Abstract
BACKGROUND
African Americans (AA) are disproportionately affected by hypertension-related health disparities. Apolipoprotein L1 (APOL1) risk variants are associated with kidney disease in hypertensive AAs.
OBJECTIVES
This study assessed the APOL1 risk alleles’ association with blood pressure traits in AAs.
METHODS
The discovery cohort included 5,204 AA participants from Mount Sinai’s BioMe biobank. Replication cohorts included additional BioMe (n = 1,623), Vanderbilt BioVU (n = 1,809), and Northwestern NUgene (n = 567) AA biobank participants. Single nucleotide polymorphisms determining APOL1 G1 and G2 risk alleles were genotyped in BioMe and imputed in BioVU/NUgene participants. APOL1 risk alleles’ association with blood pressure–related traits was tested in the discovery cohort, a meta-analysis of replication cohorts, and a combined meta-analysis under recessive and additive models after adjusting for age, sex, body mass index, and estimated glomerular filtration rate.
RESULTS
There were 14% to 16% of APOL1 variant allele homozygotes (2 copies of G1/G2) across cohorts. APOL1 risk alleles were associated under an additive model with systolic blood pressure (SBP) and age at diagnosis of hypertension, which was 2 to 5 years younger in the APOL1 variant allele homozygotes (Cox proportional hazards analysis, p value for combined meta-analysis [pcom] = 1.9 × 10−5). APOL1 risk alleles were associated with overall SBP (pcom = 7.0 × 10−8) and diastolic blood pressure (pcom = 2.8 × 10−4). After adjustment for all covariates, those in the 20- to 29-year age range showed an increase in SBP of 0.94 ± 0.44 mm Hg (pcom = 0.01) per risk variant copy. APOL1-associated estimated glomerular filtration rate decline was observed starting a decade later in life in the 30- to 39-year age range.
CONCLUSIONS
APOL1 risk alleles are associated with higher SBP and earlier hypertension diagnoses in young AAs; this relationship appears to follow an additive model.
Keywords: allele, APOL1, estimated glomerular filtration rate, genetic association studies, kidney disease
Hypertension is a leading cause of morbidity and mortality worldwide. Data from the NHANES (National Health and Nutrition Examination Survey) for 2008 demonstrated a hypertension prevalence rate of 30% in the U.S. adult population (1). There are differences in prevalence between populations, with hypertension occurring earlier in life and more frequently in African Americans (AAs) (2). Additionally, compared with European Americans (EAs), AAs have a higher mortality from, and earlier onset of, complications of hypertension, in particular stroke, heart disease, and kidney disease (3). Average systolic blood pressure (SBP) and diastolic blood pressure (DBP) are 4 and 2 mm Hg higher, respectively, in AAs compared with EAs between 18 and 39 years of age (4). In addition to socioeconomic factors (5), heritability estimates indicate that genetic factors contribute to blood pressure (BP) traits in those of African or European ancestry (6,7). Numerous genetic loci for BP and hypertension have been identified, explaining approximately 2% of variation (8).
AAs are disproportionately affected by various kidney diseases (9). Genetic association studies demonstrated that 2 distinct alleles in the apolipoprotein L1 (APOL1) gene on chromosome 22 confer substantially increased risk for a number of kidney diseases in AAs, including focal segmental glomerulosclerosis, human immunodeficiency virus–associated nephropathy, and hypertension-attributable kidney disease (10–14). APOL1 G1 (rs73885319/rs60910145; 2 missense variants in very high linkage disequilibrium) and G2 (rs71785313; 6 base pair deletion) risk alleles in the last exon of APOL1 confer resistance to lethal Trypanosoma brucei infections in Sub-Saharan Africa, resulting in their selection and considerably higher frequency in individuals of African ancestry compared with other populations (15). This difference partly accounts for health disparities in kidney disease and end-stage renal disease in individuals of African ancestry (12,16,17). APOL1 renal risk alleles were also associated with accelerated progression of chronic kidney disease (CKD) (18). The association between the APOL1 risk alleles and kidney phenotypes is believed to follow a recessive model in which APOL1 variant allele homozygotes (2 copies of G1/G2) are at increased risk. The underlying biological mechanisms, however, remain poorly understood (19).
Because of the increased prevalence and rate of progression of hypertensive kidney disease in individuals of African ancestry, we aimed to assess the association between APOL1 risk variants and BP traits. For this purpose, we utilized genomic data linkable to electronic medical record (EMR) phenotypic data available in several biobanks that contribute to the Electronic Medical Records and Genomics (eMERGE) Network (20).
METHODS
This study was conducted as part of the eMERGE Network of the National Human Genome Research Institute (20). The study was designed by 2 authors (E.E.K., E.P.B), then was reviewed and approved for eMERGE II Network-wide participation by the eMERGE Steering Committee. The institutional review boards at all participating sites approved eMERGE II Network study protocols.
The BioMe discovery cohort consisted of 5,204 self-reported AA participants who enrolled in the EMR-linked BioMe Biobank at the Icahn School of Medicine at Mount Sinai on or before January 23, 2012. An additional 1,623 AA BioMe participants who subsequently enrolled between January 24, 2012, and February 14, 2014, comprised the BioMe replication cohort. External replication cohorts consisted of 1,809 AAs enrolled in the Vanderbilt University Medical Center BioVU Biobank (21), and 567 AAs enrolled in the Northwestern University NUgene Biobank.
GENOTYPING AND PHENOTYPING
Participants of the BioMe discovery and replication cohorts were genotyped using a clinical genetic test to determine APOL1 ancestral (G0), G1, and G2 allele status. All BioVU and NUgene samples were genotyped at their respective sites using a standard genotyping array with 1 million single nucleotide polymorphisms. Following standard genotype data quality control, variants on chromosome 22 were phased and imputed using the SHAPEIT and IMPUTE2 programs, respectively (22,23). The APOL1 variant allele homozygotes were defined as carriers of 2 copies of rs73885319 (G1), 2 copies of rs71785313 (G2), or 1 copy of each. The APOL1 variant allele heterozygotes were defined as carriers of 1 copy of these alleles, and APOL1 ancestral allele homozygotes as carrying zero copies of these alleles. Under the additive model, the sum of the number of G1/G2 alleles was used, and under the recessive model, APOL1 ancestral homozygotes and heterozygotes were joined in the same group.
Age, sex, and AA/black race were extracted from participants’ EMRs for BioVU participants and from enrollment questionnaires of BioMe and NUgene participants. Concurrent Hispanic/Latino ethnicity was not considered. Time-stamped International Classification of Diseases-Ninth Revision-Clinical Modification (ICD-9-CM) codes, medication records, and laboratory values were extracted from longitudinal EMR entries. Body mass index (BMI) was calculated as the ratio between weight and the square of height in kg/m2. The estimated glomerular filtration rates (eGFR) were calculated from serum creatinine value using the Chronic Kidney Disease Epidemiology Collaboration study equation (24). Office-based SBP and DBP represented the average (mean) of all recorded BP readings (referred to as “all”) or average of BP readings prior to prescription of antihypertensive medications (referred to as “untreated”). Medications were identified from electronic prescribing systems, structured problem list entries, and use of validated natural language processing tools (25). BMI and eGFR were averaged for overall and age range–specific values, where 2 or more determinations were available. For the age group–specific analyses, SBP, DBP, BMI, and eGFR values were stratified according to the age of participant at date of determination. Therefore, observations from an individual participant were represented only once in each group analysis, but individuals could have spanned age groups. The date of first recorded entry of hypertension-related ICD-9-CM codes (401.xx) was used to determine participant age at diagnosis of hypertension.
