and discover new content about developments in the biomedical big data domain (30).
Author Contributions
S.P., P.U.I., G.J.C., R.D.M., R.C.W., I.A., and R.E.A. developed the software. S.P., P.U.I., and G.J.C. drafted the article. P.U.I., G.J.C., R.D.M., R.C.W., I.A., and R.E.A. edited the article. All the authors read and approved the final article.
Acknowledgments
This work was funded in part by the National Biomedical Computation Resource (NBCR) through National Institutes of Health (NIH) P41 GM103426 and the NIH New Innovator award OD007237 to R.E.A. This work was also supported by an NVIDIA Compute the Cure award to R.E.A.
Editor: Tamar Schlick.
Footnotes
Shweta Purawat and Pek U Ieong contributed equally to this work.
AMBER GPU Workflow User Manual is available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30552-0.
Supporting Material
References
- 1.Salomon-Ferrer R., Götz A.W., Walker R.C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh Ewald. J. Chem. Theory Comput. 2013;9:3878–3888. doi: 10.1021/ct400314y. [DOI] [PubMed] [Google Scholar]
- 2.Götz A.W., Williamson M.J., Walker R.C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 1. Generalized Born. J. Chem. Theory Comput. 2012;8:1542–1555. doi: 10.1021/ct200909j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ieong P., Amaro R.E., Li W.W. Molecular dynamics analysis of antibody recognition and escape by human H1N1 influenza hemagglutinin. Biophys. J. 2015;108:2704–2712. doi: 10.1016/j.bpj.2015.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wassman C.D., Baronio R., Amaro R.E. Computational identification of a transiently open L1/S3 pocket for reactivation of mutant p53. Nat. Commun. 2013;4:1407. doi: 10.1038/ncomms2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pronk S., Páll S., Lindahl E. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowers, K. J., H. Xu, …, D. E. Shaw. 2006. Scalable algorithms for molecular dynamics simulations on commodity clusters. In ACM/IEEE Conference on Supercomputing (SC06), Tampa, FL. ACM/IEEE, Article No. 84, http://dl.acm.org/citation.cfm?id=1188544.
- 8.Eastman P., Friedrichs M.S., Pande V.S. OpenMM 4: a reusable, extensible, hardware independent library for high performance molecular simulation. J. Chem. Theory Comput. 2013;9:461–469. doi: 10.1021/ct300857j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Case D.A., Betz R.M., Kollman P.A. University of California, San Francisco; San Francisco, CA: 2015. AMBER 2015. [Google Scholar]
- 10.Allen T.W., Kuyucak S., Chung S.H. Molecular dynamics study of the KcsA potassium channel. Biophys. J. 1999;77:2502–2516. doi: 10.1016/S0006-3495(99)77086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tai K., Shen T., McCammon J.A. Analysis of a 10-ns molecular dynamics simulation of mouse acetylcholinesterase. Biophys. J. 2001;81:715–724. doi: 10.1016/S0006-3495(01)75736-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Awasthi M., Jaiswal N., Dwivedi U.N. Molecular docking and dynamics simulation analyses unraveling the differential enzymatic catalysis by plant and fungal laccases with respect to lignin biosynthesis and degradation. J. Biomol. Struct. Dyn. 2015;33:1835–1849. doi: 10.1080/07391102.2014.975282. [DOI] [PubMed] [Google Scholar]
- 13.Walker, R. AMBER v16 GPU Acceleration Support—Benchmark. http://ambermd.org/gpus/benchmarks.htm.
- 14.Le Grand S., Gotz A.W., Walker R.C. SPFP: Speed without compromise—a mixed precision model for GPU accelerated molecular dynamics simulations. Comput. Phys. Commun. 2013;184:374–380. [Google Scholar]
- 15.Altintas, I., C. Berkley, …, S. Mock. 2004. Kepler: an extensible system for design and execution of scientific workflows. In 16th International Conference on Scientific and Statistical Database Management, Santorini Island, Greece. IEEE Xplore. http://ieeexplore.ieee.org/xpl/mostRecentIssue.jsp?punumber=9176. pp 423–424.
- 16.Altintas I., Wang J., Li W. Challenges and approaches for distributed workflow-driven analysis of large-scale biological data. In: Divesh Srivastava I.A., editor. EDBT Extending Database Technology. ACM; New York, NY: 2012. pp. 73–78. [Google Scholar]
- 17.Altintas I., Barney O., Jaeger-Frank E. Proceedings of the 2006 International Conference on Provenance and Annotation of Data. Springer; Chicago, IL: 2006. Provenance collection support in the Kepler scientific workflow system; pp. 118–132. [Google Scholar]
- 18.Ribeiro J.V., Bernardi R.C., Schulten K. QwikMD—integrative molecular dynamics toolkit for novices and experts. Sci. Rep. 2016;6:26536. doi: 10.1038/srep26536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jo S., Cheng X., Im W. CHARMM-GUI—10 years for biomolecular modeling and simulation. J. Comput. Chem. 2016;38:1114–1124. doi: 10.1002/jcc.24660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doerr S., Harvey M.J., De Fabritiis G. HTMD: high-throughput molecular dynamics for molecular discovery. J. Chem. Theory Comput. 2016;12:1845–1852. doi: 10.1021/acs.jctc.6b00049. [DOI] [PubMed] [Google Scholar]
- 21.Harrigan M.P., Sultan M.M., Pande V.S. MSMBuilder: statistical models for biomolecular dynamics. Biophys. J. 2017;112:10–15. doi: 10.1016/j.bpj.2016.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scherer M.K., Trendelkamp-Schroer B., Noé F. PyEMMA 2: a software package for estimation, validation, and analysis of Markov models. J. Chem. Theory Comput. 2015;11:5525–5542. doi: 10.1021/acs.jctc.5b00743. [DOI] [PubMed] [Google Scholar]
- 23.Roe D.R., Cheatham T.E., 3rd PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 24.Team R.C. R Foundation for Statistical Computing; Vienna, Austria: 2015. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 25.Vogelstein B., Lane D., Levine A.J. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 26.Green D.R., Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458:1127–1130. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li T., Kon N., Gu W. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell. 2012;149:1269–1283. doi: 10.1016/j.cell.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olivier M., Eeles R., Hainaut P. The IARC TP53 database: new online mutation analysis and recommendations to users. Hum. Mutat. 2002;19:607–614. doi: 10.1002/humu.10081. [DOI] [PubMed] [Google Scholar]
- 29.Duke R. AMBER Mailing List Archive; 2007. AMBER: reproducibility between software.http://archive.ambermd.org/ [Google Scholar]
- 30.Purawat S., Cowart C., Altintas I. Biomedical big data training collaborative (BBDTC): an effort to bridge the talent gap in biomedical science and research. Procedia Comput. Sci. 2016;80:1791–1800. doi: 10.1016/j.procs.2016.05.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.