Abstract
As knowledge of growth-independent functions of cancer cells is expanding, exploration into the role of chemokines in modulating cancer pathogenesis, particularly metastasis, continues to develop. However, more study into the mechanisms whereby chemokines direct the migration of cancer cells is needed before specific therapies can be generated to target metastasis. Herein, we draw attention to the longstanding conundrum in the field of chemokine biology that chemokines stimulate migration in a biphasic manner; and explore this phenomenon’s impact on chemokine function in the context of cancer. Typically, low concentrations of chemokines lead to chemotactic migration and higher concentrations halt migration. The signaling mechanisms that govern this phenomenon remain unclear. Over the last decade, we have defined a novel signaling mechanism for regulation of chemokine migration through ligand oligomerization and biased agonist signaling. We provide insight into this new paradigm for chemokine signaling and discuss how it will impact future exploration into chemokine function and biology. In the pursuit of producing more novel cancer therapies, we suggest a framework for pharmaceutical application of the principles of chemokine oligomerization and biased agonist signaling in cancer.
Keywords: G protein coupled receptor, functional selectivity, biased agonist
INTRODUCTION
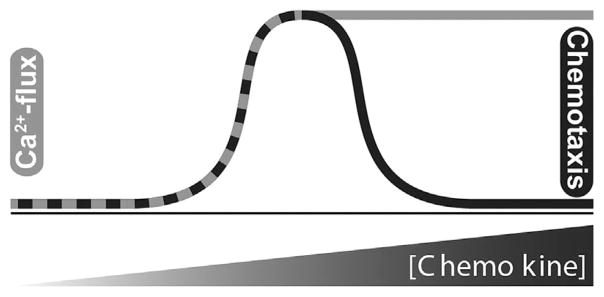
The majority of patients with advanced cancers ultimately succumb to metastasis. However, there is a paucity of therapeutics available that specifically target metastatic spread of tumor cells, providing the rationale for continuing to study chemokine biology in the context of cancer. More detailed knowledge of the molecular mechanisms that regulate cell movement is needed in order to generate viable pharmaceutical strategies for arresting cancer dissemination. Chemotactic cytokines, known as chemokines, play a fundamental role in metastasis by specifically directing the migration of cells along concentration gradients, through circulation and into specific tissues. Chemokines bind their cognate G protein-coupled seven transmembrane receptors (GPCRs) and trigger intracellular calcium flux at concentrations above a minimum threshold. In contrast, chemokines induce chemotaxis in a relatively narrow range of concentrations with migration declining as concentrations climb (Figure 1). The mechanistic origin of the biphasic or “bell-shaped” chemotaxis dose-response profile remains unclear. Our recent work with mutants of the chemokine CXCL12 has shed light on the biologic significance of ligand oligomerization on signaling and cell function. Importantly, these results suggest that regulation of chemokine function can be better understood by studying the mechanisms underlying biased agonist signaling through chemokine receptor GPCRs.
Figure 1.
Bi-modal outputs of chemokine signaling. Intracellular calcium release and cell migration display similar response profiles at lower physiological concentrations. As chemokine concentration increases, calcium flux signaling reaches a plateau (gray line) while cell migration (black line) returns to baseline levels. The “ataxic” halt of migration occurs despite maximal calcium signaling.
We have three objectives in this in-depth description and analysis of how chemokine structure regulates chemokine biology via biased agonist signaling through the cognate GPCR. First, we provide a primer to chemokine biochemistry and identify little-studied topics in cancer biology which can be addressed through chemokine biology. Second, we present a parallel mechanism to GPCR biased agonism that is chemokine protein centric and thereby establish a new model of biased agonists focused on chemokine oligomerization and receptor-ligand stoichiometry. Weanticipate that thismodel will serve to guide future exploration into the complexity of chemokine biology. Finally, we propose therapeutic options for arresting tumor malignancy through chemokine-targeted pharmaceutical intervention. Our aim is to spur additional structure-function studies into the relationship between chemokines and cancer, eventually leading to clinical use of chemokine ligand-derived therapies.
Chemokine Structure and Signaling
Chemokines are a family of ~50 proteins, ranging from 8 to 42 kDa in size, which are produced and secreted by a variety of cell types to mediate cell migration. Despite variable amino acid sequence identity, they adopt a highly conserved tertiary structure comprised of a three-stranded β-sheet and a C-terminal α-helix. Four chemokine subfamilies (CXC, CC, XC, and CX3C) are defined by the spacing of cysteine residues near the N-terminus, which is stabilized by a pair of conserved disulfide bonds [1]. The chemokine receptor family comprises ~20 distinct class A GPCRs that activate a number of intracellular signaling pathways, often in a cell type specific fashion [2]. Typical class A GPCRs bind a small molecule or peptide ligand, which then docks into a structurally complementary pocket known as the orthosteric site, within the transmembrane domain of the receptor. In contrast, chemokine receptors employ a modular two-site mechanism for ligand recognition that has been validated by mutational studies of various chemokines and receptors [3,4]. At the extracellular surface, the flexible N-terminal domain of the receptor wraps around the chemokine to create an extensive protein-protein interface (site 1). The chemokine N-terminus then docks into the orthosteric site to cause intracellular signaling (step 2) [5]. Early studies of chemokine receptors indicated that receptor dimerization is needed for activation and may be promoted by ligand binding [6–9].