STATISTICAL ANALYSIS
All association tests were performed under both a recessive model and an additive model. For overall mean outpatient SBP and DBP, linear regressions were used with year of birth, sex, and mean BMI as covariates. We performed linear regressions for overall mean eGFR with year of birth, sex, and overall mean BMI as covariates. The association test for age at hypertension onset was performed by Cox proportional hazards analysis with covariates year of birth, sex, and overall mean BMI. For the SBP and DBP age group–specific analyses, linear regressions were performed under 2 models: 1) model 1 with covariates sex and mean BMI; and 2) model 2 with covariates sex, mean BMI, and mean eGFR. For the eGFR age group–specific analyses, linear regressions were performed under 3 models: 1) model 1 with sex and mean BMI as covariates; 2) model 2 with sex, mean BMI, and mean SBP as covariates; and 3) model 3 with sex, mean BMI, mean SBP, and mean DBP as covariates.
Because some of our studied phenotypes were correlated and we replicated our results independently, Bonferroni correction was considered to be too punitive and we established a heuristic study-wide significance at p ≤ 0.01 and a test-wide significance at p ≤ 0.05. All analyses were performed with R version 3.0.3 (R Foundation for Statistical Computing, Vienna, Austria). We used METAL software for meta-analyses of replication cohorts and combined association results (26).
RESULTS
All 9,203 participants with reported African ancestry meeting quality control criteria in the BioMe discovery cohort and 3 replication cohorts (BioMe replication, Vanderbilt BioVU, and Northwestern NUgene) were included. There were 14% to 16% of APOL1 variant allele homozygotes and 40% to 47% of APOL1 variant allele heterozygotes across cohorts. Mean follow-up, based on time between first and last BP measurements, ranged from 3.6 to 7.6 years, and mean number of BP measurements ranged from 19.9 to 37.9 depending on cohort and APOL1 variant allele carrier status (Table 1). Robustness of the APOL1 genotyping/imputation was validated by replication of prior associations between APOL1 risk alleles and eGFR under a recessive model in each cohort, except in Northwestern NUgene (p = 0.14; consistent direction), which had the smallest sample size (Online Table 1).
TABLE 1.
Demographic and Clinical Characteristics
| BioMe Discovery
|
BioMe Replication
|
Vanderbilt BioVU
|
Northwestern NUgene
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
APOL1 Variant Allele Copies
|
p Value |
APOL1 Variant Allele Copies
|
p Value |
APOL1 Variant Allele Copies
|
p Value |
APOL1 Variant Allele Copies
|
p Value | |||||
| 0 or 1 (n = 4,496) | 2 (n = 708) | 0 or 1 (n = 1,356) | 2 (n = 267) | 0 or 1 (n = 1,524) | 2 (n = 285) | 0 or 1 (n = 482) | 2 (n = 85) | |||||
| Age at first blood pressure measurement, yrs | 49.4 ± 14.9 | 48.9 ± 14.6 | 0.34 | 42.5 ± 16.5 | 42.0 ± 15.7 | 0.68 | 48.3 ± 16.0 | 46.0 ± 15.1 | 0.03 | 46.8 ± 13.0 | 43.3 ± 12.6 | 0.03 |
|
| ||||||||||||
| Female | 63.4 | 65.8 | 0.22 | 61.8 | 58.4 | 0.30 | 66.0 | 68.8 | 0.38 | 70.1 | 75.3 | 0.37 |
|
| ||||||||||||
| Genetic African ancestry, % | 80.9 ± 16.0 | 86.8 ± 8.3 | 2.2 × 10−16 | NA | NA | NA | 79.5 ± 11.3 | 82.8 ± 8.8 | 8.5 × 10−8 | 78.3 ± 12.6 | 81.9 ± 9.2 | 2.3 × 10−3 |
|
| ||||||||||||
| Time between first and last blood pressure measurements, yrs | 4.8 ± 2.7 | 4.7 ± 2.6 | 0.34 | 3.5 ± 2.7 | 3.7 ± 2.8 | 0.22 | 6.0 ± 3.8 | 6.3 ± 3.8 | 0.25 | 7.5 ± 4.7 | 7.7 ± 4.9 | 0.72 |
|
| ||||||||||||
| Discrete outpatient encounters with blood pressure recordings, n | 32.1 ± 31.6 | 30.1 ± 27.9 | 0.09 | 21.5 ± 23.2 | 19.9 ± 17.4 | 0.19 | 30.9 ± 33.5 | 33.8 ± 33.0 | 0.18 | 35.6 ± 35.8 | 37.9 ± 35.0 | 0.60 |
|
| ||||||||||||
| Hypertension diagnosis* | 61.1 | 64.5 | 0.08 | 36.7 | 43.8 | 0.03 | 65.3 | 70.9 | 0.08 | 57.9 | 58.8 | 0.91 |
|
| ||||||||||||
| Antihypertensive medications† | 66.5 | 67.7 | 0.58 | 45.6 | 51.3 | 0.09 | 73.5 | 78.6 | 0.08 | 62.2 | 58.8 | 0.55 |
|
| ||||||||||||
| SBP, mm Hg | 130.8 ± 13.9 | 132.2 ± 13.9 | 0.02 | 128.0 ± 14.5 | 131.8 ± 14.8 | 1.7 × 10−4 | 131.4 ± 14.5 | 132.3 ± 13.7 | 0.32 | 126.8 ± 12.5 | 127.6 ± 13.4 | 0.61 |
|
| ||||||||||||
| DBP, mm Hg | 75.8 ± 8.0 | 76.5 ± 8.5 | 0.07 | 74.6 ± 8.5 | 75.7 ± 8.4 | 0.05 | 77.5 ± 8.0 | 79.1 ± 7.7 | 3.5 × 10−3 | 77.4 ± 7.4 | 78.3 ± 7.5 | 0.34 |
|
| ||||||||||||
| eGFR, ml/min/1.73 m2‡ | 83.9 ± 26.9 | 78.4 ± 30.9 | 1.6 × 10−5 | 90.1 ± 27.4 | 82.8 ± 38.8 | 6.0 × 10−3 | 82.1 ± 28.3 | 80.2 ± 31.4 | 0.35 | 85.8 ± 24.9 | 85.8 ± 25.5 | 0.99 |
|
| ||||||||||||
| BMI, kg/m2§ | 30.6 ± 7.9 | 31.4 ± 8.0 | 0.01 | 29.2 ± 8.0 | 30.0 ± 8.0 | 0.13 | 31.9 ± 7.9 | 32.3 ± 7.6 | 0.52 | 32.9 ± 9.5 | 35.7 ± 10.1 | 0.02 |
Values are mean ± SD or %.
Hypertension diagnosis status was assessed with presence of hypertension International Classification of Diseases-Ninth Revision-Clinical Modification diagnostic code.
Percentage of participants prescribed any antihypertensive medications.
Estimated glomerular filtration rate (eGFR) was calculated from measured serum creatinine values by the Chronic Kidney Disease Epidemiology estimating equation.
Body mass index (BMI) is the weight in kilograms divided by the square of the height in meters.
APOL1 = apolipoprotein L1; DBP = diastolic blood pressure; SBP = systolic blood pressure.
In the discovery cohort, there was an association between APOL1 risk alleles and overall SBP (p = 3.3 × 10−3) under a recessive model but no association with DBP (Online Table 1). The combined meta-analysis showed an association between APOL1 risk alleles and overall SBP (p value for the combined meta-analysis [pcom] = 1.6 × 10−7) and DBP (pcom = 4.5 × 10−4) (Online Table 2) under a recessive model.
