Main Text
Cell signaling is a cascade of events that coordinates proper responses in cells upon sensing an external stimulus available in their surroundings. The ability of cells to respond to these highly dynamic changes in their immediate environment is what enables larger, coordinated responses that range from population control in bacterial communities to tissue development and homeostasis in multicellular organisms. Conversely, deregulation of cell signaling leads to lack of this homeostasis; in humans, it can lead to severe diseases.
Intracellular interpretation of these environmental stimuli often requires the action of second messengers. These molecules act not only as translators but also as amplifiers of extracellular responses by activating a myriad of proteins within cells. Moreover, their role as amplifiers implies that their concentration must first increase for activation of cellular responses and then decrease to restore the cell’s responsiveness to future stimuli.
One of the most important secondary messengers is cyclic adenosine monophosphate (cAMP), a nucleotide that has been extensively studied over a period of 60 years due to its participation in a broad spectrum of cell responses described in many cell biology and biochemistry textbooks, including cell growth and differentiation, gene transcription, and protein expression (1). The regulation exerted by cAMP consists of two distinct phases: 1) an activation phase, where the intracellular concentration of this secondary messenger is increased through cAMP synthesis triggered after hormonal stimulation of G-protein coupled receptors, followed by binding-dependent downstream activation of specific cellular targets; and 2) a termination phase, where cAMP is hydrolyzed to 5′-AMP by phosphodiesterases (PDEs) to restore basal intracellular concentration levels of this molecule (2). As PDEs are unique in their function of hydrolyzing cAMP, several PDE inhibitors have been designed for treating diseases linked to intracellular concentrations of cAMP (3).
Protein kinase A (PKA) is one of the most prominent targets of cAMP-dependent activation. The signaling pathway defined by the interaction between these two molecules is one of the most common and versatile ones in eukaryotic cells, constituting a convergence point for several extracellular signals and performing crucial cell regulation functions in almost all tissues in mammals (4). In its inactive state, PKA is constituted by two catalytic subunits bound to a regulatory dimer (R) that contains two cyclic nucleotide-binding sites. Binding of two cAMP molecules for each R-subunit releases and activates the catalytic subunits to phosphorylate a variety of target proteins. Termination of this signaling pathway is then controlled through hydrolysis of the cAMP that is tightly bound to the R-subunits, followed by reconstitution of the inactive state (4).
Despite decades of study of this signaling pathway, two key aspects remain enigmatic. First, how do PDEs hydrolyze cAMP that is tightly bound to R-subunits, a condition required to restart the signaling pathway? Second, although it makes sense that increased cAMP synthesis leads to increased PKA activity (5), how does the increase in PDE for augmented cAMP degradation, contrary to expectation, also activate PKA (6)?
In this volume of Biophysical Journal, the excellent work by Tulsian et al. (7) provides a solution to these paradigmatic riddles using a combination of enzyme kinetics, fluorescence polarization assays, and hydrogen-deuterium exchange mass spectrometry on the R-subunit RIα of PKA and PDE8 under several conditions. Their starting point is their earlier work on PDE/PKA complexes, in which they provided compelling evidence of the ability of PDE to dissociate and hydrolyze R-subunit-bound cAMP through coupling between active sites (8, 9). Their results suggested that substrate channeling, i.e., the restricted diffusion of reaction intermediates between binding sites without their release into the cytosol, played a crucial role in this signaling pathway (8). Here, the authors describe how this substrate channeling operates to allow cAMP degradation in the PDE8/PKA-RIα complex (7).
First, they use enzyme kinetics to demonstrate how the presence of RIα bound to PDE8 enhances its hydrolytic activity against cAMP in solution compared to free PDE8. Given the low nanomolar affinity of RIα for cAMP, these results were interpreted as the consequence of the formation of a stable PDE8/RIα complex with enhanced catalytic activity. Further evidence of the steps of PDE8-dependent hydrolysis of cAMP bound to RIα was obtained using fluorescence polarization assays with two different fluorescent cAMP analogs, a non-hydrolyzable PDE8-resistant moiety working as a reporter of formation of PDE8/RIα/cAMP ternary complexes and a PDE8-hydrolyzable analog reporting cAMP-binding to PDE8 and its subsequent hydrolysis. Addition of PDE8 to RIα complexed with these fluorescent analogs led to increased polarization, reflecting the formation of ternary complexes. Strikingly, the fluorescence polarization signal of the hydrolyzable analog slowly decreased over time, thus indicating the channeling of the cAMP analog to PDE8 for its fast degradation, followed by the slow disassembly of the ternary complex. Moreover, “washing out” cAMP from RIα/cAMP-analog complexes initially incubated with an excess of this cyclic nucleotide by addition of PDE8 only allowed the reassociation of the hydrolyzable analog. Altogether, these results strongly indicated the functional importance of the association of PDE8 to RIα in the presence of cAMP, constituting a stable complex that mediates enhanced hydrolysis of cAMP via channeling of this secondary messenger between binding sites.
