Abstract
Mechanistic target of rapamycin (mTOR) plays a critical role in the development of oral squamous cell carcinoma (OSCC), but the underlying mechanisms remain poorly understood. Here we have demonstrated that the expression of osteopontin (OPN) was dramatically up-regulated in OSCC tissues and cell lines. Moreover, reduction of OPN suppressed cell proliferation, colony formation, and in vivo tumorigenic ability of OSCC cell lines Tca8113. In addition, there was a strong positive correlation between mTORC1 activity and OPN expression in OSCC tissues and cell lines. Furthermore, mTOR complex 1 (mTORC1) enhanced OPN expression through up-regulation of ERRα. Therefore, OPN is a downstream target of mTORC1 and is crucial for OSCC development. mTORC1, ERRα, and OPN may be potential targets for treatment of OSCC with aberrant mTORC1 signaling.
Keywords: mTOR, ERRα, OPN, tumorigenesis, OSCC.
Introduction
Oral squamous cell carcinoma (OSCC) ranks among the top-ten most common cancers worldwide 1. The etiology of OSCC is complicated and many factors are involved in its progression. Despite the advances in the treatment applied, the overall survival rate of patients with OSCC remains poor 2. Therefore, it is of great significance to find an effective method for diagnosis and treatment of OSCC.
Mechanistic target of rapamycin (mTOR), a serine/threonine protein kinase, exerts a critical role in cell growth, proliferation, and survival 3, 4. mTOR associates with different partners to form two complexes, mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2) 3, 5. mTORC1 is sensitive to rapamycin, whereas mTORC2 is rapamycin-insensitive. mTORC1 signaling pathway is frequently altered in many cancers 6. Recently, the critical role of mTORC1 in OSCC development has been established 7-9, but the exact mechanisms remain to be further elucidated.
Osteopontin (OPN) is a secreted phosphorylated glycoprotein and is overexpressed in a variety of cancers 10, 11. OPN plays an important role in tumor angiogenesis, apoptosis, and cancer metastasis 12. There are few characterizations on the functional regulation of PI3K/AKT/mTOR signaling on OPN expression.
In this study, we have shown that OPN expression is elevated in OSCC and positively correlated with mTORC1 activity. Depletion of OPN suppresses tumorigenic capacity of human OSCC cells. mTORC1 positively regulates OPN expression through the activation of ERRα signaling. mTORC1, ERRα, and OPN may serve as potential targets for the treatment of OSCC with dysregulated mTORC1 signaling.
Materials and Methods
Reagents and antibodies
Rapamycin and XCT-790 were purchased from Sigma; Lipofectamine 2000 and 4-12% Bis-Tris Nu-PAGE gels were obtained from Life Technologies. OPN and β-actin antibodies were purchased from Santa Cruz; p-S6 (Ser235/236), S6, mTOR, Raptor, and Rictor antibodies were obtained from Cell Signaling. GAPDH and ERRα antibodies were purchased from Abcam.
Cell cultures
The human transformed embryonic kidney cells (HEK293T) and the human OSCC cell lines KB, HSC-2, HSC-3, Tca8113, SCC-9, and SCC-15 were obtained from the ATCC. Tsc2+/+ and Tsc2-/- mouse embryonic fibroblasts (MEFs) used in this study were kindly provided by Dr. Hongbing Zhang (Peking Union Medical College) and have been described previously 13, 14. All cell lines were cultured in DMEM with 10% FBS and 1% penicillin/streptomycin in 5% CO2 at 37°C.
Clinical samples
Clinical samples of 30 OSCC and 16 paired normal mucosa tissues (adjacent normal tissues) were obtained from patients undergoing surgical resection in the affiliated hospital of stomatology, Chongqing Medical University from June 2013 to December 2013. All samples were immediately preserved in -80℃ cryogenic refrigerator after surgery. All cases of OSCC were confirmed for the first time and without prior neoadjuvant therapy. All specimens were diagnosed for primary squamous cell carcinoma of the tongue by more than two pathologists. Both oral and written informed consents were taken from every patient in the study, according to a protocol approved by ethics committees of Chongqing Medical University.
Lentivirus-mediated RNA interference
The GV248 lentiviral shRNA expression vector targeting human OPN and control scrambled (shScramble) were obtained from Genechem (Shanghai, China). Lentiviruses were produced by co-transfecting the shRNA expression vector with the Vira Power lentiviral Packaging Mix (Genechem) in HEK293T cells. Culture supernatants were collected after 48 h of transfection and then used to infect Tca8113 cells.
Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from cells using Trizol (Invitrogen) according to the protocol provided by the manufacturer. RNA was converted to cDNA using the PrimeScriptTM RT Reagent Kit (TaKaRa). qRT-PCR was performed using SYBR® Premix Ex Taq™ II (TaKaRa) as described previously 14. The primer sequences were as follows: OPN forward: 5'-CTCCATTGACTCGAACGACTC-3', reverse: 5'-CAGGTCTGCGAAACTTCTTAGAT-3'; β-actin forward: 5'-AGAAAATCTGGCACCACACC-3', reverse: 5'-AGAGGCGTACAGGGATAGCA-3'.
Western blotting analysis
Western blotting analyses were performed as described previously 13, 15 using primary antibodies against OPN, mTOR, Raptor, Rictor, p-S6, S6, ERRα, GAPDH, and β-actin.