ASSOCIATIONS OF APOL1 RISK ALLELES
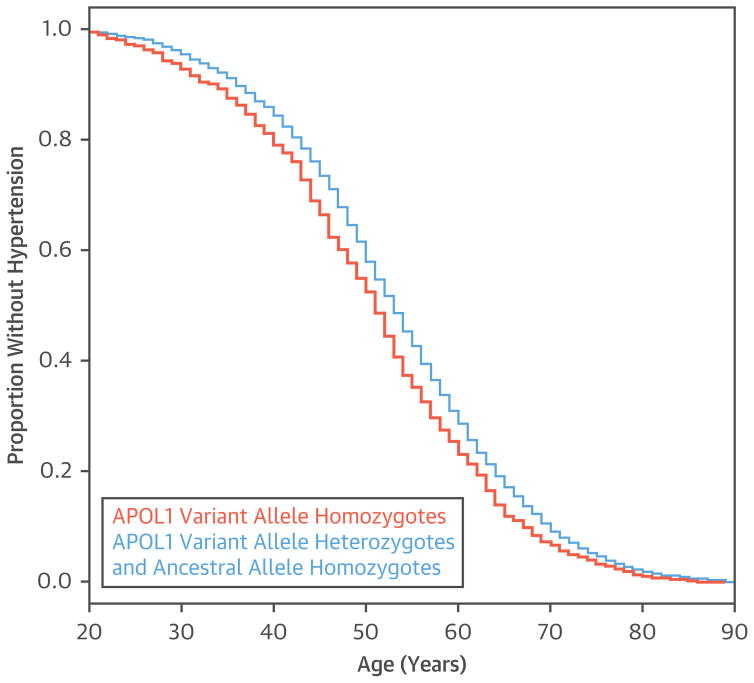
APOL1 risk allele was associated with younger age at hypertension diagnosis in a Cox proportional hazards analysis (p value for the discovery cohort [pdis] = 0.04; p value for the replication cohort [prep] = 3.2 × 10−4; pcom = 1.0 × 10−4; with age, sex, and overall mean BMI as covariates) (Figure 1). The median age of hypertension diagnosis was 2 to 5 years earlier in the APOL1 variant allele homozygotes, depending on the individual biobank cohort (Online Figure 1). APOL1 variant allele homozygote separation of age at hypertension diagnosis manifested in the 20- to 39-year age range (Figure 1).
FIGURE 1. Age at First Diagnosis of Hypertension.
Apolipoprotein L1 (APOL1) variant allele homozygotes (orange) were younger at diagnosis of hypertension than APOL1 ancestral allele homozygotes and variant allele heterozygotes (blue). A Cox proportional hazards analysis with age at hypertension diagnosis adjusted for year of birth, sex, and overall mean body mass index was significant in both the discovery cohort and replication meta-analysis.
To test age-specific associations between APOL1 risk alleles and BP, we conducted linear regression in 3 nonoverlapping 20-year age groups, including 20 to 39, 40 to 59, and 60 to 79 years, consistent with NHANES reports (4). Demographic and clinical characteristics are described for each of the 20-year age groups (Online Tables 3 to 5). Considering all BP measurements, irrespective of absence or presence of treatment in the 20- to 39-year group, APOL1 variant allele homozygosity was associated with a study-wide significantly higher SBP after adjustment for sex and BMI (model 1: discovery cohort, 2.50 ± 0.93 mm Hg, pdis = 7.3 × 10−3; replication cohort, 2.90 ± 0.83 mm Hg, prep = 4.7 × 10−4; and combined, 2.72 ± 0.62 mm Hg, pcom = 1.1 × 10−5) (Table 2), and remained associated after further adjustment for mean interval eGFR (model 2: replication cohort, 2.09 ± 0.86 mm Hg; prep = 0.01; and combined 1.91 ± 0.64 mm Hg; pcom = 2.7 × 10−3) (Table 2). We then excluded BP measurements obtained after participants were prescribed antihypertensive medications to eliminate potentially confounding effects of BP treatment and/or treatment response. APOL1 variant allele homozygosity was associated with higher SBP obtained in the absence of treatment for the 20-to 39-year age range (model 1: discovery cohort, 1.98 ± 0.96 mm Hg; pdis = 0.04; replication cohort, 2.61 ± 0.84 mm Hg, prep = 1.9 × 10−3; combined 2.34 ± 0.63 mm Hg; pcom = 2.2 × 10−4) and after further adjustment for interval eGFR (1.72 ± 0.63 mm Hg; pcom = 6.6 × 10−3) (Table 2). APOL1 variant allele homozygosity was associated with higher DBP (discovery cohort, 1.71 ± 0.68 mm Hg; pdis = 0.01; replication cohort, 1.33 ± 0.58 mm Hg; prep = 0.02; combined 1.49 ± 0.44 mm Hg; pcom = 7.1 × 10−4, respectively), but after additional adjustment for eGFR, this was attenuated to a nominal association achieved only in the combined analysis (model 2: 0.90 ± 0.45 mm Hg; p = 0.05) (Table 3). The APOL1 risk alleles were not associated with BP in the 40- to 59-year age group (Online Table 6), and an association in the 60- to 79-year age range (2.07 ± 0.75 mm Hg; pcom = 5.8 × 10−3) was attenuated after adjusting for eGFR (model 2: 1.79 ± 0.74 mm Hg; pcom = 0.02) in this age group (Online Table 7).
TABLE 2.
Association of APOL1 Risk Alleles With SBP
| BioMe Discovery | Replication Meta-Analysis | Combined Analysis | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||||
| N | 0 or 1 Copy | 0 Copies | 1 Copy | 2 Copies | β ± SE | p Value | β ± SE | p Value | β ± SE | p Value | |
|
| |||||||||||
| Model 1: Adjusted for Sex and BMI* | |||||||||||
| All patients | |||||||||||
| 20–39 yrs | |||||||||||
| Recessive | 1,132 | 122.4 ± 12.0 | 125.9 ± 14.4 | 2.50 ± 0.93 | 7.3 × 10−3 | 2.90 ± 0.83 | 4.7 × 10−4 | 2.72 ± 0.62 | 1.1 × 10−5 | ||
| Additive | 1,132 | 121.8 ± 11.5 | 123.1 ± 12.6 | 125.9 ± 14.4 | 1.40 ± 0.45 | 2.1 × 10−3 | 1.68 ± 0.43 | 9.6 × 10−5 | 1.55 ± 0.31 | 7.4 × 10−7 | |
| 20–29 yrs | |||||||||||
| Recessive | 472 | 119.7 ± 11.1 | 122.3 ± 11.4 | 2.27 ± 1.34 | 0.09 | 3.80 ± 1.02 | 1.8 × 10−4 | 3.24 ± 0.81 | 6.2 × 10−5 | ||
| Additive | 472 | 119.7 ± 10.6 | 119.8 ± 11.8 | 122.3 ± 11.4 | 1.01 ± 0.64 | 0.11 | 1.84 ± 0.53 | 4.5 × 10−4 | 1.51 ± 0.41 | 2.0 × 10−4 | |
| 30–39 yrs | |||||||||||
| Recessive | 814 | 124.0 ± 12.8 | 127.6 ± 15.2 | 2.59 ± 1.18 | 0.03 | 2.28 ± 1.11 | 0.04 | 2.42 ± 0.81 | 2.8 × 10−3 | ||
| Additive | 814 | 123.1 ± 12.3 | 125.1 ± 13.2 | 127.6 ± 15.2 | 1.64 ± 0.58 | 5 × 10−3 | 1.22 ± 0.58 | 0.04 | 1.43 ± 0.41 | 5.3 × 10−4 | |
| Untreated patients | |||||||||||
| 20–39 yrs | |||||||||||
| Recessive | 946 | 120.5 ± 10.8 | 123.1 ± 12.6 | 1.98 ± 0.96 | 0.04 | 2.61 ± 0.84 | 1.9 × 10−3 | 2.34 ± 0.63 | 2.2 × 10−4 | ||
| Additive | 946 | 120.2 ± 10.3 | 120.8 ± 11.3 | 123.1 ± 12.6 | 1.07 ± 0.46 | 0.02 | 1.54 ± 0.43 | 3.1 × 10−4 | 1.32 ± 0.31 | 2.3 × 10−5 | |
| 20–29 yrs | |||||||||||
| Recessive | 424 | 118.6 ± 10.0 | 121.8 ± 10.9 | 2.97 ± 1.30 | 0.02 | 2.69 ± 1.00 | 7.2 × 10−3 | 2.79 ± 0.79 | 4.2 × 10−4 | ||
