Abstract
Background: Proteus mirabilis is an opportunistic pathogen, commonly associated with complicated urinary tract infections (UTIs). UTIs caused by multidrug-resistant Proteus mirabilis have increased worldwide. Multidrug-resistance of Gram-negative enteric bacteria is usually associated with class 1 integrons.
Purposes: To investigate the prevalence and characterize gene cassettes of class 1 integrons in multidrug-resistant P. mirabilis
Methods: From 2006 to 2008, 314 P. mirabilis isolates from urine were collected from a regional teaching hospital. Antimicrobial resistance of the isolates was determined by disk diffusion methods. The phenotypic confirmatory test of extended-spectrum β-lactamase (ESBL) production was performed as described in the Clinical and Laboratory Standards Institute (CLSI) guideline. The genetic organization of the class 1 integron cassettes was investigated by PCR, cloning, and sequencing of the regions surrounding these genes.
Results: Seventy-nine (25%, 79/314) P. mirabilis isolates were ESBL-producing and most ESBL-producing P. mirabilis were positive for blaCTX-M. Class 1 integrons were presented in 76 isolates (24.2%, 76/314), and were more frequently found in ESBL-positive (55/79, 70%) than ESBL-negative (21/235, 8.9%) P. mirabilis isolates. The most prevalence of the cassettes encoded resistance genes were aminoglycoside (aac(6’)-Ib, aacA7, aadAl, aadA2, and aadAla), trimethoprim (dfrAl and dfrA12) and chloramphenicol (catB3 and cmlA6). The most prevalent cassette of dfr12-orfF-aadA2 was found in 49 isolates. The cassette array aadB-catB3-oxa10-aadA1 was first found in P. mirabilis. The enterobacterial repetitive intergenic consensus (ERIC)-PCR fingerprinting patterns were detected in these 76 integron positive P. mirabilis isolates and belonged to 8 profiles.
Conclusion: This study investigated the prevalence and characterized gene cassettes of class 1 integrons in MDR P. mirabilis isolates from urine samples. The frequency of gene cassettes in P. mirabilis were partially by clonal spread of the carriers and the results could provide information for effective antimicrobial therapy and infection control.
Keywords: ERIC, ESBL, Integron, Proteus mirabilis
1. Introduction
The antimicrobial genes frequently located on plasmids, transposons and integrons, lead to the rapid dissemination and treatment problem. Class 1 integrons has been proven the most common integron type present in clinical isolates of Gram-negative enteric bacteria mostly in Enterobacteriaceae, Pseudomonas aeruginosa and Acinetobacter baumannii [1]. Class 1 integrons usually associated with multidrug-resistance due to their ability to incorporate or excise one or more antimicrobial resistance gene cassettes.These genes can be integrated in the form of cassettes by three key components: the inti gene for recombination, the attl site for primary recombination known as 59 base elements, and a promoter Pc that directs expression of the cassette-encoded genes [2]. As the consequence of different insert genes, the variety of linked backbone structures of both cassettes and integrons also suggests an important role of these elements in adaptive evolution [3].
Proteus mirabilis is an opportunistic pathogen, commonly associated with complicated urinary tract infections (UTI) among patients with urolithiasis and long-term urinary catheterization in both community and healthcare settings. Infection by multidrug- resistant (MDR) P. mirabilis infections has increased worldwide in the past few years [4–7], due to its rapid acquisition and dissemination of a wide variety of antibiotic resistance genes, as well as other members of the Enterobacteriaceae family express β-lactamase. The β-lactam resistance patterns of the P. mirabilis isolates have reported production of various class extended-spectrum β-lactamases (ESBLs) and AmpC-type cephalosporinases [6, 8–10]. Previously, we reported that P. mirabilis isolates from Taiwan were susceptible to imipenem, ceftazidime, cefepime (MICs ≤ 0.5 μg/ml) but exhibited high level resistance to cefotaxime (MIC > 256 μg/ml) [11]. A high level resistance of cefotaxime is mainly mediated by co-existence of AmpC enzymes with CTX-M-type β-lactamases among P. mirabilis in Taiwan [6, 11].
