Abstract
Objectives
To characterize the microbiological, molecular and epidemiological data of an outbreak of carbapenem-resistant Enterobacteriaceae (CRE) in a tertiary-care hospital in Mexico.
Methods
From September 2014 to July 2015, all CRE clinical isolates recovered during an outbreak in the Hospital Civil "Fray Antonio Alcalde" in Jalisco, Mexico were screened for antimicrobial susceptibility, carbapenemase production, carbapenemase-encoding genes, and plasmid profiles. Horizontal transfer of imipenem resistance; and clonal diversity by pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST); as well as biofilm production and the presence of 14 virulence genes were analyzed in selected isolates.
Results
Fifty-two carbapenem-resistant isolates corresponding to 5 species were detected, i.e., Klebsiella pneumoniae (n = 46), Enterobacter cloacae (n = 3), Escherichia coli (n = 1), Providencia rettgeri (n = 1) and Citrobacter freundii (n = 1) with carbapenemase encoding genes blaNDM-1 (n = 48), blaVIM (n = 3), blaIMP (n = 1) and blaKPC (n = 1) detected in these isolates.
The blaNDM-1 gene was detected in plasmids from 130- to 170-kb in K. pneumoniae (n = 46); E. cloacae (n = 3), E. coli (n = 1) and P. rettgeri (n = 1). The transfer of plasmids harboring the blaNDM-1 gene was obtained in eight transconjugants. One plasmid restriction pattern was detected, with the blaNDM-1 identified in different restriction fragments.
Predominant clone A of K. pneumoniae isolates archived 28/46 (60%) isolates and belongs to ST392. Besides, ST307, ST309, ST846, ST2399, and ST2400 were detected for K. pneumoniae; as well as E. cloacae ST182 and E. coli ST10.
The fimA and uge genes were more likely to be identified in K. pneumoniae carbapenem-susceptible isolates (p = <0.001) and biofilm production was more liable to be observed in carbapenem-resistant isolates (p = <0.05).
Conclusions
Four Enterobacteriaceae species harboring the blaNDM-1 gene were detected in a nosocomial outbreak in Mexico; horizontal transfer and strain transmission were demonstrated for the blaNDM-1 gene. Given the variation in the size of the plasmid harboring blaNDM-1, complex rearrangements must also be occurring.
Introduction
Nosocomial infections caused by carbapenem-resistant Enterobacteriaceae (CRE) are of particular concern since they can spread rapidly worldwide, and few treatment options remain available for these diseases [1]. Several carbapenem resistance mechanisms have been described in bacteria, and one of the most important in Gram-negative species is the production of carbapenemase enzymes [1,2]. A high diversity of carbapenemases has been reported in Enterobacteriaceae, including the Ambler class A blaKPC, class B metallo-β-lactamases, blaVIM, blaIMP, and blaNDM, and class D carbapenemase blaOXA-48 type [3]. The blaNDM-1 gene is located most frequently on large conjugative plasmids of several incompatibility groups. These plasmids also harbor genes conferring resistance to almost all antibiotics that are used to treat enterobacterial infections [2,3].
In Mexico, the first report of blaNDM-1 was identified in Providencia rettgeri [4], and it was subsequently identified in a Klebsiella pneumoniae pediatric isolate [5]. Furthermore, horizontal transfer and clonal dissemination have been reported in this country, in an outbreak caused by K. pneumoniae, Escherichia coli and Enterobacter cloacae strains harboring a 101-kb IncFII plasmid carrying the blaNDM-1 gene [6].
The analysis of sixteen NDM-1-producing enterobacterial isolates from eight countries showed that the spread of the blaNDM-1 gene is not related to specific clones, specific plasmids, or a single genetic structure [3,7]. The rapid and successful spread of carbapenem-resistant NDM-1-positive organisms may be associated with other antibiotic resistance mechanisms; however, the coexistence of multidrug resistance and virulence mechanisms has also been proposed [8].
Several virulence factors have been described in K. pneumoniae, including adhesins, capsular serotype, iron-scavenging systems, lipopolysaccharide, and biofilm production [9]. The aim of this study was to characterize the epidemiological, microbiological and molecular data of an outbreak of CRE in a tertiary-care hospital in Mexico.
Materials and methods
Hospital setting and recognition of outbreak
This study was performed in the Hospital Civil de Guadalajara “Fray Antonio Alcalde” in Jalisco, Mexico. This hospital is an 899-bed tertiary-care teaching hospital located in Guadalajara, the second largest city in Mexico. This hospital provides care to adult and pediatric patients in 31 wards situated among three connected buildings.
In September 2014, resistance to carbapenem was detected in five clinical isolates of Enterobacteriaceae in the Laboratory of Bacteriology. The infection control department was alerted, and from there all isolated Enterobacteriaceae between September 2014 and July 2015 were collected for analysis. Before September 2014, we had no carbapenem-resistant isolates.
Ethics statement
The local ethics committee (Comité de Ética en Investigación del Antiguo Hospital Civil de Guadalajara “Fray Antonio Alcalde,” Jalisco, Mexico) approved this study with reference number 003/16. Informed consent was waived by the Ethics Committee because no intervention was involved and no patients identifying information was included.
Clinical isolates and patients
Enterobacteriaceae species were identified by Matrix-Assisted Laser Desorption Ionization- Time-of-Flight Mass Spectrometry (MALDI-TOF MS) using the Bruker Biotyper (Bruker Daltonics, Germany) as described previously [10].
Drug susceptibility was performed for all isolates using the VITEK automated system and confirmed by the broth microdilution method. Guidelines of the European Committee on Antimicrobial Susceptibility Testing (EUCAST) version 6.0 were used for colistin and tigecycline [11]. Guidelines of the CLSI were used for amikacin, gentamicin, ertapenem, imipenem, meropenem, ceftriaxone, trimethoprim/sulfamethoxazole, aztreonam, ampicillin, ciprofloxacin, fosfomycin, chloramphenicol [12]. Multidrug-Resistance (MDR) was defined as non-susceptibility to one or more agents of at least three different antibiotic classes [13].