| Additive | 424 | 118.5 ± 10.1 | 118.7 ± 10.2 | 121.8 ± 10.9 | 1.29 ± 0.63 | 0.04 | 1.38 ± 0.50 | 6.1 × 10−3 | 1.34 ± 0.39 | 6.0 × 10−4 | |
| 30–39 yrs | |||||||||||
| Recessive | 626 | 121.5 ± 11.1 | 123.8 ± 13.5 | 1.30 ± 1.24 | 0.30 | 2.78 ± 1.19 | 0.02 | 2.07 ± 0.86 | 0.02 | ||
| Additive | 626 | 121.0 ± 10.6 | 122.0 ± 11.6 | 123.8 ± 13.5 | 1.00 ± 0.59 | 0.09 | 1.54 ± 0.61 | 0.01 | 1.26 ± 0.42 | 3.0 × 10−3 | |
|
| |||||||||||
| Model 2: Adjusted for Sex, BMI, and eGFR† | |||||||||||
|
| |||||||||||
| All patients | |||||||||||
| 20–39 yrs | |||||||||||
| Recessive | 1,022 | 122.7 ± 12.2 | 126.4 ± 14.5 | 1.69 ± 0.95 | 0.08 | 2.09 ± 0.86 | 0.01 | 1.91 ± 0.64 | 2.7 × 10−3 | ||
| Additive | 1,022 | 122.2 ± 11.6 | 123.3 ± 12.7 | 126.4 ± 14.5 | 0.91 ± 0.47 | 0.05 | 1.22 ± 0.45 | 6.5 × 10−3 | 1.07 ± 0.32 | 9.2 × 10−4 | |
| 20–29 yrs | |||||||||||
| Recessive | 390 | 120.2 ± 11.4 | 122.1 ± 11.0 | 1.37 ± 1.45 | 0.35 | 2.65 ± 1.08 | 0.01 | 2.20 ± 0.87 | 0.01 | ||
| Additive | 390 | 120.4 ± 10.8 | 120.0 ± 12.1 | 122.1 ± 11.0 | 0.35 ± 0.70 | 0.61 | 1.32 ± 0.56 | 0.02 | 0.94 ± 0.44 | 0.03 | |
| 30–39 yrs | |||||||||||
| Recessive | 736 | 124.1 ± 12.9 | 128.3 ± 15.1 | 1.51 ± 1.21 | 0.21 | 1.93 ± 1.13 | 0.09 | 1.74 ± 0.83 | 0.04 | ||
| Additive | 736 | 123.0 ± 12.3 | 125.5 ± 13.4 | 128.3 ± 15.1 | 1.36 ± 0.59 | 0.02 | 0.91 ± 0.60 | 0.13 | 1.14 ± 0.42 | 6.9 × 10−3 | |
| Untreated patients | |||||||||||
| 20–39 yrs | |||||||||||
| Recessive | 822 | 120.2 ± 10.0 | 123.0 ± 12.3 | 1.70 ± 0.93 | 0.07 | 1.73 ± 0.86 | 0.04 | 1.72 ± 0.63 | 6.6 × 10−3 | ||
| Additive | 822 | 120.1 ± 9.8 | 120.4 ± 10.3 | 123.0 ± 12.3 | 0.74 ± 0.45 | 0.10 | 0.94 ± 0.44 | 0.03 | 0.84 ± 0.31 | 7.0 × 10−3 | |
| 20–29 yrs | |||||||||||
| Recessive | 341 | 118.8 ± 9.9 | 121.0 ± 8.9 | 1.82 ± 1.35 | 0.18 | 1.80 ± 1.13 | 0.11 | 1.81 ± 0.87 | 0.04 | ||
| Additive | 341 | 118.9 ± 9.7 | 118.7 ± 10.2 | 121.0 ± 8.9 | 0.68 ± 0.66 | 0.30 | 1.05 ± 0.56 | 0.06 | 0.89 ± 0.43 | 0.04 | |
| 30–39 yrs | |||||||||||
| Recessive | 536 | 120.8 ± 9.9 | 124.5 ± 13.6 | 1.83 ± 1.21 | 0.13 | 2.38 ± 1.17 | 0.04 | 2.12 ± 0.84 | 0.01 | ||
| Additive | 536 | 120.4 ± 9.8 | 121.3 ± 10.0 | 124.5 ± 13.6 | 1.06 ± 0.57 | 0.06 | 0.88 ± 0.60 | 0.15 | 0.97 ± 0.41 | 0.02 | |
Values are mean ± SD.
All mean values listed are for individuals with nonmissing BMI values.
All mean values listed are for individuals with nonmissing BMI and eGFR values.
β = effect size in mm Hg for APOL1 variant allele homozygotes versus APOL1 variant allele heterozygotes and ancestral allele homozygotes under the recessive model and for 1 copy of APOL1 risk allele under the additive model; SE = standard error; other abbreviations as in Table 1.
TABLE 3.
Association of APOL1 Risk Alleles With DBP
| BioMe Discovery | Replication Meta-Analysis | Combined Analysis | |||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||
| N | 0 or 1 Copy | 2 Copies | β ± SE | p Value | β ± SE | p Value | β ± SE | p Value | |
|
| |||||||||
| Model 1: Adjusted for Sex and BMI* | |||||||||
| All patients | |||||||||
| 20–39 yrs | 1,132 | 73.4 ± 8.2 | 75.7 ± 10.0 | 1.71 ± 0.68 | 0.01 | 1.33 ± 0.58 | 0.02 | 1.49 ± 0.44 | 7.1 × 10−4 |
| 20–29 yrs | 472 | 70.9 ± 7.9 | 73.6 ± 8.7 | 2.59 ± 1.03 | 0.01 | 1.31 ± 0.76 | 0.08 | 1.76 ± 0.61 | 3.9 × 10−3 |
| 30–39 yrs | 814 | 74.9 ± 8.4 | 76.8 ± 10.4 | 1.34 ± 0.84 | 0.11 | 1.48 ± 0.76 | 0.05 | 1.42 ± 0.56 | 0.01 |
| Untreated patients | |||||||||
| 20–39 yrs | 946 | 72.2 ± 7.6 | 73.5 ± 7.9 | 0.93 ± 0.71 | 0.19 | 1.13 ± 0.63 | 0.07 | 1.04 ± 0.47 | 0.03 |
| 20–29 yrs | 424 | 70.2 ± 7.2 | 72.6 ± 6.6 | 2.29 ± 0.97 | 0.02 | 0.51 ± 0.78 | 0.52 | 1.21 ± 0.61 | 0.05 |
| 30–39 yrs | 625 | 73.3 ± 7.8 | 74.1 ± 8.8 | 0.23 ± 0.92 | 0.81 | 2.06 ± 0.88 | 0.02 | 1.19 ± 0.63 | 0.06 |
|
| |||||||||
| Model 2: Adjusted for Sex, BMI, and eGFR† | |||||||||
|
| |||||||||
| All patients | |||||||||
| 20–39 yrs | 1,022 | 73.9 ± 8.1 | 75.9 ± 10.2 | 0.82 ± 0.68 | 0.23 | 0.96 ± 0.60 | 0.11 | 0.90 ± 0.45 | 0.05 |
| 20–29 yrs | 392 | 71.5 ± 8.0 | 73.5 ± 8.7 | 1.60 ± 1.08 | 0.14 | 0.93 ± 0.82 | 0.25 | 1.18 ± 0.65 | 0.07 |
| 30–39 yrs | 736 | 75.2 ± 8.5 | 77.2 ± 10.4 | 0.62 ± 0.86 | 0.47 | 1.22 ± 0.77 | 0.11 | 0.95 ± 0.57 | 0.10 |
| Untreated patients | |||||||||
| 20–39 yrs | 822 | 72.4 ± 7.1 | 73.2 ± 7.8 | 0.21 ± 0.70 | 0.77 | 1.09 ± 0.67 | 0.10 | 0.67 ± 0.49 | 0.17 |
| 20–29 yrs | 343 | 70.7 ± 7.0 | 72.1 ± 5.7 | 1.11 ± 1.02 | 0.28 | 0.54 ± 0.91 | 0.56 | 0.79 ± 0.68 | 0.24 |
| 30–39 yrs | 535 | 73.1 ± 7.2 | 74.2 ± 8.6 | 0.13 ± 0.92 | 0.88 | 1.96 ± 0.92 | 0.03 | 1.05 ± 0.65 | 0.11 |
We next tested the association of eGFR with APOL1 risk alleles in the 20- to 39-year age group, adding relevant covariates successively in 3 different models (Table 4). Compared with basic adjustment for sex and BMI (model 1: −4.83 ± 1.40 ml/min/1.73 m2; p = 5.7 × 10−4), association of APOL1 risk alleles with lower eGFR was attenuated largely by adjustment for SBP (model 2: −3.75 ± 1.39 ml/min/1.73 m2; p = 6.8 × 10−3) and to a minor degree by further adjustment for DBP (model 3: −3.67 ± 1.38 ml/min/1.73 m2; p = 7.8 × 10−3) (Table 4).