The spread of ESBLs represents a serious threat to the management of infectious diseases that restricts therapeutic options of antimicrobial uses. The prevalence of integrons and characterized gene cassettes in Gram-negative bacteria integron-associated multidrug resistance have been investigated [12–15]; however, it is seldom addressed in P. mirabilis. In this study, a comparison of integron-carrying and non-integron-carrying MDR P. mirabilis isolated from urine was made to assess the differences in their antimicrobial susceptibility and clonal dissemination.
2. Materials and methods
2.1. Organisms
From 2006 to 2008, non-duplicate P. mirabilis isolates with ampicillin resistance (n = 314) of patients with urinary tract infection were collected from a 746-bed, tertiary care regional teaching hospital in middle Taiwan (Jen-Ai hospital, Taichung). These isolates were identified on the basis of routine microbiologic methods and confirmed using the VITEK system (BioMerieux Vitek Inc, Hazelwood, MO, USA). Escherichia coli DH5a was used as the host for transformation experiments.
2.2. Antibiotic resistance
Identification the resistance phenotype of the gene cassettes within the integrons was undertaken by disc diffusion test according to the recommendations of the Clinical and Laboratory Standards Institute (CLSI). The combination-disk (Becton Dickinson Microbiology Systems, Cockeysville, MD, USA) synergy tests were performed to detect ESBL and additional AmpC β-lactamases phenotype for all the collected isolates [16, 17]. All disks were purchased from Becton Dickinson Microbiology System. The MICs of antimicrobial agents were determined by the agar gradient diffusion technique (E-test and ESBL Screen, AB BIODISK, Solna, Sweden) according to the manufacturer’s instructions. The ESBL phenotype was confirmed by a reduction of ≥ 3 doubling dilutions for MICs of either cefotaxime or ceftazidime in the presence of clavulanic acid [17]. Control strains included E. coli ATCC 25922 and Klebsiella pneumoniae 700603.
2.3. Integron detection and typing, and PCR amplification of β-lactamase-encoding genes (bla)
The genetic organization of the class 1 integron cassettes was investigated by PCR, cloning, and sequencing of the regions surrounding these genes. Primers p-1 (5′-CGGATGAAGGCACGAAC-3′) and p-2 (5′-AAGCAGACTTGACCTGA-3′) were used for class 1 integron detection [18]. Polymerase chain reaction (PCR) detection and sequencing of bla genes coding for the TEM, SHV, CTX-M and CMY enzymes were performed as described previously [11]. PCR conditions for all these genes were 3 min at 94 °C; 30 cycles of 1 min at 94 °C, 1 min at 55 °C, and 2 min at 72 °C; and finally, 7 min at 72 °C. The amplicons were revealed by electrophoresis on a 1.0% agarose gel with 0.5 × TBE (Tris-borate-EDTA) running buffer and a subsequent exposure to UV light in the presence of ethidium bromide. The amplicons were purified with PCR clean up kits (Genemark, Taichung, Taiwan) and were sequenced by an ABI PRISM 377 sequencer analyzer (Applied Biosystems, Foster City, CA, USA). Sequence analyses were performed online at the National Center for Biotechnology Information website (http:// www.ncbi.nlm.nih.gov). The phylogenetic relations of int1-positive P. mirabilis were analyzed using enterobacterial repetitive intergenic consensus (ERIC)-PCR with primer ERIC2 (5′-AAG-TAAGTGACTGGGGTGAGCG-3′) [19]. The following conditions of amplification were used: denaturation for 5 min at 94 °C; 40 cycles of 45 s at 94 °C, 1 min at 52 °C and 5 min at 72 °C; and a final extension step of 10 min at 72 °C. All PCR products were separated by electrophoresis in 1% agarose in 0.5 × Tris/acetate/ EDTA buffer for 1 h at 100 V. The generated fingerprints were compared visually.
