Abstract
Antibody–antigen interactions are governed by recognition of specific residues and structural complementarity between the antigen epitope and antibody paratope. While X-ray crystallography has provided detailed insights into static conformations of antibody–antigen complexes, factors such as conformational flexibility and dynamics, which are not readily apparent in the structures, can also have an impact on the binding event. Here we investigate the contribution of dynamics in the HIV-1 gp120 glycoprotein to antibody recognition of conserved conformational epitopes, including the CD4- and coreceptor-binding sites, and an inner domain site that is targeted by ADCC-active antibodies. Hydrogen/deuterium-exchange mass spectrometry (HDX-MS) was used to measure local structural dynamics across a panel of variable loop truncation mutants of HIV-1 gp120, including full-length gp120, ΔV3, ΔV1/V2, and extended core, which includes ΔV1/V2 and V3 loop truncations. CD4-bound full-length gp120 was also examined as a reference state. HDX-MS revealed a clear trend toward an increased level of order of the conserved subunit core resulting from loop truncation. Combined with biolayer interferometry and enzyme-linked immunosorbent assay measurements of antibody–antigen binding, we demonstrate that an increased level of ordering of the subunit core was associated with better recognition by an array of antibodies targeting complex conformational epitopes. These results provide detailed insight into the influence of structural dynamics on antibody–antigen interactions and suggest the importance of characterizing the structural stability of vaccine candidates to improve antibody recognition of complex epitopes.
Graphical abstract
Interaction of antibody and antigen involves the recognition of residues with a specific three-dimensional organization on the antigen (epitope) by a set of residues on the antibody (paratope). Understanding the structural basis of antibody recognition and immune response against antigens is important for optimization of vaccine immunogens and for understanding mechanisms of neutralization. High-resolution X-ray crystal structures have illustrated the complementary nature of the epitope–paratope coupling. This type of information has permitted epitopes to be mapped with atomic detail and suggested modifications to the antibody paratope as well as changes on the antigen that can enhance the affinity of antibody–antigen binding in optimizing potential vaccine immunogens.1,2 Structural dynamics and conformational variability of immunogens can also influence the presentation of epitopes and impact the affinity and specificity of binding. These are fundamental structural and biophysical properties that are at play essentially in all protein–protein interactions.
A correlation among structural dynamics, solvent accessibility, antigenicity, and immunogenicity has been proposed for a variety of different antigens.3–9 Directly testing the effect of structural dynamics of the antigen or antibody on the binding interaction has often proved to be challenging. Most studies that report a linkage between dynamics and antibody–antigen affinity have relied upon measurements that provide global thermodynamic parameters but little structural dynamic information for epitopes and paratopes.10–12 Fluorescence methods have examined the influence of antibody structural dynamics on recognition of relatively simple target antigens.13,14 NMR has been used for the study of antibody fragments and small antigens, providing information about structural as well as dynamic changes during binding.15–18 It remains difficult, however, to apply similar approaches to large, complex antigens, let alone to study their association with antibodies.
In the case of the majority of broadly neutralizing antibodies targeting the HIV envelope glycoprotein (Env), the target epitopes are often formed by discontinuous peptide segments, loops, and glycan chains.19,20 These types of epitopes in some cases may exhibit a high degree of flexibility in the absence of a stabilizing ligand. Indeed, in Env, conformational switching and structural flexibility appear to be key mechanisms the virus uses to evade immune recognition, particularly to hide conserved epitopes such as the chemokine coreceptor-binding sites from detection. Prior to the primary CD4 receptor binding event, the structural elements that will form the conserved coreceptor-binding site are sequestered and maintained in an organization distinct from that observed in the CD4-bound state. CD4 binding induces major structural rearrangements in Env, leading to formation of the bridging sheet and exposure of V3 that together are needed to engage the coreceptor.10,21–23 Even at equilibrium, prior to receptor binding, recent studies have demonstrated that Env can sample a range of conformational states.24,25 The frequency with which these states are sampled varies depending on specific viral isolate, with more neutralization resistant isolates remaining in the “closed” prefusion state in which the coreceptor-binding site is masked to a greater extent, while neutralization sensitive variants frequently sample more “open” states, transiently exposing the binding site as well as additional epitopes throughout Env.