TABLE 4.
Association of APOL1 Risk Alleles with eGFR*
| BioMe Discovery
|
Replication Meta-Analysis
|
Combined
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N | 0 or 1 Copy | 0 Copies | 1 Copy | 2 Copies | β ± SE | p Value | β ± SE | p Value | β ± SE | p Value | |
| 20–39 years of age | |||||||||||
| Model 1 recessive | 1,047 | 105.0 ± 22.8 | 97.2 ± 31.2 | −7.13 ± 2.10 | 7.0 × 10−4 | −2.97 ± 1.89 | 0.12 | −4.83 ± 1.40 | 5.7 × 10−4 | ||
| Model 1 additive | 1,047 | 106.0 ± 22.9 | 103.8 ± 22.6 | 97.2 ± 31.2 | −3.56 ± 1.03 | 5.6 × 10−4 | −1.88 ± 0.99 | 0.06 | −2.69 ± 0.71 | 1.6 × 10−4 | |
| Model 2 recessive | 1,022 | 105.1 ± 22.8 | 97.4 ± 30.6 | −5.84 ± 2.07 | 4.9 × 10−3 | −2.05 ± 1.87 | 0.27 | −3.75 ± 1.39 | 6.8 × 10−3 | ||
| Model 2 additive | 1,022 | 106.1 ± 23.0 | 103.9 ± 22.5 | 97.4 ± 30.6 | −2.88 ± 1.02 | 4.7 × 10−3 | −1.21 ± 0.97 | 0.21 | −2.01 ± 0.70 | 4.2 × 10−3 | |
| Model 3 recessive | 1,017 | 105.1 ± 22.8 | 97.4 ± 30.6 | −5.76 ± 2.05 | 5.2 × 10−3 | −1.95 ± 1.86 | 0.29 | −3.67 ± 1.38 | 7.8 × 10−3 | ||
| Model 3 additive | 1,017 | 106.1 ± 23.1 | 103.9 ± 22.5 | 97.4 ± 30.6 | −3.04 ± 1.01 | 2.7 × 10−3 | −1.22 ± 0.97 | 0.21 | −2.10 ± 0.70 | 2.8 × 10−3 | |
|
| |||||||||||
| 20–29 years of age | |||||||||||
| Model 1 recessive | 403 | 111.8 ± 22.9 | 111.1 ± 20.3 | −0.18 ± 3.27 | 0.96 | −0.87 ± 2.67 | 0.75 | −0.59 ± 2.07 | 0.77 | ||
| Model 1 additive | 403 | 113.0 ± 22.8 | 110.4 ± 23.0 | 111.1 ± 20.3 | −1.22 ± 1.57 | 0.44 | −1.48 ± 1.40 | 0.29 | −1.36 ± 1.04 | 0.19 | |
| Model 2 recessive | 390 | 112.0 ± 22.6 | 110.0 ± 19.9 | −0.98 ± 3.24 | 0.76 | 0.27 ± 2.66 | 0.92 | −0.24 ± 2.06 | 0.91 | ||
| Model 2 additive | 390 | 113.3 ± 21.8 | 110.3 ± 23.4 | 110.0 ± 19.9 | −1.77 ± 1.55 | 0.25 | −0.82 ± 1.39 | 0.55 | −1.25 ± 1.03 | 0.23 | |
| Model 3 recessive | 390 | 112.0 ± 22.6 | 110.0 ± 19.9 | −0.23 ± 3.18 | 0.94 | 0.14 ± 2.71 | 0.96 | −0.02 ± 2.06 | 0.99 | ||
| Model 3 additive | 390 | 113.3 ± 21.8 | 110.3 ± 23.4 | 110.0 ± 19.9 | −1.83 ± 1.52 | 0.23 | −0.89 ± 1.40 | 0.52 | −1.32 ± 1.03 | 0.20 | |
|
| |||||||||||
| 30–39 years of age | |||||||||||
| Model 1 recessive | 762 | 101.1 ± 22.8 | 91.0 ± 32.6 | −9.46 ± 2.49 | 1.6 × 10−4 | −1.80 ± 2.33 | 0.44 | −5.37 ± 1.70 | 1.6 × 10−3 | ||
| Model 1 additive | 762 | 101.4 ± 22.6 | 100.6 ± 23.0 | 91.0 ± 32.6 | −3.78 ± 1.23 | 2.3 × 10−3 | −1.03 ± 1.23 | 0.40 | −2.40 ± 0.87 | 5.9 × 10−3 | |
| Model 2 recessive | 736 | 101.1 ± 22.8 | 90.7 ± 32.0 | −8.28 ± 2.46 | 8.0 × 10−4 | −1.18 ± 2.31 | 0.61 | −4.51 ± 1.68 | 7.4 × 10−3 | ||
| Model 2 additive | 736 | 101.4 ± 22.8 | 100.7 ± 22.9 | 90.7 ± 32.0 | −2.83 ± 1.22 | 0.02 | −0.47 ± 1.22 | 0.70 | −1.65 ± 0.86 | 0.06 | |
| Model 3 recessive | 731 | 101.1 ± 22.9 | 90.7 ± 32.0 | −8.32 ± 2.46 | 7.7 × 10−4 | −1.04 ± 2.31 | 0.65 | −4.45 ± 1.69 | 8.3 × 10−3 | ||
| Model 3 additive | 731 | 101.4 ± 22.8 | 100.8 ± 22.9 | 90.7 ± 32.0 | −2.86 ± 1.23 | 0.02 | −0.41 ± 1.22 | 0.73 | −1.63 ± 0.86 | 0.06 | |
To refine further the temporal relationship between APOL1 risk alleles and higher SBP or lower eGFR, we further divided the 20- to 39-year age group into 20- to 29-year and 30- to 39year age groups. The association between APOL1 variant allele homozygosity and higher SBP was already observed in the 20- to 29-year age range replication meta-analysis and combined analysis (model 1: 3.80 ± 1.02 mm Hg replication; prep = 1.8 × 10−4; 3.24 ± 0.81 mm Hg combined; pcom = 6.2 × 10−5) (Table 2). Although attenuated, the association persisted after further adjustment for eGFR (model 2: 2.65 ± 1.08 mm Hg replication; prep = 0.01; 2.20 ± 0.87 mm Hg combined; pcom = 0.01). In contrast, we did not observe an association of lower eGFR with APOL1 variant allele homozygosity in the 20- to 29-year age group (−0.59 ± 2.07 ml/min/1.73 m2 combined; pcom = 0.77), but we did observe it in the 30- to 39-year age group (−5.37 ± 1.70 ml/min/1.73 m2 combined; pcom = 1.6 × 10−3) (Table 4).
ASSOCIATION RESULTS UNDER RECESSIVE AND ADDITIVE MODELS
As expected, the overall eGFR association with APOL1 risk alleles was stronger under a recessive model than under an additive model (recessive model: pcom = 3.0 × 10−16; additive model: pcom = 4.2 × 10−13) (Online Table 2). However, in the 20- to 39-year age range, the association with eGFR under an additive model was slightly stronger than the recessive model (recessive model: pcom = 5.7 × 10 = −4; additive model: pcom = 1.6 × 10−4) (Table 4).