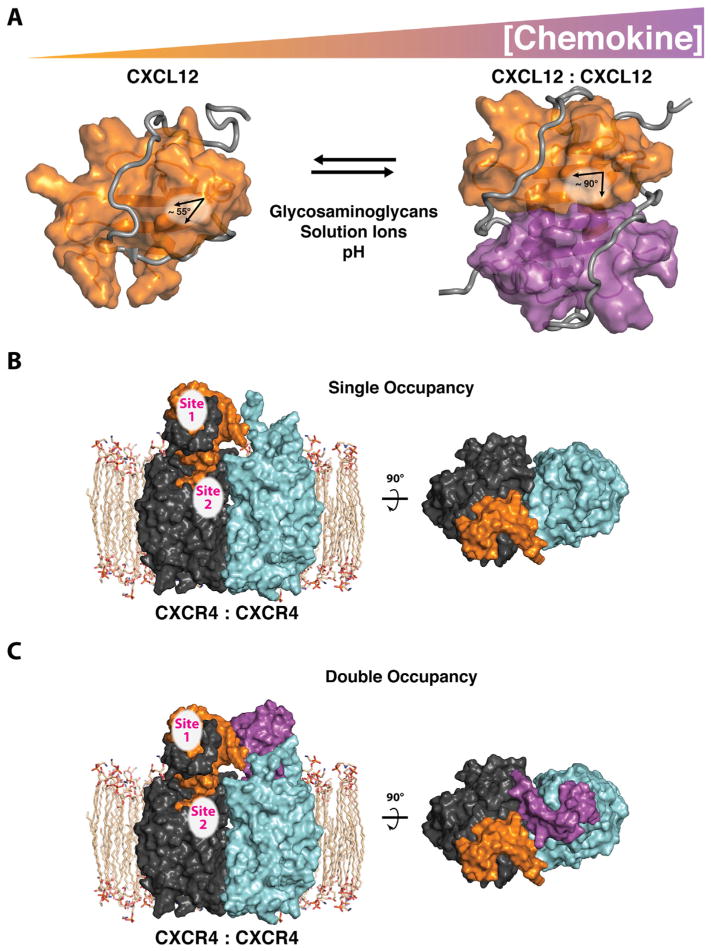
Chemokines aggregate into homodimers, heterodimers, or polymers, with oligomerization enabling key functional roles for those molecules in vivo [10]. As concentrations of the ligand increase, or in the presence of glycosaminoglycans, chemokine ligands self-associate to form dimers or higher order oligomers, with CC and CXC chemokines exhibiting distinct modes of self-association [11–14]. Indeed, CXCL8 was shown in 1994 to exist in solution as an equilibrium between monomer and dimer, each with distinct binding affinities for the chemokine’s two receptors, CXCR1 and CXCR2 [15,16]. Interestingly, Burrows noted that monomeric chemokine was the preferred conformation at lower, more physiologic concentrations. Numerous other chemokines have been shown to oligomerize, including CCL2, CCL4, CCL8, CCL7, CCL5, and CXCL10 [10–12,17]. Chemokine oligomerization states include monomers, dimers, tetramers, as evidenced by CXCL4, or larger order homo- and heteromeric polymers [18]. Most CC dimers largely involve protein–protein interactions that sequester the N-terminus and N-loop regions of two subunits within the dimer interface while the CXC chemokines are joined at the β1-strands from each monomer with the α-helices packing together against the intermolecular six-stranded β-sheet (Figure 2A) [14,18]. Rare exceptions include the CC chemokine CCL20, which crystallizes in a “CXC-type” dimer arrangement between the β1-strands [19]. Because the flexible chemokine N-terminus participates directly in GPCR activation, CC-type dimers cannot bind or activate their receptors [20]. However, in the CXC-type dimer the N-terminus remains available to interact with the receptor.
Figure 2.
Chemokine oligomerization influences receptor occupancy. (A) Chemokine concentration and environmental conditions such as glycosaminoglycans, solution ions, pH, and soluble receptor peptides (gray), regulate chemokine self-association. The C-terminal α-helix of CXCL12 is rearranged (helix angle: 55–90°) upon CXC-dimer formation at the first β-strand of each monomeric protein (shown in orange and purple). Important receptor binding epitopes (site 1) remain available on each CXCL12 monomeric subunit, irrespective of oligomeric state (PDB IDs: 2N55 and 2K05). (B) Monomeric CXCL12 forms a single-occupancy (2:1) complex with the CXCR4 receptor dimer (receptor monomers are shown as dark and light gray). (C) Monomeric CXCL12 could form a double occupancy (2:2) complex (shown). A double occupancy (2:2) complex, between dimeric CXCL12 and the CXCR4 dimer, while maintaining important sites 1 and 2 contacts, is a possible alternative mode of receptor binding (not shown).
Multiple studies have shown that chemokine receptor dimerization is needed for activation [6–9]. In some cases, ligand binding promotes dimerization, but some receptors, most notably CXCR4, are described as constitutively dimeric [21]. Assuming that receptor dimerization is obligatory for signaling, a 1:2 ligand:receptor stoichiometry, (“single occupancy”) represents the simplest functional signaling complex (Figure 2B). However, at higher chemokine concentrations, a 2:2 “double occupancy” complex could arise from the binding of two ligands to a single receptor dimer (Figure 2C). The 2:2 CXCL12:CXCR4 binding mode shown in Figure 2C was first illustrated conceptually based on the CXCR4 crystal structure [22]. Our own structural modeling of the CXCL12 dimer and dimeric receptor are compatible with 2:2 ligand–receptor binding, consistent with chemokine dimers activating CXCR4 dimers simultaneously. Differences in receptor occupancy may explain the distinct dose-dependent signaling profiles elicited by chemokines illustrated in Figure 1.
Traditional GPCR pharmacology uses maximal receptor activation (efficacy) and ligand potency as parameters to distinguish ligands that act on a common receptor with differing effects. A full agonist activates the receptor to maximal efficacy whereas a partial agonist binds and activates with sub-maximal efficacy or potency [23]. Molecular antagonists will bind to the receptor, often competing for the agonist binding site with comparable affinity, but produce no receptor activation. Additionally, ligands for some class A GPCRs (e.g., β2 adrenergic and dopamine receptors) are known to act as biased agonists. A balanced agonist activates the entire repertoire of G protein and arrestin signaling cascades for a given GPCR, while a biased agonist preferentially activates a subset of Gprotein or arrestin dependent intracellular signaling pathways for the same GPCR [24,25]. For example, morphine is a biased agonist of the μ-opioid receptor, activating only Gprotein pathways, whereas enkephalin binding to μ-opioid receptor induces balanced G protein and β-arrestin signaling [26,27]. Examples of biased agonism in existing pharmaceuticals include carvedilol and aripiprazole [28,29].
The canonical mechanism for chemokine signaling is through the heterotrimeric G protein complex. Typically, chemokine ligand–receptor binding results in activation of the Gαi subunit of the heterotrimeric complex, which in turn leads to mobilization of intracellular calcium from the endoplasmic reticulum [30]. Additionally, signaling pathways are activated in response to G protein coupled receptor kinase phosphorylation and subsequent binding of β-arrestin to the receptor C-terminus, which leads to internalization of the active chemokine-receptor-arrestin complex [31–33]. The active receptor complex ultimately stimulates calmodulin kinases, phospholipase C, and monomeric GTPase signaling in order to coordinate the cytoskeletal rearrangements necessary for cell movement [34]. However, the mechanisms behind differential regulation and coordination of chemokine-mediated G protein and arrestin signaling cascades are largely unknown.
Chemokine receptor activation has also been associated with migration-independent and cell-type specific functions, including proliferation, cell–cell adhesion, cell-matrix adhesion, and apoptosis [35]. Phosphatidylinositide 3-kinase, mitogen-activated protein kinases, and a variety of tyrosine kinases have been shown to be activated downstream of multiple chemokine receptors [34,36–41]. By stimulating several cell functions simultaneously, chemokines regulate whole-organ and -body physiologic processes such as immune or hematopoetic cell homing, angiogenesis, embryogenesis, and wound healing. Our recent work suggests that chemokine dimerization and biased agonism of chemokine receptors contributes to their functional diversity [37,42,43], but the role of biased chemokine receptor signaling in both physiologic processes and disease has yet to be fully explored.