Carbapenem-resistant isolates were screened to detect carbapenemase production using the CarbaNP test [12], and to detect the carbapenemase-encoding genes for blaKPC, MBL (blaVIM, blaIMP, and blaNDM) and blaOXA-48 by PCR [14–16]. PCR products were sequenced using the chain termination method with a Big-Dye Terminator kit (Applied Biosystems Foster City, CA) and ABI PRISM 3130 (Applied Biosystems). ESBL encoding genes (blaTEM, blaSHV, blaCTX-M and blaCYM), mcr-1 and mcr-2 genes were screened by PCR [17–19].
We recovered demographic and clinical data from patients infected with a carbapenem resistant-carbapenemase producer isolates. From each enterobacterial species, one isolate per patient was analyzed.
Plasmid analysis
Plasmid profiles were obtained from all carbapenem-resistant isolates according to the method described by Kieser [20]. In isolates that presented different plasmid profile according to Kieser method, the S1 nuclease assay was performed [21].
Horizontal transfer of carbapenem resistance by bacterial conjugation with E. coli J53-2 as the recipient strain was performed by liquid and solid-phase mating as described [22,23] in isolates that presented a different plasmid profile according to S1 nuclease assay. Transconjugants were selected on Luria-Bertani (LB) agar supplemented with rifampin (100 μg/ml) plus imipenem (2 μg/ml) when the conjugation was unsuccessful the assay was performed in LB agar supplemented with rifampin (100 μg/ml) plus cefoxitin (30 μg/ml). Enzymatic digestion with HinIII (Invitrogen, California, USA) was performed in transconjugants with only one plasmid present. The incompatibility groups were detected by PCR replicon typing in these isolates [24].
Additionally, Southern hybridization with a non-radioactive probe (ECL direct nucleic acid labeling and detection system; GE Healthcare, Piscataway, NJ) of the blaNDM-1 gene was performed in transconjugants and blaNDM-1 positive isolates.
Clonal diversity studies
Clonal diversity was performed by pulsed-field gel electrophoresis (PFGE) and Multilocus sequence typing (MLST) analysis of selected isolates. For PFGE, chromosomal DNA was prepared using the methodology described by Kaufmann [25] with some modifications. Chromosomal DNA from the isolates was digested with 10 U of XbaI (Takara Bio Inc., Shiga, Japan) with following conditions: temperature of 14°C, the voltage of 6 V/cm, run time of 23 h, and switch time of 1–30 s. PFGE patterns were analyzed visually, and when the restriction patterns presented 100% similarity, the isolates were classified as a clone. When two or three difference in the restriction pattern were detected the isolates were considered as subtypes as suggested by Tenover et al. [26].
MLST was performed on selected isolates harboring blaNDM-1 gene according to species, PFGE pattern and plasmid analysis using the MLST websites: http://bigsdb.pasteur.fr, http://mlst.warwick.ac.uk and http://pubmlst.org [27–29].
Virulence factors
Detection of genes encoding virulence factors and determination of biofilm formation were conducted only for the K. pneumoniae isolates. For a comparison of virulence factors of carbapenem-resistant (blaNDM-1 positive) and carbapenem-susceptible isolates, a group of twenty-three carbapenem-susceptible isolates was randomly selected for analysis. The susceptible isolates were obtained in the same period of the carbapenem-resistant isolates (September 2014-June 2015) and were collected from similar specimens and hospital wards to carbapenem-resistant isolates.
Virulence genes from K. pneumoniae [serotypes K1 and K2, rmpA, rmpA2 (regulator of mucoid phenotype), uge (uridine diphosphate galacturonate-4 epimerase), ureA (urease), entB (enterobactin), iroB (salmochelin), irp2 (yersiniabactin), iucA (aerobactin), fimA (fimbrial), fimH (fimbrial), mrkA (fimbrial), and mrkD (fimbrial)] were screened by PCR [30].
Furthermore, semi-quantitative determination of biofilm formation was performed in these isolates (both carbapenem-resistant and carbapenem susceptible) by crystal violet staining as previously described by Bandeira et al., with modifications described by Burmølle et al. [9,31]. The biofilm index (OD595/OD600) was used to normalize the amount of biofilm formed to the total cell content of each sample tested. The biofilm production was classified using the biofilm index as non-adherent (<0.90), weakly adherent (>0.90–<1.20) and strongly adherent (>1.20) The cut-off values were defined according to a comparison in the classification with others methodologies previously reported [32]. Staphylococcus aureus ATCC 29213 (a high biofilm producer) and E. coli ATCC 25922 (a low biofilm producer) were used as quality control organisms.
Statistical analysis
The similarity coefficients were generated from a similarity matrix calculated using the Jaccard’s coefficient.
Percentages of biofilm production and from each virulence factor were compared using Mann-Whitney test. A P value less than 0.05 was considered statistically significant. Statistical analysis was performed in the SPSS Statistics 22 software (IBM Corporation, Somers, NY, USA).
Results
Species and resistance genes
During the eleven months of study, 3044 isolates of Enterobacteriaceae were recovered; 86/3044 (2.83%) of them were carbapenem-resistant and carbapenemase-producers. From each enterobacterial species, one isolate per patient was selected for the study (52 isolates).
Five species were found to produce a carbapenemase: K. pneumoniae (n = 46, 88%), E. cloacae (n = 3, 6%), E. coli, P. rettgeri and Citrobacter freundii (n = 1, 2% each).
The blaNDM-1 gene was detected in 48/52 (92.3%) isolates belonging to K. pneumoniae (n = 43, 90%), E. cloacae (n = 3, 6%), E. coli and P. rettgeri (n = 1, 2%, each).
The blaVIM gene was detected in 3/52 (5.6%) isolates belonging to K. pneumoniae (n = 2) and C. freundii (n = 1). The blaKPC gene was detected in one isolate of K. pneumoniae, and the P. rettgeri isolate harbored two carbapenemase genes (blaNDM-1 and blaIMP). The blaOXA-48 gene was not detected in any of the isolates.
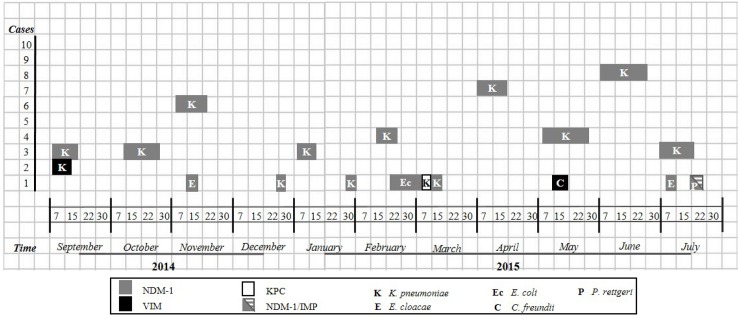
The first carbapenem-resistance isolate was an NDM-1-producing K. pneumoniae (Fig 1). Isolates harboring the blaNDM-1 gene were detected throughout the study.