RNA interference
Cells were seeded in 12-well plates and transfected with siRNAs using Lipofectamine 2000 following the manufacturer's instructions. All siRNA were synthesized by GenePharma (Shanghai, China). siRNA targeted sequences were as follows: mTOR: 5'-CCCTGCTTCTGTCATGCCT-3'; Raptor: 5'-GGACAACGGCCACAAGTAC-3'; Rictor: 5'-ACUTGTGAAGAATCGTATC-3'; ERRα: 5'-GGACCTATGAGACCTTCAA-3'; negative control: 5'-TTCTCCGAACGTGTCACGT-3'.
Reporter constructs and luciferase reporter assay
A 834-bp human OPN promoter fragment was cloned into the kpn I / xho I sites of the luciferase reporter plasmid pGL3-basic (Promega) and named as OPN-luc. The primer sequences were as follows: forward, 5'-CGGGGTACCCATGGATGAGGGAACAAGG-3'; reverse, 5'-CCGCTCGAGTACCTTGGTCGGCGTTTGG-3'. The putative ERRα binding sites in the OPN promoter were mutated using the Quick Change site-directed mutagenesis kit (Promega). The primer sequences were as follows: Mut1, forward 5'-GCCCAAGGTTGCACATATTTGCAGTGACACAGCGGA-3'; reverse 5'-TCCGCTGTGTCACTGCAAATATGTGCAACCTTGGGC-3'. Mut2, forward 5'-AAAGCTAAGCTTGAGTAGTAGACCAGTGAGGCAAGTTTTCTG-3'; reverse 5'-CAGAAAACTTGCCTCCATGGTCTACTACTCAAGCTTAGCTTT-3'. Cells were incubated in triplicate in 24-well plates and transfected with the promoter constructs (200 ng) in combination with the plasmid pRL-TK (20 ng) as an internal control. Luciferase activity was examined with the Dual-Luciferase Reporter Assay System (Promega).
Chromatin immunoprecipitation assay
Immunoprecipitation assay with an anti-ERRα antibody was performed with a SimpleChIP® Enzymatic Chromatin IP kit (Cell Signaling) according to the manufacturer's protocol. The immunoprecipitated DNA was purified for the analysis by qRT-PCR. The primer sequences used were as follows: the putative ERRα-binding site region 1 (PBR1) of human OPN forward, 5′-TGGATGAGGGAACAAGGATAGGTAGG-3′, reverse, 5′-GACCGTGGTTCTGAATTCCGCTGT-3′; the putative ERRα-binding site region 2 (PBR2) of human OPN forward, 5′-TGCATACTCGAAATCACAAAGCTAAG-3′, reverse, 5′-AATGGATTTTTGTTTCTTTCGGTTTA-3′; a nonspecific ERRα-binding region (NBR) of human OPN forward, 5′-AAGAAATAGCAATCACCTGGGCAGTC-3′, reverse, 5′-CTCCCGGCCTCCTGAATTAAACGTCT-3′.
Cell proliferation assay
Cells were seeded in 96-well plates at a density of 2×103 per well and the cell proliferation was evaluated with MTT assay as described previously 16.
Soft agar assay
A 0.6% (w/v %) bottom layer of low melting point agarose in common culture medium was placed in 6-well plates. On top, a layer of 0.3% agarose containing 1×103 cells was placed, and the colony number in the soft agar was counted after three weeks 14.
Induction of subcutaneous tumors in nude mice
Subcutaneous tumors were established in nude mice (BALB/c, 5 week old) as described previously 17, 18. Six male mice were used in each cohort. 2×106 Tca8113 cells expressing shOPN1 or shScramble in 100 μl of DMEM were s.c. inoculated into the right posterior back region. Mice were sacrificed when tumor size was greater than 1,000 mm3, there was ulceration over the tumor, or weight loss of more than 10% occurred. All animals were maintained and used in accordance with the guidelines of the Animal Center of Chongqing Medical University.
Enzyme-linked immunosorbent assay (ELISA)
The presence of the OPN released by the OSCC cell lines into the culture medium was measured using a human OPN Quantikine ELISA kit (R&D Systems) according to the manufacturer's instructions. In brief, Cells (6 × 105 cells/well) were plated into 6-well culture plates. After 12 h, the culture medium was replaced with an equivalent volume of fresh serum-free medium with or without rapamycin (50 nM). Cell culture supernatants were collected 24 h later and the OPN levels were determined using an ELISA kit.
Statistical analysis
The tumor development and survival data were analyzed with Kaplan-Meier log-rank test and other data were analyzed using a 2-tailed paired Student's t-test as described previously 13, 14. It is statistically significant when P < 0.05.