Chemokines in Cancer Progression
In the last decade, cancer research has expanded beyond a focus on unregulated tumor growth and now explores multiple cellular characteristics altered during tumorigenesis that lead to malignancy [44]. Malignant tumors were historically defined as abnormal growths that contain cells with an enhanced ability to proliferate and survive. Conventionally, the governing elements for tumorigenesis were thought to be oncogenes or tumor suppressors. However, as numerous investigators have clearly emphasized [44,45], the contributing factors to tumor malignancy are far more varied and complex than originally outlined. Growth-independent characteristics of malignant tumor cells include altered adherence, upregulated ability to invade surrounding tissue, and enhanced migratory capacity, each of which combine to confer cell entry into circulation and eventual metastasis [46]. Several additional intra- and extra-tumoral interactions regulate tumorigenesis and malignancy, including tumor stroma deposition, tumor associated inflammation, and tumor angiogenesis [44].
To the detriment of patients with advanced cancers, relatively few pharmaceutical efforts have been directed at the growth-independent functions that promote tumor malignancy. Though the vast majority of patients with advanced solid-tumor malignancies will eventually succumb to metastasis, there remains a remarkable lack of novel pharmaceuticals entering late phase clinical trials that specifically target metastasis-driving signaling mechanisms. As regulators of cell migration and homing, the chemokine family is a rational target for further discovery of metastasis specific pharmaceutical interventions. Since the seminal study by Muller et al. [47], elevated chemokine receptor expression has been associated with a variety of human cancers. The receptors CXCR4, CCR6, and CCR7 have each been connected with cancer metastasis [35]. Expression of each of these receptors is thought to sensitize cancer cells’ ability to metastasize to organs rich in production of their respective ligands. CXCR4 and CCR6 mediate metastasis to the liver, an organ with high production of their respective cognate ligands CXCL12 and CCL20 [47,48]. CXCR4 is also linked to metastasis to the lung and bone, two additional organs that produce CXCL12. CCR7 expression facilitates spread to lymph nodes, which secrete high levels of the cognate ligands CCL19 and CCL21 [49,50]. Despite the linkage between tumor cell expression of chemokine receptors, and the implicit understanding that chemokine-based migration and metastasis is dependent on concentration gradients of chemokine ligands, relatively little information is known with regard to the status or role of chemokine ligand expression in cancers.
More recently, we have demonstrated a tumor suppressive role for the chemokine ligand CXCL12 in colorectal, mammary, and pancreatic cancers [51–53]. In each of these cancers, pathologic epigenetic silencing of CXCL12 expression in tumor cells concomitantly expressing CXCR4 permits metastatic spread. Additionally, in models of colorectal and pancreatic cancer, cells that stably express CXCL12 were susceptible to detachment-based apoptosis and slower growth rates, respectively [53,54]. The concurrent ability of CXCL12 to regulate both cancer cell movement and cancer cell viability in those tumor models reflects the complex role of chemokines in cell function. Despite those emerging roles for CXCL12 in cancer, the expression or functional roles of the ligands CCL19, CCL20, or CCL21 have yet to be mechanistically explored in the context of cancer.
Current chemokine-targeted therapeutic options are conventionally focused on antagonizing chemokine receptors. This approach in cancer biology omits key nuances in the regulation of chemokine ligand-driven function. The ability of chemokines, such as CXCL12, to regulate multiple functions within malignant tumors mirrors the current movement toward development of multifunctional pharmaceutical targets in cancer. The current paradigm for the role of chemokines in cancer largely ignores the emerging concept of biased agonism. Herein, we first describe the role of biased agonism in regulation of chemokine function, and then explore the options for exploiting chemokine biased agonism in order to generate more effective cancer therapeutics.
Chemokine Oligomerization
When free in solution, CXCL12 is predominantly a monomeric species, even at concentrations well above the likely physiological range. However, glycosaminoglycans or a soluble fragment of its receptor CXCR4 are known to induce CXCL12 self-association [55,56]. The helix angle and histidine side chain orientation of CXCL12 are distinguishing structural features for such self-association or oligomerization. The monomeric CXCL12 molecule has an acute α-helix angle relative to the β-sheet that swings to ~90° upon dimer formation (Figure 2A) [43,57]. The charged histidine residue at position 25 and lysine at position 27 discourage dimer formation unless solution ions, glycosaminoglycans, or CXCR4 N-terminal peptide are available to buffer the charge–charge repulsion and help promote dimer formation [37,55,58].
Depending on specific environmental conditions, a monomeric or dimeric CXCL12 molecule will bind and activate the receptor in the canonical chemokine binding model. As illustrated in Figure 2A, the N-loop and third β-strand of CXCL12 define a receptor binding region that recognizes a sulfotyrosine in the CXCR4 N-terminal domain and may be conserved across the chemokine family [59]. This binding site is distant from the CXC-type β-sheet dimer interface but adjacent to the CC-type dimer interface. A hydrophobic cleft at the junction of the β-sheet and C-terminal helix exposed in the CXCL12 monomer but buried at the dimer interface also binds residues at CXCR4’s extreme N-terminus while maintaining comparable binding at the sulfotyrosine pocket. Taken together, the β-sheet dimer conformation of CXCL12 does not preclude receptor binding but alters the interaction surface with the potential to influence receptor activation.
The Model
A central focus of our collaborative efforts has been to address an important and longstanding conundrum in chemokine biology. All chemokines stimulate migration in a dose-dependent fashion. However, migration is only stimulated over a specific concentration range for each axis and cell type. Beyond that range, chemokines do not stimulate active migration of cells and instead produce little to no migration (Figure 1). This phenomenon may function as a self-regulating “stop signal” for chemokine function as a cell approaches its final destination near the source of chemokine secretion within a particular tissue. Our continuing goal has been to understand the underlying mechanisms behind this signal that occurs at elevated doses of chemokine. Our recent work has described a biochemical mechanism whereby elevated concentrations of CXCL12 directly evoke a lack of cell movement, which we have termed “ataxis” [60]. The clinical implications of uncovering such knowledge are clear. For patients with early metastatic cancer, eliciting a specific signal to stop migration would be a powerful chemotherapeutic tool.
