Abstract
Metastatic castration resistant prostate cancer (CRPC) is associated with substantial clinical, pathologic, and molecular heterogeneity. Most tumors remain driven by androgen receptor (AR) signaling, which has clinical implications for patient selection for AR-directed approaches. However, histologic and clinical resistance phenotypes can emerge after AR-inhibition, in which the tumors become less dependent on the AR. In this review we discuss prostate cancer variants including neuroendocrine (NEPC) and aggressive variant (AVPC) prostate cancers and their clinical implications. Improvements in the understanding of the biologic mechanisms and molecular features underlying prostate cancer variants may help prognostication and facilitate the development of novel therapeutic approaches for subclasses of patient with CRPC.
Keywords: castration-resistant prostate cancer, neuroendocrine prostate cancer, small cell prostate cancer, aggressive variant, androgen indifference, androgen independent prostate cancer
Introduction
Prostate cancer arises as an androgen driven disease [1]. Therefore the mainstay of systemic therapy for patients with advanced disease is androgen deprivation therapy [2]. However despite significant responses, nearly all patients ultimately progress and castration resistance ensues [3]. Castration resistant prostate cancer (CRPC) therefore represents a stage in the continuum of the disease. CRPC is defined by consecutive rises in serum prostate specific antigen (PSA) levels or/and progression of metastatic spread in the setting of castrate levels of testosterone [4, 5]. Despite recent advances in the treatment of patients progressing with metastatic CRPC, average survival remains approximately three years [6]. Identification of more effective and/or precise therapeutic approaches is a clinical unmet need.
Disease progression in CRPC is most commonly driven by reactivation of AR signaling [7]. This has led to the clinical development and approval of potent AR-targeted therapies (ie., the cytochrome P450 17 inhibitor abiraterone acetate (ABI) and the AR antagonist enzalutamide (ENZ)) and several others in clinical development [8] for patients with metastatic CRPC. Disease progression after potent AR-therapies is still mediated by activated AR signaling in the majority of cases, and this can occur through secondary alterations involving the AR gene (eg., amplification or activating point mutations, AR splice variants), AR bypass or crosstalk mechanisms (eg,. glucocorticoid receptor, PI3K/AKT pathway activation), or other means (Figure). It has been increasingly recognized that a subset of CRPC tumors evade therapy through another mechanism in which the tumors become less dependent on AR signaling (ie., “androgen indifferent”), and this is associated with lineage plasticity, loss of luminal markers, and acquisition of neuroendocrine features (Figure).
Figure 1. Schematic illustrating the three main classes of treatment resistance to AR targeted therapies in prostate cancer.
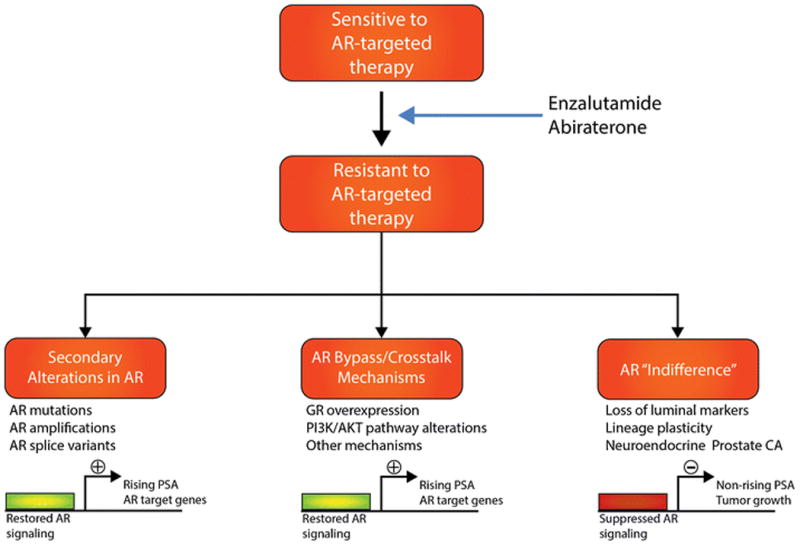
Other approved therapeutic options for metastatic CRPC with demonstrated survival benefit in randomized Phase 3 trials include taxane chemotherapies (docetaxel and cabazitaxel), autologous cell-based immunotherapy with sipuleucel-T, and the radioisotope 223radium [9]. Although these agents have significantly improved outcomes and quality of life for patients, there can be wide clinical variability in individual response between patients. Therefore there is a growing need for biomarkers to help improve patient selection and inform sequencing of systemic therapies [9]. Further, emerging therapeutics such as PARP inhibitors for CRPC have been shown to benefit only subsets of patients in early phase trials (eg., DNA repair alterations in the case of PARP [10]), highlighting the need for predictive biomarkers.
One challenge in the development of molecular biomarkers for advanced prostate cancer is the availability of tumor tissue [11]. Metastatic biopsies have been challenged by a predominance of sclerotic bone lesions, which are technically challenging and harbor limited viable tissues for extensive analyses. Nonetheless, in recent years through collaborative efforts and clinical trials, biopsies in metastatic prostate cancer have become feasible [12]. Tumor biopsy programs including those from the Stand Up To Cancer (SU2C)- Prostate Cancer Foundation (PCF) Dream Team clinical trials [13, 14] have revealed 1) genomic diversity between patients with CRPC; 2) acquisition and/or enrichment of molecular alterations in CRPC compared to primary prostate cancer; 3) emergence of pathologic variants including atypical and neuroendocrine phenotypes. These studies have provided insights into mechanisms of treatment resistance, intra- and inter-patient heterogeneity, and clonal evolution [15]. This review focuses on recent advances in our understanding of emerging clinical and pathologic variants in CRPC. Based on a growing number of biopsies being performed clinically, refinement of pathologic criteria for histological subtyping [16], and an expanded use of high-throughput technologies [11], the clinical, pathologic and molecular features CRPC variants have become better characterized. Their full characterization is a subject of ongoing investigation.
Neuroendocrine prostate cancer
The diagnosis of neuroendocrine prostate cancer (NEPC) is histologically defined, encompasses morphologies that overlap with neuroendocrine tumors arising from other primary sites (eg., lung, bladder), and may occur with mixed or overlapping features with prostate adenocarcinoma. The recent refinement of diagnostic terminology for NEPC came after accumulation of new clinical data and a revised pathologic consensus for defining NEPC subtypes [16]. These subtypes include: a) usual prostate adenocarcinoma with neuroendocrine differentiation; b) adenocarcinoma with Paneth cell neuroendocrine differentiation; c) carcinoid tumor; d) small cell carcinoma; e) large cell neuroendocrine carcinoma; and f) mixed neuroendocrine carcinoma - acinar adenocarcinoma [7]. Immunohistochemical detection of neuroendocrine markers such as synaptophysin, chromogranin, and CD56 may help support the diagnosis of NEPC but are not required in the presence of morphologic characteristics. Pure small-cell neuroendocrine prostate carcinoma is a subtype of NEPC that has the most aggressive biologic behavior and poor outcomes. The clinical significance of other NEPC subtypes especially mixed or hybrid phenotypes is not well defined.
Although NEPC rarely arises de novo and accounts for less than 2% of patients at the time of diagnosis of prostate cancer [17], NEPC is enriched in the advanced disease setting and reported as high as 10-20% in CRPC [14, 18]. Clinically, NEPC is often suspected in patients that progress in the setting of low or moderately rising PSA levels (suggesting less AR driven disease) with a predominance of visceral and/or lytic bone metastases. Morphologically, NEPC shares features with other high grade neuroendocrine cancers, including presence of small cells with ‘salt and pepper’ chromatin, high mitotic count and nuclear molding [19]. In the majority of cases at least one neuroendocrine immunohistochemical (IHC) marker, such as neuron-specific enolase, synaptophysin, chromogranin, or CD56 stains positive. NEPC is often negative for classical luminal markers of prostatic glandular differentiation (eg., prostate-specific antigen (PSA) and prostatic acid phosphatase). Uncommonly, polypeptide hormones may be present (including ACTH, antidiuretic hormone (ADH), and corticotropin-releasing factor).