The Env gp120 receptor-binding subunit provides a highly suitable construct for investigating links between epitope structural dynamics and antibody recognition. gp120 bears a number of epitopes that are targets for neutralizing antibodies against HIV, and it has been intensively investigated as a component in HIV vaccines.26–29 It has been suggested via past thermodynamic measurements that the intrinsic conformational flexibility of the gp120 subunit can impact antibody recognition. Calorimetry experiments revealed that unliganded gp120 subunits exhibit a high degree of conformational flexibility.11 This led to the hypothesis that epitope disorder and structural dynamics in gp120 may hinder antibody binding and serve as a mechanism of immune evasion.10,30,31 Subunit flexibility becomes constrained when CD4 binds and orders the conserved subunit core.11,31,32 In a recent study, it was shown that gp120 variable loop truncation can modulate the antigenicity and structure of the gp120 core to the extent that the extended core lacking variable loops (Coree) crystallized in a conformation nearly identical to that of the CD4-bound state.33 Kwon and colleagues reported that the loop truncation mutants displayed stronger binding affinities than full-length gp120 for antibodies recognizing the receptor-bound state. While the previous study used antibody binding to covalently cross-linked gp120 constructs to assess the structure of the panel of truncation mutants, strong antibody–antigen interactions, cross-linking, and crystallization can themselves impose significant influences on antigen structure, rendering such approaches rather invasive probes of structure.
While a number of studies have examined the effect of the overall conformational stability of HIV Env on antibody binding,34,35 to date, it has been difficult to directly demonstrate the link between local structural dynamics and the affinity and specificity of antibody binding to those local structural features.25,30 Here we selected HIV-1 gp120 as a prime example of a complex antigen and used a combination of solution-phase biophysical methods to investigate the underlying relationship among local epitope structural order, structural dynamics, and antigenicity under native solution conditions. Hydrogen/deuterium-exchange mass spectrometry (HDX-MS) was used to probe the local structural dynamics in the panel of variable loop truncation mutants of gp120, revealing significant changes in structural dynamics of epitopes resulting from variable loop truncation. We show that antibody recognition of conserved conformational epitopes, such as the CD4- and coreceptor-binding sites, correlates inversely with the degree of local dynamics. The findings suggest that it may be beneficial to optimize the dynamic profile of antigens to enhance the presentation of complex epitopes and improve their immune recognition.
MATERIALS AND METHODS
Reagents
The expression vectors for HIV-1 clade B YU2 gp120 glycoproteins, including FL, ΔV1/V2, ΔV3, and Coree, were kindly supplied by P. D. Kwong.33 Antibodies 17b,21,36 48d,36 A32,37 VRC01,38 NIH45–46,1 VRC03,38 F105,39 and b1240 were obtained from the NIH AIDS Reagents Program. sCD4 and CD4-IgG2 were obtained from the NIH AIDS Reagents Program.41 Antibodies N5i5 and B18 were generously provided by Y. Guan and G. Lewis (University of Maryland, College Park, MD).42,43
High-Density Transfection and Expression of YU2 gp120 Envelope Glycoproteins
293F cells were grown in HyClone SFM4Transfx-293 medium (Fisher Scientific). The various HIV-1 Clade BYU2 gp120 glycoproteins were expressed in human embryonic kidney cell line HEK293F by high-density (HD) transient transfection as previously described.44 Briefly, 293F cells were resuspended at a density of 2 × 107 cells/mL in fresh SFM4Transfx-293 medium. Sterile filtered plasmid DNA (20 μg/mL) was added into the cells directly, immediately followed by PEI MAX (40 μg/mL, Polysciences, Inc.) (1:2 DNA:PEI MAX). After being gently mixed, the cells were incubated at 37 °C for 4 h. The culture volume was expended 20-fold by adding fresh medium containing 20 mM Hepes (pH 7.3) (Life Technologies) and 1× GlutaMAX (Life Technologies). Seventy-two hours after transfection, the supernatant was collected and filtered by a 0.45 μm filter (Fisher Scientific). A cocktail of protease inhibitors (Roche Diagnostics) was added. The supernatant was concentrated 20-fold by being passed through a KvickLab ultrafiltration flat sheet cassette (30 kDa, GE Healthcare) using a Masterflex L/S Easy-Load II system (Cole-parmer). Then the concentrated supernatant was diluted 30-fold with Galanthus nivalis lectin affinity (GNA) binding buffer [20 mM Tris-HCl (pH 7.4), 100 mM NaCl, 1 mM EDTA, and 1 mM EGTA]. Finally, the sample was concentrated to 50 mL before being loaded on a GNA affinity column.