Results
The expression of OPN is significantly up-regulated in OSCC and positively correlated with mTORC1 signaling
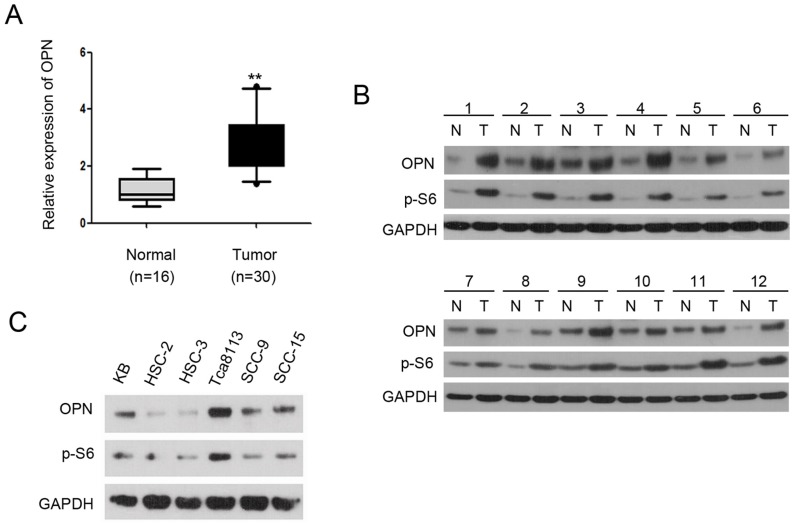
OPN plays a critical role in tumorigenesis and is frequently overexpressed in various types of cancers 11, 19, 20. However, the role of OPN in the development of OSCC remains largely unknown. To investigate the expression abundance of OPN in OSCC, we performed qRT-PCR with cDNA from 30 OSCC tissues and 16 normal mucosa tissues. As shown in Figure 1A, the mRNA level of OPN was significantly higher in OSCC tissues compared with that of normal tissues. To determine whether the protein levels of OPN were also elevated in OSCC tissues, we measured the expression level of OPN in OSCC tissues by western blotting. The result revealed that OPN was markedly up-regulated in OSCC tissues compared to adjacent normal tissues. Interestingly, there were consistent increase of p-S6 level with OPN expression in the tumor tissues (p-S6 is an indicator of mTORC1 activity) 21 (Figure 1B). To further explore the relationship between mTORC1 and OPN, we used six human OSCC cell lines (KB, HSC-2, HSC-3, Tca8113, SCC-9, and SCC-15) to analyze the expression levels of p-S6 and OPN. As shown in Figure 1C, the expression level of p-S6 was positively correlated with OPN in these cell lines. Taken together, these data indicate that OPN is up-regulated in OSCC and the expression of OPN is positively correlated with mTORC1 activity in OSCC.
Figure 1.
OPN expression is elevated in OSCC and positively correlated with mTORC1 activity. (A) qRT-PCR analysis of OPN expression in 30 OSCC tissues compared to 16 normal mucosa tissues. **P<0.01, compared with the respective controls. Box plots display the median, 25th and 75th percentiles. Whiskers represent 5-95 percentiles and dots the outliers. (B) Western blotting analysis of OPN and p-S6 levels in 12 paired of OSCC tissues and adjacent normal/pre-cancerous tissues and GAPDH was used as a control. (C) Cell lysates from KB, HSC-2, HSC-3, Tca8113, SCC-9, and SCC-15 cells were subjected to immunoblotting with the indicated antibodies.
mTORC1 positively regulates OPN in OSCC cell lines
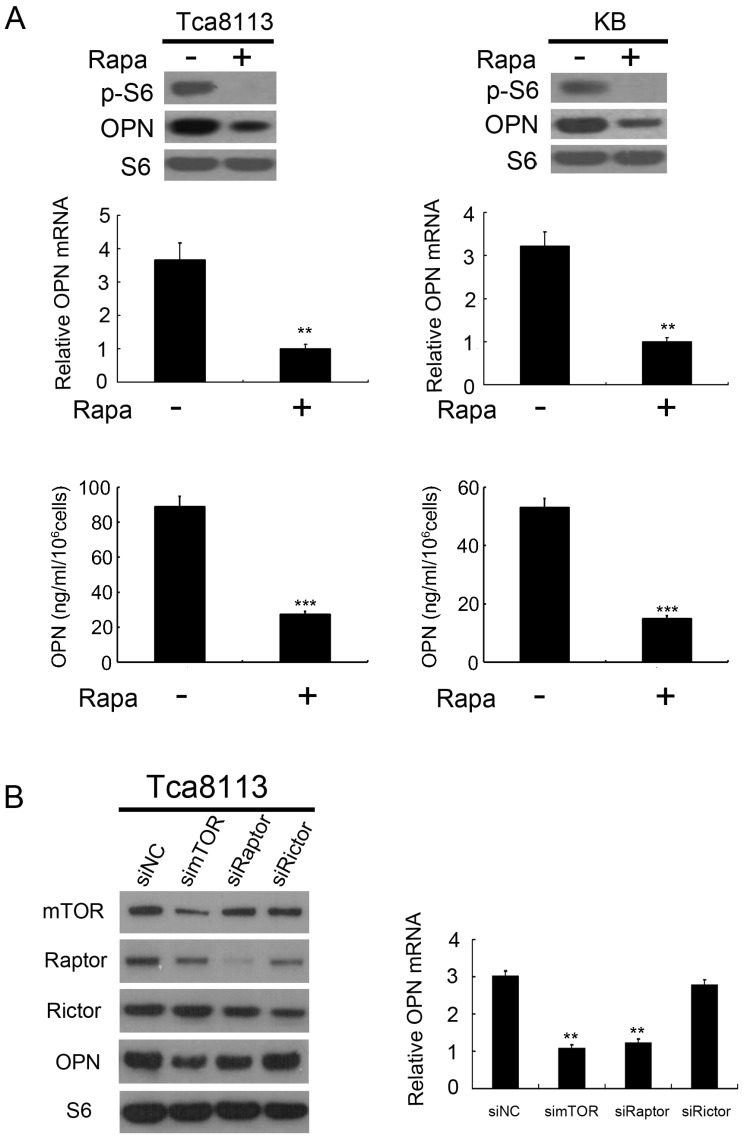
mTOR exists in two multi-protein complexes (rapamycin-sensitive mTORC1 and rapamycin-insensitive mTORC2). To determine whether OPN is controlled by mTORC1, we evaluated the effect of rapamycin, specific inhibitor of mTORC1 on OPN expression. As depicted in Figure 2A, the expression and secretion of OPN were decreased in response to rapamycin treatment in Tca8113 cells and KB cells. In order to confirm the regulation effect of mTORC1 on OPN expression, we examined the expression level of OPN in mTOR-, Raptor (a specific component of mTORC1)-, or Rictor (a specific component of mTORC2)-knockdown cells. Knockdown of mTOR or Raptor with siRNAs dramatically decreased the expression level of OPN in Tca8113 cells, whereas the level of OPN in cells transfected with Rictor siRNAs was no change with that in the control (Figure 2B). Collectively, these data suggest that it is mTORC1, but not mTORC2, that up-regulates OPN expression.