In ambiguous cases or in cases of small cell carcinoma of unknown primary, ERG break-apart fluorescence in situ hybridization (FISH) may be used clinically to evaluate for the presence of the prostate cancer-specific ERG gene rearrangement. ERG rearrangement is present in approximately 50% of NEPC and is diagnostic of prostate cancer [20, 21], though a negative test does not rule out prostate origin. Patients with NEPC are often treated like other high-grade neuroendocrine tumors with platinum-based chemotherapy [22], though this treatment decision may depend on the clinical context especially in cases with mixed features. Although there are few published clinical series reporting clinical outcomes of patients with NEPC, median survival of small cell NEPC has been reported at 7 months [23] with patients with mixed adenocarcinoma-NEPC tumors having a more variable prognosis [24].
Early insights into the molecular background of NEPC came with the use of next generation RNA-sequencing comparing 7 NEPC tumors, 30 PC adenocarcinomas (PCA) and 5 benign prostate tissue samples [25]. The transcriptome of NEPC was distinct from PCA, with 936 genes differentially expressed. One notable finding was overexpression and amplification of the oncogenic transcription factor N-myc (MYCN). In multiple preclinical models N-myc has been found to be a key driver of the NEPC phenotype [25-27]. Overexpression of N-myc in the context of either PTEN loss or AKT overexpression results in the development of aggressive features, including histologic features of NEPC, low AR and low AR signaling, up-regulation of NEPC markers, and resistance to AR therapies [26, 27]. Similar to what has been observed in N-myc-amplified neuroblastoma [28, 29], N-myc may be inhibited indirectly by targeting its allosteric protein partner Aurora kinase A (AURKA) using several of the available aurora kinase inhibitors (eg., alisertib, CD532) [28-30]. AURKA is a mitotic kinase and oncogene [31], also significantly overexpressed in NEPC [25]. A mechanistic link has also been demonstrated between mutated TP53 and elevated levels of AURKA, through increased expression of miR25 and resultant inhibition of E3 ubiquitin ligase FBXW7-mediated AURKA degradation [32].
In an extended cohort evaluating metastatic biopsies from 81 patients with CRPC, including 51 with clinical and pathologic features of AR-driven adenocarcinoma and 30 with features of AR-indifferent NEPC [33], the two ends of the spectrum were compared in order to elucidate the molecular features that distinguish NEPC. Unexpectedly, despite distinct pathologic and clinical differences, whole exome sequencing (WES) revealed significant overlap in the mutational and copy number landscape between castration resistant adenocarcinoma and NEPC tumors, supporting clonal evolution of NEPC from an adenocarcinoma precursor. There was significant enrichment in RB1 loss and TP53 mutations and deletions in NEPC, alterations commonly observed in other high-grade neuroendocrine tumors [34]. In another study reported by Tan et al, RB1 alterations were found in 90% of small cell NEPC (26 of 29 patients) [35]. In prostate cancer preclinical models, the combination of RB1 and TP53 drives the development of NEPC [36-39], suggesting that RB1 and TP53 contribute to the pathogenesis of NEPC. Extending upon these findings, Mu et al (38) and Ku et al (39) recently described in two elegant studies how loss of p53 and Rb1 can drive resistance to AR-antagonists in prostate cancer by promoting lineage plasticity and progression towards an androgen indifferent state – a state in which AR protein is still present, but tumors are less dependent on androgen signaling - mediated in part by SOX2, a lineage transcription factor also capable of reprogramming fibroblasts to pluripotent stem cells.
As opposed to their largely similar genomic profiles, the epigenomic landscape of NEPC is distinct compared to castration resistant adenocarcinoma [33]. Combined analysis of genome-wide CpG methylation with transcript data revealed epigenetically dysregulated pathways including those involved in epithelial mesenchymal transition, neuronal, stem cell like, and developmental pathways, suggesting a key role of the epigenome in modulating the NEPC phenotype. The histone methyltransferase EZH2 is also up-regulated in NEPC and EZH2-supressed target genes down-regulated. Treatment with EZH2 inhibitors demonstrated antitumor activity in NEPC models and modulated NEPC and AR signaling pathways (33, 39), pointing to a cellular plasticity that may be exploited therapeutically. A mechanistic link between N-myc and EZH2 has also been elucidated in that they cooperate to drive histone methylation, suppress AR signaling, and drive a NEPC phenotype [27].
Development of preclinical models is another important tool for in vitro and in vivo replication of disease subtypes, identification of biomarkers, and evaluation of novel treatments. In addition to the N-myc/PTEN [27], N-myc/AKT [26], and the TP53/RB1 model [36-39] systems that drive NEPC, enzalutamide resistant models [40, 41], patient-derived xenograft (PDX) models [42, 43] and patient-derived organoids [44, 45] may be used to study drug resistance and the development of NEPC [42, 43]. Preclinical observations using these model systems combined with evaluation of additional patient samples have elucidated additional NEPC associated biologic pathways. The RE1-silencing transcription factor (REST), a neuronal signaling transcriptional repressor, is significantly downregulated in NEPC, mediated by the splicing regulator SRRM4. Overexpression of SRRM4 occurring in 50% of NEPC as opposed to only 3% of PCA cases (N=218) [43, 46, 47] and is a central regulator of alternative splicing events [48]. SRRM4 was found to be a potent inducer of neuroendocrine differentiation under conditions of AR pathway inhibition, accentuated by RB1 and p53 loss, acting upstream of several neuronal signaling and chromatin remodeling genes, including REST [48]. De-repression of the placental gene PEG10 also occurs during the NEPC transition, and is regulated by AR and the E2F/RB pathway. PEG10 promotes cell-cycle progression in the context of TP53 loss and regulates Snail expression via TGF-β signaling to promote invasion [49]. Loss of the androgen regulated ERG overexpression in the context of TMPRSS2-ERG fusion- positive NEPC has also been implicated in promoting de-differentiation, proliferation, resistance to AR therapies, and the NEPC phenotype [50]. In addition to AURKA, up-regulation of UBE2C and other mitotic genes as well as cyclin D1 (CCND1) overexpression also occurs in NEPC [51, 52]. Recently the neural transcription factor BRN2 was identified as a key driver of the NEPC phenotype, significantly overexpressed in NEPC, and inversely correlated with AR signaling in CRPC. Mechanistic studies have revealed that BRN2 is directly repressed by the AR and this can be lost under selective pressure of enzalutamide, thereby promoting the development of NEPC both in vitro and in vivo [40].