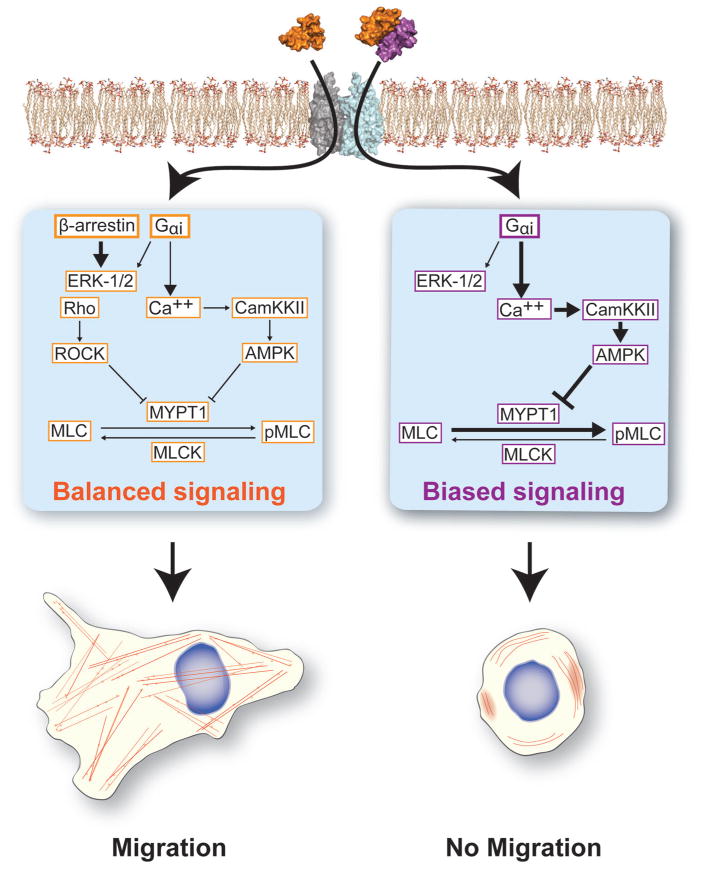
Historically, the proposed mechanism for halted migration at elevated doses of chemokine has been through receptor internalization and desensitization [61]. The notion has been that at supraphysiological ligand concentrations cells exhaust unbound membranous receptor and therefore lack the ability to continue binding to extracellular chemokine. Other mechanisms for the change in chemotactic migration as ligand concentration have been observed, including changes in cell polarity, dynamic filopodia formation in migrating, or ataxic cells [62], as well as receptor tyrosine kinase signaling [63]. In particular, at elevated concentrations CCL5 binding to CCR5 stimulated cell proliferation, and not migration, through transactivation of receptor tyrosine kinases [63]. Our collaborative findings over the last decade suggest a new and complementary mechanism for the ability of chemokines to halt cell migration. As mentioned above, in Veldkamp et al., we first showed that the CXCL12 protein free in solution exists in an equilibrium where the monomeric state is favored at lower concentrations and the dimeric complex is favored at higher concentrations (Figure 2A) [55]. Through the generation of mutant CXCL12 proteins, we demonstrated that a preferential monomeric mutant of CXCL12 stimulated migration over a wider concentration range than the wild-type protein, initially supporting our hypothesis that oligomerization regulates chemokine function [64]. Veldkamp et al. showed that CXCL12 dimer induces calcium signaling but no chemotaxis [57]. Further, the engineered CXCL12 dimer prevented migration to subsequent stimulation with wild-type CXCL12, consistent with the dimer functioning as a competitive inhibitor of the receptor CXCR4 [57]. Drury et al. subsequently demonstrated that CXCL12 was capable of binding to the cognate receptors CXCR4 and, with much weaker affinity, CXCR7 [37]. The dimeric mutant showed a blunted recruitment of β-arrestin and led to more transient extracellular signal-regulated kinase 1/2 signaling compared to the monomeric variant of CXCL12. Finally, the engineered dimeric mutant of CXCL12 most effectively blocked liver and lung metastases in mouse models of colorectal cancer and melanoma, respectively [37,65]. Based on these observations, we proposed that oligomerization provided the mechanistic foundation for a biased agonist model of chemokine ligand regulation of cell migration (Figure 3).
Figure 3.
Receptor occupancy dictates cell migratory fate. Single receptor occupancy (i.e., 1:2 ligand–receptor binding) with a CXCL12 monomer promotes activation of both Gαi and β-arrestin signaling leading to the balanced cycling of phosphorylated myosin light chain (MLC). Single occupancy balanced signaling promotes the formation and cycling of actin stress fibers needed for cell motility. When double, or 2:2, receptor occupancy occurs, β-arrestin signaling remains basally activate while the G protein signaling arm is activated. This leads to a G protein and calcium mobilization dependent increase in AMPK activity and unbalanced cycling of MLC phosphorylation, therefore preventing the organization of actin stress fibers and limited cell migration.
We then further explored the downstream biochemical signals that lead to halted migration at elevated concentrations of CXCL12 (Figure 3). Using a pancreatic cancer model in Roy et al., we demonstrated that elevated concentrations of CXCL12 led to increased bioenergetic stress and prolonged AMP-Kinase (AMPK) activity compared with lower, migratory doses of CXCL12 [60]. CXCL12 activation of AMPK was dependent on CXCR4 G protein and calcium signaling. Further, the sustained AMPK activity driven by elevated levels of CXCL12 led to downstream disruption of cycling of myosin light chain phosphorylation and thereby prevented the formation of actin stress fibers (Figure 3). The cumulative result of our studies was a novel biophysical and biochemical mechanism for regulation of chemokine migration—wherein the oligomerization state of the chemokine influences whether balanced β-arrestin/G protein signaling will occur, triggering chemotactic migration, or G protein biased signaling will occur, leading to lack of migration. As opposed to receptor inhibition, elevated levels of ligand or a dimeric chemokine variant actively signal to prevent migration.
Chemokine Therapeutics
At the clinical level, therapeutics designed to target chemokine signaling and function in disease have remained relatively few in number and narrow in scope of application. Chemokine pharmaceutical efforts were initiated by the discovery of small molecule antagonists for the co-receptors of HIV, CCR5, and CXCR4. Most notably, this lead to the clinical development of Maraviroc, a CCR5 inhibitor, and Plerixafor, a bicyclam derivative CXCR4 inhibitor. While Maraviroc was eventually approved for use in patients with HIV, the advancement of Plerixafor as an antiviral was halted after it demonstrated poor efficacy and notable off-target side effects [66,67]. More recently, both inhibitors have seen clinical use as acute regulators of host cell circulation in patients with hematologic malignancies. Influenced by the plethora of small molecule binding partners available for several related GPCR sub-families [68], subsequent approaches for targeting chemokine signaling have been to search for and test small-molecules that antagonize specific chemokine receptors. However, the contrasting fates of Maraviroc and Plerixafor underscore the potential pitfalls of broadly inhibiting constitutively expressed chemokine receptors that have a wide variety of roles in normal physiology.