Figure 2.
mTORC1 positively regulates OPN expression. (A) Tca8113 and KB cells were treated with or without 50 nM rapamycin (Rapa) for 24 h. Expression levels of OPN, p-S6 and S6 were detected by western blot analysis (top). The mRNA levels of OPN were analyzed by qRT-PCR (middle). OPN levels in the cell supernatants were detected by an ELISA (bottom). Data indicate mean ± SD of triplicate samples. **P<0.01; ***P<0.001, compared with the respective controls. (B) Tca8113 cells were transfected with mTOR, Raptor or Rictor siRNAs for 48 h. Cell lysates were subjected to immunoblotting with the indicated antibodies (right). The mRNA levels of OPN were analyzed by qRT-PCR (left). Data indicate mean ± SD of triplicate samples. **P<0.01, compared with the respective controls.
mTORC1 up-regulates OPN through activation of ERRa signaling
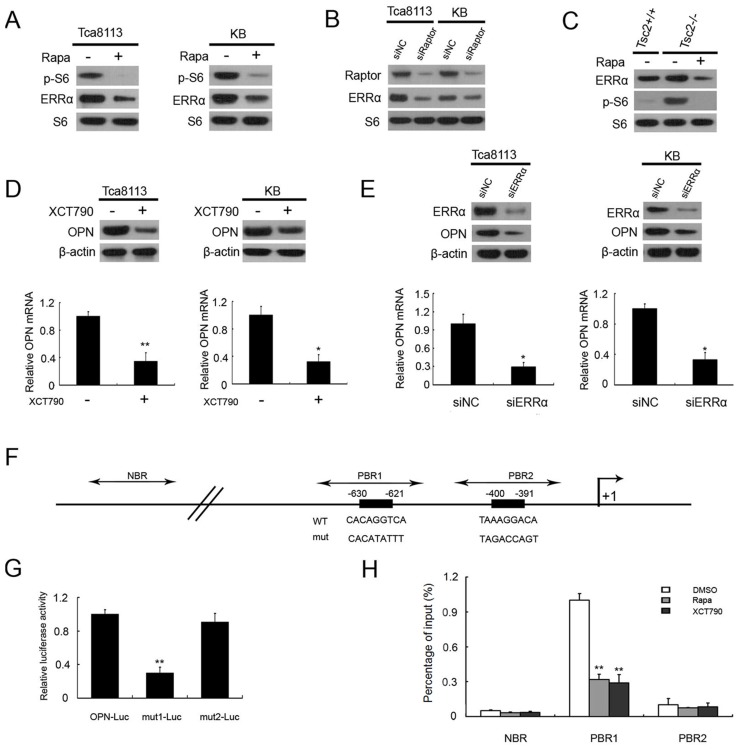
Estrogen-related receptor α (ERRα) is an orphan nuclear receptor that plays a crucial role in the transcriptional regulation of mitochondrial function and energy metabolism 22, 23. Next we investigated whether there was a contribution of ERRα in mTOR-mediated up-regulation of OPN. We first determined whether ERRα is controlled by mTORC1. As shown in Figure 3A, the protein levels of ERRα were dramatically decreased after treatment with rapamycin in KB and Tca8113 cells. Moreover, knockdown of Raptor remarkably reduced ERRα expression in Tca8113 cells and KB cells (Figure 3B). Given that TSC2 is the key negative regulator of mTORC1, Tsc2-deficient MEFs have been widely used as cell models for the study on mTOR-related diseases 24-26. The ERRα expression was markedly increased in Tsc2-/- MEFs compared to WT MEFs, and impaired by the treatment with rapamycin (Figure 3C). Therefore, these results suggested that ERRα is a downstream effector of mTORC1.
Figure 3.