2.4. Amplicon cloning and sequence
Isolates yielding two and three amplicons of difference sizes were inserted into a cloning vector (pGEM-T easy) according to the instructions for a pGEM-T easy cloning kit (Promega, cooperation USA), and E. coli DH5a was transformed with the recombinant plasmid. The sequence of the insert was verified by nucleotide sequencing.
3. Results and discussion
3.1. Antimicrobial susceptibility and ESBL survey
MIC distributions and resistance rates of tested antimicrobial agents are shown in Table 1. Except ceftazidime and meropenem, integron-positive isolates have significantly higher resistance rate (p < 0.05) than integron-negative isolates. Eighty-seven isolates (27.7%) presented a multi-resistant phenotype (resistance to three or more antimicrobial families) and 25 isolates (8.0%) exhibited resistance to at least five different families of antimicrobial agents (data not shown). Moreover, 79 (25.1%) of the 314 isolates exhibited a positive ESBL test. Among 79 ESBL-producing P. mirabilis isolates, 55 (69.6%) were class 1 integron positive (Table 2). By PCR and nucleotide sequencing, we detected the presence of only CTX-M-14 in 70 isolates, only CTX-M-3 in 2 isolates, and both CTX-M-14 and CTX-M-3 in 7 isolates. Furthermore, all of the 79 isolates confirmed 100% identity with blaTEM-1 and no amplicons were observed for blaSHV and plasmid-mediated AmpC β-lactamases blaCMY genes. CTX-M-3 and CTX-M-14 were the most common CTX-M variants in this study as were reported previously in a survey in 2009 in middle Taiwan [20], but different as Enterobacteriaceae coding CTX-M-15 in Asia-Pacific region [21]. Although ESBL-producing P. mirabilis isolates coproducing CTX-M (27.7% in this study) have been shown to be major pathogens for urine infection, the trends of CTX-M variants or other ESBLs resistance should be carefully monitored.
Table 1.
Susceptibility testing results of 76 intll-positive and 238 intll-negative Proteus mirabilis isolates.
| intl1-positive isolates (n = 76) ESBL positive n = 55 |
intl1-negative isolates (n = 238) ESBL positive n = 24 |
|||
|---|---|---|---|---|
| Antibiotic | MIC (μg/ml) (no. of isolates) | Resistance | MIC (μg/ml) (no. of isolates) | Resistance |
| Amoxicillin-clavulanic acida | 16 (4), ≤ 8 (72) | 5.2% | 16 (2), ≤ 8 (236) | 0.1% |
| Piperacillina | ≥ 256 (75), 64 (1) | 98.7% | ≥ 256 (35), 32 (203) | 14.7% |
| Cefotaximea | ≥ 256 (29), 64 (13), 16–32 (12), ≤ 2 (22) | 71.1% | ≥ 256 (10), 64 (7), 16-32 (7), ≤ 2 (214) | 10.1% |
| Ceftazidime | 2(1), ≤ 0.5 (75) | 1.3% | ≤ 0.5 (238) | 0% |
| Cefepimea | 32 (1), 4 (3), ≤ 2 (72) | 5.3% | 4 (1), ≤ 2 (237) | ~0% |
| Cefoxitina | 16–32 (3), ≤ 8 (73) | 3.9% | 16 (1), ≤ 8 (237) | ~0% |
| Meropenem | ≤ 0.5 (76) | 0% | ≤ 0.5 (238) | 0% |
| Gentamicina | ≥ 16 (66), ≤ 2 (10) | 86.8% | ≥ 16 (21), ≤ 2 (217) | 8.8% |
| Amikacina | ≥ 16 (65), ≤ 2 (11) | 85.5% | ≥ 16 (17), ≤ 2 (221) | 7.1% |
| Ciprofloxacina | ≥ 4 (28), ≤ 1 (48) | 36.8% | ≥ 4 (11), ≤ 1 (227) | 4.6% |
The chi-square test was employed to compare the antimicrobial resistance rate between intll-positive and intll-negative P. mirabilis isolates. Susceptibility was determined according to the interpretive criteria of the Clinical and Laboratory Standards Institute 2005.
p-value is less than 0.05.