Fig 1. Temporal distribution of recovered carbapenem-resistant Enterobacteriaceae isolates from September 2014 to July 2015.
Grey squares represent blaNDM-1; black squares represent blaVIM; white squares represent blaKPC; grey and lined squares represent the blaNDM-1/blaIMP. K represents K. pneumoniae; E represents E. cloacae; Ec represents E. coli; C represents C. freundii; and P represents P. rettgeri.
The blaSHV gene was detected in 45/52 (86.5%) isolates, the blaCTX-M gene was detected in 39/52 (75%) isolates, the blaTEM gene was detected in 30/52 (57.7%) isolates and, blaCYM, mcr-1, and mcr-2 genes were not detected in any isolate.
Demographic and clinical data
The 52 isolates were recovered from 51 patients, with one of them was co-infected with two blaNDM-1 species (E. cloacae and E. coli). The mean age was 44 (0–81) years; 66.7% were male, and the mean length of stay (LOS) before positive culture was 26 (range 3–76) days. Clinical characteristics and outcome of the patients are shown in S1 Table. The most frequent cause of hospitalization was brain injury (20% of patients), followed by lower respiratory tract infection (15%), chronic renal failure, and burn injuries (7.5% each).
Antimicrobial susceptibility patterns
Regarding all the 52 CRE, resistance to ampicillin (100%), ceftriaxone (100%), trimethoprim/sulfamethoxazole (98%), ertapenem (96%), fosfomycin (92%), meropenem (90%), imipenem (88%), ciprofloxacin (87%), gentamicin (83%), amikacin (79%), and aztreonam (73%) was detected. Lower resistance to chloramphenicol (52%), tigecycline (19%), and colistin (4%) was observed. The two colistin-resistant isolates corresponded to K. pneumoniae positives for blaNDM-1 and the patients were treated with carbapenems and colistin before the CRE isolation, and after the CRE isolation, a combined therapy of colistin and tigecycline was used.
From the CRE isolates, 51/52 (98%) were classified as MDR and the single non-MDR isolate corresponded to C. freundii positive for blaVIM carbapenemase. The C. freundii isolate was susceptible to the three carbapenems tested by microdilution but resistant to meropenem and ertapenem using the VITEK system. The other K. pneumoniae isolates that harbored blaVIM presented only resistance to imipenem.
Considering the E. cloacae isolates, all were resistant to all antibiotics tested except for tigecycline (one strain resistant).
Regarding only the NDM-1-producing K. pneumoniae, a high drug resistance was detected. The minimum inhibitory concentration (MIC) range, MIC90, and MIC50, as well as the percentage of resistant and susceptible isolates to each of the antimicrobial agents tested, are indicated in Table 1.
Table 1. Antimicrobial susceptibility of blaNDM-1 producing K. pneumoniae isolates (n = 43).
| Antibiotic | MIC (mg/L) | Isolates n (%) | |||
|---|---|---|---|---|---|
| Range | 50% | 90% | Susceptible | Resistant | |
| Amikacin | ≤4–≥ 128 | ≥ 128 | ≥ 128 | 6 (14) | 37 (86) |
| Gentamicin | ≤1- ≥ 32 | ≥ 32 | ≥ 32 | 5 (11.6) | 38 (88.4) |
| Ertapenem | 4–≥ 128 | 16 | 32 | 0 (0) | 43 (100) |
| Imipenem | 2–≥256 | 8 | 16 | 0 (0) | 38 (88.4) |
| Meropenem | 2–256 | 8 | 16 | 1 (2.3) | 40 (93) |
| Ceftriaxone | ≥ 64 | ≥ 64 | ≥ 64 | 0 (0) | 43 (100) |
| Trimethoprim/ Sulfamethoxazole | 8/152–16/304 | 16/304 | 16/304 | 0 (0) | 43 (100) |
| Aztreonam | ≤2- ≥128 | 16 | 32 | 7 (16.3) | 32 (74.4) |
| Ampicillin | 64- ≥128 | ≥ 128 | ≥ 128 | 0 (0) | 43 (100) |
| Ciprofloxacin | ≤0.5–≥16 | ≥ 16 | ≥ 16 | 3 (7) | 38 (88.4) |
| Fosfomycin | 256–≥512 | 512 | ≥ 512 | 0 (0) | 43 (100) |
| Chloramphenicol | ≤4- ≥128 | 16 | 128 | 6 (14) | 20 (46.5) |
| Colistin | ≤0.5- ≥16 | ≤0.5 | 2 | 41 (95.3) | 2 (4.7) |
| Tigecycline | ≤0.5–8 | 2 | 4 | 7 (16.3) | 9 (21) |
Classification of resistance and susceptibility to amikacin, gentamicin, ertapenem, imipenem, meropenem, ceftriaxone, trimethoprim/sulfamethoxazole, aztreonam, ampicillin, ciprofloxacin, fosfomycin, chloramphenicol was based on CLSI interpretive criteria. Classification of resistance and susceptibility to colistin and tigecycline was based on EUCAST interpretative criteria.
Plasmid pattern, hybridization and transfer of carbapenem resistance
The plasmid profiles of all the 52 CRE clinical isolates harbored between one and five plasmids with sizes of 40 to170 kb. The plasmid profile of NDM-harboring Enterobacteriaceae isolates (n = 48) was heterogeneous with 15 different sizes of plasmids according to Kieser method and eight different sizes by S1 nuclease assay.