Purification of HIV Envelope Glycoproteins
The initial purification was conducted by using the affinity technique with G. nivalis lectin (GNA) (Sigma). Subsequent purification of HIV envelope glycoproteins was achieved by gel filtration using a HiLoad 16/60 Superdex 200 column (GE Healthcare) equilibrated with phosphate-buffered saline (1× PBS, pH 7.4, 1 mM EDTA, 0.02% NaN3). Fractions (500 μL) were collected and analyzed by using sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and Blue-native PAGE (BN-PAGE). Quantification of proteins was conducted by checking the absorbance at 280 nm. The purified HIV envelope glycoproteins were pooled and stored in appropriate buffer for further experiments (Figure S1).
SDS–PAGE and BN-PAGE
For SDS–PAGE, the gp120 envelope glycoprotein samples were loaded on a NuPAGE Novex 4–12% Bis-Tris gel (Invitrogen) and run at 180 V for 50 min. The gel was fixed with 50% methanol and 7% acetic acid for 10 min, stained with SYPRO Ruby protein gel stain (Invitrogen), and washed with 10% methanol and 7% acetic acid. Benchmark protein ladder (Invitrogen) was used for SDS–PAGE. For BN-PAGE, the protein samples were loaded on a 3–12% Bis-Tris Native PAGE gel (Invitrogen). The gel was run at 150 V for 1 h and 250 V for 2 h, fixed for 15 min with 50% methanol and 10% acetic acid, and destained using dH2O. Native Mark protein standard (Invitrogen) was used for BN-PAGE.
Light Scattering Measurements
Dynamic and static scattering of gp120 envelope glycoproteins were measured using a Dynapro Nanostar (Wyatt Technology, Santa Barbara, CA) at 20 °C. A total of 30 acquisitions of 5 s were collected for each sample. The data were analyzed by using the Dynamics analysis software package (Wyatt Technology), assuming a spherical model. The theoretical expected molecular mass for each gp120 envelope glycoprotein was calculated as the sum of the molecular mass of the polypeptide and the mass of the glycans (Man8, 1.7 kDa, for the high-mannose type and fucosylated monosialylated biantennary, 2.1 kDa, for the complex type).
Hydrogen/Deuterium-Exchange Mass Spectrometry
As previously described,31 hydrogen/deuterium (H/D) exchange was initiated by mixing 100 μg/mL gp120 envelope glycoprotein in phosphate buffer [10 mM phosphate and 144 mM NaCl (pH 7.4)] with 85% D2O (Cambridge Isotope Laboratories). The solution was maintained at room temperature for 3 s, 1 min, 30 min, and 20 h for the H/D exchange to occur. At each time point, the solution was quenched by a 1:1 dilution in 1 M tris(2-carboxyethyl)phosphine hydrochloride (TCEP) in 0.02% formic acid with a final pH of 2.5. The quenched sample was immediately digested with pepsin (Worthington Laboratories) (2:1 by mass) on ice for 5 min. The samples were frozen in liquid nitrogen and stored at −80 °C for future data collection. To correct for H/D exchange during pepsin digestion, a zero time point sample was prepared by mixing the gp120 with prequenched PBS and D2O. To correct for back exchange, a fully deuterated sample was prepared by mixing denatured gp120 in 50 mM Tris-HCl (pH 8.0), 4 M GndHCl, and 50 mM dithiothreitol with 85% D2O at 40 °C for 4 h. The FL+sCD4 complex was prepared by mixing FL with a 3-fold molar excess of sCD4 at room temperature for 1 h. The final concentration for FL in the FL+sCD4 complex was 100 μg/mL. The FL+sCD4 complex was deuterated under the same conditions as those for the unliganded gp120 samples.
All spectra were obtained by liquid chromatography and mass spectrometry (LC–MS) on a Synapt HDMS system (Waters, Milford, MA) integrated with a Waters Acquity Ultra-Performance LC (UPLC). The frozen samples were thawed on ice for 5 min and directly injected into a BEH C18 trap column [1.7 μm, 2.1 mm × 5 mm (Waters)] in 0.1% trifluoroacetic acid (TFA) at a rate of 100 μL/min. After being washed for 3 min, the peptides were resolved on a Hypersil C18 column [1.9 μm, 50 mm × 1 mm (Thermo Scientific)] with a linear gradient of 5 to 100% solvent B for 15 min [solvent A, 5% acetonitrile (ACN) and 0.05% TFA; solvent B, 80% ACN and 0.05% TFA]. Between injections, the syringe, loop, and trap column were washed with 10% formic acid, 80% methanol, a 2:1 2-propanol/ACN mixture, and 80% ACN sequentially.