In contrast, outside of the chemokine field, biased-agonist targeted strategies have already demonstrated significant progress toward the development of next-generation therapies. Biased small molecule agonists, eliciting either arrestin- or G protein-biased signaling, exist for β adrenergic receptors, opioid receptors, angiotensin receptors, dopaminergic, and serotonergic receptors [69,70]. Development of therapies targeted at biased chemokine-signaling can be approached from two different perspectives. A more classical approach, similar to those employed for other GPCR classes, would be to screen for small molecule ligands that are capable of recapitulating the biased signaling previously observed. However, our prior reports suggest that tailored recombinant proteins may offer a superior approach to targeting cancer malignancy. Our studies demonstrate that recombinant wild-type CXCL12, along with oligomeric variants, are capable of blocking metastasis in a preclinical model of colorectal cancer [37]. In both colorectal cancer and melanoma models, we further showed that a CXCL12 locked dimer mutant had increased efficacy compared to wild-type protein. Significantly, in the melanoma study, the locked dimer form of CXCL12 had greater efficacy than Plerixafor [65]. Thus, chemokine ligand targeted therapies provide a potential for greater efficacy and specificity in their ability to inhibit the malignant characteristics of tumors by eliciting biased agonist signaling.
The role of other chemokines in cancer progression warrants further study. Though the biased agonism conferred by CCL19 or CCL21 binding to CCR7 has been examined previously [71], the distinct functions of these two ligands has not been fully explored in the context of cancer. Nonetheless, CCL19 and CCL21 are promising targets for pharmaceutical intervention of not only lymph node metastasis, but also host adaptive immune reaction to tumor malignancy [39,72]. Preclinical studies into the role of CCL20, a chemokine capable of directing liver metastasis [48] and which dimerizes in a similar fashion to CXC chemokines rather than CC chemokines, may parallel findings made in CXCL12.
Future Directions
With the rapid evolution of techniques for solving protein structures and the simultaneous burgeoning exploration of G protein-independent signaling of GPCRs, the chemokine field is poised to undergo substantial evolution in the coming decade. The seminal work of Lefkowitz et al. delineating β-arrestin dependent, G protein independent signaling of small molecule binding class A GPCRs opened the door to question whether chemokines, as much larger molecules, were capable of similar signaling. In parallel, the multitude of available chemokine ligands, compared to the number of existing chemokine receptors, suggested that individual chemokines are potentially capable of inducing distinct signaling events. Thus, the concept of receptor selectivity, wherein a single chemokine receptor can elicit distinct signaling outputs from different chemokines, was born. We have focused on a related question—how can a single soluble chemokine elicit distinct functional outcomes, such as the biphasic migration curve or cell proliferation, at different concentrations? Our initial studies demonstrating that chemokines such as CXCL12 can exist as dimers under varying environmental conditions suggested that unique oligomeric states of CXCL12 could provide ligand selectivity for distinct cellular functions. Indeed, our subsequent work demonstrated that dimeric CXCL12 actively elicits a non-migratory, or “ataxic,” phenotype in cancer cells through a G protein biased signaling mechanism that ultimately locks cell migration machinery for that ligand. Thus, our collective work suggests not only that biased agonist signaling plays a crucial role in regulating chemokine function, but that these biased signals can then specifically regulate the function of cancer cells. Given the growing implication of a variety of chemokines in cancer progression, further study into biased agonist chemokine signaling will likely yield promising and novel targets for therapies in cancer.
Many aspects of biased agonist chemokine signaling have yet to be examined rigorously. Though we have now demonstrated chemokine “ligand selectivity” through CXCL12, exploration into additional chemokines axes, particularly CXC-type chemokines that form functionally active oligomers, is necessary. We predict that given certain biophysical criteria (i.e., “CXC dimer” conformation in a monogamous ligand–receptor pair) other chemokines, such as CCL5, may also demonstrate ligand selective biased agonism. Conversely, in polygamous chemokine ligand–receptor relationships, independent of oligomerization, we propose that individual chemokine ligands may access distinct, biased agonist signaling while binding to the same receptor. Exploration into the role of receptor selectivity, or receptor biased chemokine signaling is emerging, but to date has been relatively limited [71]. Emerging work, reviewed by Kleist et al. [73], is revealing the key architectural features of the chemokine-receptor interface likely to dictate ligand and receptor biased signaling events. Closer examination into the role of ligand biased, receptor biased, or even tissue biased signaling in the association and activation of each chemokine ligand with each chemokine receptor in distinct cells and tissues or cancers is necessary before the complete impact of this novel mechanism for chemokine signaling can be truly understood.
Our recent work detailing biased agonist signaling in CXCL12-driven tumor cell migration raises several additional key biochemical and biophysical questions that should be addressed. While we have now described a G protein biased mechanism, the independent role of β-arrestin biased signaling in chemokine function has yet to be defined. Further, the stoichiometry of how dimeric, or oligomeric, chemokine ligands bind to dimeric receptors has yet to be elucidated and could shed important light on the structural mechanisms underpinning biased agonist signaling of GPCRs. Similarly, examination of the intracellular loops of the receptor should yield more detailed information regarding the structural mechanisms guiding the activity and recruitment of G protein coupled receptor kinase and β-arrestin proteins that dictate biased signaling [73].
At the level of cellular function, previous biased agonist studies have primarily focused on differing signaling events. To more fully understand potential roles for chemokine biased signaling in cancer, future studies should increasingly link biased biochemical signaling events with chemokine specific and cell type specific functions. While our collective work has demonstrated a role for biased agonist signaling in chemotaxis, more study into the role of this mechanism in chemokine driven non-migratory events is needed. For example, while CXCL12 is known to regulate cell proliferation and apoptosis in a variety of cellular or disease contexts, the mechanisms underlying those cell and disease selective phenotypes remains unknown. We would speculate that ligand biased signaling may account, at least in part, for this selectivity.
As tumors remain a highly heterogeneous population of cancer cells within a cancer-unique microenvironment, it remains likely that biased agonist signaling of chemokine or other GPCRs may play key roles in tumor development and progression.
Given the prior pharmaceutical application of biased agonist principles to small molecule GPCRs such as β-blockers and dopaminergic agents [28,74], there is substantial therapeutic potential for the application of chemokine biased agonists to precisely modulate cancer pathogenesis. While additional study is needed before proceeding to clinical trials, our preclinical work indicates that engineered chemokine proteins provide a novel, biologically driven avenue for generating therapies that can regulate cancer cell function, particularly the motor machinery that dictates tumor metastasis. Further preclinical trials of chemokine biased agonist-targeted drugs will explore physiologic effects of this novel chemokine strategy to uncover tumor autonomous versus tumor parenchyma effects. In particular changes in innate and adaptive immune responses or organ development remain to be examined.
In sum, we are now entering an exciting era in the field of chemokines. Incorporating the novel concepts generated from further study of chemokine biased signaling into our understanding of cancer biology will be essential to strengthening our perception of tumors as a diseased organ. In particular, investigation into chemokine biased signaling will have significant impact on our understanding of cancer metastasis and cancer immunity. The eventual development of chemokine biased agonist directed therapies may yet offer new hope for addressing both cancer metastasis and cancer progression overall.