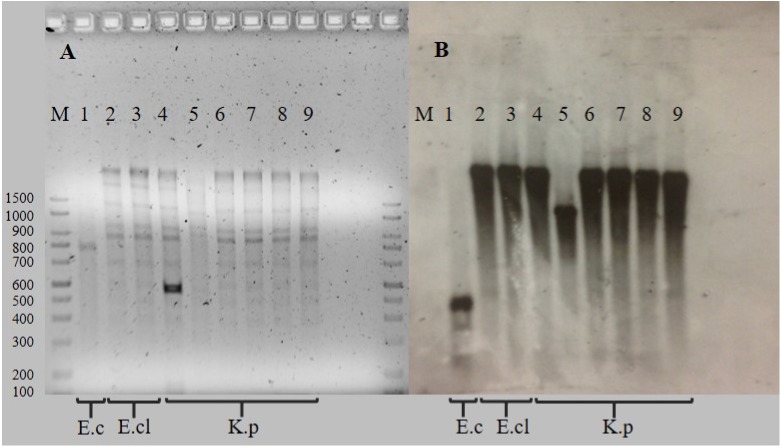
Characteristics of 15 representative isolates of Enterobacteriaceae harboring blaNDM-1 are shown in Table 2. Thirteen isolates (K. pneumoniae, n = 11; and E. cloacae, n = 2) were successful in transfer the resistance with eight transconjugants receiving only one plasmid, (from 130 to 150 kb) and contained the blaNDM-1 gene. Enzymatic digestion revealed one unique restriction pattern, with the blaNDM-1 gene present on a >1.5 kb fragment in seven transconjugants and a smaller 1.0 Kb fragment in one transconjugant (Fig 2). The IncFIIk and IncFIIy incompatibility groups were identified in the eight transconjugants and the E. coli isolate that presented a single plasmid, in 88.9% (8/9) and 22.2% (2/9) of the plasmids, respectively.
Table 2. Characteristics of representative isolates of Enterobacteriaceae harboring blaNDM-1.
| Clinical isolate data | |||||||||||||||
| Strain ID | 14–3335 | 14–3337 | 14–3338 | 14–3423 | 14–3424 | 14–3425 | 14–3442 | 15–0026* | 15–1880 | 15–1327* | 15–1363 | 15–1362 | 15–1372 | 15–1887 | 15–1941 |
| Species | K. pn | K. pn | K. pn | K. pn | K. pn | K. pn | E. cl | E. cl | K. pn | E. coli | K. pn | K. pn | K. pn | K. pn | P. re |
| Susceptibility. MIC mg/L (Interpretation) | |||||||||||||||
| AMP | >128 | 64 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 |
| CRO | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 |
| ETP | >128 | 4 | 8 | 16 | 32 | 16 | 32 | 128 | 32 | 8 | 32 | 32 | 16 | 16 | 4 |
| IMP | 256 | 8 | 8 | 8 | 8 | 8 | 8 | 32 | 256 | 4 | 8 | 4 | 8 | 8 | 4 |
| MEM | 256 | 2 | 4 | 8 | 16 | 8 | 16 | 16 | 128 | 4 | 16 | 8 | 4 | 8 | 8 |
| ATM | 16 | <2 | 8 | 16 | 16 | 16 | 16 | 16 | 16 | 8 | 16 | <2 | 16 | 16 | <2 |
| SXT | 16/304 | 16/304 | 16/304 | 16/304 | 16/304 | 16/304 | 16/304 | 16/304 | 16/304 | 16/304 | 16/304 | 16/304 | 16/304 | 16/304 | 16/304 |
| GEN | >32 | <1 | 16 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | <1 | >32 | >32 | <1 |
| AMK | >128 | <4 | 16 | >128 | >128 | >128 | >128 | >128 | 128 | 8 | >128 | 16 | >128 | >128 | 16 |
| CIP | >16 | <0.5 | 2 | >16 | >16 | >16 | >16 | >16 | >16 | 2 | >16 | 8 | >16 | >16 | 16 |
| FOF | >512 | 512 | 256 | 512 | 512 | 512 | 512 | >512 | >512 | 32 | >512 | 256 | 512 | >512 | 128 |
| CHL | >128 | <4 | 16 | 32 | 16 | 16 | >128 | >128 | 32 | >128 | 128 | 64 | 32 | 32 | 128 |
| TGC | 2 | <0.5 | 1 | 2 | 2 | 2 | 1 | <0.5 | <0.5 | <0.5 | 2 | 2 | 2 | 8 | 1 |
| CST | <0.5 | <0.5 | <0.5 | <0.5 | <0.5 | <0.5 | <0.5 | <0.5 | <0.5 | <0.5 | <0.5 | 2 | <0.5 | <0.5 | >16 |
| Molecular characterization | |||||||||||||||
| Clone | A (n = 28) | D (n = 1) | E (n = 1) | A1 (n = 2) | A2 (n = 1) | A (n = 28) | A1 (n = 1) | A (n = 2) | A5 (n = 1) | N/A | A3 (n = 2) | G (n = 1) | A4 (n = 1) | A6 (n = 1) | N/A |
| ST | 392 | 309 | 846 | 307 | N/D | 392 | 182 | N/D | N/D | 10 | N/D | 2400 | N/D | N/D | N/A |
| Plasmid profile by Kieser (No. of isolates) |
130, 160, 165 (21) |
130, 145, 155 (1) | 130, 145, 155 (1) | 130, 150, 160, 170 (1) | 130, 162, 166 (3) | 130, 164, 166 (7) | 135, 142, 150 (1) | 130, 142, 150 (2) | 130, 162, 166 (1) |
140 (1) | 70, 78, 130, 142, 166 (2) | 160, 170 (4) |
130,163, 166 (1) |
130, 163, 166 (1) |
128, 165 (1) |
| Plasmid profile by S1 nuclease assay | 82 | 82, 90, 96 | 52, 77, 90, 96 | 82 | 82, 90, 96 | 82 | 82, 86, 117, 122 | 82, 100, 117, 142 | 82 | 77 | 82, 102, 133, 142 | 82 | 82 | 82 | 59 |
| Plasmid carrying NDM-1 (kb) | 130 | 130 | 130 | 130, 150 | 130 | 130 | 150 | 150 | 130 | 130 | 130 | 170 | 130 | 130 | - |
| Positive conjugation (presented a single plasmid) | Yes (Yes) | Yes (Yes) | Yes (No) | Yes (No) | Yes (Yes) | Yes (Yes) | Yes (Yes) | Yes (Yes) | Yes (No) | No | Yes (Yes) | Yes (No) | Yes (Yes) | Yes (No) | No |
| Incompatibility groups | IIIk | FIIy | N/D | N/D | FIIy | FIIy | FIIy | FIIy | N/D | FIIy, FIIk | FIIy | N/D | FIIy | N/D | No |
(*): same patient
K. pn: K. pneumoniae; E. cl: E. cloacae; N/D: Not detected; N/A: Not applicable; AMP: ampicillin; CRO: ceftriaxone; ETP: ertapenem; IMP: imipenem; MEM: meropenem; ATM: aztreonam; SXT: trimethoprim/sulfamethoxazole; GEN: gentamicin; AMK: amikacin; CIP: ciprofloxacin; FOF: fosfomycin; CHL: chloramphenicol; TGC: tigecycline; CST: colistin.