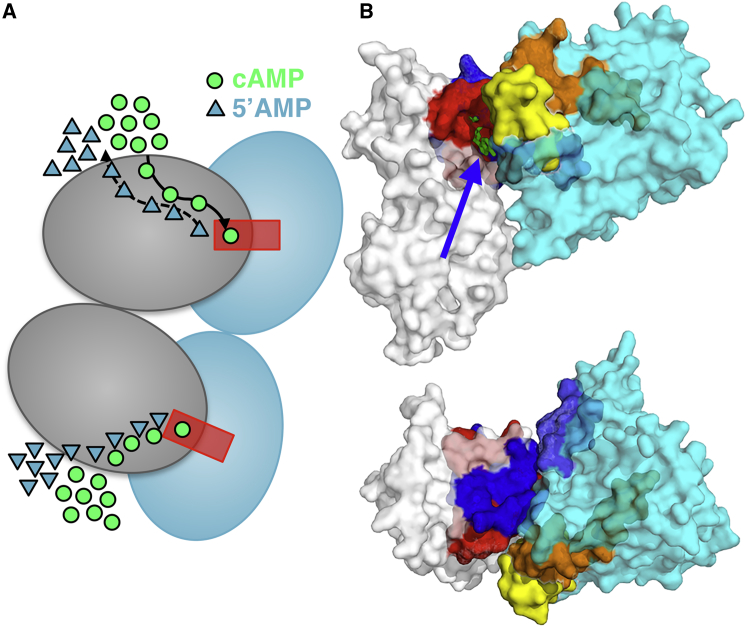
Structural information of the features of the complex and its substrate channel was obtained from hydrogen-deuterium exchange mass spectrometry experiments on the PDE8/RIα/cAMP complex, in which the extent of deuterium exchange over localized regions of a protein is used as a probe of their flexibility. These studies revealed that the flexibility of the cAMP binding site in RIα increased over time after addition of PDE8, demonstrating that cAMP was hydrolyzed from RIα only upon active association of the PDE8. Moreover, the interaction surfaces between PDE8 and RIα were identified based on their decreased deuterium exchange in comparison with their free forms in solution. For RIα and PDE8, the protein interaction surfaces span loop regions in the vicinity of the two cyclic nucleotide binding sites of the R-subunit and the catalytic site of PDE8, such that a channel-like complex that encloses the flexible cAMP binding sites is formed, whereas excess cAMP increases the stability of these protein-protein interactions (Fig. 1). Finally, comparison of the extent of deuterium exchange of these complexes in limiting and excess-cAMP conditions allowed the authors to demonstrate the effective hydrolysis of cAMP in the context of a stable PDE8/RIα.
Figure 1.
Substrate channeling between RIα and PDE8. (A) Substrate channeling between the nucleotide binding sites CNB:A and CNB:B of RIα (gray) and PDE8 (cyan). PDE8 preferentially hydrolyzes RIα-bound cAMP by forming a substrate channel (red) between their active and binding sites, respectively, releasing 5′AMP as a result. (B) Side and top views of a potential structure of the RIα CNB:A/PDE8 complex generated using the protein interfaces predicted by hydrogen-deuterium exchange mass spectrometry in combination with the ZDOCK server. RIα (PDB: 4MX3) is shown in white, with the PDE8-binding region and the CNB:A binding site in blue and red, respectively. PDE8 (PDB: 3ECN) is shown in cyan, and the RIα interaction surface and substrate-recognition and catalytic sites are in light blue, yellow, and orange, respectively. The arrow indicates the cAMP binding site of RIα. The cartoon in (A) was adapted from Tulsian et al. (7). To see this figure in color, go online.