Acknowledgments
Grant sponsor: NIH; Grant numbers: U01 CA178960; R01 AI058072; T32 GM080202; Grant sponsor: the Bobbie Nick Voss Charitable Foundation
Supported in part by U01 CA178960 (MBD), R01 AI058072 (BFV), as well as a grant from the MCW Cancer Center and philanthropic donations from the Bobbie Nick Voss Charitable Foundation (MBD). Ishan Roy is a member of the NIH supported (T32 GM080202) Medical Scientist Training Program at MCW. Dr. Dwinell and Dr. Volkman are co-founders and majority stakeholders in Protein Foundry, LLC, a producer of molecular grade chemokines for use in biomedical research and have patents issued and pending regarding engineered chemokine variants and there use in cancer.
Footnotes
Disclosure: B.F. Volkman and M.B. Dwinell have ownership interests in Protein Foundry, LLC. The other authors have no conflict of interest to disclose.
References
- 1.Zlotnik A, Yoshie O. Chemokines: A new classification system and their role in immunity. Immunity. 2000;12:121–127. doi: 10.1016/s1074-7613(00)80165-x. [DOI] [PubMed] [Google Scholar]
- 2.Murphy PM, Baggiolini M, Charo IF, et al. International union of pharmacology. XXII. Nomenclature for chemokine receptors. Pharmacol Rev. 2000;52:145–176. [PubMed] [Google Scholar]
- 3.Ohnishi Y, Senda T, Nandhagopal N, et al. Crystal structure of recombinant native SDF-1alpha with additional mutagenesis studies: An attempt at a more comprehensive interpretation of accumulated structure-activity relationship data. J Interferon Cytokine Res. 2000;8:691–700. doi: 10.1089/10799900050116390. [DOI] [PubMed] [Google Scholar]
- 4.Crump MP, Gong J, Loetscher P, et al. Solution structure and basis for functional activity of stromal cell-derived factor-1; dissociation of CXCR4 activation from binding and inhibition of HIV-1. EMBO J. 1997;16:6996–7007. doi: 10.1093/emboj/16.23.6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monteclaro FS, Charo IF. The amino-terminal extracellular domain of the MCP-1 receptor, but not the RANTES/MIP-1a receptor, confers chemokine selectivity: Evidence for a two-step mechanism for MCP-1 receptor activation. J Biol Chem. 1996;271:19084–19092. doi: 10.1074/jbc.271.32.19084. [DOI] [PubMed] [Google Scholar]
- 6.Vila-Coro AJ, Rodriguez-Frade JM, Martin De Ana A, Moreno-Ortiz MC, Martinez-A C, Mellado M. The chemokine SDF-1a triggers CXCR4 receptor dimerization and activates the JAK/STAT pathway. FASEB J. 1999;13:1699–1710. [PubMed] [Google Scholar]
- 7.Rodríguez-Frade JM, Vila-Coro AJ, Martín de Ana A, Albar JP, Martínez-A C, Mellado M. The chemokine monocyte chemo-attractant protein-1 induces functional responses through dimerization of its receptor CCR2. Proc Natl Acad Sci USA. 1999;96:3628–3633. doi: 10.1073/pnas.96.7.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trettel F, Di Bartolomeo S, Lauro C, Catalano M, Ciotti MT, Limatola C. Ligand-independent CXCR2 dimerization. J Biol Chem. 2003;278:40980–40988. doi: 10.1074/jbc.M306815200. [DOI] [PubMed] [Google Scholar]
- 9.Hernanz-Falcon P, Rodriguez-Frade J, Serrano A, et al. Identification of amino acid residues crucial for chemokine receptor dimerization. Nat Immunol. 2004;5:216–223. doi: 10.1038/ni1027. [DOI] [PubMed] [Google Scholar]
- 10.Proudfoot AEI, Handel TM, Johnson Z, et al. Glycosaminoglycan binding and oligomerization are essential for the in vivo activity of certain chemokines. Proc Natl Acad Sci. 2003;100:1885–1890. doi: 10.1073/pnas.0334864100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lowman HB, Fairbrother WJ, Slagle PH, et al. Monomeric variants of IL-8: Effects of side chain substitutions and solution conditionsupon dimer formation. Protein Sci. 1997;6:598–608. doi: 10.1002/pro.5560060309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Rollins BJ. A dominant negative inhibitor indicates that monocyte chemoattractant protein 1 functions as a dimer. Mol Cell Biol. 1995;15:4851–4855. doi: 10.1128/mcb.15.9.4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Czaplewski LG, McKeating J, Craven CJ, et al. Identification of amino acid residues critical for aggregation of human CC chemokines macrophage inflammatory protein (MIP)-1a, MIP-1β, and RANTES: Characterization of active dissaggregated chemokine variants. J Biol Chem. 1999;274:16077–16084. doi: 10.1074/jbc.274.23.16077. [DOI] [PubMed] [Google Scholar]
- 14.Clore GM, Gronenborn AM. Three-dimensional structures of alpha and beta chemokines. FASEB J. 1995;9:57–62. doi: 10.1096/fasebj.9.1.7821760. [DOI] [PubMed] [Google Scholar]
- 15.Schnitzel W, Monschein U, Besemer J. Monomer-dimer equilibria of interleukin-8 and neutrophil-activating peptide 2. Evidence for IL-8 binding as a dimer and oligomer to IL-8 receptor B. J Leukoc Biol. 1994;55:763–770. doi: 10.1002/jlb.55.6.763. [DOI] [PubMed] [Google Scholar]
- 16.Burrows SD, Doyle ML, Murphy KP, et al. Determination of the monomer-dimer equilibrium of interleukin-8 reveals it is a monomer at physiological concentrations. Biochemistry. 1994;33:12741–12745. doi: 10.1021/bi00209a002. [DOI] [PubMed] [Google Scholar]
- 17.Crown SE, Yu Y, Sweeney MD, Leary JA, Handel TM. Heterodimerization of CCR2 chemokines and regulation by glycosaminoglycan binding. J Biol Chem. 2006;281:25438–25446. doi: 10.1074/jbc.M601518200. [DOI] [PubMed] [Google Scholar]
- 18.Kufareva I, Salanga CL, Handel TM. Chemokine and chemokine receptor structure and interactions: Implications for therapeutic strategies. Immunol Cell Biol. 2015;93:372–383. doi: 10.1038/icb.2015.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malik ZA, Tack BF. Structure of human MIP-3[alpha] chemokine. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2006;62:631–634. doi: 10.1107/S1744309106006890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan JHY, Ludeman JP, Wedderburn J, et al. Tyrosine sulfation of chemokine receptor CCR2 enhances interactions with both monomeric and dimeric forms of the chemokine monocyte chemoattractant protein-1 (MCP-1) J Biol Chem. 2013;288:10024–10034. doi: 10.1074/jbc.M112.447359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Babcock GJ, Farzan M, Sodroski J. Ligand-independent dimerization of CXCR4, a principal HIV-1 coreceptor. J Biol Chem. 2003;278:3378–3385. doi: 10.1074/jbc.M210140200. [DOI] [PubMed] [Google Scholar]