mTORC1 up-regulates OPN through activation of ERRα signaling.(A) Cell lysates from Tca8113 and KB cells treated with or without 50 nM Rapa for 24 h were subjected to immunoblotting with the indicated antibodies. (B) Cell lysates from Tca8113 or KB cells transfected with Raptor or the control (siNC) siRNAs for 48 h were subjected to immunoblotting with the indicated antibodies.(C) Cell lysates from Tsc2+/+ MEFs and Tsc2-/- MEFs treated with or without 20 nM Rapa for 24 h were subjected to immunoblotting with the indicated antibodies. (D) Tca8113 and KB cells were treated with or without 50 nM Rapa for 24 h. The proteins were detected by immunoblotting with the indicated antibodies (top). The mRNA levels of OPN were analyzed by qRT-PCR (bottom). Data indicate mean ± SD of triplicate samples. **P<0.01, compared with the respective controls. (E) Tca8113 and KB cells were transfected with ERRα or the control siRNAs for 48 h. The proteins were detected by immunoblotting with the indicated antibodies (top). The mRNA levels of OPN were analyzed by qRT-PCR (bottom). Data indicate mean ± SD of triplicate samples. **P<0.01, compared with the respective controls. (F) Schematic representation of the putative wild-type (WT) and mutated (mut) ERRα-binding sites within the promoter region of human OPN gene. Dark rectangles indicate the predicted ERRα binding sites; two-way arrows indicate the fragments amplified in ChIP real-time PCR analysis. The transcription start site is indicated by an arrow above the gene. PBR1, putative binding region 1; PBR2, putative binding region 2; NBR, nonspecific binding region. (G) Tca8113 cells were co-transfected with pOPN-Luc, pmut1-Luc, or pmut2-Luc reporter plasmid and pRL-TK plasmid. Relative luciferase activity was examined 48 h after transfection. Data indicate mean ± SD of triplicate samples. **P<0.01, compared with the respective controls. (H) Tca8113 cells treated with Rapamycin, XCT790, or DMSO were subjected to ChIP assay using an anti-ERRα antibody. Normal rabbit IgG antibody served as the negative control. qRT-PCR were performed to amplify a fragment surrounding PBR1, PBR2, or NBR. The data were plotted as the ratio of immunoprecipitated DNA subtracting nonspecific binding to IgG vs. total input DNA. Data indicate mean ± SD of triplicate samples. **P<0.01, compared with the respective controls.
To investigate whether mTORC1 up-regulates OPN via ERRα, Tca8113 and KB cells were treated with XCT-790, a specific inhibitor of ERRα. As shown in Figure 3D, the levels of OPN were markedly decreased in the presence of XCT-790. Moreover, knockdown of ERRα with siRNAs led to decrease of OPN expression in Tca8113 and KB cells (Figure 3E). Taken together, these data indicated that mTORC1 up-regulates OPN through activation of ERRα signaling.
To explore the mechanism by which ERRα up-regulates OPN expression, we analyzed the promoter region of OPN gene and found that there were 2 putative ERRα-binding sequence (-400/-391 TAAAGGACA, -630/-621 CACAGGTCA) in the promoter of OPN gene (Figure 3F). We inserted a 834-bp fragment of the promoter sequence of human OPN gene containing the putative ERRα-binding sequences, into the luciferase reporter plasmid to evaluate the transcriptional activity of ERRα on this region. The transcriptional activity was significantly impaired when one of the putative ERRα-binding sites was mutated (Figure 3G). ChIP assay revealed that the direct binding of ERRα to one of the putative ERRα-binding sites within the promoter of OPN gene were significantly attenuated by the treatment of rapamycin or XCT790 (Figure 3H). Taken together, ERRα transcriptionally regulates OPN expression downstream of mTORC1 signaling.
Depletion of OPN suppresses tumorigenic capacity of human OSCC cells
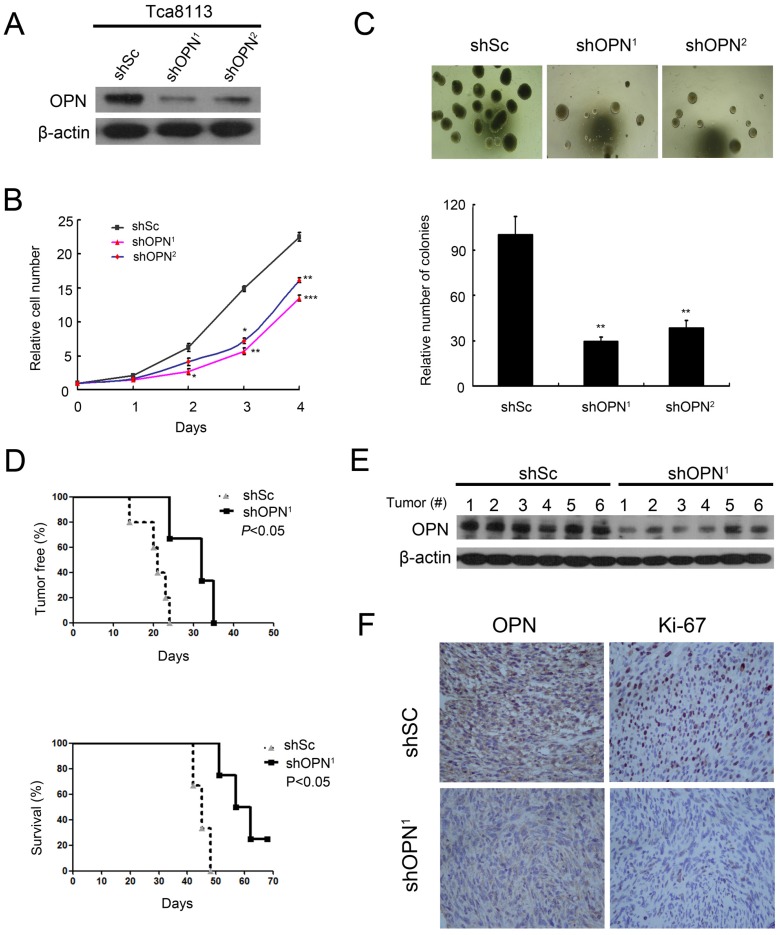
To investigate the role of OPN in the tumorigenesis of human OSCC cells, we knocked down OPN expression with two different shRNAs for OPN (shOPN1 and shOPN2) in Tca8113 cells. Both shOPN1 and shOPN2 dramatically decreased the OPN expression (Figure 4A). Reduction of OPN expression led to a decrease of cell proliferation and suppression of colony formation in Tca8113 cells (Figure 4B and C). To determine whether depletion of OPN influences the tumorigenic ability of Tca8113 cells in vivo, we inoculated Tca8113 cells expressing shOPN1 or shScramble subcutaneously into nude mice. Depletion of OPN substantially inhibited the tumorigenesis of Tca8113 cells in nude mice (Figure 4D). Impaired OPN expression in the tumor tissues derived from these nude mice was confirmed by western blotting (Figure 4E). Moreover, IHC analysis showed that reduction of OPN expression led to a decrease of proliferation marker Ki-67 expression in vivo (Figure 4F). Taken together, these data indicate that OPN is critical for the proliferation, colony formation, and tumor growth of OSCC cells.