The conclusions drawn by these experiments are a major contribution to our understanding of how the cAMP-dependent PKA signaling pathway can briefly amplify extracellular stimuli within the cell interior and rapidly return to responsive conditions. First, it rigorously provides an elaborate solution to the paradigm of PDE-dependent signal amplification of PKA. The enhanced catalytic activity of PDE is due to its ability to sequester the R-subunit via a stable complex formed for serial hydrolysis of all available cAMP molecules, even under low concentrations of this secondary messenger (Fig. 1), which in turn blocks the reassociation of the catalytic subunit into the inactive PKA. This mechanism explains how an increase in the concentration of PDE leads to the activation of PKA for cAMP (6) and positions PDE as a key component of both the activation and termination phases of cAMP-dependent signaling. This behavior allows dynamic cellular responses for subsequent stimulus, as in a precise oscillator, which could ultimately lead to cellular adaptation to the ever-changing environment (5). Second, it constitutes yet another remarkable example of the fundamental role of protein-protein interactions in controlling cell-signaling processes and maintaining cellular homeostasis (10), as they provide specificity and diversity simultaneously. For the PKA signaling pathway, restricted cAMP diffusion due to the formation of the PDE8/RIα complex enables compartmentalization (specificity) and partitioning (diversity) of the different signaling pathways within cells (4). Finally, the work of these authors demonstrates how the right combination of biophysical approaches can reveal paradigmatic cellular behaviors by careful observation of the system dynamics, which should be enlightening for other researchers challenged by similar riddles.
Acknowledgments
C.A.R.S. is supported by Fondo Nacional de Desarrollo Científico y Tecnológico (Fondecyt grant 11140601) and the Chilean Antarctic Institute (INACH grant RG_47-16).
Editor: Elizabeth Komives.
References
- 1.Beavo J.A., Brunton L.L. Cyclic nucleotide research—still expanding after half a century. Nat. Rev. Mol. Cell Biol. 2002;3:710–718. doi: 10.1038/nrm911. [DOI] [PubMed] [Google Scholar]
- 2.Skroblin P., Grossmann S., Klussmann E. Mechanisms of protein kinase A anchoring. Int. Rev. Cell Mol. Biol. 2010;283:235–330. doi: 10.1016/S1937-6448(10)83005-9. [DOI] [PubMed] [Google Scholar]
- 3.Yan K., Gao L.N., Zhou X. The cyclic AMP signaling pathway: exploring targets for successful drug discovery (Review) Mol. Med. Rep. 2016;13:3715–3723. doi: 10.3892/mmr.2016.5005. (review) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taskén K., Aandahl E.M. Localized effects of cAMP mediated by distinct routes of protein kinase A. Physiol. Rev. 2004;84:137–167. doi: 10.1152/physrev.00021.2003. [DOI] [PubMed] [Google Scholar]
- 5.Houslay M.D. Adaptation in cyclic AMP signalling processes: a central role for cyclic AMP phosphodiesterases. Semin. Cell Dev. Biol. 1998;9:161–167. doi: 10.1006/scdb.1997.0221. [DOI] [PubMed] [Google Scholar]
- 6.Leiser M., Fleischer N., Erlichman J. Enhanced activation of cAMP-dependent protein kinase by rapid synthesis and degradation of cAMP. J. Biol. Chem. 1986;261:15486–15490. [PubMed] [Google Scholar]
- 7.Tulsian N.K., Krishnamurthy S., Anand G.S. Channeling of cAMP in PDE-PKA complexes promotes signal adaptation. Biophys. J. 2017;112:2552–2566. doi: 10.1016/j.bpj.2017.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krishnamurthy S., Moorthy B.S., Anand G.S. Active site coupling in PDE:PKA complexes promotes resetting of mammalian cAMP signaling. Biophys. J. 2014;107:1426–1440. doi: 10.1016/j.bpj.2014.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krishnamurthy S., Tulsian N.K., Anand G.S. Parallel allostery by cAMP and PDE coordinates activation and termination phases in cAMP signaling. Biophys. J. 2015;109:1251–1263. doi: 10.1016/j.bpj.2015.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pawson T., Nash P. Protein-protein interactions define specificity in signal transduction. Genes Dev. 2000;14:1027–1047. [PubMed] [Google Scholar]