Aggressive variant or anaplastic prostate cancer
As metastatic biopsies are invasive and may be challenging to perform, clinical features suggestive of small cell/NEPC may be useful to select patients for platinum-based chemotherapy and potentially other NEPC-directed approaches. The aggressive variant prostate cancer (AVPC), formally termed as anaplastic prostate cancer, is clinically defined as CRPC with at least 1 of the following: 1) histologic evidence of small-cell NEPC (pure or mixed), 2) the presence of exclusively visceral metastases, 3) radiographically predominant lytic bone metastases by plain x-ray or CT scan, 4) bulky (≥ 5 cm) lymphadenopathy or bulky (≥5 cm) high-grade (Gleason ≥ 8) tumor mass in prostate/pelvis. 5) low PSA (≤ 10 ng/mL) at initial presentation (prior to ADT or at symptomatic progression in the castrate setting) plus high volume (≥ 20) bone metastases, 6) presence of neuroendocrine markers on histology (positive staining of chromogranin A or synaptophysin) or in serum (abnormal high serum levels for chromogranin A or gastrin-releasing peptide (GRP)) at initial diagnosis or at progression, plus any of the following in the absence of other causes: (a) elevated serum LDH (≥ 2 × ULN), (b) malignant hypercalcemia, (c) elevated serum CEA (≥ 2 × ULN), 7) short interval (≤ 6 months) to androgen-independent progression following the initiation of hormonal therapy with or without the presence of neuroendocrine markers [53]. These features have been used as inclusion criteria for patient enrollment in clinical trials incorporating platinum chemotherapy. In a prospective Phase 2 clinical trial of carboplatin and docetaxel (CD) with cisplatin-etoposide (EP) upon progression, 120 patients with at least one out of the seven clinical features were enrolled [53]. The primary objectives were to estimate the response rate and time to progression while secondary objectives included the collection of tissues for investigating the underlying biology of this subset, the toxicity of the regimens and overall survival [53]. The high response rates (65.4% and 33.8% after four cycles of CD and EP, respectively) and the significant associations of certain clinical criteria with outcomes (ie., bulky disease, serum CEA) led to the exploration of a molecular signature within this cohort [54]. Targeted genomic analysis in 36 trial participants and 8 patient tumor xenografts (PDX) revealed 8q amplification (MYC), and RB1 and PTEN copy number losses (CNL) as the most frequent events in approximately 50% of samples [54]. More recently the combination cabazitaxel/carboplatin (CAB/CARB) in clinically defined AVPC patients demonstrated a significant 3-month PFS benefit compared to cabazitaxel (CAB) alone (3.8 months with CAB vs. 5.6 months with CAB/CARB) and a trend towards survival improvement (17.4 months with CAB vs. 19.2 with CAB/CARB). Approximately half (42/79) of CAB patients and a quarter (19/81) of CAB/CARB patients received additional salvage platinum-based therapy after progression [55]. Combined alterations in RB1, TP53, and/or PTEN were more frequent in the AVPC cohort as compared to an unselected CRPC cohort [13] and The Cancer Genome Atlas (TCGA) datasets [54]. Overall these data suggest that a subset of CRPC defined by aggressive clinical features may be more amenable to platinum, potentially based on the molecular overlap with NEPC. These studies as well as the high frequency of germline and somatic DNA repair alterations in CRPC [55, 56], warrant further exploration of platinum chemotherapy for a biomarker -selected or clinically enriched subset of patients with CRPC.
Intermediate Atypical prostate cancer
The West Coast Stand Up To Cancer (SU2C)- Prostate Cancer Foundation (PCF) Dream Team recently presented data from a prospective metastatic biopsy trial of patients with CRPC with primary and acquired resistance to abiraterone and/or enzalutamide. One unexpected finding after evaluation of the first 226 cases was a high frequency of histologic variants including small cell NEPC, mixed NEPC/adenocarcinoma, intermediate atypical prostate cancer (IAC) variants [57]. The molecular classification and prognostic role of these variants are yet to be fully reported, but there is some suggestion that there may be a transition state in CRPC in which patients develop mixed or overlapping pathologic and molecular features with NEPC.
In a related study, a 91 basal stem cell gene signature, termed CD49f Hi, was generated after purification and transcriptional profiling of basal and luminal epithelial cells from primary prostate tumors and adjacent benign tissue [58]. The signature, comprising E2F target genes, was particularly enriched in small-cell NEPC when applied to the TCGA, Weill Cornell and West Coast SU2C-PCF datasets, as compared to adenocarcinoma and IAC, implying the presence of a conserved transcriptional program between normal prostate basal stem cells and small-cell NEPC [58]. Overall, it is expected that as molecular data accumulate, clinical and pathologic recommendations regarding classification of tumors may be refined to address variability in reporting and areas of uncertainty regarding morphologies with focal neuroendocrine differentiation of primary prostate cancer, treatment-induced changes, pure small cell or large cell carcinoma of the prostate, and other variant or hybrid phenotypes [16, 59].
Other rare histological variants
Another histologic variant with aggressive clinical behavior is ductal adenocarcinoma. Its histologic appearance is similar to endometrial carcinoma [60]. Clinical characteristics involve presentation with a high Gleason score, local symptoms including intractable urinary symptoms, often atypical visceral metastases, and poor response to hormonal and radiotherapy [60, 61]. TMPRSS2:ERG fusion is reported at similar frequency (50%) as typical prostate acinar adenocarcinomas [62] supporting a same cell of origin. Notably there is an enrichment in mismatch repair alterations and a hyper-mutated phenotype in ductal adenocarcinoma [63], which has both biologic and potential treatment implications and warrants further investigation. Other rare histologic variants in prostate cancer include adenosquamous, mucinous, prostate leiyomyosarcomas, cystosarcoma phyllodes, carcinosarcoma, radiation associated sarcoma, or primary lymphoma of the prostate [64]. Clinical (survival), histological and molecular characteristics of the most common CRPC variants are summarized in Table 1.
Table 1. Clinical, histological and molecular characteristics of most common CRPC variants.
| CRPC variant | Survival | Histology | Molecular aberrations | Ref |
|---|---|---|---|---|
|
| ||||
| Neuroendocrine (NEPC) | 1-2 years | chromogranin (+/-),synaptophysin (+/-), AR low/absent | low AR signaling, RB1 loss, TP53 loss, REST, high MYCN, AURKA, EZH2, PEG10, SRRM4, DLL3, epigenetic | [19, 23-27, 33, 49, 50, 52, 60] |
| Small cell carcinoma | 7 months | Small blue/oat cells | ||
|
| ||||
| “AR indifferent” castration resistant adenocarcinoma | To be defined | Adenocarcinoma, AR (+), | AR signaling low, RB1 loss, TP53 loss, high EZH2 | [33, 38, 39] |
|
| ||||
| Intermediate atypical | To be defined | To be defined | Mixed/overlapping molecular features with adenocarcinoma and NEPC | [14, 57] |
|
| ||||
| Aggressive variant | 9-17 months | Heterogeneous (Adenocarcinoma or NEPC) | RB1, TP53, PTEN loss | [51, 53-55] |
|
| ||||
| Ductal | 84 months | High Gleason, endometrioid | MMR gene alterations/hypermutation | [60, 61, 63] |
Other clinical phenotypes of aggressiveness
There are other clinical characteristics with prognostic value in CRPC, such as circulating tumor cell count and sites of metastases. For instance, lymph node disease has been associated with longer median survival compared to bone and visceral (particularly liver) metastases (31.6 months vs. 21.3 months vs. 13.5 months, respectively) [65]. Other clinical characteristics that have independently been associated with worse outcomes are anemia, pain not responding to treatment, less than 30 or 50% PSA decline with therapy, and elevated lactate dehydrogenase (LDH) [66]. Recently, an updated prognostication for CRPC patients has identified three risk groups, low, intermediate and high, on the basis of 8 prognostic factors, including Eastern Cooperative Oncology Group performance status, disease site, LDH, opioid analgesic use, albumin, hemoglobin, prostate-specific antigen, and alkaline phosphatase [67]. The molecular features underlying these risk groups and their respective drivers of tumor aggressiveness require further characterization.
