ABSTRACT
Fecal microbiota transplantation is best understood as an effective and inexpensive therapy for recurrent Clostridium difficile infection but fecal donor selection and screening should be periodically revised. Here, we review current recommendations for selection and screening of fecal donors for fecal microbiota transplantation. We recommend considering diabetes mellitus, prior cardiovascular events, and clinical healthcare exposure as fecal donor exclusion criteria until more is known about the association of these conditions with the human gut microbiome. We review the non-bacterial members of the human gut microbiome, associations of the gut microbiome with colorectal malignancies, the human gut resistome and how these may impact future donor screening recommendations. Collaboration between clinicians, clinical laboratory scientists, industry and regulatory agencies will be critically important for continued improvement in donor selection and screening.
KEYWORDS: antibiotic resistance, Clostridium difficile, FMT, fecal microbiota transplantation, gut microbial therapeutics, microbiome, resistome, stool donor screening
Introduction
The last decade has seen a dramatic expansion in understanding of the composition and function of the microbiota of the human gut. Advances in genetic sequencing techniques and bioinformatics approaches to analyze large amounts of data have advanced our capacity to query the relationship of the microbiome, the sum genetic material of the microbiota, to human health and disease. As more is understood about the influence of the human gut microbiota, basic science and clinical trial data should be rapidly incorporated to translate benefits and limit risk to patients through updated donor and stool screening for fecal microbiota transplantation (FMT).
FMT is best understood as an effective, inexpensive and apparently safe method for clinical enrichment of human gut microbiota to treat recurrent Clostridium difficile infection (RCDI). Risk of Clostridium difficile infection (CDI) is thought to be driven by loss of diversity of the gut microbiota, or dysbiosis, as a complication of antibiotic use. The negative impacts of CDI in the United States include an epidemiologic burden of disease of 434,000 infections and 29,000 deaths in 2011 and may contribute up to 4.8 billion in 2008-value adjusted US dollars.1,2 CDI has a high likelihood of recurrence, which can have diminishing cure rates when successive episodes are treated with additional antibiotics. The reported 90% cure rate for RCDI treated with FMT, with few reported adverse effects, underscores the need for rapid and continued improvement in understanding of this treatment.3-6
While the practice of preparing human stool to treat human disease has been traced back 1700 years, the benefits of FMT have only recently begun to be clarified in modern structured clinical trials.7 And though FMT is best understood as treatment of RCDI, interest in microbiota enrichment for other clinical indications is expanding. As of September 1, 2016, there are 197 clinical trials registered under the terms “FMT,” “fecal microbiota,” and “fecal microbiota transplantation” on www.clinicaltrials.gov for indications as varied as primary sclerosing cholangitis, non-alcoholic steatohepatitis (NASH), type II diabetes mellitus, irritable bowel syndrome, inflammatory bowel disease, hepatic encephalopathy and eradication of colonization by multi-drug resistant organisms (MDRO).8 Increasing numbers of academic medical centers have established FMT programs in an attempt to meet the demand for an effective, inexpensive treatment of RCDI. Still, expansion of these programs, not to mention home preparations of FMT guided by resources like YouTube instructional videos, has greatly outpaced evaluation of safety parameters and long-term treatment outcomes, which prompted the Food and Drug Administration (FDA) to regulate its use. While the FDA has released statements regarding donor screening and FMT for treatment of RCDI, they continue to exercise enforcement discretion, and some guidance remains in draft form.
There have been very few structured clinical trials and no published long-term safety data to inform optimal donor selection beyond expert opinion. In addition, there may be future changes in FMT regulation, including which indications require Investigational New Drug (IND) applications, oversight of donor selection, screening and stool banking practices.9,10 As an example of the importance of donor selection, outcomes for FMT for Inflammatory Bowel Disease (IBD) in particular may be sensitive to donor selection and the donor stool microbiota diversity.11-13
Here we review the various current recommendations for stool donor selection and screening for FMT product, donor screening test performance characteristics and validation issues of tests that were designed and validated for diarrheal disease. We also consider how donor testing may change to reflect evolving understanding of microbiome diversity indices and implications of associations with metabolism and the gut microbiome, cardiovascular disease, colorectal cancer and multi-drug resistant organisms on donor selection and screening.
The FDA and regulation in flux
The FDA announced in May of 2013 its intent to classify stool processed for FMT as a biologic agent and regulate fecal material as a drug used for treatment of human disease. Subsequently, the FDA issued revised guidance in response to feedback, indicating a decision to exercise enforcement discretion, which allows licensed healthcare professionals to administer FMT without an IND only for RCDI. By request of the FDA, a joint society recommendation for selection and screening of donors was released in 2013 as an open letter from the presidents of the Infectious Diseases Society of America (IDSA), American Society for Gastrointestinal Endoscopy (ASGE), North American Society for Pediatric Gastroenterology, Hepatology and Nutrition (NASPGHAN), American Gastroenterological Association (AGA) and American College of Gastroenterology (ACG).14 These joint society recommendations were largely informed by the foundational recommendations of the FMT Workgroup, which were published in 2011.15 FDA guidance for FMT remains in flux but ideally, their ultimate regulatory decisions would standardize donor selection and screening to reduce risk without overly limiting access.
Guidance from professional societies and the FDA suggest a preference for selecting stool donors who are known to the patient or providers. On the other hand, clinical trials and case reports show a general trend toward sourcing stool (which is often banked) from screened donors unknown to patients. Thus, overarching recommendations are not fully keeping in step with the apparent trend of clinical practice. Indeed, the most recent draft guidance for industry from the FDA seems to suggest that stool banking organizations like OpenBiome would not be included in the IND enforcement discretion exception and that they would be required to submit an IND for their operations. If this draft guidance is enforced and stool banks are not prepared, or if there is not a viable commercially manufactured alternative FMT product available, the burden of donor screening may be shifted to local institutions by default. Indeed, multiple authors have argued that instead of pursuit of a centralized manufacturing facility as would be seen in small molecule and non-FMT biologic therapeutics that FMT material should instead be regulated as a tissue more analogous to decentralized blood banking practices.16-18 Close collaboration between the FDA and leaders of infectious disease and gastroenterology professional societies is needed to ensure appropriate and relevant regulatory oversight while ensuring access to FMT.
Current donor selection and screening guidance
In their initial 2011 recommendations, Bakken et al outlined several key issues to consider when selecting donors. These recommendations and subsequent updates have used blood donor screening as a robust medicolegal analogy to fecal donor screening with donor questionnaires and laboratory testing.16,17,19 This informs much of current published guidance to use questionnaires to identify risk for diseases for which there are no valid laboratory tests (or for which there may be an undetectable early-stage or window-period) and to use appropriate diagnostics for diseases which can be tested.15 Since the FDA's announcement in 2013, there has been incremental revision in donor selection and screening practices with input from experts and donor screening data from public stool banking organizations.20,21 A summary of published donor screening protocols is shown in Table 1. Still, the relative merits of selecting donors known or unknown to patients, though a subject of much speculation, are largely undefined.22-24 Further research on the relative merits of donor selection and screening is needed.