- 22.Wu B, Chien EY, Mol CD, et al. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010;330:1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rajagopal S, Rajagopal K, Lefkowitz RJ. Teaching old receptors new tricks: Biasing seven-transmembrane receptors. Nat Rev Drug Discov. 2010;9:373–386. doi: 10.1038/nrd3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kenakin T. Agonist-receptor efficacy II: Agonist trafficking of receptor signals. Trends Pharmacol Sci. 1995;16:232–238. doi: 10.1016/s0165-6147(00)89032-x. [DOI] [PubMed] [Google Scholar]
- 25.Urban JD, Clarke WP, von Zastrow M, et al. Functional selectivity and classical concepts of quantitative pharmacology. J Pharmacol Exp Ther. 2007;320:1–13. doi: 10.1124/jpet.106.104463. [DOI] [PubMed] [Google Scholar]
- 26.Keith DE, Murray SR, Zaki PA, et al. Morphine activates opioid receptors without causing their rapid internalization. J Biol Chem. 1996;271:19021–19024. doi: 10.1074/jbc.271.32.19021. [DOI] [PubMed] [Google Scholar]
- 27.Groer CE, Tidgewell K, Moyer RA, et al. An opioid agonist that does not induce μ-opioid receptor–Arrestin interactions or receptor internalization. Mol Pharmacol. 2007;71:549–557. doi: 10.1124/mol.106.028258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wisler JW, DeWire SM, Whalen EJ, et al. A unique mechanism of β-blocker action: Carvedilol stimulates β-arrestin signaling. Proc Natl Acad Sci. 2007;104:16657–16662. doi: 10.1073/pnas.0707936104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Urban JD, Vargas GA, von Zastrow M, Mailman RB. Aripiprazole has functionally selective actions at dopamine D2 receptor-mediated signaling pathways. Neuropsychopharmacology. 2006;32:67–77. doi: 10.1038/sj.npp.1301071. [DOI] [PubMed] [Google Scholar]
- 30.Sozzani S, Luini W, Molino M, et al. The signal transduction pathway involved in the migration induced by a monocyte chemotactic cytokine. J Immunol. 1991;147:2215–2221. [PubMed] [Google Scholar]
- 31.Barlic J, Khandaker MH, Mahon E, et al. β-Arrestins regulate interleukin-8-induced CXCR1 internalization. J Biol Chem. 1999;274:16287–16294. doi: 10.1074/jbc.274.23.16287. [DOI] [PubMed] [Google Scholar]
- 32.Cheng Z, Zhao J, Sun Y, et al. β-Arrestin differentially regulates the chemokine receptor CXCR4-mediated signaling and receptor internalization, and this implicates multiple interaction sites between β-arrestin and CXCR4. J Biol Chem. 2000;275:2479–2485. doi: 10.1074/jbc.275.4.2479. [DOI] [PubMed] [Google Scholar]
- 33.Fong AM, Premont RT, Richardson RM, Yu YA, Lefkowitz RJ, Patel DD. Defective lymphocyte chemotaxis in β-arrestin2- and GRK6-deficient mice. Proc Natl Acad Sci. 2002;99:7478–7483. doi: 10.1073/pnas.112198299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moyer RA, Wendt MK, Johanesen PA, Turner JR, Dwinell MB. Rho activation regulates CXCL12 chemokine stimulated actin rearrangement and restitution in model intestinal epithelia. Lab Invest. 2007;87:807–817. doi: 10.1038/labinvest.3700595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roy I, Evans DB, Dwinell MB. Chemokines and chemokine receptors: Update on utility and challenges for the clinician. Surgery. 2014;155:961–973. doi: 10.1016/j.surg.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drury LJ, Wendt MK, Dwinell MB. CXCL12 chemokine expression and secretion regulates colorectal carcinoma cell anoikis through bim-mediated intrinsic apoptosis. PLoS ONE. 2010;5:e12895. doi: 10.1371/journal.pone.0012895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Drury LJ, Ziarek JJ, Gravel S, et al. Monomeric and dimeric CXCL12 inhibit metastasis through distinct CXCR4 interactions and signaling pathways. Proc Natl Acad Sci USA. 2011;108:17655–17660. doi: 10.1073/pnas.1101133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Agle KA, Vongsa RA, Dwinell MB. Calcium mobilization triggered by the chemokine CXCL12 regulates migration in wounded intestinal epithelial monolayers. J Biol Chem. 2010;285:16066–16075. doi: 10.1074/jbc.M109.061416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murakami T, Cardones AR, Finkelstein SE, et al. Immune evasion by murine melanoma mediated through CC chemokine receptor-10. J Exp Med. 2003;198:1337–1347. doi: 10.1084/jem.20030593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wong M, Uddin S, Majchrzak B, et al. Rantes activates Jak2 and Jak3 to regulate engagement of multiple signaling pathways in T cells. J Biol Chem. 2001;276:11427–11431. doi: 10.1074/jbc.M010750200. [DOI] [PubMed] [Google Scholar]
- 41.Mellado M, Rodríguez-Frade JM, Mañes S, Martínez-A C. Chemokine signaling and functional responses: The role of receptor dimerization and TK pathway activation. Annu Rev Immunol. 2001;19:397–421. doi: 10.1146/annurev.immunol.19.1.397. [DOI] [PubMed] [Google Scholar]
- 42.Ziarek JJ, Veldkamp CT, Zhang F, et al. Heparin oligosaccharides inhibit chemokine (CXC motif) ligand 12 (CXCL12) cardioprotection by binding orthogonal to the dimerization interface, promoting oligomerization, and competing with the chemokine (CXC motif) receptor 4 (CXCR4) N terminus. J Biol Chem. 2013;288:737–746. doi: 10.1074/jbc.M112.394064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Veldkamp CT, Ziarek JJ, Su J, et al. Monomeric structure of the cardioprotective chemokine SDF-1/CXCL12. Protein Sci. 2009;18:1359–1369. doi: 10.1002/pro.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hanahan D, Weinberg R. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 45.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 46.Fidler IJ. The pathogenesis of cancer metastasis: The ‘seed and soil’ hypothesis revisited. Nat Rev Cancer. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 47.Muller A, Homey B, Soto H, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 48.Ghadjar P, Rubie C, Aebersold DM, Keilholz U. The chemokine CCL20 and its receptor CCR6 in human malignancy with focus on colorectal cancer. Int J Cancer. 2009;125:741–745. doi: 10.1002/ijc.24468. [DOI] [PubMed] [Google Scholar]
- 49.Yoshida R, Imai T, Hieshima K, et al. Molecular cloning of a novel human CC chemokine EBI1-ligand chemokine that is a specific functional ligand for EBI1, CCR7. J Biol Chem. 1997;272:13803–13809. doi: 10.1074/jbc.272.21.13803. [DOI] [PubMed] [Google Scholar]