Fig 2. Restriction pattern and Southern hybridization of selected plasmids.
(A): Restriction pattern; (B): Southern hybridization; M: Molecular weight marker of 1.5 kb; 1: 15–1327; 2: TT 14–3442 E. cloacae; 3: TT 15–0026; 4: TT 14–3335; 5: TT 14–3337; 6: TT 14–3424; 7: TT 14–3425; 8: TT 15–1363; 9: TT 15–1372. TT represents transconjugants; E.cl represents E. cloacae; E.c represents E. coli; and K.p. represents K. pneumoniae.
The Southern hybridization experiments showed the presence of the blaNDM-1 gene in four different plasmids, with sizes from 130 to 170 kb. One blaNDM-1 gene copy was identified in two different plasmids (130 and 150 kb) harbored in K. pneumoniae (14–3423) isolate. Conjugation was unsuccessful for the E. coli, and P. rettgeri clinical isolates. The Southern hybridization experiment was unsuccessful for the P. rettgeri isolate.
Clonal diversity
K. pneumoniae (n = 46) and E. cloacae (n = 3) isolates were subjected to PFGE assays. Regarding K. pneumoniae, 14 distinct patterns were detected. The percentage of similarity ranged from 75% to 100%, with restriction patterns of 15–20 bands and the 60.9% (28/46) of the isolates corresponding to clone A; 4.3% (2/46) to clones B and C; and 2.2% (1/46) to clones D to H.
Clone A harbored blaNDM-1, Clone B, and F strains harbored blaVIM and blaKPC genes, respectively. The clone A presented six subtypes (19.5%, 9/46) with two or three differences in the restriction pattern in comparison with clone A restriction pattern.
Regarding E. cloacae isolates, they were classified as closely related with only two different bands in the restriction pattern and are considered as subtypes. The Clone A and A1 presented one and two isolates, respectively.
Regarding MLST assays, selected isolates were K. pneumoniae (n = 6), E. coli and E. cloacae (n = 1 each). Regarding K. pneumoniae, only isolates harboring blaNDM-1 gene and with different plasmid profile were selected. Four ST previously reported were detected (ST392, strain 14–3335; ST309, strain 14–3337; ST846, strain 14–3338; and ST307, strain 14–3423). Furthermore, two new ST were identified (ST2400, strain 15–1362; and ST2399, strain 15–1600). The clone A corresponded to ST392 and was detected during the 11 months of surveillance.
E. coli (15–1327) isolate corresponded to ST10, and E. cloacae (14–3442) isolate corresponded to ST182.
Virulence factors
Virulence factors analyzed in both carbapenem-resistant and carbapenem susceptible isolates showed that the presence of fimA and uge genes was more likely to be detected in carbapenem-susceptible isolates (P = <0.001) (Table 3).
Table 3. Distribution of virulence factors in blaNDM-1 producing K. pneumoniae isolates and carbapenem susceptible isolates.
| Group | Virulence genes | Biofilm production | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| entB | iroB, K2 | irp2 | fimA | fimH | mrkA | mrkD | uge | ureA | rmpA | non-adherent | weakly adherent | strongly adherent | |
| CR | 97.7% (42/43) |
0% (0/43) |
60.5% (26/43) |
18.6% (8/43) |
100% (43/43) |
74.4% (32/43) |
0% (0/43) |
27.9% (12/43) |
76.7% (33/43) |
0% (0/43) |
69.8% (30/43) |
4.6% (2/43) |
25.6% (11/43) |
| CS | 100% (23/23) |
13% (3/23) |
47.8% (11/23) |
65.2% (15/23) |
95.7% (22/23) |
100% (23/23) |
4.3% (1/23) |
82.6% (19/23) |
100% (23/23) |
8.7% (2/23) |
100% (23/23) | 0% (0/23) | 0% (0/23) |
| P-value | NS | NS | 0.467 | <0.001 | NS | NS | NS | <0.001 | NS | NS | NS | NS | NS |
CR: Carbapenem-resistant isolates; CS: Carbapenem-susceptible isolates. NS: Not significant. iucA, K1, and rmpA2 genes were not detected.
Regarding biofilm production, the 81.8% (54/66) of the isolates were classified as non-adherent, 1.5% (1/66) as weakly adherent, and 16.7% (11/66) as strongly adherent. All strongly and weakly adherent isolates were carbapenem-resistant (Table 3). Biofilm production (strong plus weak) was found to be associated with carbapenem resistance (P = <0.05).
Discussion
In this study, we characterized the epidemiological, microbiological, and molecular data of an outbreak of CRE in a tertiary-care hospital in western Mexico and detected that the most commonly gene identified was blaNDM-1 in a predominant clone A of K. pneumoniae. The presence of only one carbapenemase type (blaNDM-1) involving K. pneumoniae, E. cloacae, and E. coli in four epidemiologically related patients was reported in a tertiary care hospital in Mexico City in 2015 [6], but not comprising the high number of species and isolates reported in this study.
In addition to being detected in K. pneumoniae, E. cloacae, and E. coli, the blaNDM-1 gene has been reported in P. rettgeri isolates [4]. In this species, resistance to carbapenems is rarely described, and when is reported, it is mainly associated with blaNDM-1 [4,33]. In this study, the P. rettgeri isolate presented both blaNDM-1 and the blaIMP gene. The presence of blaIMP in P. rettgeri has been reported only in Japan [34,35], and to the best of our knowledge, the presence of blaNDM-1 and blaIMP in the same isolate has not been reported to date. Unfortunately, the P. rettgeri isolate lost the plasmid carrying blaNDM-1 during experiments, and we were unable to characterize it. Due to the presence of the blaIMP gene, the isolate remained carbapenem-resistant.