The Cox proportional hazards analysis of age at hypertension diagnosis was also more significant under an additive model (recessive model: pcom = 1.0 × 10−4; additive model: pcom = 1.9 × 10−5), with APOL1 variant allele heterozygotes in 3 of the 4 cohorts having a younger median age of hypertension diagnosis than APOL1 ancestral allele homozygotes.
Both the overall associations with SBP (recessive model: pcom = 1.6 × 10−7; additive model: pcom = 7.0 × 10−8) and DBP (recessive model: pcom = 4.5 × 10−4; additive model: pcom = 2.8 × 10−4) with APOL1 risk alleles were slightly stronger under an additive model (Online Table 2). In the 20- to 39-year age range, the association between the APOL1 risk alleles with SBP was stronger under the additive model compared with the recessive model, except for untreated patients when eGFR was added to the model (Table 2). For DBP, the additive model was not more associated in any age range (Online Table 8). Testing for the additive model in the other age ranges did not uncover additional associations.
AGE-RELATED APOL1 RISK STATUS
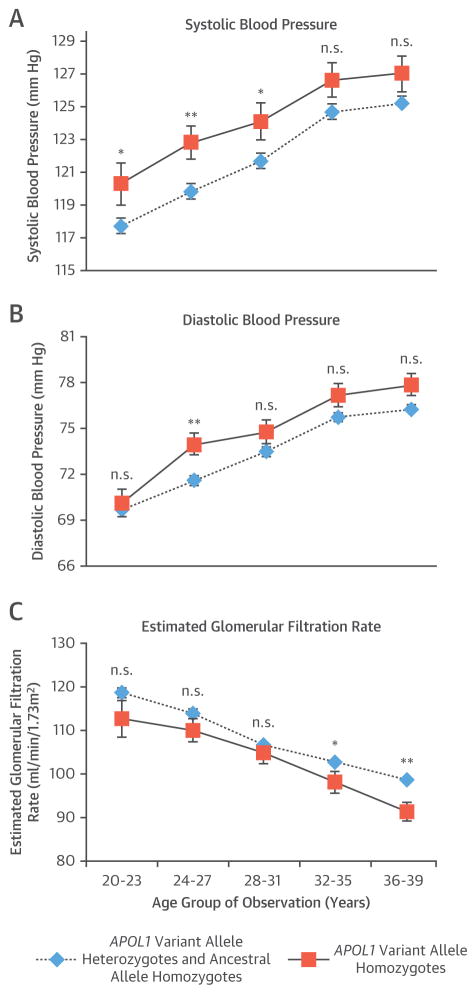
Next, we divided the 20- to 39-year age group in consecutive, nonoverlapping 4-year groups with average SBP, DBP, and eGFR measurements across all cohorts (Figure 2). SBP was significantly increased in APOL1 variant allele homozygotes in 3 consecutive 4-year age groups between 20 and 31 years, respectively (Figure 2A). However, DBP was not significantly higher in APOL1 variant allele homozygotes in any of the 4-year intervals, except for the 24- to 27-year interval (Figure 2B). In contrast, eGFR did not significantly decrease in APOL1 variant allele homozygotes in 3 age groups between 20 and 31 years, but significantly decreased in 2 subsequent age groups between 32 and 39 years (Figure 2C).
FIGURE 2. Change of Blood Pressure and eGFR.
Variations in (A) systolic blood pressure, (B) diastolic blood pressure, and (C) estimated glomerular filtration rate (eGFR) were seen in apolipoprotein L1 (APOL1) ancestral allele homozygotes and APOL1 variant allele heterozygotes versus APOL1 variant allele homozygotes for all cohorts combined between the ages of 20 and 39 years. Line graphs show mean values with 95% confidence intervals. *p < 0.05; **p < 0.01. n.s. = not significant.
Because urine albumin/protein laboratory data were largely missing in EMRs, we could only summarize the available results. In the 20- to 39-year age range, 23% of APOL1 variant allele homozygotes and 19% of APOL1 variant allele heterozygotes and APOL1 ancestral allele homozygotes had a urine albumin/protein test recorded. Among those, 42% versus 55% manifested albuminuria or proteinuria at 29.9 ± 6.1 versus 30.8 ± 5.3 years in APOL1 variant allele homozygotes versus combined APOL1 variant allele heterozygotes and ancestral allele homozygotes, respectively (differences not significant). Thus, albuminuria/proteinuria was first documented nearly a decade after higher SBP and prior to significantly accelerated decline of eGFR in APOL1 variant allele homozygotes.
DISCUSSION
Our findings demonstrated that APOL1 risk alleles are associated with increased SBP and earlier onset of hypertension in young adults of African ancestry (Central Illustration). This association seems to be stronger under an additive model. After adjusting for major covariates such as BMI and eGFR, each copy of an APOL1 risk allele accounted for 1.1-mm Hg higher SBP considering all BP measurements in the 20- to 39-year age group. When analysis was further limited to BP measured in the absence of antihypertensive medications, each copy of an APOL1 risk allele accounted for a 0.8-mm Hg higher SBP, demonstrating that the association was independent of antihypertensive therapy.
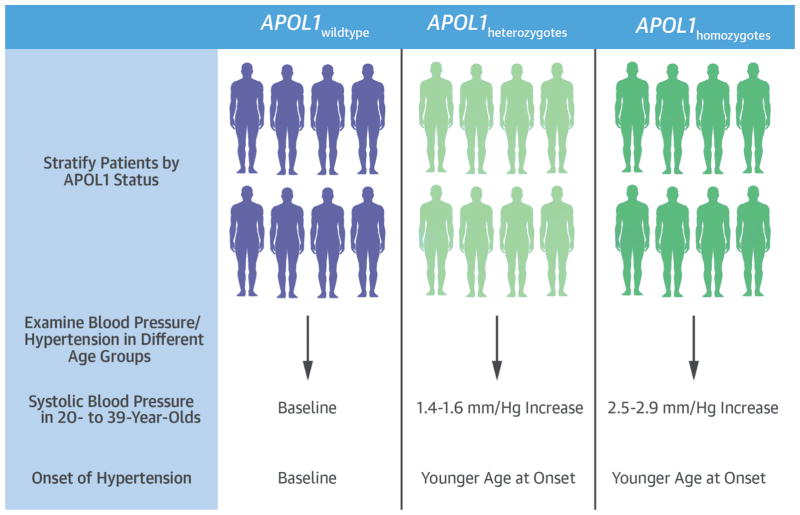
CENTRAL ILLUSTRATION. APOL1 Genotypes and Phenotypic Data From EMRs.
Hypertension-related health problems affect African Americans (AAs) to a greater degree than other groups, and apolipoprotein L1 (APOL1) risk variants are associated with kidney disease in hypertensive AAs. Analyzing data from electronic medical records (EMRs) from 3 major health systems in discovery and replication cohorts using recessive and additive models demonstrated that APOL1 risk alleles were associated with higher systolic blood pressure in AAs of 20 to 39 years of age and younger at diagnosis of hypertension.
APOL1 risk alleles could account for a significant proportion of higher SBP in AAs compared with EAs between 18 and 39 years of age, although the entire difference in quantitative SBP cannot be explained by APOL1 risk alleles. Significantly higher average SBP in APOL1 variant allele homozygotes was already apparent in the 20- to 23-year age group, the youngest in our study. Future studies in adolescent and pediatric populations will be needed to identify an age range at which the effect of APOL1 risk alleles on BP traits first manifests. Because an estimated 14% of AAs are APOL1 variant allele homozygotes (27), and approximately 40% to 47% are APOL1 variant allele heterozygotes, APOL1 genetic testing in young AAs might define a new area to study health disparities associated with increased BP burden (28). The APOL1 variant allele homozygotes subpopulation is also at increased risk for progressive renal (18) and possibly cardiovascular complications of hypertension (27).