Table 1.
Summarized Donor Screening Recommendations14,15,19,21
| Initial Screen | • Health questionnaire similar to that of AABB for blood donors |
| Consider: | |
| • Provider visit for history and physical examination | |
| Inclusion Criteria | • Age > 18 |
| • Feeling well at time of donation | |
| • Able to provide informed consent | |
| • Children may donate with parental consent and child's assent | |
| Exclusion Criteria | • High-risk behaviors: |
| o Injection or other illicit drug use | |
| o Known exposure with HIV, HBV or HCV infection in previous 12 months | |
| o Unprotected intercourse or intercourse with sex worker in previous 12 months | |
| o Tattoo or piercing within previous 6 months | |
| o Incarceration or history of incarceration | |
| • Risk factors for variant Cruetzfeldt-Jakob disease | |
| • Known current communicable disease | |
| • Any of the following in the previous 4 weeks: fever, vomiting, diarrhea or other symptoms of infection | |
| • Any of the following in the previous 8 weeks: vaccinations, injections or contact with a recipient of the smallpox vaccine | |
| • Any of the following in the previous 12 months: blood transfusion, accidental needle stick or blood exposure | |
| • International travel within previous 6 months to areas of high risk of travelers' diarrhea | |
| • Household members with active gastrointestinal infection | |
| • Receipt of antibiotics in the previous 3 months | |
| • History of intrinsic gastrointestinal disease, including inflammatory bowel disease, irritable bowel syndrome, chronic constipation, gastrointestinal malignancy or prior major gastrointestinal surgery or procedure | |
| • Strong family history of colorectal cancer involving 2 or more first-degree relatives | |
| • History of autoimmune or atopic illness | |
| • History of ongoing immunomodulatory therapy | |
| • History of other malignancy or ongoing chemotherapy | |
| • History of metabolic syndrome obesity, or moderate to severe malnutrition | |
| • History of chronic pain syndromes, neurologic or neurodevelopmental disorders | |
| Consider: | |
| • Diabetes mellitus | |
| • History of cardiovascular event or stroke | |
| Blood Testing | • HAV IgM |
| • HBV surface antigen | |
| • HCV antibody | |
| • HIV 1and2 immunoassay | |
| • Testing for syphilis (classical sequence or reverse sequence per local laboratory) | |
| Consider: | |
| • Human T-cell Lymphotrophic Virus (HTLV 1and2) | |
| • Epstein-Barr virus, JC Virus, BK Virus | |
| • Hepatitis B virus core antibody, Hepatitis B virus surface antibody | |
| • Strongyloides stercoralis, Schistosoma spp serologies | |
| Stool Testing | • Clostridium difficile* |
| • Routine bacterial culture for enteric pathogens (Salmonella, Shigella, Campylobacter and Escherichia coli serotype O157) | |
| • Ova and parasites examination, if travel history suggests | |
| Consider by donor or recipient characteristics: | |
| • Giardia | |
| • Cryptosporidium | |
| • Isospora and Cyclospora | |
| • Rotavirus | |
| • Norovirus | |
| • Listeria spp | |
| • Vibrio spp | |
| Possibly Consider: | |
| • Vancomycin-resistant Enterococcus culture | |
| • Methicillin-resistant Staphylococcus aureus culture | |
| • Helicobacter pylori | |
| • Dientamoeba fragilis | |
| • Blastocystis hominis | |
| • Entamoeba histolytica | |
| Frequency of Testing | • Initial laboratory screening should be within 4 weeks of stool donation |
| Consider: | |
| • Repeat screening if donor is still active after 4–8 weeks | |
| Additional Considerations | • Complete blood count, liver function tests, electrolytes and creatinine |
Note. HIV - human immunodeficiency virus, HAV - hepatitis A virus, HBV - hepatitis B virus, HCV - hepatitis C virus.
Off-label C. difficile toxin gene polymerase chain reaction testing is most commonly recommended, although off-label antigen, toxin enzyme immunoassay or toxigenic culture are also likely appropriate.
As concerns about transmission of infectious and classically non-infectious diseases related to the gut microbiome remain, mitigating harm to recipients has remained the primary driver of the current framework of strict exclusion criteria for donors. For example, many screening recommendations include testing asymptomatic donors for pathogens that have not been documented to be transmitted via the fecal-oral route. However, the limited number of harmful effects directly attributed to FMT to date may be an encouraging signal that current screening is effective for avoiding short-term harm. Reported serious adverse effects in published trials are most commonly due to procedures related to FMT rather than the transplanted fecal material itself. Wang et al recently published a systematic review of adverse events of FMT. They identified 50 relevant articles reporting outcomes for 1089 patients who were treated with FMT.25 The majority of articles reviewed included case series and case reports (and thus were without a control group for comparison), although the article did cite 4 randomized controlled trials. They found a 28.5% (310/1089) overall incidence of adverse effects after FMT for all indications, the majority of which were minor and included abdominal discomfort, diarrhea, transient fever, nausea, vomiting and constipation. Slightly higher rates of adverse effects were reported in upper routes of administration compared with lower, though there have been no clinical head-to-head trials to evaluate the effect of route of administration. Recent randomized controlled studies have also shown that rates of these minor adverse effects were similar in treatment and control groups, suggesting that they are not necessarily intrinsic to the FMT material.3,26
Currently, clinical trials that are approved by the FDA under an Investigational New Drug (IND) application are chiefly submitted as Phase I clinical trials and as their results are published, understanding of safety and attributable adverse effects of FMT will improve. Further, a national registry for FMT outcomes data was recently announced, which will also enhance short- and long-term safety end point data collection and feedback into appropriate donor selection and screening.27
Limitations of screening asymptomatic donors
There is consensus on the importance of screening donors for potentially transmissible conditions related to the gut microbiome. However, the use of laboratory tests beyond the indications for which they were clinically validated raises important issues. For example, use of tests in populations with lower prevalence than the original reference population could increase the number of false positive results. On the other hand, use of nucleic acid based diagnostics on formed stool instead of watery stool may decrease the sensitivity by decreasing the number of true carriers detected if there are inadequate primers for the sample being tested.28 Two recent reports of donor screening practice outcomes indicated that only 6–10% of screened donors were ultimately found to be eligible for stool donation.20,21 Detection of viruses (namely norovirus and rotavirus) and parasites of unclear or variable pathogenicity (Endolimax nana, Blastocystis hominis and Dientamoeba fragilis) were among the most common reasons for exclusion based on stool testing.20,21
Notably, protozoa are frequently found in stool of healthy individuals. In one study, 32.5% of healthy fecal donors were found to have Blastocystis by triple feces test (TFT), compared with 13.3% of patients with active ulcerative colitis.29 Much higher rates have been reported in other studies ranging from 50–100% Blastocystis colonization in healthy individuals in industrialized countries and Senegalese children respectively.30 Detection of Blastocystis by PCR has been associated with higher microbiota diversity and lower abundance of Enterobacteriaceae, and has inconsistently been associated with irritable bowel syndrome (IBS).31-33 However, the literature on this topic is inconsistent.