Of the several carbapenemases previously described worldwide, the blaKPC has been reported as the predominant carbapenemase gene associated with CRE intrahospital infections [36]. Nevertheless, in the recent years, blaNDM-1 gene has frequently been associated with outbreaks, particularly with strains of K. pneumoniae and E. coli [3].
We detected more than one CRE species and multiple carbapenemases genes in the same hospital in the same period. This diversity has been previously reported, but in countries geographically distant from Mexico (China and Kuwait) [37,38]. Because of this, our report underscores the importance of active surveillance in all enterobacterial species.
The transfer of plasmids of 130 to170 kb carrying blaNDM-1 was demonstrated for K. pneumoniae, and E. cloacae and these experiments partially explain, the high dissemination inter and intra-species observed during the outbreak. In contrast, the inability of conjugation of E. coli and P. rettgeri could explain the lack of dissemination of these species/plasmids during the outbreak.
Similar restriction patterns were detected in eight plasmids of similar size (130 to150 kb) with the blaNDM-1 gene present on the same restriction fragment in 7/8 plasmids. These results and the different size of plasmids, strongly suggest rearrangements of plasmids during the short period of this outbreak. Rearrangements of plasmids have been previously reported for plasmids harboring blaVIM gene [39], but to our knowledge, rearrangements in plasmids encoding blaNDM-1 have not been previously described.
One of the most significant findings of our study was the high attributable mortality detected for any CRE (35%, 14/40), even that this percentage was lower than previous reports (57.4%, 54/94) in clinical isolates of K. pneumoniae, E. coli, E. cloacae, C. freundii, Enterobacter aerogenes, Klebsiella oxytoca, Raoultella ornithinolytica and Raoultella planticola obtained from 2010 to 2014 in China [37]. Furthermore, a high attributable mortality related to CRE blaNDM-1 producers was observed, regarding that lower values have been reported (28.6%, 6/21) in Kuwait for K. pneumoniae, E. coli, E. cloacae, M. morganii and P. stuartii recovered in 2014 [38].
In our study, the 19.6% isolates were resistant to tigecycline. The high resistance detected is a point of concern because the SENTRY Antimicrobial Surveillance Program reported in 2016 that only 2.6% of CRE from Latin America presented tigecycline resistance [40], and by these new findings, we may infer that tigecycline resistance is increasing.
Colistin resistance was also detected (4%), and this is now a serious global menace as previous reports from India and UK showed percentages of resistance of 6% and 11%, respectively [41]. This study is the second report of colistin-resistant Enterobacteriaceae in Mexico [42]. The molecular mechanism of resistance to colistin presented in these colistin-resistant isolates was not acquired due to mcr gene was not detected, and additional analysis is ongoing in our group to evaluate the mechanism(s) involved.
We detected K. pneumoniae ST307, ST392, and ST846 which have been reported as harboring blaKPC, blaOXA-48 and blaNDM-1 genes [43–45]. In contrast, the K. pneumoniae ST309 detected has not been related to carbapenemases genes.
E. cloacae ST182 harboring blaNDM-1 has been reported in Mexico and Finland [6,46]; the ST10 of E. coli has only been reported in isolates harboring ESBL genes and not carbapenemase genes [47].
The combination of clonal expansion and horizontal gene transfer demonstrated in this study has been described in Mexico, UK, and Chennai, India [6,41]. In contrast, isolates from Haryana, India, showed an apparent clonal expansion by the demonstration of two types of predominant plasmids [41].
In our study, we evaluated 14 virulence genes in all carbapenem-resistant and in selected carbapenem-susceptible K. pneumoniae isolates, and we found that the frequency of virulence genes was similar to reported when 6 K pneumoniae KPC +, ST258 were analyzed [30].
The virulence gene distribution between both groups was similar except for the fimA and uge genes. The presence of fimA and uge genes was more frequently detected in carbapenem-susceptible K. pneumoniae isolates (P <0.001). The type 1 fimbriae, encoded by fimA, contribute to the invasion of bladder cells and biofilm formation. Despite the low frequency of the fimA gene in the carbapenem-resistant isolates, these strains were capable of high biofilm production.
In this study, we confirmed that biofilm production is higher in the carbapenem-resistant isolates than in the carbapenem-susceptible isolates as previously reported [8], and the presence of biofilm may contribute to drug resistance. However, more studies are required to define the impact of virulence genes.
This studies had several limitations. First, there was a lack of information about the travel of patients or healthcare workers to places where NDM-1-producing strains are endemic. However, blaNDM-1 strains been reported in Mexico previously, and the dissemination could be the result of regional transmission. Another significant limitation of our study is the absence of analysis of CRE carriers in the hospital. The above limitations could be resolved in future studies of CRE in this region.
The results obtained in this study indicate that blaNDM-1 was disseminated horizontally among different species in a tertiary care Hospital in Mexico, also with proof of strain spread predominantly of K. pneumoniae ST392. We have provided evidence of plasmid transfer but, given the variation in plasmid sizes, complex rearrangements must also be occurring. In this analysis, the presence of other carbapenemase genes encoding blaVIM, blaKPC and blaIMP were sporadic.
Supporting information
(DOC)
(DOC)
Acknowledgments
The authors thank Alejandro Sanchez-Perez and Teresa Rojas for their assistance in the laboratory. We are grateful to the team of the curators of the Institut Pasteur MLST system (Paris, France) for importing novel alleles, profiles, and/or isolates at http://bigsdb.pasteur.fr/. This work was partially supported by grants 136339, 130224 and 256927 from CONACyT (Mexican Council for Science and Technology).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by grants 136339 and 130224 from Mexican Council for Science and Technology (http://conacyt.gob.mx/) to JSS.