Molecular tools for patient selection
The clinical utility of molecular biomarkers to identify emerging variants and improve patient selection for therapy is an area of active investigation. The use of molecular criteria to identify NEPC and other clinical or pathologic variants may help overcome some of the current issues with pathologic heterogeneity and pave the way for non-invasive testing. As with any biomarker, application and validation of tissue-based and potentially liquid biomarkers will require prospective testing and extensive clinical validation. Circulating tumor cells (CTCs) have shown promise beyond enumeration in metastatic CRPC [68, 69] and have demonstrated feasibility for molecular investigation across various platforms. For instance, the presence of nuclear AR expression and active AR signaling (“AR-mixed” CTCs and “AR-on” cells) despite treatment with ARPI therapies (ABI, ENZ) has demonstrated prognostic value [70, 71]. At the gene level, differential expression of 282 genes between samples with ≥5 CTCs versus <5 CTCs was demonstrated, with significant prognostic ability of a two-gene model [72]. Concordance of PTEN status between CTCs and fresh tumor tissue is promising of the ability of CTC profiling to reflect tumor mutational status [73]. In addition, AR-V7 splice variant detection in CTCs may predict better response to taxanes compared to ARPIs [74, 75]. A NEPC classifier composed of protein and morphological characteristics including low or absent AR expression, lower cytokeratin expression, and smaller morphology, has also been proposed [76]. Circulating tumor DNA (ctDNA), including evaluation of gene copy number and mutation status is also feasible and has shown utility in clinical cohorts for predicting response to either ABT or ENZ [77-80]. Investigation of ctDNA as a platform to evaluate NEPC genomic changes [33] for non-invasive detection is also an area of ongoing research. Urine is another source of tumor cell DNA and was also recently exploited for whole genome sequencing at different stages, before or after ADT in hormone sensitive disease as well as post-docetaxel chemotherapy in CRPC patients [81]. Urine [82] or blood exosome, oncosomes [83], or microvesicles (extracellular vesicles) have also been introduced as a non-invasive platform for studying gene expression changes reflective of primary or metastatic prostate tissue. How these alternative technologies may apply to the detection of CRPC variants and/or other resistance pathways and the best timing and cohorts to evaluate warrant further investigation.
Novel therapeutic targets for CRPC variants
During the time of initial molecular data accumulation on the implication of AURKA and MYCN in the pathogenesis of NEPC, a phase II trial evaluated the Aurora kinase inhibitor danusertib in 88 metastatic CRPC patients but failed to achieve its primary end point of PSA response and showed minimal objective tumor responses with stable disease rates of 18.6% and 34.2% respectively, in the two different dose arms of the study [84]. However, the unselected population of the study and the non-surprising fact of poor performance of PSA as a biomarker in the androgen-independent NEPC subtype of the disease encouraged the initiation of an ongoing phase II trial of alisertib, a selective AURKA inhibitor in an enriched population of NEPC (NCT01799278). Given the physical interaction between AURKA and MYCN and their consistent and concurrent up-regulation in NEPC [25-27], it is anticipated that novel strategies to target MYCN or the AURKA/MYCN complex may also be effective. Pharmacologic inhibition of MYC family transcription factors is challenging, however a direct inhibition strategy with DCR-MYC, which is a Dicer-substrate small, interfering double-stranded RNA (si-RNA) targeted to the MYC oncogene is currently being tested (NCT02110563). Other approaches of targeting MYC family genes have focused on inhibiting bromodomains of putative co-activator proteins implicated in its transcriptional initiation and elongation (BET) [85]. Inhibition of the N-terminal bromodomain of bromodomain-containing protein 4 (BRD4) has also implications for AR signaling given BRD4 interaction with the N-terminal domain of AR [86]. Approaches for targeting of other emerging key drivers of NEPC such as BRN2 [40] warrant further investigation.
The epigenomic landscape is a critical area of change in NEPC, particularly, but not exclusively, pertaining to polycomb group (PcG) protein-mediated epigenetic silencing, leading to a de-differentiated phenotype [27, 33, 87]. Two such PcG family members, CBX2 and EZH2 were the most overexpressed in comparative gene expression analysis in an in vivo model of ADT-induced NEPC with clinical NEPC cohort validation, and found to be associated with a NE-associated repression signature (NEARS) of 185 genes, conferring a more aggressive clinical phenotype and progression [88]. Selective inhibition of EZH2 in in vitro and in vivo models also resulted in modulation of AR signaling and/or down-regulation of NEPC-associated genes [26, 33, 39, 49]. DNA methylation profiles are also distinct between castration resistant adenocarcinoma and NEPC, associated with differentially methylated promoters and downstream transcriptional changes [33, 89]. As a result, epigenetic strategies with use of EZH2 inhibitors (EPZ-6438, GSK2816126) and DNA methyltransferase (DNMT) inhibitors (azacitidine) are currently under phase I/II testing.
As patients with NEPC and AVPC are often treated with similar regimens as small cell lung cancer (ie. platinum), emerging targets for small cell lung cancer (SCLC) may have relevance for CRPC variants. The antibody drug target SC16LD6.5 (rovalpituzumab) targets the notch ligand delta-like 3 (DLL3), a cell surface protein aberrantly expressed in SCLC [90]. Recent early phase studies have demonstrated antitumor activity of rovalpituzumab in patients with recurrent SCLC [91]. A Phase 1 trial investigating rovalpituzumab in other neuroendocrine tumors, including a dedicated NEPC arm, is now underway (NCT02709889). Based on the emerging role of anti-PD1 therapy in SCLC [92, 93], high frequency of mismatch repair defects in ductal adenocarcinoma [63], and new data suggesting a subset of enzalutamide resistant CRPC may benefit from the addition of checkpoint inhibition [94], further investigation of immunotherapy for CRPC variants is also warranted.
Conclusions
The identification and characterization of clinical and pathologic variants of CRPC have provided new insights into the spectrum of CRPC, have facilitated preclinical studies exploring the biology of AR-indifferent CRPC, and have led to the development of new biomarker-enriched clinical trials [95]. A collective effort and use of common language in terms of standardization of methods applied and for the clinical or/and molecular categorization of metastatic CRPC patients is required and will help facilitate the design of future prospective studies. These efforts may ultimately help define the most appropriate choice and sequencing of treatments in order to maximize patient responses and improve clinical outcomes.
Acknowledgments
Himisha Beltran has received research support through grants from Millennium/Takeda, Stemcentrx, Astellas/Medivation, and Janssen.
Footnotes
Compliance with Ethics Guidelines: Conflict of Interest: Panagiotis J. Vlachostergios declares that he has no conflict of interest. Loredana Puca declares that she has no conflict of interest.
Human and Animal Rights and Informed Consent: This article does not contain any studies with human or animal subjects performed by any of the authors.