Figure 4.
Depletion of OPN suppresses cell proliferation, colony formation, and tumorigenesis. (A) Two independent shRNAs that target OPN (shOPN1 and shOPN2) or a control shRNA (shSc) were stably expressed in Tca8113 cells. The expression of OPN was examined by western blotting and β-actin was used as a control. (B) The proliferation of Tca8113 cells expressing shSc, shOPN1, or shOPN2 was examined by MTT assay. Data indicate mean ± SD of triplicate samples. *P<0.05, **P<0.01, ***P<0.001, compared with the respective controls. (C) The number of colonies formed in soft agar by Tca8113 cells expressing shSc, shOPN1, or shOPN2. Representative images were presented. Data indicate mean ± SD of triplicate samples. **P<0.01, compared with the respective controls. (D) Tca8113 cells expressing shSc or shOPN1 were inoculated subcutaneously into nude mice, and followed for tumor development (top panel) and survival (bottom panel). (E) Tumor tissues from these nude mice were subjected to immunoblotting for OPN expression and β-actin was used as a control. (F) Tumor tissues were subjected to immunohistochemical staining. Representative images were presented.
Discussion
The underlying mechanisms of OSCC mediated by hyperactivated mTOR remain largely elusive. In this study, we have demonstrated that OPN expression is significantly elevated in OSCC. Moreover, depletion of OPN inhibits tumorigenic capacity of human OSCC cells. In addition, OPN levels positively correlated with mTORC1 activity in OSCC. Furthermore, mTORC1 positively regulates OPN via activation of ERRα signaling.
The level of OPN is obviously increased in the serum of patients with tumor metastasis, and it may be a diagnostic indicator of tumor metastasis 27. It has also been reported that OPN is overexpressed in many cancers, including oral squamous cell carcinoma and early invasive tongue squamous cell carcinoma 28, 29. Aberrant OPN expression was associated with the malignization of oral squamous epithelium 30. In this study, we also found that OPN was overexpressed in OSCC tissues. We demonstrated that OPN was required for the growth of OSCC cells in vitro and in vivo through knockdown of OPN expression in Tca8113 cells, MTT assay, clone formation assay, and human OSCC xenografts in nude mice. Therefore, OPN may be a candidate target for the treatment against OSCC.
The aberrant PI3K/AKT/mTOR signaling pathway plays a critical role in the development of cancers 31-33. The PI3K/AKT/mTOR signaling is reported to be activated in OSCC 8, 9. mTOR exists in two different complexes: mTOR complex 1 (mTORCl) and mTOR complex 2 (mTORC2). mTORC1 is sensitive to rapamycin and composed of mTOR, Raptor, and mLST8/GβL. mTORC2 is insensitive to rapamycin and composed of mTOR, Rictor, mLST8/ GβL, and Sin1 34, 35. mTORC2 plays an important role in phosphorylation of AKT. Here we demonstrated that the mTOR signaling was activated in OSCC tissues examined, which suggest that there may be a putative link between activation of PI3K/AKT/mTOR signaling pathway and OPN overexpression in OSCC. Subsequent research revealed that OPN was positively regulated by mTORC1, but not mTORC2, in OSCC cell lines Tca8113 and KB. This work established that OPN is a novel effector downstream of mTORC1, and there is a strong link between mTORC1-OPN axis and OSCC development.
Cell- and tissue-specific OPN expression is regulated by hormones, growth factors, and oncogenes. The sequence of OPN promoter is highly conservative and can be combined with a variety of transcription factors 36-38. The majority of studies have shown that OPN expression is frequently regulated at the level of transcription 38. ERRα is an orphan nuclear receptor, which is a member of the nuclear receptor superfamily 39. ERRα is able to combine with DNA to activate transcription 40, 41. OPN is reported to be a target gene of ERRα in osteoblast differentiation 41. Although there is no classic estrogen responsive element in the OPN promoter, estrogen can selectively bind to 7 steroid factor response elements of OPN promoter via ERα and ERRα to activate OPN transcription 42. In this study, we demonstrated that mTORC1 is a positive regulator of ERRα, and ERRα regulate OPN transcription through directly binding to the promoter region of OPN gene. The newly identified mTORC1-ERRα-OPN signaling cascade contributes to the mechanism of OSCC.
In conclusion, hyperactive mTORC1 signaling contributes to OSCC via activation of ERRα-OPN signaling. This study provides new insight into the mechanism of OSCC mediated by hyperactive mTOR. mTORC1, ERRα, and OPN may be candidate targets for treatment of OSCC with abnormal mTORC1 signaling.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grants 81371778 to Y.Y) and Chongqing Science and Technology Commission Grant for Distinguished Young Scholar of Chongqing (grant cstc2014jcyjjq10002 to J.C.).