Screening stool for potential pathogens is a critically important step in reducing risk to FMT recipients. However, for C. difficile and other gastrointestinal pathogens more generally, polymerase chain reaction (PCR) based tests have not been validated for this purpose. Ruling out asymptomatic toxin-forming C. difficile colonization is of particular interest when identifying potential donors for FMT as a therapy for CDI. Studies of rates of asymptomatic colonization with C. difficile have suggested overall rates of 7.6% in otherwise healthy community-dwelling adults in Japan using culture followed by PCR toxin assay.36 In a more recent study from 2013–2015 in Montreal, 4.8% of patients screened were identified as carriers by PCR toxin assay performed on rectal swabs at time of hospitalization.37 While it is apparent that several healthy individuals will be asymptomatically colonized with C. difficile, efficient screening for this is not well described and there is no FDA approved test for this indication. Most assays currently in use were validated on loose or watery stools and not for the well-formed stools donors would be anticipated to produce.
Screening by toxigenic culture would be the primary alternative test of choice for screening of asymptomatic carriers, however it is impractical due to cost and processing times and is no longer performed by many clinical laboratories. Commercial C. difficile PCR assays are approximately 90% sensitive and 95% specific as compared with toxigenic culture in validation studies of patients with loose stool, with higher reported sensitivities compared with antigen testing.38,39 Notably, PCR diagnostics for symptomatic CDI may result in over diagnosis compared with toxin enzyme immunoassay (EIA) tests, though in the case of screening, this is likely an acceptable bias.40,41 Off-label rectal swab PCR testing for asymptomatic carriers has been used in at least one study.37 However, further clinical validation of C. difficile PCR testing is urgently needed to evaluate its use for screening of asymptomatic individuals.37
Similarly, PCR-based multiplex panels that have been developed for rapid identification of other bacterial, viral and parasitic pathogens in infectious diarrhea have been developed compared with culture of loose/watery stool specimens.42 While most donor screening recommendations suggest stool culture, antigen testing and microscopic examination for ova and parasites as a means of screening for asymptomatic carriage, a study to validate gastrointestinal pathogen panels for asymptomatic pathogen colonization could potentially reduce costs to stool banking programs and improve efficiency of screening.
Using non-toxigenic C. difficile strains as a means to outcompete toxigenic strains is being tested in clinical trials.26 Validated commercial assays testing alternative PCR targets that identify donors colonized with non-pathogenic strains of C. difficile, while ruling out asymptomatic carriage of other viral, bacterial and protozoal pathogens would be a helpful inclusion criterion for ideal stool donors. Understanding how to effectively screen donors will help to streamline logistical processes, while protecting the health of FMT recipients.
Antibiotic resistance screening and the resistome
Where the microbiome is the sum of microbiota genetic information, the sum of antibiotic resistance genes in a sample has been referred to as the resistome. Rapid molecular diagnostics for bacterial antibiotic susceptibility is an area of intense interest and active investigation. Many questions remain about the relative utility and most appropriate combination of phenotypic and genotypic assays for antibiotic resistance and integration with healthcare “big data” systems.43,44 Case reports and case series have shown that FMT can effectively reduce the overall number of antibiotic resistance genes as well as eradicate colonic colonization by MDRO.45,46 Some have advocated for a whole genome sequencing approach to screening for antibiotic resistance while others have indicated that the field is changing too rapidly to establish actionable/understandable platforms and that this approach is much more costly.47,48 One of the most attractive features of FMT for CDI is that it is a non-antibiotic based approach to treatment of an antibiotic-associated illness. Clinical trials are underway to evaluate an extension of this premise to decolonize patients with MDRO colonization for whom antibiotics may have selected for multi-drug resistance. Further study, perhaps including integration with microbiology laboratory phenotypic testing results, is needed to identify an ideal complement of molecular and phenotypic diagnostic screening; however, we recommend screening donors for carbapenem-resistant Enterobacteriaceae, vancomycin-resistant Enterobacteriaceae and frequent contact with healthcare as additional donor exclusion criteria.
Microbiome diversity indices
Beyond screening for infectious diseases and specific taxa associated with classically non-infectious diseases, clarification of normal gut microbiome diversity may allow for tests of deviation from this norm through microbiome diversity indices.49 As antibiotic use is a known risk factor for CDI, there is interest in characterizing the effects of specific antibiotics on microbiome diversity to more fully inform their clinical use. For example, fidaxomicin has been reported to have a microbiome-sparing aspect when used for treatment of C. difficile.50 Further, recent studies have investigated the specific impact of antibiotics on subsequent C. difficile infection.51-53 Microbiome diversity indices may provide a way to evaluate unexpected impacts of a drug and how they modulate risk of subsequent MDRO infection or colonization. Whether such measures of diversity will be operationalized with nucleic acid sequencing techniques, or simple surveillance culture results as a surrogate for dysbiosis, is still unclear.
Screening for non-bacterial members of the gut microbiota
Much of our understanding of the human gut microbiome has grown from the simultaneous emergence of increasingly affordable and efficient sequencing technology and the evaluation of the clinical application of FMT for CDI as it relates to dysbiosis. However, our grasp of roles that non-bacterial microbes like viruses, Archaea, fungi and parasites may play in human health and disease is still very limited.
Most work to date evaluating the viral constituents of the gut microbiome suggests that phages make up the majority of viruses in both healthy and dysbiotic individuals.54-56 Yet outcomes from these studies suggest differing models of the roles of viruses and range of normal diversity. Some work suggests that there are components of a shared healthy virome, while others suggest that increased phage diversity was seen in patients with IBD.56,57 It bears mention that many of these viral sequence reads correspond to lysogenic and temperate phages, which could theoretically be harbored in bacteria in a manner that is challenging to describe as distinct from the bacterial microbiota. Further, these studies may provide more questions than answers in clarifying whether the role of phages is that of predator to prey, engine of genetic recombination and diversity generation, or still some other purpose. Still, as none of these studies of the viromes of healthy individuals identified viruses known to infect human cells, and human gut viromes are not yet defined well enough to establish universally desirable profiles if they do exist, screening for asymptomatic shedding of known pathogenic viruses is likely the most appropriate manner to proceed.
Eukaryotes are also frequently found in stool of healthy individuals. Further complicating understanding of burden and occurrence of Blastocystis and Dientamoeba is their intermittent shedding in the gut, which limits testing validity.34,35 Neither Blastocystis nor Dientamoeba are clearly or consistently established as direct pathogens, though it is likely a reasonable practice to exclude donors with Blastocystis and Dientamoeba while the topic continues to be investigated, (including the possibility that the presence of Blastocystis may be beneficial) if alternative healthy donors are available.30 The investigational use of helminths with intent to modulate immunological pathways of the gut for IBD and other indications is beyond the scope of this review. Further study of the significance of protozoal detection in stool is needed. The investigational use of helminths with intent to modulate immunological pathways of the gut for IBD and other indications is beyond the scope of this review.