References
Papers of particular interest, published recently, have been highlighted as:
• Of importance,
•• Of major importance
- 1.Weischenfeldt J, Simon R, Feuerbach L, Schlangen K, Weichenhan D, Minner S, et al. Integrative genomic analyses reveal an androgen-driven somatic alteration landscape in early-onset prostate cancer. Cancer Cell. 2013;23(2):159–70. doi: 10.1016/j.ccr.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 2.Huggins C, Hodges CV. Studies on prostatic cancer. I. The effect of castration, of estrogen and androgen injection on serum phosphatases in metastatic carcinoma of the prostate. CA Cancer J Clin. 1972;22(4):232–40. doi: 10.3322/canjclin.22.4.232. [DOI] [PubMed] [Google Scholar]
- 3.Kirby M, Hirst C, Crawford ED. Characterising the castration-resistant prostate cancer population: a systematic review. Int J Clin Pract. 2011;65(11):1180–92. doi: 10.1111/j.1742-1241.2011.02799.x. [DOI] [PubMed] [Google Scholar]
- 4.Cookson MS, Roth BJ, Dahm P, Engstrom C, Freedland SJ, Hussain M, et al. Castration-resistant prostate cancer: AUA Guideline. J Urol. 2013;190(2):429–38. doi: 10.1016/j.juro.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 5.Heidenreich A, Bastian PJ, Bellmunt J, Bolla M, Joniau S, van der Kwast T, et al. EAU guidelines on prostate cancer. Part II: Treatment of advanced, relapsing, and castration-resistant prostate cancer. Eur Urol. 2014;65(2):467–79. doi: 10.1016/j.eururo.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 6.Roviello G, Sigala S, Sandhu S, Bonetta A, Cappelletti MR, Zanotti L, et al. Role of the novel generation of androgen receptor pathway targeted agents in the management of castration-resistant prostate cancer: A literature based meta-analysis of randomized trials. Eur J Cancer. 2016;61:111–21. doi: 10.1016/j.ejca.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 7*.Watson PA, Arora VK, Sawyers CL. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat Rev Cancer. 2015;15(12):701–11. doi: 10.1038/nrc4016. Comprehensive review of mechanisms of resistance in CRPC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graham L, Schweizer MT. Targeting persistent androgen receptor signaling in castration-resistant prostate cancer. Med Oncol. 2016;33(5):44. doi: 10.1007/s12032-016-0759-3. [DOI] [PubMed] [Google Scholar]
- 9•.Crawford ED, Higano CS, Shore ND, Hussain M, Petrylak DP. Treating patients with metastatic castration resistant prostate cancer: A comprehensive review of available therapies. J Urol. 2015;194(6):1537–47. doi: 10.1016/j.juro.2015.06.106. Comprehensive review of currently approved therapies in CRPC. [DOI] [PubMed] [Google Scholar]
- 10••.Mateo J, Carreira S, Sandhu S, Miranda S, Mossop H, Perez-Lopez R, et al. DNA-repair defects and olaparib in metastatic prostate Cancer. N Engl J Med. 2015;373(18):1697–708. doi: 10.1056/NEJMoa1506859. First biomarker-driven phase 2 trial in mCRPC patients showing significant responses in patients with BRCA2 and ATM DNA repair defects. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fraser M, Berlin A, Bristow RG, van der Kwast T. Genomic, pathological, clinical heterogeneity as drivers of personalized medicine in prostate cancer. Urol Oncol. 2015;33(2):85–94. doi: 10.1016/j.urolonc.2013.10.020. [DOI] [PubMed] [Google Scholar]
- 12.McKay RR, Zukotynski KA, Werner L, Voznesensky O, Wu JS, Smith SE, et al. Imaging, procedural and clinical variables associated with tumor yield on bone biopsy in metastatic castration-resistant prostate cancer. Prostate Cancer Prostatic Dis. 2014;17(4):325–31. doi: 10.1038/pcan.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13••.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161(5):1215–28. doi: 10.1016/j.cell.2015.05.001. The first multi-institutional, comprehensive assessment of whole-exome and transcriptome sequencing from mCRPC lesions revealed the most common gene aberrations, involving AR, ETS, TP53, and PTEN and DNA repair genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14••.Small EJ, Aggarwal RR, Huang J, Sokolov A, Zhang L, Alumkal JJ, et al. Clinical and genomic characterization of metastatic small cell/neuroendocrine prostate cancer (SCNC) and intermediate atypical prostate cancer (IAC): Results from the SU2C/PCF/AACRWest Coast Prostate Cancer Dream Team (WCDT) J Clin Oncol. 2016;34 suppl; abstr 5019. The identification and characterization of a new pathologic subtype, IAC, in mCRPC patients resistant to ABT or ENZ The majority of CRPC metastases were found to display non-adenocarcinoma features, associated with a shortened survival IAC was found to have a distinct transcriptional signature compared to classic adenocarcinoma and SCPC and an intermediate median survival. [Google Scholar]
- 15.Zong Y, Goldstein AS. Adaptation or selection–mechanisms of castration-resistant prostate cancer. Nat Rev Urol. 2013;10(2):90–8. doi: 10.1038/nrurol.2012.237. [DOI] [PubMed] [Google Scholar]
- 16•.Epstein JI, Amin MB, Beltran H, Lotan TL, Mosquera JM, Reuter VE, et al. Proposed morphologic classification of prostate cancer with neuroendocrine differentiation. Am J Surg Pathol. 2014;38(6):756–67. doi: 10.1097/PAS.0000000000000208. Refined diagnostic terminology describing the full spectrum of NE differentiation in 5 different pathological entities. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parimi V, Goyal R, Poropatich K, Yang XJ. Neuroendocrine differentiation of prostate cancer: a review. Am J Clin Exp Urol. 2014;2(4):273–85. [PMC free article] [PubMed] [Google Scholar]
- 18.Nadal R, Schweizer M, Kryvenko ON, Epstein JI, Eisenberger MA. Small cell carcinoma of the prostate. Nat Rev Urol. 2014;11(4):213–9. doi: 10.1038/nrurol.2014.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang W, Epstein JI. Small cell carcinoma of the prostate. A morphologic and immunohistochemical study of 95 cases. Am J Surg Pathol. 2008;32(1):65–71. doi: 10.1097/PAS.0b013e318058a96b. [DOI] [PubMed] [Google Scholar]
- 20.Scheble VJ, Braun M, Wilbertz T, Stiedl AC, Petersen K, Schilling D, et al. ERG rearrangement in small cell prostatic and lung cancer. Histopathology. 2010;56(7):937–43. doi: 10.1111/j.1365-2559.2010.03564.x. [DOI] [PubMed] [Google Scholar]
- 21.Lotan TL, Gupta NS, Wang W, Toubaji A, Haffner MC, Chaux A, et al. ERG gene rearrangements are common in prostatic small cell carcinomas. Mod Pathol. 2011;24(6):820–8. doi: 10.1038/modpathol.2011.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papandreou CN, Daliani DD, Thall PF, Tu SM, Wang X, Reyes A, et al. Results of a phase II study with doxorubicin, etoposide, and cisplatin in patients with fully characterized small-cell carcinoma of the prostate. J Clin Oncol. 2002;20(14):3072–80. doi: 10.1200/JCO.2002.12.065. [DOI] [PubMed] [Google Scholar]
- 23.Wang HT, Yao YH, Li BG, Tang Y, Chang JW, Zhang J. Neuroendocrine Prostate Cancer (NEPC) progressing from conventional prostatic adenocarcinoma: factors associated with time to development of NEPC and survival from NEPC diagnosis-a systematic review and pooled analysis. J Clin Oncol. 2014;32(30):3383–90. doi: 10.1200/JCO.2013.54.3553. [DOI] [PubMed] [Google Scholar]
- 24.Deorah S, Rao MB, Raman R, Gaitonde K, Donovan JF. Survival of patients with small cell carcinoma of the prostate during 1973-2003: a population-based study. BJU Int. 2012;109(6):824–30. doi: 10.1111/j.1464-410X.2011.10523.x. [DOI] [PubMed] [Google Scholar]