Table 2.
Resistance phenotype of class 1 integrons and their gene cassettes in76 intl1-positive Proteus mirabilis isolates.
| Integrons group (no. of isolates) | Amplicon size of integrons (Kb) | Gene cassette(s) | Resistance phenotype | ERIC type (number) |
|---|---|---|---|---|
| A single amplicon produced | ||||
| 1 (1) | 3.5 | aadB-cat-oxa10-aadA1 | KmCAmSmSp | G (1) |
| 2 (1) | 3.2 | aac(6’)-Ib-aacA7-cmlA | GmCTpSmSp | H (1) |
| 3 (38) | 2.1 | dfr12-orfF-aadA2 | TpSmSp | C (9), D (4), E (2), G(21), H (1), ND(1) |
| 4 (8) | 1.8 | aadB-aadA2 | GmSmSp | B (2), G (4), ND (2) |
| 5 (5) | 1.8 | dfrA1-aadA1a | TpSmSp | A (1), B (1), G (3) |
| 6 (3) | 1.0 | aadA1 | SmSp | E (1), G (2) |
| 7 (5) | 0.9 | dfr12 | Tp | C (2), D (1), ND(2) |
| Two or more amplicons produced | ||||
| 8 (5) | 2.1/0.9 | dfr12-orfF-aadA2/dfr12 | TpSmSp | C (2), D (2), G (1) |
| 9 (3) | 2.1/1.8 | dfr12-orfF-aadA2/aadB-aadA2 | TpSmSp | C (1), F (1), G(1) |
| 10 (2) | 2.1/1.8/0.9 | dfr12-orfF-aadA2/aadB-aadA2/ dfr12 | TpSmSpGm | C (1), G (1) |
| 11 (1) | 3.1/2.1/1.8 | aadB-cat-aadA1a/dfr12-orfF-aadA2/dfrA1- aadA1a | KmTpSmSp | G (1) |
| Negative for cassette regions | ||||
| 12 (4) | 0.15 | – | – | C (1), D (2), ND(1) |
ND, not detected.
Am, ampicillin; C, chloramphenicol; Gm, gentamicin; Km, kenamycin; Tp, trimethoprim; Sm, streptomycin; Sp, spectinomycin.
3.2. Prevalence of classes 1 integrons in the urinary isolates
Class 1 integrons were presented in 76 (24.2%) isolates, from 314 urinary isolates of P. mirabilis (Table 1). Among 76 isolates, 63 yielded one amplicon, 9 yielded 2 amplicons, and 4 yielded 3 amplicons of different sizes. The amplicon lengths, corresponding to the approximate sizes of the cassette regions, varied from 0.15 to 3.5 kb (Table 2). The presence of class 1 integrons was classified 12 groups by the length and the numbers of amplicons in a single isolate (Table 2). An integron carrying 2.1-kb insert in length was presented in 38 (60.3%) of the 63 isolates yielded one amplicon. This result demonstrated that a widespread distribution of class 1 integrons and their antimicrobial-resistant gene cassettes existed among urinary P. mirabilis isolates with ampicillin resistance in the middle Taiwan.