- 50.Yoshida R, Nagira M, Kitaura M, Imagawa N, Imai T, Yoshie O. Secondary lymphoid-tissue chemokine is a functional ligand for the CC chemokine receptor CCR7. J Biol Chem. 1998;273:7118–7122. doi: 10.1074/jbc.273.12.7118. [DOI] [PubMed] [Google Scholar]
- 51.Wendt MK, Johanesen PA, Kang-Decker N, Binion DG, Shah V, Dwinell MB. Silencing of epithelial CXCL12 expression by DNA hypermethylation promotes colonic carcinoma metastasis. Oncogene. 2006;25:4986–4997. doi: 10.1038/sj.onc.1209505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wendt MK, Cooper AN, Dwinell MB. Epigenetic silencing of CXCL12 increases the metastatic potential of mammary carcinoma cells. Oncogene. 2008;27:1461–1471. doi: 10.1038/sj.onc.1210751. [DOI] [PubMed] [Google Scholar]
- 53.Roy I, Zimmerman NP, Mackinnon AC, Tsai S, Evans DB, Dwinell MB. CXCL12 chemokine expression suppresses human pancreatic cancer growth and metastasis. PLoS ONE. 2014;9:e90400. doi: 10.1371/journal.pone.0090400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wendt MK, Drury LJ, Vongsa RA, Dwinell MB. Constitutive CXCL12 expression induces anoikis in colorectal carcinoma cells. Gastroenterology. 2008;135:508–517. doi: 10.1053/j.gastro.2008.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Veldkamp CT, Peterson FC, Pelzek AJ, Volkman BF. The monomer-dimer equilibrium of stromal cell-derived factor-1 (CXCL 12) is altered by pH, phosphate, sulfate, and heparin. Protein Sci. 2005;14:1071–1081. doi: 10.1110/ps.041219505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Veldkamp CT, Seibert C, Peterson FC, Sakmar TP, Volkman BF. Recognition of a CXCR4 sulfotyrosine by the chemokine stromal cell-derived factor-1alpha (SDF-1alpha/CXCL12) J Mol Biol. 2006;359:1400–1409. doi: 10.1016/j.jmb.2006.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Veldkamp CT, Seibert C, Peterson FC, et al. Structural basis of CXCR4 sulfotyrosine recognition by the chemokine SDF-1/CXCL12. Sci Signal. 2008;1:ra4. doi: 10.1126/scisignal.1160755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ziarek JJ, Veldkamp CT, Zhang F, et al. Heparin oligosaccharides inhibit chemokine (CXC motif) ligand 12 (CXCL12) cardioprotection by binding orthogonal to the dimerization interface, promoting oligomerization, and competing with the chemokine (CXC motif) receptor 4 (CXCR4) N terminus. J Biol Chem. 2013;288:737–746. doi: 10.1074/jbc.M112.394064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ziarek JJ, Heroux MS, Veldkamp CT, Peterson FC, Volkman BF. Sulfotyrosine recognition as marker for druggable sites in the extracellular space. Int J Mol Sci. 2011;12:3740–3756. doi: 10.3390/ijms12063740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roy I, McAllister DM, Gorse E, et al. Pancreatic cancer cell migration and metastasis is regulated by chemokine-biased agonism and bioenergetic signaling. Cancer Res. 2015;75:3529–3542. doi: 10.1158/0008-5472.CAN-14-2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oberlin E, Amara A, Bachelerie F, et al. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature. 1996;382:833–835. doi: 10.1038/382833a0. [DOI] [PubMed] [Google Scholar]
- 62.Meyen D, Tarbashevich K, Banisch TU, et al. Dynamic filopodia are required for chemokine-dependent intracellular polarization during guided cell migration in vivo. eLife. 2015;4:e05279. doi: 10.7554/eLife.05279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bacon K, Premack B, Gardner P, Schall T. Activation of dual T cell signaling pathways by the chemokine RANTES. Science. 1995;269:1727–1730. doi: 10.1126/science.7569902. [DOI] [PubMed] [Google Scholar]
- 64.Veldkamp CT, Seibert C, Peterson FC, et al. Structural basis of CXCR4 sulfotyrosine recognition by the chemokine SDF-1/CXCL12. Sci Signal. 2008;1:ra4. doi: 10.1126/scisignal.1160755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Takekoshi T, Ziarek JJ, Volkman BF, Hwang ST. A locked, dimeric CXCL12 variant effectively inhibits pulmonary metastasis of CXCR4-expressing melanoma cells due to enhanced serumstability. MolCancer Therapeutics. 2012;11:2516–2525. doi: 10.1158/1535-7163.MCT-12-0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Horuk R. Chemokine receptor antagonists: Overcoming developmental hurdles. Nat Rev Drug Discov. 2009;8:23–33. doi: 10.1038/nrd2734. [DOI] [PubMed] [Google Scholar]
- 67.Hendrix CW, Collier AC, Lederman MM, et al. Safety, pharmacokinetics, and antiviral activity of AMD3100, a selective CXCR4 receptor inhibitor, in HIV-1 infection. J Acquir Immune Defic Syndr. 2004;37:1253–1262. doi: 10.1097/01.qai.0000137371.80695.ef. [DOI] [PubMed] [Google Scholar]
- 68.Kobilka BK. G protein coupled receptor structure and activation. Biochim Biophys Acta. 2007;1768:794–807. doi: 10.1016/j.bbamem.2006.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Luttrell LM. Minireview: More than just a hammer: Ligand “Bias” and pharmaceutical discovery. Mol Endocrinol. 2014;28:281–294. doi: 10.1210/me.2013-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shonberg J, Lopez L, Scammells PJ, Christopoulos A, Capuano B, Lane JR. Biased agonism at G protein-coupled receptors: The promise and the challenges–A medicinal chemistry perspective. Med Res Rev. 2014;34:1286–1330. doi: 10.1002/med.21318. [DOI] [PubMed] [Google Scholar]
- 71.Zidar DA, Violin JD, Whalen EJ, Lefkowitz RJ. Selective engagement of G protein coupled receptor kinases (GRKs) encodes distinct functions of biased ligands. Proc Natl Acad Sci. 2009;106:9649–9654. doi: 10.1073/pnas.0904361106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zlotnik A. Chemokines and cancer. Int J Cancer. 2006;119:2026–2029. doi: 10.1002/ijc.22024. [DOI] [PubMed] [Google Scholar]
- 73.Kleist AB, Getschman AE, Ziarek JJ, et al. New paradigms in chemokine receptor signal transduction: Moving beyond the two-site model. Biochem Pharmacol. 2016;114:53–68. doi: 10.1016/j.bcp.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mailman RB. GPCR functional selectivity has therapeutic impact. Trends Pharmacol Sci. 2007;28:390–396. doi: 10.1016/j.tips.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]