References
- 1.Miyahara M, Tanuma J, Sugihara K, Semba I. Tumor lymphangiogenesis correlates with lymph node metastasis and clinicopathologic parameters in oral squamous cell carcinoma. Cancer. 2007;110:1287–94. doi: 10.1002/cncr.22900. [DOI] [PubMed] [Google Scholar]
- 2.Kademani D, Bell RB, Schmidt BL, Blanchaert R, Fernandes R, Lambert P. et al. Oral and maxillofacial surgeons treating oral cancer: a preliminary report from the American Association of Oral and Maxillofacial Surgeons Task Force on Oral Cancer. Journal of oral and maxillofacial surgery: official journal of the American Association of Oral and Maxillofacial Surgeons. 2008;66:2151–7. doi: 10.1016/j.joms.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 3.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 4.Yang Q, Guan KL. Expanding mTOR signaling. Cell research. 2007;17:666–81. doi: 10.1038/cr.2007.64. [DOI] [PubMed] [Google Scholar]
- 5.Gulhati P, Bowen KA, Liu J, Stevens PD, Rychahou PG, Chen M. et al. mTORC1 and mTORC2 regulate EMT, motility, and metastasis of colorectal cancer via RhoA and Rac1 signaling pathways. Cancer research. 2011;71:3246–56. doi: 10.1158/0008-5472.CAN-10-4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lane HA, Breuleux M. Optimal targeting of the mTORC1 kinase in human cancer. Current opinion in cell biology. 2009;21:219–29. doi: 10.1016/j.ceb.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 7.Ji WT, Yang SR, Chen JY, Cheng YP, Lee YR, Chiang MK. et al. Arecoline downregulates levels of p21 and p27 through the reactive oxygen species/mTOR complex 1 pathway and may contribute to oral squamous cell carcinoma. Cancer science. 2012;103:1221–9. doi: 10.1111/j.1349-7006.2012.02294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chakraborty S, Mohiyuddin SM, Gopinath KS, Kumar A. Involvement of TSC genes and differential expression of other members of the mTOR signaling pathway in oral squamous cell carcinoma. BMC cancer. 2008;8:163. doi: 10.1186/1471-2407-8-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naruse T, Kawasaki G, Yanamoto S, Mizuno A, Umeda M. Immunohistochemical study of VEGF expression in oral squamous cell carcinomas: correlation with the mTOR-HIF-1alpha pathway. Anticancer research. 2011;31:4429–37. [PubMed] [Google Scholar]
- 10.Shevde LA, Metge BJ, Mitra A, Xi Y, Ju J, King JA, Samant RS. Spheroid-forming subpopulation of breast cancer cells demonstrates vasculogenic mimicry via hsa-miR-299-5p regulated de novo expression of osteopontin. J Cell Mol Med. 2010 Jun;14(6B):1693–706. doi: 10.1111/j.1582-4934.2009.00821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rittling SR, Chambers AF. Role of osteopontin in tumour progression. British journal of cancer. 2004;90:1877–81. doi: 10.1038/sj.bjc.6601839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denhardt DT, Guo X. Osteopontin: a protein with diverse functions. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 1993;7:1475–82. [PubMed] [Google Scholar]
- 13.Ma J, Meng Y, Kwiatkowski DJ, Chen X, Peng H, Sun Q. et al. Mammalian target of rapamycin regulates murine and human cell differentiation through STAT3/p63/Jagged/Notch cascade. The Journal of clinical investigation. 2010;120:103–14. doi: 10.1172/JCI37964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zha X, Hu Z, Ji S, Jin F, Jiang K, Li C. et al. NFkappaB up-regulation of glucose transporter 3 is essential for hyperactive mammalian target of rapamycin-induced aerobic glycolysis and tumor growth. Cancer letters. 2015;359:97–106. doi: 10.1016/j.canlet.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 15.Zhang H, Cicchetti G, Onda H, Koon HB, Asrican K, Bajraszewski N. et al. Loss of Tsc1/Tsc2 activates mTOR and disrupts PI3K-Akt signaling through downregulation of PDGFR. The Journal of clinical investigation. 2003;112:1223–33. doi: 10.1172/JCI17222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zha X, Wang F, Wang Y, He S, Jing Y, Wu X. et al. Lactate dehydrogenase B is critical for hyperactive mTOR-mediated tumorigenesis. Cancer research. 2011;71:13–8. doi: 10.1158/0008-5472.CAN-10-1668. [DOI] [PubMed] [Google Scholar]
- 17.Zhang H, Bajraszewski N, Wu E, Wang H, Moseman AP, Dabora SL. et al. PDGFRs are critical for PI3K/Akt activation and negatively regulated by mTOR. The Journal of clinical investigation. 2007;117:730–8. doi: 10.1172/JCI28984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun Q, Chen X, Ma J, Peng H, Wang F, Zha X. et al. Mammalian target of rapamycin up-regulation of pyruvate kinase isoenzyme type M2 is critical for aerobic glycolysis and tumor growth. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:4129–34. doi: 10.1073/pnas.1014769108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rangaswami H, Bulbule A, Kundu GC. Osteopontin: role in cell signaling and cancer progression. Trends in cell biology. 2006;16:79–87. doi: 10.1016/j.tcb.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 20.Shevde LA, Samant RS. Role of osteopontin in the pathophysiology of cancer. Matrix biology: journal of the International Society for Matrix Biology. 2014;37:131–41. doi: 10.1016/j.matbio.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iwenofu OH, Lackman RD, Staddon AP, Goodwin DG, Haupt HM, Brooks JS. Phospho-S6 ribosomal protein: a potential new predictive sarcoma marker for targeted mTOR therapy. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2008;21:231–7. doi: 10.1038/modpathol.3800995. [DOI] [PubMed] [Google Scholar]