Fungi are frequently found in stool culture as well and are thought to also play important roles in gut microbiome function and disease. Similar to the observed diversity from bacterial sequencing reads compared with culture methods, several fungal genera and species are identifiable by molecular tests but are challenging or impossible to culture. Unfortunately, while rapid sequencing approaches for bacterial genetic reads have matured relatively quickly, fungal sequencing techniques are still in varied stages of development.58 Further, interpreting sequence reads is complicated because databases with known fungal sequence reads for assignment to operational taxonomic units (OTU), are not as robust as those for bacterial reads and may have more errors.58 Further study is needed before recommendations can be made about routine stool screening for fungal organisms.
Screening for obesity and metabolic diseases
In addition to screening for known pathogens, most fecal donor screening programs exclude donors above a body mass index (BMI) of > 30 kg/m2, metabolic syndrome or patients with moderate to severe malnutrition. A particularly striking example of the impact of donor selection was reported by Alang and Kelly, in which a patient was effectively treated for RCDI but became obese after FMT from an obese but otherwise healthy donor known to the patient.59 Though donors are screened for malnutrition and metabolic syndrome, it may be appropriate to exclude donors for other metabolic disorders as well, since studies have suggested that even some disorders not previously considered to be infectious are associated with the gut microbiome.
The influence of the microbiome on nutrition and metabolic disease is suggested by proximity of the microbiota to the intestinal tract, but only recently beginning to become more clear. Key influences and associations of the gut microbiome in health and disease are shown in Figure 1. In particular, associations with bacterial metabolites like blood levels of trimethylamine N-oxide (TMAO) and thrombosis, atherosclerosis and cardiovascular events appear to suggest a mechanism of cardiovascular risk modulation after animal protein ingestion.60,61 Further, insulin sensitivity may be improved after FMT and microbiome differences between malnourished children in Bangladesh may account for differences in growth rates after therapeutic food interventions.60,62 Many of these studies found strong associations between bacterial microbiota and objective clinical outcomes, however, parameters like TMAO levels are not routinely screened in most laboratories. Nor is it likely that testing for novel microbiota metabolites identified in the future will be feasible to scale up. Thus, routine laboratory screening for these factors is unlikely to be a practical or cost-effective endeavor. As the risk of transmission of these classically non-communicable diseases through donor feces is unclear, it may be prudent to exclude donors with known cardiovascular disease, stroke or history of diabetes mellitus in addition to exclusion of donors with obesity, metabolic syndrome or malnutrition.
Figure 1.
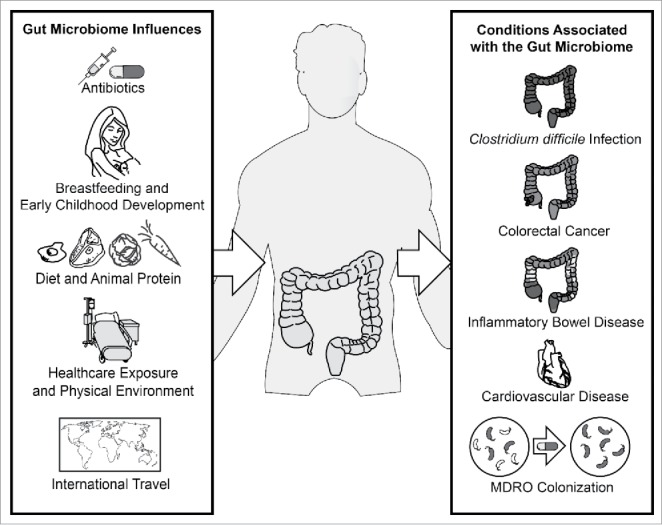
Established influences and health conditions associated with the human gut microbiome. MDRO denotes multi-drug resistant organism.
Future studies to evaluate FMT to reduce risk of these conditions should clearly describe their donor recruitment and screening strategies and ideally indicate if donors adhere to a vegetarian, vegan or other specific diet to aid evaluation of long-term outcomes given the time period over which many of these conditions are thought to develop.
Screening for oncogenic potential
Multiple pathways have been described by which the human gut microbiome could potentially influence the development of colorectal cancer, including virulence factors, local immunoregulatory interactions and toxic metabolites.63,64 The influence of the human gut microbiome in development of colorectal cancer echoes a theme of its influence in other conditions in apparent redundancy in operational taxonomic units (OTUs) that can result in similar functional results. Despite this redundancy in function in the microbiome, multiple studies have shown significant association of Fusobacterium spp with the development of colorectal cancer and others have suggested that Fusobacterium nucleatum is associated with clinical features included in staging and outcomes of colorectal cancer.65,66
In addition, Zackular et al developed a composite stool screen for colon lesions that improved pre-test probability testing and sensitivity for detection of colon adenomas and carcinomas. Still, it is not entirely clear if routine screening of stool will correlate with mucosal biofilm colonies, which have been associated with right-sided colorectal cancers in particular.67-69
There are currently insufficient data to operationalize a screening recommendation for microbiota-related oncogenic potential. For example, an argument could be made to consider restricting the age of fecal donors to below 50 y of age, beyond which colon cancer is known to increase in incidence. On the other hand, it is conceivable that a well-screened donor over this age may have a less oncogenically-risky microbiota and may be favorable for transplantation compared with a younger donor of unknown colorectal malignancy risk. As understanding of associations with malignancy and the gut microbiome improve, donor screening recommendations should continue to be updated.
Future directions for donor screening
Although there has been much speculation and enthusiasm about the influence of the gut microbiome on human health and disease, ongoing collaboration between clinical specialists, clinical laboratories, stool banking organizations, pharmaceutical companies and regulatory agencies will be of primary import. Much work remains on describing which components of the gut microbiome constitute keystone members. Alternatively, it may be the case that ecological principles of microbial community organization and signaling may be more important than specific keystone taxa. Still, improved understanding in healthy microbiome indices may allow opportunity for clinical interventions to improve microbiota resilience against insults. Further study of conditions that have been associated with the human gut microbiome will clarify which appear more likely to be causal, which should be considered in donor selection and screening and which may appropriate for further clinical trials of safety and efficacy.
Beyond selecting ideal donors for FMT for RCDI, in the future, it may be relevant to select specific donors for specific FMT indications by relative diversity in keystone populations. Long-term outcomes databases and centralized adverse effect reporting may inform best practices for donor selection, means of manufacturing fecal material for transplantation and optimal routes of administration. Long-term follow up in patients with more diverse comorbidities, including patients with cancer or who are otherwise immunocompromised may elucidate pharmacomicrobial interactions. Further clarification of healthy microbiome indices may streamline stool screens, lowering cost of screening and FMT overall, ultimately increasing access. Ongoing collaboration with clinicians, industry and regulatory agencies and feedback to donor selection and screening processes will likely prove to be the most important factors in optimizing outcomes while reducing risk to patients.