- 25••.Beltran H, Rickman DS, Park K, Chae SS, Sboner A, MacDonald TY, et al. Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Discov. 2011;1(6):487–95. doi: 10.1158/2159-8290.CD-11-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26••.Lee JK, Phillips JW, Smith BA, Park JW, Stoyanova T, McCaffrey EF, et al. N-Myc drives neuroendocrine prostate cancer initiated from human prostate epithelial cells. Cancer Cell. 2016;29(4):536–47. doi: 10.1016/j.ccell.2016.03.001. Identification of N-Myc and activated AKT1 as oncogenic transformers of CRPC cells to NEPC with foci of divergent differentiation from adenocarcinoma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27••.Dardenne E, Beltran H, Benelli M, Gayvert K, Berger A, Puca L, et al. N-Myc induces an EZH2-mediated transcriptional program driving neuroendocrine prostate cancer. Cancer Cell. 2016;30(4):563–577. doi: 10.1016/j.ccell.2016.09.005. Functional characterization of the role of N-myc as a driver of NE differentiation via EZH2-mediated repression of AR signaling, and sensitization to EZH2 and AURKA inhibitors, in N-myc overexpressing isogenic cell lines, xenografts, genetically engineered mice and tumor organoid cultures. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Otto T, Horn S, Brockmann M, Eilers U, Schüttrumpf L, Popov N, et al. Stabilization of N-Myc is a critical function of Aurora A in human neuroblastoma. Cancer Cell. 2009;15(1):67–78. doi: 10.1016/j.ccr.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 29.Brockmann M, Poon E, Berry T, Carstensen A, Deubzer HE, Rycak L, et al. Small molecule inhibitors of aurora-a induce proteasomal degradation of N-myc in childhood neuroblastoma. Cancer Cell. 2013;24(1):75–89. doi: 10.1016/j.ccr.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gustafson WC, Meyerowitz JG, Nekritz EA, Chen J, Benes C, Charron E, et al. Drugging MYCN through an allosteric transition in Aurora kinase A. Cancer Cell. 2014;26(3):414–27. doi: 10.1016/j.ccr.2014.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dominguez-Brauer C, Thu KL, Mason JM, Blaser H, Bray MR, Mak TW. Targeting mitosis in cancer: emerging strategies. Mol Cell. 2015;60(4):524–36. doi: 10.1016/j.molcel.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 32.Li Z, Sun Y, Chen X, Squires J, Nowroozizadeh B, Liang C, et al. p53 mutation directs AURKA overexpression via miR-25 and FBXW7 in prostatic small cell neuroendocrine carcinoma. Mol Cancer Res. 2015;13(3):584–91. doi: 10.1158/1541-7786.MCR-14-0277-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33••.Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med. 2016;22(3):298–305. doi: 10.1038/nm.4045. Integrative molecular analysis of NEPC. Whole exome sequencing provided evidence of divergent clonal evolution of adenocarcinoma to NEPC. In contrast, significant epigenetic differences between the 2 states were shown, including upregulation of the histone methyltransferase EZH2 and downregulation of EZH2-repressed target genes in NEPC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.George J, Lim JS, Jang SJ, Cun Y, Ozretid L, Kong G, et al. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524(7563):47–53. doi: 10.1038/nature14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan HL, Sood A, Rahimi HA, Wang W, Gupta N, Hicks J, et al. Rb loss is characteristic of prostatic small cell neuroendocrine carcinoma. Clin Cancer Res. 2014;20(4):890–903. doi: 10.1158/1078-0432.CCR-13-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou Z, Flesken-Nikitin A, Corney DC, Wang W, Goodrich DW, Roy-Burman P, et al. Synergy of p53 and Rb deficiency in a conditional mouse model for metastatic prostate cancer. Cancer Res. 2006;66(16):7889–98. doi: 10.1158/0008-5472.CAN-06-0486. [DOI] [PubMed] [Google Scholar]
- 37.Bianchi-Frias D, Hernandez SA, Coleman R, Wu H, Nelson PS. The landscape of somatic chromosomal copy number aberrations in GEM models of prostate carcinoma. Mol Cancer Res. 2015;13(2):339–47. doi: 10.1158/1541-7786.MCR-14-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38••.Mu P, Zhang Z, Benelli M, Karthaus WR, Hoover E, Chen CC, et al. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science. 2017;355(6320):84–8. doi: 10.1126/science.aah4307. Combined Rb1 and TP53 loss is associated with loss of luminal features, lineage plasticity, and enzalutamide-resistance, and this is mediated by the reprogramming factor SOX2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39••.Ku SY, Rosario S, Wang Y, Mu P, Seshadri M, Goodrich ZW, et al. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science. 2017;355(6320):78–83. doi: 10.1126/science.aah4199. Loss of Rb1 and Trp53 is associated with propagation of lineage plasticity and emergence of NE phenotypes mediated by epigenetic reprogramming through EZH2 which confers antiandrogen therapy resistance and can be reversed by EZH2 inhibition in CRPC and NEPC mouse models. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40••.Bishop JL, Thaper D, Vahid S, Jama R, Ketola K, Kim S, et al. The master neural transcription factor BRN2 is an androgen receptor suppressed driver of neuroendocrine differentiation in prostate cancer. Cancer Discov. 2017;7(1):54–71. doi: 10.1158/2159-8290.CD-15-1263. Mechanistic link between AR and the master regulator of neuronal differentiation, POU-domain transcription factor BRN2. In context of enzalutamide resistance and an AR-low phenotype, suppression of BRN2 is lost and BRN2 thereby drives neuroendocrine differentiation and aggressive disease. [DOI] [PubMed] [Google Scholar]
- 41.Arora VK, Schenkein E, Murali R, Subudhi SK, Wongvipat J, Balbas MD, et al. Glucocorticoid receptor confers resistance to antiandrogens by bypassing androgen receptor blockade. Cell. 2013;155(6):1309–22. doi: 10.1016/j.cell.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin D, Wyatt AW, Xue H, Wang Y, Dong X, Haegert A, et al. High fidelity patient-derived xenografts for accelerating prostate cancer discovery and drug development. Cancer Res. 2014;74(4):1272–83. doi: 10.1158/0008-5472.CAN-13-2921-T. [DOI] [PubMed] [Google Scholar]
- 43.Zhang X, Coleman IM, Brown LG, True LD, Kollath L, Lucas JM, et al. SRRM4 expression and the loss of REST activity may promote the emergence of the neuroendocrine phenotype in castration-resistant prostate cancer. Clin Cancer Res. 2015;21(20):4698–708. doi: 10.1158/1078-0432.CCR-15-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao D, Vela I, Sboner A, Iaquinta PJ, Karthaus WR, Gopalan A, et al. Organoid cultures derived from patients with advanced prostate cancer. Cell. 2014;159(1):176–87. doi: 10.1016/j.cell.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pauli C, Puca L, Mosquera JM, Robinson BD, Beltran H, Rubin MA, et al. An emerging role for cytopathology in precision oncology. Cancer Cytopathol. 2016;124(3):167–73. doi: 10.1002/cncy.21647. [DOI] [PubMed] [Google Scholar]
- 46.Lapuk AV, Wu C, Wyatt AW, McPherson A, McConeghy BJ, Brahmbhatt S, et al. From sequence to molecular pathology, and a mechanism driving the neuroendocrine phenotype in prostate cancer. J Pathol. 2012;227(3):286–97. doi: 10.1002/path.4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18(1):11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48••.Li Y, Donmez N, Sahinalp C, Xie N, Wang Y, Xue H, et al. SRRM4 drives neuroendocrine transdifferentiation of prostate adenocarcinoma under androgen receptor pathway inhibition. Eur Urol. 2016 May 11; doi: 10.1016/j.eururo.2016.04.028. pii: S0302-2838(16)30138-5. Identification of a NEPC-specific mRNA splicing signature driven by SRRM4, inducing NE transdifferentiation through REST1 alternative splicing-associated downregulation, in 2 independent CRPC-NEPC patient cohorts. [DOI] [PubMed] [Google Scholar]