3.3. Characteristic of integrons and arrangement of integron gene cassettes
The PCR products of one amplicon were sequenced directly and more than one amplicons were cloned and subjected to DNA sequence and alignment. Primers of p-1 or p-2 and the second or third in cassettes were used mapping the gene cassettes. Most integrons contained 1 or 2 gene cassettes with various configurations and different sizes as indicated in Table 2. We identified 10 different gene cassettes, including 9 cassettes pertaining to antibiotic resistance and 1 cassettes (orfF) encoding non-antimicrobial resistance products. The antibiotic resistance genes included those encoding β-lactamase resistance to ampicillin (bla0XA_10), dihydrofolate reductase family resistance to trimethoprim (dfr12), chloramphenicol resistance genes, (cmlA, catB), and aminoglycoside-modifying enzymes family (aadA1, aadA1a, aadA2, aadB and aac(6’)-Ib) resistance to aminoglycosides. Eight distinct kinds of gene cassette arrays were identified. Of these, the 2.1-kb insert with dfr12-orfF-aadA2 and 1.8-kb insert with aadB-aadA2 were found most prevalent. This 2.1-kb insert was also present in isolates yielding two or more amplicons. Sequence analysis of the 150-bp PCR amplicon had not encoded additional antibiotic resistance genes except the basic genetic elements of class 1 integron. The observation that different bacterial species harboured class 1 integrons with aadA2 and dfr12 cassettes is a common finding in clinical medicine [22, 23]. Most of the identical drug- resistant gene cassettes in this study were commonly distributed in pathogenic bacteria isolated from human. The cassette array aadB-cat-oxa10-aadA1 has not been reported previously in P. mirabilis, although it is present in A. baumannii (accession: DQ288250), Klebsiella pneumoniae (accession:HQ880271), Proteus sp. IICAZ2 (accession: HQ386837) and Providencia rettgeri (accession: KJ488989).
3.4. Clonal spread of functional class 1 integron-harbouring P. mirabilis
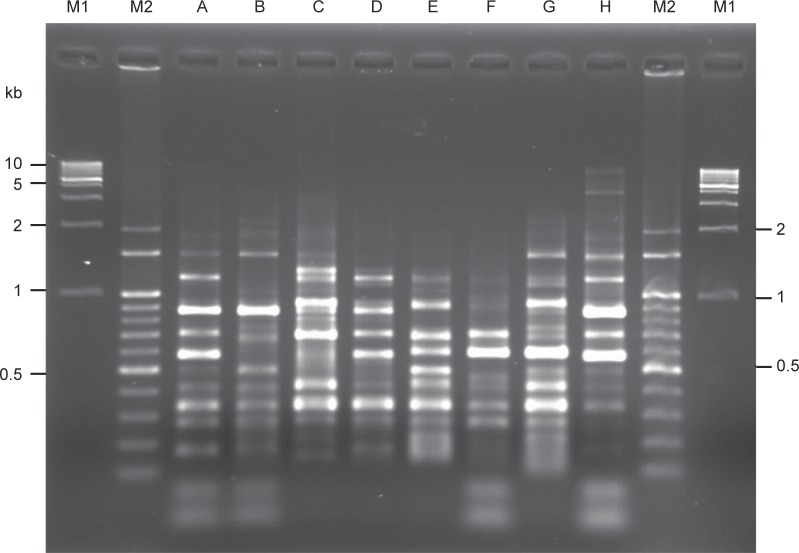
ERIC-PCR was used to type the phylogenetic relations of 76 class 1integron-positive P. mirabilis isolates. Eight ERIC types were obtained according to the electrophoresis patterns. As shown in fig. 1, products ranging from 200-900 bp were encountered more routinely. From the ERIC-PCR fingerprints results, we found 35 (46.1%, 35/76) integron-positive P. mirabilis isolates belonged to ERIC type G. Most of these ERIC type G P. mirabilis isolates (71.4%, 25/35) were also positive for class 1 integrons with the dfr12-orfF-aadA2 gene cassette arrays. This pattern was also reported in urinary E. coli isolates both in Taiwan and Korea study [23, 24], and conferring a kind of gene cassettes with stable integration and predominant in multidrug-resistant S. Choleraesuis isolates [25]. These results indicated the clonal dissemination of functional class 1 integron harbouring P. mirabilis in our hospital.
Fig. 1.
Fingerprinting patterns of eight ERIC types of class 1 integron-positive P. mirabilis isolates in this study. Lane M1, 1 Kb molecular size marker. Lane M2, 100 bp molecular size marker. Lanes A-H, fingerprinting patterns of ERIC types A-H, respectively.