Since the original report attributing substantial risk for focal segmental glomerulosclerosis and human immunodeficiency virus-associated nephropathy to the G1 and G2 APOL1 risk alleles in a recessive model (12), the spectrum of APOL1-related nephropathies has expanded to a range of glomerular conditions (13,29–31), albuminuria and renal function decline (32), CKD and end-stage renal disease (33,34), and accelerated CKD progression (18). The intense search for how APOL1 risk alleles contribute mechanistically to a range of distinct glomerulopathies has focused on their potential to exert direct toxicity and cell death in kidney cells. Indeed, findings of the trypanolytic activity of APOL1, forming pores in lysosomal membranes causing lysosomal permeabilization and cell death, have been observed following overexpression of APOL1 ancestral (G0) and risk alleles (G1 and G2) in cultured human podocytes (35). However, APOL1 is broadly expressed in human tissues, raising the question of whether the APOL1 risk allele toxicity is restricted to renal cells or whether other tissues manifest adverse phenotypes yet to be identified.
Our results raised the possibility that the effect of APOL1 risk variants on hypertensive nephropathy could result, at least in part, from an increased BP burden demonstrated here, although subclinical nephropathy due to APOL1 variant allele carrier status could also manifest itself as increased BP early in life (26). First, the SBP effect of APOL1 risk alleles was already manifest in 20- to 29-year-old patients and persisted after adjustment for kidney function. In contrast, the APOL1 association with reduced kidney function was not detectable in this age group but was evident in the 30- to 39-year-old group. Second, a kidney disease–independent, BP-dependent effect in APOL1 variant allele homozygotes might underlie its previously reported association with cardiovascular disease burden (27), although this finding remains controversial (36). Third, APOL1 protein expression was observed in vascular media of preglomerular resistance vessels in diseased kidneys, but not in normal kidneys (37,38). It will be interesting to see whether vascular smooth muscle expression of APOL1 is associated with BP levels.
The results suggesting an additive model for the genetic association between APOL1 risk alleles and SBP, and possibly with eGFR, in 20- to 39-year-olds were surprising. Further studies, including participants of all ages, will be needed to clarify whether the underlying mechanism behind the association between APOL1 and kidney phenotypes could follow an additive model where heterozygosity would not be sufficient to lead to kidney disease. Nonetheless, young APOL1 variant allele heterozygotes showed elevated SBP, which increased the APOL1-related public health burden to the majority of young AAs.
STUDY LIMITATIONS
We were unable to adjust for albuminuria or proteinuria because of missing data in EMRs. Interestingly, in a community-based cohort of EA and AA young adults, albuminuria was first documented after age 30 years in APOL1 variant allele homozygotes and combined APOL1 variant heterozygotes and ancestral allele homozygotes, and excess incidence of albuminuria in AAs was largely explained by traditional risk factors including obesity, diabetes, and BP levels (32). However, in light of our results demonstrating an association between APOL1 risk alleles and SBP, caution is warranted to apply adjustment for BP levels as independent covariate in association testing with APOL1 risk alleles.
Although the effect size of an APOL1 risk allele on SBP was larger than any previous common variant on BP, it is quantitatively low, was attenuated after exclusion of BP measurements obtained after participants were prescribed antihypertensive medications, and cannot explain the entire difference in BP between AAs and EAs ages 20 to 39 years. We did not find a significant independent association with DBP; the association of APOL1 risk alleles and cardiovascular risk might not be mediated by BP alone. Due to the observational nature of the study, residual confounding cannot be excluded. Additionally, BP measurements were recorded during clinical care encounters, not as part of a research protocol; therefore, exact details of measurements methods were not available, and we could not use more robust measures such as 24-h ambulatory BP monitoring, because such monitoring is not routinely measured. However, there was no significant difference in number of BP measurements and lengths of observation period between APOL1 variant allele carrier status in individual bio-bank cohorts. Although administrative codes could have limitations, the hypertension ICD-9-CM codes have excellent sensitivity and specificity.
Ascertainment bias for some phenotypes, such as eGFR, for which patients with clinical signs/symptoms or family history of kidney disease would have been more likely to be tested more frequently and at a younger age, cannot be ruled out. We used self-reported AA race as inclusion criteria leading to a possibility of selection bias; however, because participants had predominantly African genetic ancestry, any bias was likely small and associations remained significant after correcting for principal components in cohorts for whom genome-wide genotyping data was available. Finally, because we only investigated the effect of APOL1 risk alleles in AAs, it is unclear whether these findings are generalizable to Africans, Afro Caribbeans, and other populations of African descent.
CONCLUSIONS
In young AA adults, APOL1 variant allele homozygotes manifested elevated SBP prior to eGFR decline and earlier hypertension diagnosis. Resolving whether APOL1 initially affects vascular or renal cell biology requires further studies, including in adolescent and childhood populations. APOL1 genetic testing can identify young individuals of African ancestry with increased BP burden and increased risk for hypertension-attributable cardiovascular and renal health disparities.
Supplementary Material
PERSPECTIVES.
COMPETENCY IN MEDICAL KNOWLEDGE
APOL1 alleles are strongly associated with elevated SBP and onset of hypertension in young AA adults before renal impairment develops.
TRANSLATIONAL OUTLOOK
Additional studies should examine whether the mechanisms underlying the association between APOL1 and hypertensive kidney disease are additive and whether APOL1 heterozygosity is associated with kidney disease.
Acknowledgments
The BioMe health care delivery cohort at Mount Sinai was established and maintained with a generous gift from the Andrea and Charles Bronfman Philanthropies. The eMERGE Network was initiated and funded by National Human Genome Research Institute through the following grants: U01HG006389 (Essentia Institute of Rural Health, Marshfield Clinic Research Foundation, and Pennsylvania State University), U01HG006382 (Geisinger Clinic), U01HG006375 (Group Health Cooperative/University of Washington), U01HG006379 (Mayo Clinic), U01HG006380 (Icahn School of Medicine at Mount Sinai), U01HG006388 (Northwestern University), U01HG006378 (Vanderbilt University Medical Center), and U01HG006385 (Vanderbilt University Medical Center serving as the Coordinating Center); U01HG004438 (CIDR) and U01HG004424 (the Broad Institute) served as Genotyping Centers. Research reported in this publication was supported by the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health under Award Number K23DK107908 (to Dr. Nadkarni).
This work was supported in part through the computational resources and staff expertise provided by the Department of Scientific Computing at the Icahn School of Medicine at Mount Sinai.
ABBREVIATIONS AND ACRONYMS
- APOL1
apolipoprotein L1
- BMI
body mass index
- CKD
chronic kidney disease
- DBP
diastolic blood pressure
- eGFR
estimated glomerular filtration rate
- EMR
electronic medical record
- SBP
systolic blood pressure
Footnotes
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Dr. Galarneau is a recipient of a Canadian Institute of Health Research postdoctoral fellowship award (MFE-140913).
All other authors have reported that they have no relationships relevant to the contents of this paper to disclose.
APPENDIX For an expanded Methods section as well as a supplemental figure and tables, please see the online version of this article.
References
- 1.Egan BM, Zhao Y, Axon RN. US trends in prevalence, awareness, treatment, and control of hypertension, 1988–2008. JAMA. 2010;303:2043–50. doi: 10.1001/jama.2010.650. [DOI] [PubMed] [Google Scholar]
- 2.Hajjar I, Kotchen TA. Trends in prevalence, awareness, treatment, and control of hypertension in the United States, 1988–2000. JAMA. 2003;290:199–206. doi: 10.1001/jama.290.2.199. [DOI] [PubMed] [Google Scholar]
- 3.Mozaffarian D, Benjamin EJ, Go AS, et al. Heart disease and stroke statistics—2016 update: a report from the American Heart Association. Circulation. 2016;133:e38–360. doi: 10.1161/CIR.0000000000000350. [DOI] [PubMed] [Google Scholar]
- 4.Wright JD, Hughes JP, Ostchega Y, Yoon SS, Nwankwo T for the Division of Health and Nutrition Examination Surveys, National Center for Health Statistics. Mean systolic and diastolic blood pressure in adults aged 18 and over in the United States, 2001–2008. [Accessed April 1, 2016];National Health Statistics Reports. Available at: http://www.cdc.gov/nchs/data/nhsr/nhsr035.pdf.