- 22.Villena JA, Kralli A. ERRalpha: a metabolic function for the oldest orphan. Trends in endocrinology and metabolism: TEM. 2008;19:269–76. doi: 10.1016/j.tem.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schreiber SN, Emter R, Hock MB, Knutti D, Cardenas J, Podvinec M. et al. The estrogen-related receptor alpha (ERRalpha) functions in PPARgamma coactivator 1alpha (PGC-1alpha)-induced mitochondrial biogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:6472–7. doi: 10.1073/pnas.0308686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nature cell biology. 2002;4:648–57. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 25.Smith EM, Finn SG, Tee AR, Browne GJ, Proud CG. The tuberous sclerosis protein TSC2 is not required for the regulation of the mammalian target of rapamycin by amino acids and certain cellular stresses. The Journal of biological chemistry. 2005;280:18717–27. doi: 10.1074/jbc.M414499200. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Nicholatos J, Dreier JR, Ricoult SJ, Widenmaier SB, Hotamisligil GS. et al. Coordinated regulation of protein synthesis and degradation by mTORC1. Nature. 2014;513:440–3. doi: 10.1038/nature13492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kita Y, Natsugoe S, Okumura H, Matsumoto M, Uchikado Y, Setoyama T. et al. Expression of osteopontin in oesophageal squamous cell carcinoma. British journal of cancer. 2006;95:634–8. doi: 10.1038/sj.bjc.6603296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogbureke KU, Nikitakis NG, Warburton G, Ord RA, Sauk JJ, Waller JL. et al. Up-regulation of SIBLING proteins and correlation with cognate MMP expression in oral cancer. Oral oncology. 2007;43:920–32. doi: 10.1016/j.oraloncology.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 29.Matsuzaki H, Shima K, Muramatsu T, Ro Y, Hashimoto S, Shibahara T. et al. Osteopontin as biomarker in early invasion by squamous cell carcinoma in tongue. Journal of oral pathology & medicine: official publication of the International Association of Oral Pathologists and the American Academy of Oral Pathology. 2007;36:30–4. doi: 10.1111/j.1600-0714.2006.00493.x. [DOI] [PubMed] [Google Scholar]
- 30.Jin H, Valverde P, Chen J. Cloning of hamster osteopontin and expression distribution in normal tissues and experimentally induced oral squamous-cell carcinoma. Archives of oral biology. 2006;51:236–45. doi: 10.1016/j.archoralbio.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 31.Morgensztern D, McLeod HL. PI3K/Akt/mTOR pathway as a target for cancer therapy. Anti-cancer drugs. 2005;16:797–803. doi: 10.1097/01.cad.0000173476.67239.3b. [DOI] [PubMed] [Google Scholar]
- 32.Porta C, Paglino C, Mosca A. Targeting PI3K/Akt/mTOR Signaling in Cancer. Frontiers in oncology. 2014;4:64. doi: 10.3389/fonc.2014.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Populo H, Lopes JM, Soares P. The mTOR signalling pathway in human cancer. International journal of molecular sciences. 2012;13:1886–918. doi: 10.3390/ijms13021886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H. et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Current biology: CB. 2004;14:1296–302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 35.Jacinto E, Facchinetti V, Liu D, Soto N, Wei S, Jung SY. et al. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell. 2006;127:125–37. doi: 10.1016/j.cell.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 36.Sodek J, Ganss B, McKee MD. Osteopontin. Critical reviews in oral biology and medicine: an official publication of the American Association of Oral Biologists. 2000;11:279–303. doi: 10.1177/10454411000110030101. [DOI] [PubMed] [Google Scholar]
- 37.El-Tanani MK, Campbell FC, Kurisetty V, Jin D, McCann M, Rudland PS. The regulation and role of osteopontin in malignant transformation and cancer. Cytokine & growth factor reviews. 2006;17:463–74. doi: 10.1016/j.cytogfr.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 38.Denhardt DT, Noda M. Osteopontin expression and function: role in bone remodeling. Journal of cellular biochemistry Supplement. 1998;30-31:92–102. [PubMed] [Google Scholar]
- 39.Giguere V, Yang N, Segui P, Evans RM. Identification of a new class of steroid hormone receptors. Nature. 1988;331:91–4. doi: 10.1038/331091a0. [DOI] [PubMed] [Google Scholar]
- 40.Johnston SD, Liu X, Zuo F, Eisenbraun TL, Wiley SR, Kraus RJ. et al. Estrogen-related receptor alpha 1 functionally binds as a monomer to extended half-site sequences including ones contained within estrogen-response elements. Molecular endocrinology. 1997;11:342–52. doi: 10.1210/mend.11.3.9897. [DOI] [PubMed] [Google Scholar]
- 41.Vanacker JM, Delmarre C, Guo X, Laudet V. Activation of the osteopontin promoter by the orphan nuclear receptor estrogen receptor related alpha. Cell growth & differentiation: the molecular biology journal of the American Association for Cancer Research. 1998;9:1007–14. [PubMed] [Google Scholar]
- 42.El-Tanani M, Fernig DG, Barraclough R, Green C, Rudland P. Differential modulation of transcriptional activity of estrogen receptors by direct protein-protein interactions with the T cell factor family of transcription factors. The Journal of biological chemistry. 2001;276:41675–82. doi: 10.1074/jbc.M103966200. [DOI] [PubMed] [Google Scholar]