- 49••.Akamatsu S, Wyatt AW, Lin D, Lysakowski S, Zhang F, Kim S, et al. The Placental Gene PEG10 Promotes Progression of Neuroendocrine Prostate Cancer. Cell Rep. 2015;12(6):922–36. doi: 10.1016/j.celrep.2015.07.012. Identification of PEG10 involvement in NEPC transdifferentiation and invasion through transcriptional re-expression and translational stabilization of PEG10 via AR-, p53- and RB1/E2F1-dependent mechanisms in patient-derived xenografts. [DOI] [PubMed] [Google Scholar]
- 50.Mounir Z, Lin F, Lin VG, Korn JM, Yu Y, Valdez R, et al. TMPRSS2:ERG blocks neuroendocrine and luminal cell differentiation to maintain prostate cancer proliferation. Oncogene. 2015;34(29):3815–25. doi: 10.1038/onc.2014.308. [DOI] [PubMed] [Google Scholar]
- 51.Tzelepi V, Zhang J, Lu JF, Kleb B, Wu G, Wan X, et al. Modeling a lethal prostate cancer variant with small-cell carcinoma features. Clin Cancer Res. 2012;18(3):666–77. doi: 10.1158/1078-0432.CCR-11-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52•.Tsai H, Morais CL, Alshalalfa M, Tan HL, Haddad Z, Hicks J, et al. Cyclin D1 loss distinguishes prostatic small-cell carcinoma from most prostatic adenocarcinomas. Clin Cancer Res. 2015;21(24):5619–29. doi: 10.1158/1078-0432.CCR-15-0744. A high ratio of CDKN2A to CCND1 expression and loss of cyclin D1 protein expression are associated with underlying Rb functional loss and distinguish morphologically identified SCPC from adenocarcinoma in 2 distinct rapid autopsy mCRPC patient cohorts and PDX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53••.Aparicio AM, Harzstark AL, Corn PG, Wen S, Araujo JC, Tu SM, et al. Platinum-based chemotherapy for variant castrate-resistant prostate cancer. Clin Cancer Res. 2013;19(13):3621–30. doi: 10.1158/1078-0432.CCR-12-3791. First phase 2 evidence of platinum-based chemotherapy efficacy in AVPC with 7 “anaplastic” clinically defined criteria, based on SCPC characteristics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54••.Aparicio AM, Shen L, Tapia EL, Lu JF, Chen HC, Zhang J, et al. Combined Tumor Suppressor Defects Characterize Clinically Defined Aggressive Variant Prostate Cancers. Clin Cancer Res. 2016;22(6):1520–30. doi: 10.1158/1078-0432.CCR-15-1259. Combined alterations in RB1, Tp53, and/or PTEN copy number losses in AVPC clinical trial patients and AVPC PDX model were identified as the strongest discriminators between AVPC and unselected mCRPC patients from other cohorts. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55•.Corn PG, Li-Ning-Tapia E, Xiao L, Heath EI, Chancoco H, Hoang A, et al. Confirmatory analysis to determine associations between platinum-sensitivity, molecular signature of combined tumor suppressor defects and aggressive variant prostate carcinomas (AVPC) J Clin Oncol. 2016;34 suppl; abstr 5020. Activity of cabazitaxel-carboplatin combination with evidence of PFS- but not OS-benefit in AVPC patients. [Google Scholar]
- 56.Pritchard CC, Mateo J, Walsh MF, De Sarkar N, Abida W, Beltran H, et al. Inherited DNA Repair Gene Mutations in Men with Metastatic Prostate Cancer, New England Journal of Medicine. N Engl J Med. 2016;375(5):443–53. doi: 10.1056/NEJMoa1603144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Small EJ, Huang J, Youngren J, Sokolov A, Aggarwal RR, Thomas G, et al. Characterization of neuroendocrine prostate cancer (NEPC) in patients with metastatic castration resistant prostate cancer (mCRPC) resistant to abiraterone (Abi) or enzalutamide (Enz): Preliminary results from the SU2C/PCF/AACR West Coast Prostate Cancer Dream Team (WCDT) J Clin Oncol. 2015;33 suppl; abstr 5003. [Google Scholar]
- 58••.Smith BA, Sokolov A, Uzunangelov V, Baertsch R, Newton Y, Graim K, et al. A basal stem cell signature identifies aggressive prostate cancer phenotypes. Proc Natl Acad Sci U S A. 2015;112(47):E6544–52. doi: 10.1073/pnas.1518007112. First evidence of a conserved, E2F-driven, stemness transcriptional program between primary prostate basal cells and small cell NEPC cells after comparison with independent mCRPC patient cohorts. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beltran H, Tomlins S, Aparicio A, Arora V, Rickman D, Ayala G, et al. Aggressive variants of castration-resistant prostate cancer. Clin Cancer Res. 2014;20(11):2846–50. doi: 10.1158/1078-0432.CCR-13-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marcus DM, Goodman M, Jani AB, Osunkoya AO, Rossi PJ. A comprehensive review of incidence and survival in patients with rare histological variants of prostate cancer in the United States from 1973 to 2008. Prostate Cancer Prostatic Dis. 2012;15(3):283–8. doi: 10.1038/pcan.2012.4. [DOI] [PubMed] [Google Scholar]
- 61.Tu SM, Lopez A, Leibovici D, Bilen MA, Evliyaoglu F, Aparicio A, et al. Ductal adenocarcinoma of the prostate: clinical features and implications after local therapy. Cancer. 2009;115(13):2872–80. doi: 10.1002/cncr.24326. [DOI] [PubMed] [Google Scholar]
- 62.Han B, Mehra R, Suleman K, Tomlins SA, Wang L, Singhal N, et al. Characterization of ETS gene aberrations in select histologic variants of prostate carcinoma. Mod Pathol. 2009;22(9):1176–85. doi: 10.1038/modpathol.2009.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63•.Schweizer MT, Cheng HH, Tretiakova MS, Vakar-Lopez F, Klemfuss N, Konnick EQ, et al. Mismatch repair deficiency may be common in ductal adenocarcinoma of the prostate. Oncotarget. 2016 Oct 15; doi: 10.18632/oncotarget.12697. [Epub ahead of print] Description of clinical and genomic characteristics of patient series with ductal prostate adenocarcinoma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fine SW. Variants and Unusual Patterns of Prostate Cancer. Surg Pathol Clin. 2008;1(1):77–104. doi: 10.1016/j.path.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 65.Halabi S, Kelly WK, Ma H, Zhou H, Solomon NC, Fizazi K, et al. Meta-Analysis evaluating the impact of site of metastasis on overall survival in men with castration-resistant prostate cancer. J Clin Oncol. 2016;34(14):1652–9. doi: 10.1200/JCO.2015.65.7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang T, Armstrong AJ. Clinical phenotypes of castration-resistant prostate cancer. Clin Adv Hematol Oncol. 2013;11(11):707–18. [PubMed] [Google Scholar]