- 5.Grotto I, Huerta M, Sharabi Y. Hypertension and socioeconomic status. Curr Opin Cardiol. 2008;23:335–9. doi: 10.1097/HCO.0b013e3283021c70. [DOI] [PubMed] [Google Scholar]
- 6.Salfati E, Morrison AC, Boerwinkle E, Chakravarti A. Direct Estimates of the Genomic Contributions to Blood Pressure Heritability within a Population-Based Cohort (ARIC) PLoS ONE. 2015;10:e0133031. doi: 10.1371/journal.pone.0133031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Snieder H, Harshfield GA, Treiber FA. Heritability of Blood Pressure and Hemodynamics in African- and European-American Youth. Hypertension. 2003;41:1196–201. doi: 10.1161/01.HYP.0000072269.19820.0D. [DOI] [PubMed] [Google Scholar]
- 8.Kaufman JS, Dolman L, Rushani D, Cooper RS. The contribution of genomic research to explaining racial disparities in cardiovascular disease: a systematic review. Am J Epidemiol. 2015;181:464–72. doi: 10.1093/aje/kwu319. [DOI] [PubMed] [Google Scholar]
- 9.Klag MJ, Whelton PK, Randall BL, Neaton JD, Brancati FL, Stamler J. End-stage renal disease in African-American and white men. 16-year MRFIT findings. JAMA. 1997;277:1293–8. [PubMed] [Google Scholar]
- 10.Genovese G, Friedman DJ, Ross MD, et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010;329:841–5. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lipkowitz MS, Freedman BI, Langefeld CD, et al. Apolipoprotein L1 gene variants associate with hypertension-attributed nephropathy and the rate of kidney function decline in African Americans. Kidney Int. 2013;83:114–20. doi: 10.1038/ki.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kopp JB, Nelson GW, Sampath K, et al. APOL1 genetic variants in focal segmental glomerulo-sclerosis and HIV-associated nephropathy. J Am Soc Nephrol. 2011;22:2129–37. doi: 10.1681/ASN.2011040388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomson R, Genovese G, Canon C, et al. Evolution of the primate trypanolytic factor APOL1. Proc Natl Acad Sci U S A. 2014;111:E2130–9. doi: 10.1073/pnas.1400699111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tzur S, Rosset S, Shemer R, et al. Missense mutations in the APOL1 gene are highly associated with end stage kidney disease risk previously attributed to the MYH9 gene. Hum Genet. 2010;128:345–50. doi: 10.1007/s00439-010-0861-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosset S, Tzur S, Behar DM, Wasser WG, Skorecki K. The population genetics of chronic kidney disease: insights from the MYH9-APOL1 locus. Nat Rev Nephrol. 2011;7:313–26. doi: 10.1038/nrneph.2011.52. [DOI] [PubMed] [Google Scholar]
- 16.Parsa A, Kao WHL, Xie D, et al. APOL1 risk variants, race, and progression of chronic kidney disease. N Engl J Med. 2013;369:2183–96. doi: 10.1056/NEJMoa1310345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Genovese G, Friedman DJ, Pollak MR. APOL1 variants and kidney disease in people of recent African ancestry. Nat Rev Nephrol. 2013;9:240–4. doi: 10.1038/nrneph.2013.34. [DOI] [PubMed] [Google Scholar]
- 18.Gottesman O, Kuivaniemi H, Tromp G, et al. The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet Med Off J Am Coll Med Genet. 2013;15:761–71. doi: 10.1038/gim.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roden DM, Pulley JM, Basford MA, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther. 2008;84:362–9. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delaneau O, Marchini J, Zagury J-F. A linear complexity phasing method for thousands of genomes. Nat Methods. 2012;9:179–81. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 21.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levey AS, Stevens LA, Schmid CH, et al. A new equation to estimate glomerular filtration rate. Ann Intern Med. 2009;150:604–12. doi: 10.7326/0003-4819-150-9-200905050-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu H, Stenner SP, Doan S, Johnson KB, Waitman LR, Denny JC. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc. 2010;17:19–24. doi: 10.1197/jamia.M3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinforma Oxf Engl. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedman DJ, Kozlitina J, Genovese G, Jog P, Pollak MR. Population-based risk assessment of APOL1 on renal disease. J Am Soc Nephrol. 2011;22:2098–105. doi: 10.1681/ASN.2011050519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freedman BI, Murea M. Target organ damage in African American hypertension: role of APOL1. Curr Hypertens Rep. 2012;14:21–8. doi: 10.1007/s11906-011-0237-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ito K, Bick AG, Flannick J, et al. Increased burden of cardiovascular disease in carriers of APOL1 genetic variants. Circ Res. 2014;114:845–50. doi: 10.1161/CIRCRESAHA.114.302347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wong MD, Shapiro MF, Boscardin WJ, Ettner SL. Contribution of major diseases to disparities in mortality. N Engl J Med. 2002;347:1585–92. doi: 10.1056/NEJMsa012979. [DOI] [PubMed] [Google Scholar]
- 29.Ashley-Koch AE, Okocha EC, Garrett ME, et al. MYH9 and APOL1 are both associated with sickle cell disease nephropathy. Br J Haematol. 2011;155:386–94. doi: 10.1111/j.1365-2141.2011.08832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freedman BI, Langefeld CD, Andringa KK, et al. End-stage renal disease in African Americans with lupus nephritis is associated with APOL1. Arthritis Rheumatol. 2014;66:390–6. doi: 10.1002/art.38220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larsen CP, Beggs ML, Saeed M, Walker PD. Apolipoprotein L1 risk variants associate with systemic lupus erythematosus-associated collapsing glomerulopathy. J Am Soc Nephrol. 2013;24:722–5. doi: 10.1681/ASN.2012121180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peralta CA, Bibbins-Domingo K, Vittinghoff E, et al. APOL1 genotype and race differences in incident albuminuria and renal function decline. J Am Soc Nephrol. 2016;27:887–93. doi: 10.1681/ASN.2015020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Foster MC, Coresh J, Fornage M, et al. APOL1 variants associate with increased risk of CKD among African Americans. J Am Soc Nephrol. 2013;24:1484–91. doi: 10.1681/ASN.2013010113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tzur S, Rosset S, Skorecki K, Wasser WG. APOL1 allelic variants are associated with lower age of dialysis initiation and thereby increased dialysis vintage in African and Hispanic Americans with non-diabetic end-stage kidney disease. Nephrol Dial Transplant. 2012;27:1498–505. doi: 10.1093/ndt/gfr796. [DOI] [PubMed] [Google Scholar]
- 35.Lan X, Jhaveri A, Cheng K, et al. APOL1 risk variants enhance podocyte necrosis through compromising lysosomal membrane permeability. Am J Physiol Renal Physiol. 2014;307:F326–36. doi: 10.1152/ajprenal.00647.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langefeld CD, Divers J, Pajewski NM, et al. Apolipoprotein L1 gene variants associate with prevalent kidney but not prevalent cardiovascular disease in the Systolic Blood Pressure Intervention Trial. Kidney Int. 2015;87:169–75. doi: 10.1038/ki.2014.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Madhavan SM, O’Toole JF, Konieczkowski M, Ganesan S, Bruggeman LA, Sedor JR. APOL1 localization in normal kidney and nondiabetic kidney disease. J Am Soc Nephrol. 2011;22:2119–28. doi: 10.1681/ASN.2011010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Monajemi H, Fontijn RD, Pannekoek H, Horrevoets AJG. The apolipoprotein L gene cluster has emerged recently in evolution and is expressed in human vascular tissue. Genomics. 2002;79:539–46. doi: 10.1006/geno.2002.6729. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.