In conclusion, the class 1 integron-borne gene cassette dfr12-orfF-aadA2 had been found widely disseminated among P. mirabilis isolates from urine samples. The high prevalence of class 1 integrons harboring different arrays of gene cassettes in CTX-M-type ESBLs, including CTX-M-3 and CTX-M-14, within P. mirabilis from urine samples which indicates that class 1 integrons were more commonly associated with the blaCTX-M gene than non-ESBLs-producing isolates. Furthermore, these functional class 1 integron-harbouring P. mirabilis isolates were likely to be the result of clonal spread in our hospital. Additional investigations into class 1 integrons associated ESBL bla genes are needed to employ effective means to avoid dissemination of multidrug- resistant bacteria.
Conflicts of interest statement
The authors declare that there are no conflicts of interest.
References
- 1. Poirel L, Naas T, Nordmann P. Genetic support of extended- spectrum beta-lactamases. Clin Microbiol Infect. 2008; 14 Suppl 1: 75–81. [DOI] [PubMed] [Google Scholar]
- 2. Hall RM, Collis CM. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol Microbiol. 1995; 15: 593–600. [DOI] [PubMed] [Google Scholar]
- 3. Baquero F.. Environmental stress and evolvability in microbial systems. Clin Microbiol Infect. 2009; 15 Suppl 1: 5–10. [DOI] [PubMed] [Google Scholar]
- 4. Luzzaro F, Perilli M, Amicosante G, Lombardi G, Belloni R, Zollo A, et al. Properties of multidrug-resistant, ESBL-producing Proteus mirabilis isolates and possible role of beta-lactam/beta-lactamase inhibitor combinations. Int J Antimicrob Agents. 2001; 17: 131–135. [DOI] [PubMed] [Google Scholar]
- 5. Adler A, Baraniak A, Izdebski R, Fiett J, Gniadkowski M, Hryniewicz W, et al. A binational cohort study of intestinal colonization with extended-spectrum beta-lactamase-producing Proteus mirabilis in patients admitted to rehabilitation centres. Clin Microbiol Infect. 2013; 19: E51–E58. [DOI] [PubMed] [Google Scholar]
- 6. Wang JT, Chen PC, Chang SC, Shiau YR, Wang HY, Lai JF, et al. Antimicrobial susceptibilities of Proteus mirabilis: a longitudinal nationwide study from the Taiwan surveillance of antimicrobial resistance (TSAR) program. BMC Infect Dis. 2014; 14: 486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yang Q, Zhang H, Cheng J, Xu Z, Xu Y, Cao B, et al. In vitro activity of flomoxef and comparators against Escherichia coli, Klebsiella pneumoniae and Proteus mirabilis producing extended-spectrum beta-lactamases in China. Int J Antimicrob Agents. 2015; 45: 485–490. [DOI] [PubMed] [Google Scholar]
- 8. Tibbetts R, Frye JG, Marschall J, Warren D, Dunne W. Detection of KPC-2 in a clinical isolate of Proteus mirabilis and first reported description of carbapenemase resistance caused by a KPC beta-lactamase in P. mirabilis. J Clin Microbiol. 2008; 46: 30803083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tsakris A, Ikonomidis A, Poulou A, Spanakis N, Pournaras S, Markou F. Transmission in the community of clonal Proteus mirabi- lis carrying VIM-1 metallo-beta-lactamase. J Antimicrob Chemother. 2007; 60: 136–139. [DOI] [PubMed] [Google Scholar]
- 10. Luzzaro F, Brigante G, D’Andrea MM, Pini B, Giani T, Mantengoli E, et al. Spread of multidrug-resistant Proteus mirabilis isolates producing an AmpC-type beta-lactamase: epidemiology and clinical management. Int J Antimicrob Agents. 2009; 33: 328–333. [DOI] [PubMed] [Google Scholar]