Abbreviations
- ACG
American College of Gastroenterology
- AGA
American Gastroenterological Association
- ASGE
American Society for Gastrointestinal Endoscopy
- CDI
Clostridium difficile Infection
- EIA
Enzyme immunoassay
- FDA
US. Food and Drug Administration
- FMT
Fecal Microbiota Transplant or Transplantation
- IBD
Inflammatory Bowel Disease
- IBS
Irritable Bowel Syndrome
- IDSA
Infectious Diseases Society of America
- IND
Investigational New Drug
- MDRO
Multi-Drug Resistant Organisms
- NASH
Non-Alcoholic Steatohepatitis
- NASPGHAN
North American Society for Pediatric Gastroenterology, Hepatology and Nutrition
- OTU
Operational Taxonomic Unit
- PCR
Polymerase Chain Reaction
- PFGE
Pulsed-field gel electrophoresis
- RCDI
Recurrent Clostridium difficile Infection
- TFT
Triple Feces Test
- TMAO
Trimethylamine-N-Oxide
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Funding
Supported by the National Center for Advancing Translational Sciences of the National Institutes of Health under Award number UL1TR000454.
References
- [1].Dubberke ER, Olsen MA. Burden of clostridium difficile on the healthcare system. Clin Infect Dis 2012; 55:88-92; http://dx.doi.org/ 10.1093/cid/cis335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, Farley MM, Holzbauer SM, Meek JI, Phipps EC, et al.. Burden of Clostridium difficile infection in the United States. N Engl J Med [Internet] 2015; 372:825-34. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25714160; PMID:25714160; http://dx.doi.org/ 10.1056/NEJMoa1408913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kelly CR, Khoruts A, Staley C, Sadowsky MJ, Abd M, Alani M, Bakow B, Curran P, McKenney J, Tisch A, et al.. Effect of fecal microbiota transplantation on recurrence in multiply recurrent clostridium difficile infection. Ann Intern Med [Internet] 2016; 165(9):609-616; Available from: http://annals.org/article.aspx?; PMID:27547925; http://dx.doi.org/ 10.7326/M16-0271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, Visser CE, Kuijper EJ, Bartelsman JFWM, Tijssen JGP, et al.. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med [Internet] 2013; 368:407-15. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23323867; PMID:23323867; http://dx.doi.org/ 10.1056/NEJMoa1205037 [DOI] [PubMed] [Google Scholar]
- [5].Cammarota G, Masucci L, Ianiro G, Bibbò S, Dinoi G, Costamagna G, Sanguinetti M, Gasbarrini A. Randomised clinical trial: faecal microbiota transplantation by colonoscopy vs. vancomycin for the treatment of recurrent Clostridium difficile infection. Aliment Pharmacol Ther [Internet] 2015; 41(9):835-43; [Epub ahead of print]. Available from: http://doi.wiley.com/10.1111/apt.13144; PMID:25728808; http://dx.doi.org/ 10.1111/apt.13144 [DOI] [PubMed] [Google Scholar]
- [6].Gough E, Shaikh H, Manges AR. Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent clostridium difficile infection. Clin Infect Dis 2011; 53:994-1002; PMID:22002980; http://dx.doi.org/ 10.1093/cid/cir632 [DOI] [PubMed] [Google Scholar]
- [7].Zhang F, Luo W, Shi Y, Fan Z, Ji G. Should we standardize the 1,700-year-old fecal microbiota transplantation? Am J Gastroenterol [Internet] 2012; 107:1755; author reply p.1755-p.6. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23160295%5Cnhttp://dx.doi.org/10.1038/ajg.2012.251; PMID:23160295; http://dx.doi.org/ 10.1038/ajg.2012.251 [DOI] [PubMed] [Google Scholar]
- [8].National Library of Medicine (US). ClinicalTrials.gov [Internet]. 2016 [cited 2016 Sep 1]; Available from: https://clinicaltrials.gov/ct2/results?termDfecalCtransplant&SearchDSearch
- [9].FDA Guidance for industry enforcement policy regarding investigational new drug requirements for use of fecal microbiota for transplantation to treat clostridium difficile infection not responsive to standard therapies [Internet]. 2014. [cited 2016 Sep 1]; Available from: http://www.fda.gov/downloads/BiologicsBloodVaccines/GuidanceComplianceRegulatoryInformation/Guidances/Vaccines/UCM387255.pdf
- [10].FDA Enforcement policy regarding investigational new drug requirements for use of fecal microbiota for transplantation to treat clostridium difficile infection not responsive to standard therapies. U.S Food and Drug Administration [Internet]. 2016. Available from: http://www.fda.gov/BiologicsBloodVaccines/GuidanceComplianceRegulatoryInformation/Guidances/Vaccines/ucm361379.html [Google Scholar]
- [11].Vermeire S, Joossens M, Verbeke K, Wang J, Machiels K, Sabino J, Ferrante M, Van Assche G, Rutgeerts P, Raes J. Donor species richness determines faecal microbiota transplantation success in inflammatory bowel disease. J Crohns Colitis [Internet] 2016; 10:387-94. Available from: http://ecco-jcc.oxfordjournals.org/; PMID:26519463; http://dx.doi.org/ 10.1093/ecco-jcc/jjv203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Moayyedi P, Surette MG, Kim PT, Libertucci J, Wolfe M, Onischi C, Armstrong D, Marshall JK, Kassam Z, Reinisch W, et al.. Fecal microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled trial. Gastroenterology [Internet] 2015; 149:102-109.e6. Available from: http://dx.doi.org/ 10.1053/j.gastro.2015.04.001; PMID:25857665; http://dx.doi.org/ 10.1053/j.gastro.2015.04.001 [DOI] [PubMed] [Google Scholar]
- [13].Rossen NG, Fuentes S, van der Spek MJ, Tijssen JG, Hartman JHA, Duflou A, Löwenberg M, van den Brink GR, Mathus-Vliegen EMH, de Vos WM, et al.. Findings from a randomized controlled trial of fecal transplantation for patients with Ulcerative Colitis. Gastroenterology [Internet] 2015; 149:110-118.e4. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25836986; PMID:25836986; http://dx.doi.org/ 10.1053/j.gastro.2015.03.045 [DOI] [PubMed] [Google Scholar]
- [14].Relman D, Vender RJ, Rustgi AK, Wang KK, Bousvaros A. Current consensus guidance on donor screening and stool testing for FMT. Bethesda, MD: American Gastroenterological Association; 2013. Available from: https://www.gastro.org/research/Joint_Society_FMT_Guidance.pdf [Google Scholar]