References
- 1.Rozales FP, Ribeiro VB, Magagnin CM, Pagano M, Lutz L, et al. (2014) Emergence of NDM-1-producing Enterobacteriaceae in Porto Alegre, Brazil. Int J Infect Dis 25: 79–81. doi: 10.1016/j.ijid.2014.01.005 [DOI] [PubMed] [Google Scholar]
- 2.Wei WJ, Yang HF, Ye Y, Li JB (2015) New Delhi Metallo-beta-Lactamase-Mediated Carbapenem Resistance: Origin, Diagnosis, Treatment and Public Health Concern. Chin Med J (Engl) 128: 1969–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dortet L, Poirel L, Nordmann P (2014) Worldwide dissemination of the NDM-type carbapenemases in Gram-negative bacteria. Biomed Res Int 2014: 249856 doi: 10.1155/2014/249856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrios H, Garza-Ramos U, Reyna-Flores F, Sanchez-Perez A, Rojas-Moreno T, et al. (2013) Isolation of carbapenem-resistant NDM-1-positive Providencia rettgeri in Mexico. J Antimicrob Chemother 68: 1934–1936. doi: 10.1093/jac/dkt124 [DOI] [PubMed] [Google Scholar]
- 5.Barrios H, Silva-Sanchez J, Reyna-Flores F, Sanchez-Perez A, Sanchez-Francia D, et al. (2014) Detection of a NDM-1-producing Klebsiella pneumoniae (ST22) clinical isolate at a pediatric hospital in Mexico. Pediatr Infect Dis J 33: 335 doi: 10.1097/INF.0000000000000173 [DOI] [PubMed] [Google Scholar]
- 6.Torres-Gonzalez P, Bobadilla-Del Valle M, Tovar-Calderon E, Leal-Vega F, Hernandez-Cruz A, et al. (2015) Outbreak caused by Enterobacteriaceae harboring NDM-1 metallo-beta-lactamase carried in an IncFII plasmid in a tertiary care hospital in Mexico City. Antimicrob Agents Chemother 59: 7080–7083. doi: 10.1128/AAC.00055-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poirel L, Dortet L, Bernabeu S, Nordmann P (2011) Genetic features of blaNDM-1-positive Enterobacteriaceae. Antimicrob Agents Chemother 55: 5403–5407. doi: 10.1128/AAC.00585-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuursted K, Scholer L, Hansen F, Dam K, Bojer MS, et al. (2012) Virulence of a Klebsiella pneumoniae strain carrying the New Delhi metallo-beta-lactamase-1 (NDM-1). Microbes Infect 14: 155–158. doi: 10.1016/j.micinf.2011.08.015 [DOI] [PubMed] [Google Scholar]
- 9.Bandeira M, Carvalho PA, Duarte A, Jordao L (2014) Exploring Dangerous Connections between Klebsiella pneumoniae Biofilms and Healthcare-Associated Infections. Pathogens 3: 720–731. doi: 10.3390/pathogens3030720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levesque S, Dufresne PJ, Soualhine H, Domingo MC, Bekal S, et al. (2015) A Side by Side Comparison of Bruker Biotyper and VITEK MS: Utility of MALDI-TOF MS Technology for Microorganism Identification in a Public Health Reference Laboratory. PLoS One 10: e0144878 doi: 10.1371/journal.pone.0144878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.European Committee on Antimicrobial Susceptibility Testing EUCAST. Clinical breakpoints—bacteria (v 60) 2016. [DOI] [PMC free article] [PubMed]
- 12.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. Performance standards for antimicrobial susceptibility testing: Twenty-six informational supplement M100S CLSI, Wayne, PA, USA, 2016.
- 13.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, et al. (2012) Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18: 268–281. doi: 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- 14.Nordmann P, Poirel L (2002) Emerging carbapenemases in Gram-negative aerobes. Clin Microbiol Infect 8: 321–331. [DOI] [PubMed] [Google Scholar]
- 15.Ellington MJ, Kistler J, Livermore DM, Woodford N (2007) Multiplex PCR for rapid detection of genes encoding acquired metallo-beta-lactamases. J Antimicrob Chemother 59: 321–322. doi: 10.1093/jac/dkl481 [DOI] [PubMed] [Google Scholar]
- 16.Baroud M, Dandache I, Araj GF, Wakim R, Kanj S, et al. (2013) Underlying mechanisms of carbapenem resistance in extended-spectrum beta-lactamase-producing Klebsiella pneumoniae and Escherichia coli isolates at a tertiary care centre in Lebanon: role of OXA-48 and NDM-1 carbapenemases. Int J Antimicrob Agents 41: 75–79. doi: 10.1016/j.ijantimicag.2012.08.010 [DOI] [PubMed] [Google Scholar]
- 17.Chang FY, Siu LK, Fung CP, Huang MH, Ho M (2001) Diversity of SHV and TEM beta-lactamases in Klebsiella pneumoniae: gene evolution in Northern Taiwan and two novel beta-lactamases, SHV-25 and SHV-26. Antimicrob Agents Chemother 45: 2407–2413. doi: 10.1128/AAC.45.9.2407-2413.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perez-Perez FJ, Hanson ND (2002) Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol 40: 2153–2162. doi: 10.1128/JCM.40.6.2153-2162.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bontron S, Poirel L, Nordmann P (2016) Real-time PCR for detection of plasmid-mediated polymyxin resistance (mcr-1) from cultured bacteria and stools. J Antimicrob Chemother 71: 2318–2320. doi: 10.1093/jac/dkw139 [DOI] [PubMed] [Google Scholar]
- 20.Kieser T (1984) Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid 12: 19–36. [DOI] [PubMed] [Google Scholar]
- 21.Barton BM, Harding GP, Zuccarelli AJ (1995) A general method for detecting and sizing large plasmids. Anal Biochem 226: 235–240. doi: 10.1006/abio.1995.1220 [DOI] [PubMed] [Google Scholar]
- 22.Miller JH (1972) Experiments in molecular genetics. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory; xvi, 466 p. p. [Google Scholar]
- 23.Hamprecht A, Poirel L, Gottig S, Seifert H, Kaase M, et al. (2013) Detection of the carbapenemase GIM-1 in Enterobacter cloacae in Germany. J Antimicrob Chemother 68: 558–561. doi: 10.1093/jac/dks447 [DOI] [PubMed] [Google Scholar]
- 24.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, et al. (2005) Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63: 219–228. doi: 10.1016/j.mimet.2005.03.018 [DOI] [PubMed] [Google Scholar]
- 25.Kaufmann ME (1998) Pulsed-field gel electrophoresis. Methods Mol Med 15: 33–50. doi: 10.1385/0-89603-498-4:33 [DOI] [PubMed] [Google Scholar]