- 11. Wu LT, Wu HJ, Chung JG, Chuang YC, Cheng KC, Yu WL. Dissemination of Proteus mirabilis isolates harboring CTX-M-14 and CTX-M-3 beta-lactamases at 2 hospitals in Taiwan. Diagn Microbiol Infect Dis. 2006; 54: 89–94. [DOI] [PubMed] [Google Scholar]
- 12. Fluit AC, Schmitz FJ. Resistance integrons and super-integrons. Clin Microbiol Infect. 2004; 10: 272–288. [DOI] [PubMed] [Google Scholar]
- 13. Mazel D. Integrons: agents of bacterial evolution. Nat Rev Microbiol. 2006; 4: 608–620. [DOI] [PubMed] [Google Scholar]
- 14. Partridge SR, Tsafnat G, Coiera E, Iredell JR. Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol Rev. 2009; 33: 757–784. [DOI] [PubMed] [Google Scholar]
- 15. Domingues S, da Silva GJ, Nielsen KM. Global dissemination patterns of common gene cassette arrays in class 1 integrons. Microbiology. 2015; 161: 1313–1337. [DOI] [PubMed] [Google Scholar]
- 16. Tzelepi E, Giakkoupi P, Sofianou D, Loukova V, Kemeroglou A, Tsakris A. Detection of extended-spectrum beta-lactamases in clinical isolates of Enterobacter cloacae and Enterobacter aerogenes. J Clin Microbiol. 2000; 38: 542–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Clinical and Laboratory Standards Institute (CLSI. Performance Standards for Antimicrobial Susceptibility Testing; 15th Informational Supplement M100-S15., Wayne, PA: Clinical and Laboratory Standards Institute, 2005. [Google Scholar]
- 18. Iyobe S, Kusadokoro H, Ozaki J, Matsumura N, Minami S, Haruta S, et al. Amino acid substitutions in a variant of IMP-1 metallo-beta- lactamase. Antimicrob Agents Chemother. 2000; 44: 2023–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Versalovic J, Koeuth T, Lupski JR. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991; 19: 6823–6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang CW, Chien JH, Peng RY, Tsai DJ, Li MH, Lee HM, et al. Molecular epidemiology of CTX-M-type extended-spectrum beta-lactamase-producing Proteus mirabilis isolates in Taiwan. Int J Antimicrob Agents. 2015; 45: 84–85. [DOI] [PubMed] [Google Scholar]
- 21. Sheng WH, Badal RE, Hsueh PR, Program S. Distribution of extended-spectrum beta-lactamases, AmpC beta-lactamases, and carbapenemases among Enterobacteriaceae isolates causing intraabdominal infections in the Asia-Pacific region: results of the study for Monitoring Antimicrobial Resistance Trends (SMART). Antimicrob Agents Chemother. 2013; 57: 2981–2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gestal AM, Stokes HW, Partridge SR, Hall RM. Recombination between the dfrA12-orfF-aadA2 cassette array and an aadA1 gene cassette creates a hybrid cassette, aadA8b. Antimicrob Agents Chemother. 2005; 49: 4771–4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chang LL, Chang TM, Chang CY. Variable gene cassette patterns of class 1 integron-associated drug-resistant Escherichia coli in Taiwan. Kaohsiung J Med Sci. 2007; 23: 273–280. [DOI] [PubMed] [Google Scholar]
- 24. Kang HY, Jeong YS, Oh JY, Tae SH, Choi CH, Moon DC, et al. Characterization of antimicrobial resistance and class 1 integrons found in Escherichia coli isolates from humans and animals in Korea. J Antimicrob Chemother. 2005; 55: 639–644. [DOI] [PubMed] [Google Scholar]
- 25. Lee MF, Chen YH, Peng CF. Molecular characterisation of class 1 integrons in Salmonella enterica serovar Choleraesuis isolates from southern Taiwan. Int J Antimicrob Agents. 2009; 33: 216–222. [DOI] [PubMed] [Google Scholar]