- 67.Halabi S, Lin CY, Kelly WK, Fizazi KS, Moul JW, Kaplan EB, et al. Updated prognostic model for predicting overall survival in first-line chemotherapy for patients with metastatic castration-resistant prostate cancer. J Clin Oncol. 2014;32(7):671–7. doi: 10.1200/JCO.2013.52.3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008;14(19):6302–9. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 69.Scher HI, Heller G, Molina A, Attard G, Danila DC, Jia X, et al. Circulating tumor cell biomarker panel as an individual-level surrogate for survival in metastatic castration-resistant prostate cancer. J Clin Oncol. 2015;33(12):1348–55. doi: 10.1200/JCO.2014.55.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Miyamoto DT, Lee RJ, Stott SL, Ting DT, Wittner BS, Ulman M, et al. Androgen receptor signaling in circulating tumor cells as a marker of hormonally responsive prostate cancer. Cancer Discov. 2012;2(11):995–1003. doi: 10.1158/2159-8290.CD-12-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Crespo M, van Dalum G, Ferraldeschi R, Zafeiriou Z, Sideris S, Lorente D, et al. Androgen receptor expression in circulating tumour cells from castration-resistant prostate cancer patients treated with novel endocrine agents. Br J Cancer. 2015;112(7):1166–74. doi: 10.1038/bjc.2015.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marín-Aguilera M, Reig Ò, Lozano JJ, Jiménez N, García-Recio S, Erill N, et al. Molecular profiling of peripheral blood is associated with circulating tumor cells content and poor survival in metastatic castration-resistant prostate cancer. Oncotarget. 2015;6(12):10604–16. doi: 10.18632/oncotarget.3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Punnoose EA, Ferraldeschi R, Szafer-Glusman E, Tucker EK, Mohan S, Flohr P, et al. PTEN loss in circulating tumour cells correlates with PTEN loss in fresh tumour tissue from castration-resistant prostate cancer patients. Br J Cancer. 2015;113(8):1225–33. doi: 10.1038/bjc.2015.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Antonarakis ES, Lu C, Luber B, Wang H, Chen Y, Nakazawa M, et al. Androgen receptor splice variant 7 and efficacy of taxane chemotherapy in patients with metastatic castration-resistant prostate cancer. JAMA Oncol. 2015;1(5):582–91. doi: 10.1001/jamaoncol.2015.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Scher HI, Lu D, Schreiber NA, Louw J, Graf RP, Vargas HA, et al. Association of AR-V7 on circulating tumor cells as a treatment-specific biomarker with outcomes and survival in castration-resistant prostate cancer. JAMA Oncol. 2016 Jun 4; doi: 10.1001/jamaoncol.2016.1828. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76••.Beltran H, Jendrisak A, Landers M, Mosquera JM, Kossai M, Louw J, et al. The initial detection and partial characterization of circulating tumor cells in neuroendocrine prostate cancer. Clin Cancer Res. 2016;22(6):1510–9. doi: 10.1158/1078-0432.CCR-15-0137. Development of a NEPC CTC classifier based on low or absent AR expression, lower cytokeratin expression, and smaller cell morphology compared to typical CRPC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Romanel A, Gasi Tandefelt D, Conteduca V, Jayaram A, Casiraghi N, Wetterskog D, et al. Plasma AR and abiraterone-resistant prostate cancer. Sci Transl Med. 2015;7(312) doi: 10.1126/scitranslmed.aac9511. 312re10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Carreira S, Romanel A, Goodall J, Grist E, Ferraldeschi R, Miranda S, et al. Tumor clone dynamics in lethal prostate cancer. Sci Transl Med. 2014;6(254) doi: 10.1126/scitranslmed.3009448. 254ra125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Azad AA, Volik SV, Wyatt AW, Haegert A, Le Bihan S, Bell RH, et al. Androgen receptor gene aberrations in circulating cell-free DNA: biomarkers of therapeutic resistance in castration-resistant prostate cancer. Clin Cancer Res. 2015;21(10):2315–24. doi: 10.1158/1078-0432.CCR-14-2666. [DOI] [PubMed] [Google Scholar]
- 80.Wyatt AW, Azad AA, Volik SV, Annala M, Beja K, McConeghy B, et al. Genomic alterations in cell-free DNA and enzalutamide resistance in castration-resistant prostate cancer. JAMA Oncol. 2016 May 5; doi: 10.1001/jamaoncol.2016.0494. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xia Y, Huang CC, Dittmar R, Du M, Wang Y, Liu H, et al. Copy number variations in urine cell free DNA as biomarkers in advanced prostate cancer. Oncotarget. 2016;7(24):35818–35831. doi: 10.18632/oncotarget.9027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Motamedinia P, Scott AN, Bate KL, Sadeghi N, Salazar G, Shapiro E, et al. Urine exosomes for non-invasive assessment of gene expression and mutations of prostate cancer. PLoS One. 2016 May 4;11(5):e0154507. doi: 10.1371/journal.pone.0154507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Minciacchi VR, You S, Spinelli C, Morley S, Zandian M, Aspuria PJ, et al. Large oncosomes contain distinct protein cargo and represent a separate functional class of tumor-derived extracellular vesicles. Oncotarget. 2015;6(13):11327–41. doi: 10.18632/oncotarget.3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Meulenbeld HJ, Bleuse JP, Vinci EM, Raymond E, Vitali G, Santoro A, et al. Randomized phase II study of danusertib in patients with metastatic castration-resistant prostate cancer after docetaxel failure. BJU Int. 2013;111(1):44–52. doi: 10.1111/j.1464-410X.2012.11404.x. [DOI] [PubMed] [Google Scholar]
- 85.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146(6):904–17. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Asangani IA, Dommeti VL, Wang X, Malik R, Cieslik M, Yang R, et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014;510(7504):278–82. doi: 10.1038/nature13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bracken AP, Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer. 2009;9(11):773–84. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- 88.Clermont PL, Lin D, Crea F, Wu R, Xue H, Wang Y, et al. Polycomb-mediated silencing in neuroendocrine prostate cancer. Clin Epigenetics. 2015;7:40. doi: 10.1186/s13148-015-0074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kleb B, Estécio MR, Zhang J, Tzelepi V, Chung W, Jelinek J, et al. Differentially methylated genes and androgen receptor re-expression in small cell prostate carcinomas. Epigenetics. 2016;11(3):184–93. doi: 10.1080/15592294.2016.1146851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Saunders LR, Bankovich AJ, Anderson WC, Aujay MA, Bheddah S, Black K, et al. A DLL3-targeted antibody-drug conjugate eradicates high-grade pulmonary neuroendocrine tumor-initiating cells in vivo. Sci Transl Med. 2015;7(302) doi: 10.1126/scitranslmed.aac9459. 302ra136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rudin CM, Pietanza MC, Bauer TM, Spigel DR, Ready N, Morgensztern D, et al. Safety and efficacy of single-agent rovalpituzumab tesirine (SC16LD6.5), a delta-like protein 3 (DLL3)-targeted antibody-drug conjugate (ADC) in recurrent or refractory small cell lung cancer (SCLC) J Clin Oncol. 2016;34 suppl; abstr LBA8505. [Google Scholar]
- 92.Ott PA, Elez Fernandez ME, Hiret S, Kim DW, Moss RA, Winser T, et al. Pembrolizumab (MK-3475) in patients (pts) with extensive-stage small cell lung cancer (SCLC): Preliminary safety and efficacy results from KEYNOTE-028. J Clin Oncol. 2015;33 suppl; abstr 7502. [Google Scholar]
- 93.Antonia SJ, López-Martin JA, Bendell J, Ott PA, Taylor M, Eder JP, et al. Nivolumab alone and nivolumab plus ipilimumab in recurrent small-cell lung cancer (CheckMate 032): a multicentre, open-label, phase 1/2 trial. Lancet Oncol. 2016;17(7):883–95. doi: 10.1016/S1470-2045(16)30098-5. [DOI] [PubMed] [Google Scholar]
- 94.Graff JN, Alumkal JJ, Drake CG, Thomas GV, Redmond WL, Farhad M, et al. Early evidence of anti-PD-1 activity in enzalutamide-resistant prostate cancer. Oncotarget. 2016 Jul 12; doi: 10.18632/oncotarget.10547. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Geethakumari PR, Cookson MS, Kelly WK, Prostate Cancer Clinical Trials Working Group 3 The Evolving Biology of Castration-Resistant Prostate Cancer: Review of Recommendations From the Prostate Cancer Clinical Trials Working Group 3. Oncology (Williston Park) 2016;30(2):187–95. 199. [PubMed] [Google Scholar]