- 26.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, et al. (1995) Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 33: 2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S (2005) Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol 43: 4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wirth T, Falush D, Lan R, Colles F, Mensa P, et al. (2006) Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60: 1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyoshi-Akiyama T, Hayakawa K, Ohmagari N, Shimojima M, Kirikae T (2013) Multilocus sequence typing (MLST) for characterization of Enterobacter cloacae. PLoS One 8: e66358 doi: 10.1371/journal.pone.0066358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garza-Ramos U, Moreno-Dominguez S, Hernandez-Castro R, Silva-Sanchez J, Barrios H, et al. (2016) Identification and Characterization of Imipenem-Resistant Klebsiella pneumoniae and Susceptible Klebsiella variicola Isolates Obtained from the Same Patient. Microb Drug Resist 22: 179–184. doi: 10.1089/mdr.2015.0181 [DOI] [PubMed] [Google Scholar]
- 31.Burmolle M, Norman A, Sorensen SJ, Hansen LH (2012) Sequencing of IncX-plasmids suggests ubiquity of mobile forms of a biofilm-promoting gene cassette recruited from Klebsiella pneumoniae. PLoS One 7: e41259 doi: 10.1371/journal.pone.0041259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stepanovic S, Vukovic D, Hola V, Di Bonaventura G, Djukic S, et al. (2007) Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS 115: 891–899. doi: 10.1111/j.1600-0463.2007.apm_630.x [DOI] [PubMed] [Google Scholar]
- 33.Carvalho-Assef AP, Pereira PS, Albano RM, Beriao GC, Chagas TP, et al. (2013) Isolation of NDM-producing Providencia rettgeri in Brazil. J Antimicrob Chemother 68: 2956–2957. doi: 10.1093/jac/dkt298 [DOI] [PubMed] [Google Scholar]
- 34.Nishio H, Komatsu M, Shibata N, Shimakawa K, Sueyoshi N, et al. (2004) Metallo-beta-lactamase-producing gram-negative bacilli: laboratory-based surveillance in cooperation with 13 clinical laboratories in the Kinki region of Japan. J Clin Microbiol 42: 5256–5263. doi: 10.1128/JCM.42.11.5256-5263.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shiroto K, Ishii Y, Kimura S, Alba J, Watanabe K, et al. (2005) Metallo-beta-lactamase IMP-1 in Providencia rettgeri from two different hospitals in Japan. J Med Microbiol 54: 1065–1070. doi: 10.1099/jmm.0.46194-0 [DOI] [PubMed] [Google Scholar]
- 36.Jean SS, Lee WS, Lam C, Hsu CW, Chen RJ, et al. (2015) Carbapenemase-producing Gram-negative bacteria: current epidemics, antimicrobial susceptibility and treatment options. Future Microbiol 10: 407–425. doi: 10.2217/fmb.14.135 [DOI] [PubMed] [Google Scholar]
- 37.Wang Q, Zhang Y, Yao X, Xian H, Liu Y, et al. (2016) Risk factors and clinical outcomes for carbapenem-resistant Enterobacteriaceae nosocomial infections. Eur J Clin Microbiol Infect Dis. [DOI] [PubMed] [Google Scholar]
- 38.Jamal WY, Albert MJ, Rotimi VO (2016) High Prevalence of New Delhi Metallo-beta-Lactamase-1 (NDM-1) Producers among Carbapenem-Resistant Enterobacteriaceae in Kuwait. PLoS One 11: e0152638 doi: 10.1371/journal.pone.0152638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aschbacher R, Doumith M, Livermore DM, Larcher C, Woodford N (2008) Linkage of acquired quinolone resistance (qnrS1) and metallo-beta-lactamase (blaVIM-1) genes in multiple species of Enterobacteriaceae from Bolzano, Italy. J Antimicrob Chemother 61: 515–523. doi: 10.1093/jac/dkm508 [DOI] [PubMed] [Google Scholar]
- 40.Sader HS, Castanheira M, Farrell DJ, Flamm RK, Mendes RE, et al. (2016) Tigecycline antimicrobial activity tested against clinical bacteria from Latin American medical centres: results from SENTRY Antimicrobial Surveillance Program (2011–2014). Int J Antimicrob Agents 48: 144–150. doi: 10.1016/j.ijantimicag.2016.04.021 [DOI] [PubMed] [Google Scholar]
- 41.Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, et al. (2010) Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis 10: 597–602. doi: 10.1016/S1473-3099(10)70143-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garza-Ramos U, Barrios H, Reyna-Flores F, Sanchez-Perez A, Tamayo-Legorreta E, et al. (2014) Characteristics of KPC-2-producing Klebsiella pneumoniae (ST258) clinical isolates from outbreaks in 2 Mexican medical centers. Diagn Microbiol Infect Dis 79: 483–485. doi: 10.1016/j.diagmicrobio.2014.05.010 [DOI] [PubMed] [Google Scholar]
- 43.Xie L, Dou Y, Zhou K, Chen Y, Han L, et al. (2017) Coexistence of blaOXA-48 and Truncated blaNDM-1 on Different Plasmids in a Klebsiella pneumoniae Isolate in China. Front Microbiol 8: 133 doi: 10.3389/fmicb.2017.00133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rojas LJ, Wright MS, De La Cadena E, Motoa G, Hujer KM, et al. (2016) Initial Assessment of the Molecular Epidemiology of blaNDM-1 in Colombia. Antimicrob Agents Chemother 60: 4346–4350. doi: 10.1128/AAC.03072-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu J, Sun L, Ding B, Yang Y, Xu X, et al. (2016) Outbreak of NDM-1-producing Klebsiella pneumoniae ST76 and ST37 isolates in neonates. Eur J Clin Microbiol Infect Dis 35: 611–618. doi: 10.1007/s10096-016-2578-z [DOI] [PubMed] [Google Scholar]
- 46.Osterblad M, Kirveskari J, Hakanen AJ, Tissari P, Vaara M, et al. (2012) Carbapenemase-producing Enterobacteriaceae in Finland: the first years (2008–11). J Antimicrob Chemother 67: 2860–2864. doi: 10.1093/jac/dks299 [DOI] [PubMed] [Google Scholar]
- 47.Bado I, Gutierrez C, Garcia-Fulgueiras V, Cordeiro NF, Araujo Pirez L, et al. (2016) CTX-M-15 in combination with aac(6')-Ib-cr is the most prevalent mechanism of resistance both in Escherichia coli and Klebsiella pneumoniae, including K. pneumoniae ST258, in an ICU in Uruguay. J Glob Antimicrob Resist 6: 5–9. doi: 10.1016/j.jgar.2016.02.001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.