- [15].Bakken JS, Borody T, Brandt LJ, Brill JV, Demarco DC, Franzos MA, Kelly C, Khoruts A, Louie T, Martinelli LP, et al.. Treating Clostridium difficile infection with fecal microbiota transplantation. Clin Gastroenterol Hepatol [Internet] 2011; 9:1044-9. Available from: http://dx.doi.org/ 10.1016/j.cgh.2011.08.014; PMID:21871249; http://dx.doi.org/ 10.1016/j.cgh.2011.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Sachs RE, Edelstein CA. Ensuring the safe and effective FDA regulation of fecal microbiota transplantation. J Law Biosci [Internet] 2015; 2:396-415. Available from: http://jlb.oxfordjournals.org/lookup/doi/10.1093/jlb/lsv032; PMID:27774199; http://dx.doi.org/ 10.1093/jlb/lsv032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Edelstein CA, Kassam Z, Daw J, Smith MB, Kelly CR. The regulation of fecal microbiota for transplantation: An international perspective for policy and public health. Clin Res Regul Aff [Internet] 2015; 32:101-9. Available from: http://ovidsp.ovid.com/ovidweb.cgi?T=JS&CSC=Y&NEWS=N&PAGE=fulltext&D=emed13&AN=2015341771; http://dx.doi.org/ 10.3109/10601333.2015.1046602 [DOI] [Google Scholar]
- [18].Smith MB, Kelly C, Alm EJ. Policy: How to regulate faecal transplants. Nature [Internet] 2014; 506:290-1. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24558658; PMID:24558658; http://dx.doi.org/ 10.1038/506290a [DOI] [PubMed] [Google Scholar]
- [19].Kelly CR, Kahn S, Kashyap P, Laine L, Rubin D, Atreja A, Moore T, Wu G. Update on Fecal Microbiota Transplantation 2015: Indications, methodologies, mechanisms, and outlook. Gastroenterology [Internet] 2015; 149:223-37. Available from: http://dx.doi.org/ 10.1053/j.gastro.2015.05.008; PMID:25982290; http://dx.doi.org/ 10.1053/j.gastro.2015.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Burns LJ, Dubois N, Smith MB, Mendolia GM, Burgess J, Edelstein C, Noh A, Alm E, Kassam Z. 499 donor recruitment and eligibility for fecal microbiota transplantation: results from an international public stool bank. Gastroenterology [Internet] 2015; 148:S-96-S-97. Available from: http://linkinghub.elsevier.com/retrieve/pii/S0016508515303310 [Google Scholar]
- [21].Paramsothy S, Borody TJ, Lin E, Finlayson S, Walsh AJ, Samuel D, van den Bogaerde J, Leong RWL, Connor S, Ng W, et al.. Donor Recruitment for Fecal Microbiota Transplantation. Inflamm Bowel Dis [Internet] 2015; 21:1600-6. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26070003; PMID:26070003; http://dx.doi.org/ 10.1097/MIB.0000000000000405 [DOI] [PubMed] [Google Scholar]
- [22].Drekonja D, Reich J, Gezahegn S, Greer N, Shaukat A, MacDonald R, Rutks I, Wilt TJ. Fecal microbiota transplantation for clostridium difficile infection: A systematic review. Ann Intern Med [Internet] 2015; 162:630-8. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25938992; PMID:25938992; http://dx.doi.org/ 10.7326/M14-2693 [DOI] [PubMed] [Google Scholar]
- [23].Gathe JC, Diejomaoh EM, Mayberry CC, Clemmons JB. Fecal transplantation for Clostridium Difficile-“All stool may not be created equal.” J Int Assoc Provid AIDS Care [Internet] 2016; 15:107-8. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26821578; PMID:26821578; http://dx.doi.org/ 10.1177/2325957415627695 [DOI] [PubMed] [Google Scholar]
- [24].Merenstein D, El-nachef N, Lynch S V. Fecal microbial therapy: promises and pitfalls. J Pediatr Gastroenterol Nutr [Internet]. 2014 Aug; 59(2):157-61. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24796803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wang S, Xu M, Wang W, Cao X, Piao M, Khan S, Yan F, Cao H, Wang B. Systematic review: Adverse events of fecal microbiota transplantation. PLoS One [Internet] 2016; 11:e0161174. Available from: http://dx.plos.org/10.1371/journal.pone.0161174; PMID:27529553; http://dx.doi.org/ 10.1371/journal.pone.0161174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Gerding DN, Meyer T, Lee C, Cohen SH, Murthy UK, Poirier A, Van Schooneveld TC, Pardi DS, Ramos A, Barron MA, et al.. Administration of Spores of Nontoxigenic Clostridium difficile Strain M3 for prevention of recurrent C difficile infection: A randomized clinical trial. Jama [Internet] 2015; 313:1719-27. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25942722; PMID:25942722; http://dx.doi.org/ 10.1001/jama.2015.3725 [DOI] [PubMed] [Google Scholar]
- [27].American Gastroenterological Association. AGA Establishes NIH-Funded Registry to Track Fecal Microbiota Transplants. [Internet]. Bethesda (MD): American Gastroenterological Association; 2016 Aug 4 [cited 2016 Aug 26]
- [28].Guh A, McDonald L. Containing what lies under thewaterline invited commentary invited commentary active surveillance and isolation of asymptomatic carriers of Clostridium difficile at hospital admission. JAMA Intern Med 2016; 176:1-2; PMID:27479247; http://dx.doi.org/ 10.1001/jamainternmed.2016.1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Rossen NG, Bart A, Verhaar N, van Nood E, Kootte R, de Groot PF, D'Haens GR, Ponsioen CY, van Gool T. Low prevalence of Blastocystis sp. in active ulcerative colitis patients. Eur J Clin Microbiol Infect Dis [Internet] 2015; 34:1039-44. Available from: http://ecco-jcc.oxfordjournals.org/cgi/doi/10.1016/S1873-9946(14)60787-X; PMID:25680316; http://dx.doi.org/ 10.1007/s10096-015-2312-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Andersen LOB. Blastocystis in Health and Disease Are we moving from a clinical to a public health perspective? PubMed Commons. J Clin Microbiol 2016; 54:524/8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Audebert C, Even G, Cian A, Blastocystis Investigation Group, Loywick A, Merlin S, Viscogliosi E, Chabé M. Colonization with the enteric protozoa Blastocystis is associated with increased diversity of human gut bacterial microbiota. Sci Rep [Internet] 2016; 6:25255. Available from: http://www.nature.com/articles/srep25255; PMID:27147260; http://dx.doi.org/ 10.1038/srep25255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Nourrisson C, Scanzi J, Pereira B, NkoudMongo C, Wawrzyniak I, Cian A, Viscogliosi E, Livrelli V, Delbac F, Dapoigny M, et al.. Blastocystis is associated with decrease of fecal microbiota protective bacteria: comparative analysis between patients with irritable bowel syndrome and control subjects. PLoS One [Internet] 2014; 9:e111868. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4218853&tool=pmcentrez&rendertype=abstract; PMID:25365580; http://dx.doi.org/ 10.1371/journal.pone.0111868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Krogsgaard LR, Engsbro AL, Stensvold CR, Nielsen HV, Bytzer P. The prevalence of intestinal parasites is not greater among individuals with irritable bowel syndrome: A population-based case-control study. Clin Gastroenterol Hepatol 2015; 13:507-513.e2; PMID:25229421; http://dx.doi.org/ 10.1016/j.cgh.2014.07.065 [DOI] [PubMed] [Google Scholar]
- [34].Vennila GD, Suresh Kumar G, Khairul Anuar A, Rajah S, Saminathan R, Sivanandan S, Ramakrishnan K. Irregular shedding of Blastocystis hominis. Parasitol Res [Internet] 1999. [cited 2016 Nov 10]; 85:162-4. Available from: http://www.ncbi.nlm.nih.gov/pubmed/9934969; PMID:9934969; http://dx.doi.org/ 10.1007/s004360050528 [DOI] [PubMed] [Google Scholar]
- [35].Stark D, Barratt J, Roberts T, Marriott D, Harkness J, Ellis J. A review of the clinical presentation of dientamoebiasis. Am J Trop Med Hyg [Internet] 2010; 82:614-9. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2844584&tool=pmcentrez&rendertype=abstract; PMID:20348509; http://dx.doi.org/ 10.4269/ajtmh.2010.09-0478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Furuya-Kanamori L, Marquess J, Yakob L, Riley T V, Paterson DL, Foster NF, Huber CA, Clements ACA. Asymptomatic Clostridium difficile colonization: epidemiology and clinical implications. BMC Infect Dis [Internet] 2015; 15:516. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4647607&tool=pmcentrez&rendertype=abstract; PMID:26573915; http://dx.doi.org/ 10.1186/s12879-015-1258-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Longtin Y, Paquet-Bolduc B, Gilca R, Garenc C, Fortin E, Longtin J, Trottier S, Gervais P, Roussy J-F, Lévesque S, et al.. Effect of Detecting and Isolating Clostridium difficile Carriers at Hospital Admission on the Incidence of C difficile Infections: A Quasi-Experimental Controlled Study. JAMA Intern Med [Internet] 2016; 176:796-804. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27111806; PMID:27111806; http://dx.doi.org/ 10.1001/jamainternmed.2016.0177 [DOI] [PubMed] [Google Scholar]
- [38].Stamper PD, Alcabasa R, Aird D, Babiker W, Wehrlin J, Ikpeama I, Carroll KC. Comparison of a commercial real-time PCR assay for tcdB detection to a cell culture cytotoxicity assay and toxigenic culture for direct detection of toxin-producing Clostridium difficile in clinical samples. J Clin Microbiol 2009; 47:373-8; PMID:19073875; http://dx.doi.org/ 10.1128/JCM.01613-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Babady NE, Stiles J, Ruggiero P, Khosa P, Huang D, Shuptar S, Kamboj M, Kiehn TE. Evaluation of the Cepheid Xpert Clostridium difficile Epi assay for diagnosis of Clostridium difficile infection and typing of the NAP1 strain at a cancer hospital. J Clin Microbiol [Internet] 2010; 48:4519-24. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20943860; PMID:20943860; http://dx.doi.org/ 10.1128/JCM.01648-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Polage CR, Gyorke CE, Kennedy MA, Leslie JL, Chin DL, Wang S, Nguyen HH, Huang B, Tang YW, Lee LW, et al.. Overdiagnosis of Clostridium difficile Infection in the Molecular Test Era. JAMA Intern Med [Internet] 2015; 175:1-10. Available from: http://archinte.jamanetwork.com/article.aspx?articleid=2434732; http://dx.doi.org/ 10.1001/jamainternmed.2015.4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Koo HL, Van JN, Zhao M, Ye X, Revell PA, Jiang Z-D, Grimes CZ, Koo DC, Lasco T, Kozinetz CA, et al.. Real-time polymerase chain reaction detection of asymptomatic Clostridium difficile colonization and rising C. difficile-associated disease rates. Infect Control Hosp Epidemiol [Internet] 2014; 35:667-73. Available from: http://search.ebscohost.com/login.aspx?direct=true&db=jlh&AN=2012573261&site=ehost-live%5Cnhttp://www.jstor.org.ezp2.lib.umn.edu/stable/pdfplus/10.1086/676433.pdf; PMID:24799643; http://dx.doi.org/ 10.1086/676433 [DOI] [PubMed] [Google Scholar]
- [42].Buss SN, Leber A, Chapin K, Fey PD, Bankowski MJ, Jones MK, Rogatcheva M, Kanack KJ, Bourzac KM. Multicenter evaluation of the BioFire FilmArray gastrointestinal panel for etiologic diagnosis of infectious gastroenteritis. J Clin Microbiol 2015; 53:915-25; PMID:25588652; http://dx.doi.org/ 10.1128/JCM.02674-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Evans SR, Hujer AM, Jiang H, Hujer KM, Hall T, Marzan C, Jacobs MR, Sampath R, Ecker DJ, Manca C, et al.. Rapid molecular diagnostics, antibiotic treatment decisions, and developing approaches to inform empiric therapy: PRIMERS I and II. Clin Infect Dis [Internet] 2016; 62:181-9. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26409063; PMID:26409063; http://dx.doi.org/ 10.1093/cid/civ837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Pak TR, Kasarskis A. How Next-generation sequencing and multiscale data analysis will transform infectious disease management. Clin Infect Dis [Internet] 2015; 61:civ670 Available from: http://cid.oxfordjournals.org/lookup/doi/10.1093/cid/civ670; http://dx.doi.org/ 10.1093/cid/civ670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Crum-Cianflone NF, Sullivan E, Ballon-Landa G. Fecal microbiota transplantation and successful resolution of multidrug-resistant-organism colonization. J Clin Microbiol 2015; 53:1986-9; PMID:25878340; http://dx.doi.org/ 10.1128/JCM.00820-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Millan B, Park H, Hotte N, Mathieu O, Burguiere P, Tompkins TA, Kao D, Madsen KL. Fecal microbial transplants reduce antibiotic-resistant genes in patients with recurrent Clostridium difficile infection. Clin Infect Dis [Internet] 2016:1-9. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27025836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Lesho EP, Clifford RJ. For rapid molecular detection, Why not a whole genome approach? Clin Infect Dis [Internet] 2016; 63:570-1. Available from: http://cid.oxfordjournals.org/lookup/doi/10.1093/cid/ciw332; PMID:27225237; http://dx.doi.org/ 10.1093/cid/ciw332 [DOI] [PubMed] [Google Scholar]
- [48].Evans S, Kreiswirth B, Fowler V, Chambers H, Patel R, Hujer AM, Perez F, Bonomo RA. Reply to Lesho and Clifford. Clin Infect Dis [Internet] 2016; 63:571-2. Available from: http://cid.oxfordjournals.org/lookup/doi/10.1093/cid/ciw336; PMID:27225238; http://dx.doi.org/ 10.1093/cid/ciw336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Halpin AL, McDonald LC. Editorial Commentary: The dawning of microbiome remediation for addressing antibiotic resistance. Clin Infect Dis [Internet] 2016; 62:1487-8. Available from: http://cid.oxfordjournals.org/lookup/doi/10.1093/cid/ciw187; PMID:27025827; http://dx.doi.org/ 10.1093/cid/ciw187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Louie TJ, Cannon K, Byrne B, Emery J, Ward L, Eyben M, Krulicki W. Fidaxomicin preserves the intestinal microbiome during and after treatment of clostridium difficile infection (CDI) and reduces both toxin reexpression and recurrence of CDI. Clin Infect Dis 2012; 55:132-42; http://dx.doi.org/ 10.1093/cid/cis338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Brown KA, Khanafer N, Daneman N, Fisman DN. Meta-analysis of antibiotics and the risk of community-associated Clostridium difficile infection. Antimicrob Agents Chemother 2013; 57:2326-32; PMID:23478961; http://dx.doi.org/ 10.1128/AAC.02176-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Langdon A, Crook N, Dantas G. The effects of antibiotics on the microbiome throughout development and alternative approaches for therapeutic modulation. Genome Med [Internet] 2016; 8:39. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4831151&tool=pmcentrez&rendertype=abstract; PMID:27074706; http://dx.doi.org/ 10.1186/s13073-016-0294-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Knecht H, Neulinger SC, Heinsen FA, Knecht C, Schilhabel A, Schmitz RA, Zimmermann A, dos Santos VM, Ferrer M, Rosenstiel PC, et al.. Effects of β-lactam antibiotics and fluoroquinolones on human gut microbiota in relation to Clostridium difficile associated diarrhea. PLoS One [Internet] 2014; 9:e89417. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24586762; PMID:24586762; http://dx.doi.org/ 10.1371/journal.pone.0089417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F, Gordon JI. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature [Internet] 2010; 466:334-8. Available from: http://dx.doi.org/ 10.1038/nature09199; PMID:20631792; http://dx.doi.org/ 10.1038/nature09199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Minot S, Bryson A. Rapid evolution of the human gut virome. Proc Natl Acad Sci U S A [Internet] 2013; 110:12450-5. Available from: http://www.pnas.org/content/110/30/12450.short [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Manrique P, Bolduc B, Walk ST, van der Oost J, de Vos WM, Young MJ. Healthy human gut phageome. Proc Natl Acad Sci [Internet] 2016; 113(37):10400-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Norman JM, Handley SA, Baldridge MT, Droit L, Liu CY, Keller BC, Kambal A, Monaco CL, Zhao G, Fleshner P, et al.. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell [Internet] 2015; 160:447-60. Available from: http://dx.doi.org/ 10.1016/j.cell.2015.01.002; PMID:25619688; http://dx.doi.org/ 10.1016/j.cell.2015.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Cui L, Morris A, Ghedin E. The human mycobiome in health and disease. Genome Med 2013; 5:1-12; PMID:23311897; http://dx.doi.org/ 10.1186/gm467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Alang N, Kelly CR. Weight gain after fecal microbiota transplantation. Open forum Infect Dis [Internet] 2015; 2:ofv004. Available from: http://ofid.oxfordjournals.org/content/early/2014/06/01/ofid.ofu038.short; PMID:26034755; http://dx.doi.org/ 10.1093/ofid/ofv004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Senthong V, Wang Z, Li XS, Fan Y, Wu Y, Wilson Tang WH, Hazen SL. Intestinal Microbiota-Generated Metabolite Trimethylamine- N- Oxide and 5-year mortality risk in stable coronary artery disease: The contributory role of intestinal microbiota in a COURAGE-like patient cohort. J Am Heart Assoc [Internet] 2016; 5:e002816. Available from: http://jaha.ahajournals.org/lookup/doi/10.1161/JAHA.115.002816; PMID:27287696; http://dx.doi.org/ 10.1161/JAHA.115.002816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Zhu W, Gregory JC, Org E, Buffa JA, Gupta N, Wang Z, Li L, Fu X, Wu Y, Mehrabian M, et al.. Gut Microbial Metabolite TMAO Enhances Platelet Hyperreactivity and Thrombosis Risk. Cell [Internet] 2016; 165:111-24. Available from: http://dx.doi.org/ 10.1016/j.cell.2016.02.011; PMID:26972052; http://dx.doi.org/ 10.1016/j.cell.2016.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Vrieze A, Van Nood E, Holleman F, Salojärvi J, Kootte RS, Bartelsman JFWM, Dallinga-Thie GM, Ackermans MT, Serlie MJ, Oozeer R, et al.. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology [Internet] 2012; 143:913-916.e7. Available from: http://dx.doi.org/ 10.1053/j.gastro.2012.06.031; PMID:22728514; http://dx.doi.org/ 10.1053/j.gastro.2012.06.031 [DOI] [PubMed] [Google Scholar]
- [63].Sears CL, Garrett WS. Microbes, microbiota, and colon cancer. Cell Host Microbe [Internet] 2014; 15:317-28. Available from: http://dx.doi.org/ 10.1016/j.chom.2014.02.007; PMID:24629338; http://dx.doi.org/ 10.1016/j.chom.2014.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Dejea CM, Wick EC, Hechenbleikner EM, White JR, Mark Welch JL, Rossetti BJ, Peterson SN, Snesrud EC, Borisy GG, Lazarev M, et al.. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc Natl Acad Sci U S A [Internet] 2014; 111:18321-6. Available from: http://www.pnas.org/content/111/51/18321; PMID:25489084; http://dx.doi.org/ 10.1073/pnas.1406199111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Zackular JP, Rogers MAM, Ruffin MT, Schloss PD. The human gut microbiome as a screening tool for colorectal cancer. Cancer Prev Res 2014; 7:1112-21; http://dx.doi.org/ 10.1158/1940-6207.CAPR-14-0129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Drewes JL, Housseau F, Sears CL. Sporadic colorectal cancer: microbial contributors to disease prevention, development and therapy. Br J Cancer [Internet] 2016; 115:273-80. Available from: http://www.nature.com/doifinder/10.1038/bjc.2016.189; PMID:27380134; http://dx.doi.org/ 10.1038/bjc.2016.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Huse SM, Young VB, Morrison HG, Antonopoulos DA, Kwon J, Dalal S, Arrieta R, Hubert NA, Shen L, Vineis JH, et al.. Comparison of brush and biopsy sampling methods of the ileal pouch for assessment of mucosa-associated microbiota of human subjects. Microbiome [Internet] 2014; 2:5. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3931571&tool=pmcentrez&rendertype=abstract; PMID:24529162; http://dx.doi.org/ 10.1186/2049-2618-2-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science [Internet] 2005; 308:1635-8. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15831718; PMID:15831718; http://dx.doi.org/ 10.1126/science.1110591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Mira-Pascual L, Cabrera-Rubio R, Ocon S, Costales P, Parra A, Suarez A, Moris F, Rodrigo L, Mira A, Collado MC. Microbial mucosal colonic shifts associated with the development of colorectal cancer reveal the presence of different bacterial and archaeal biomarkers. J Gastroenterol [Internet] 2015; 50:167-79. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24811328; PMID:24811328; http://dx.doi.org/ 10.1007/s00535-014-0963-x [DOI] [PubMed] [Google Scholar]