Data Analysis
Mass shifts were analyzed by using HX Express,45 and the percent deuteration was calculated by46
where m is the peptide mass centroid at a given time point, m0% is the zero time point peptide mass centroid, and m100% is the fully deuterated peptide mass centroid.
No obvious differences were observed between different glycoforms of the same peptide. The centroids for the different glycoforms of each peptide were averaged. Binomial distribution fitting was applied for overlapped and noisy H/D-exchange data.45,47
Small Angle X-ray Scattering (SAXS)
SAXS measurements were conducted on beamline 4–2 at the Stanford Synchrotron Radiation Laboratory.48 The data collection procedure was as described previously.30,31 Briefly, each sample (3–5 mg/mL, 100 μL) was injected into a Sepharose 200 column (GE Healthcare) pre-equilibrated with PBSE [1× PBS, 1 mM EDTA, and 0.02% NaN3 (pH 7.4)] at a flow rate of 50 μL/min. The eluted sample was passed through in a thin-wall quartz capillary cell and irradiated by focused 11 keV X-ray. One s exposures were recorded by an MX 225HE detector (Rayonix, Evanston, IL) placed 1.7 m upstream of the quartz cell. The detector pixel numbers were converted into the momentum transfer (Q), Q = 4π × sin θ/λ, where 2θ is the scattering angle and λ is the X-ray wavelength of 1.127 Å, calibrated with silver behenate powder at the capillary position. Protein scattering data were processed by using MarParse, scaled for the transmitted beam intensity integrated for each exposure, and azimuthally averaged.48 Buffer scattering data were averaged from the first 100 data points (before the void volume) and subtracted from the corresponding protein scattering data. The ATSAS software suite was used to analyze SAXS parameters and patterns.49,50 The approximate radius of gyration (Rg) was determined from the Guinier plot (qRg < 1.3)51 in Primus.52 AutoGNOM was used to estimate Dmax for the P(r) plot, the pairwise distance distribution histogram.53 The final P(r) plot was generated by GNOM version 4.5.50 Ab initio shape reconstruction was conducted with the generated P(r) plot using DAMMIN version 5.3i.54 The bead models were aligned using SUPCOMB13 with enantiomers considered55 and averaged using DAMAVER.56 Sites of low bead occupancy in the averaged model were filtered using DAMFILT.57 The final models were converted into a volume envelope using pdb2vol in the SITUS2.2 package.58 Molecular weights were calculated with SAXSMoW.59
Enzyme-Linked Immunosorbent Assays (ELISAs)
For ELISAs, the gp120 envelope glycoproteins (50 ng/well) in phosphate-buffered saline (PBS) were immobilized onto an Immulon 2HB plate (Thermo Scientific) at 4 °C overnight. The following day, the plates were blocked by PBS with 2% bovine serum albumin (BSA) and 0.01% Tween 20 for 2 h at room temperature and washed with Tris-buffered saline (TBS) with 0.02% Tween 20 subsequently. The primary antibodies with a starting concentration of 10 μg/mL for human antibodies or 5 μg/mL for mouse antibodies were added and then diluted into 4-fold dilution series by antibody dilution buffer (PBS with 1% BSA and 0.01% Tween 20) and incubated for 1 h at room temperature. After several washes, the horseradish peroxidase (HRP)-conjugated goat anti-human or goat anti-mouse secondary antibodies (Jackson ImmunoResearch) (diluted 1:5000) were added to the wells for 1 h at room temperature. After a final set of washes, the plates were developed with SureBlue Reserve trimethylbenzidine (KPL, Inc.) and stopped with 1 M HCl. The absorbance at 450 nm was measured using a Tecan Infinite M200 plate reader.
Kinetic Assay by Biolayer Interferometry (BLI)
A FortéBio (Menlo Park, CA) Octet RED biolayer interferometry system was used to measure the binding of each gp120 (FL, ΔV3, ΔV1/V2, and Coree) to different antibodies. Anti-Human IgG Fc Capture (AHC) biosensors were hydrated for at least 10 min prior to antibody loading. Human anti-HIV antibody (10 μg/mL) in HBS-EP+ buffer [10 mM HEPES, 150 mM NaCl, 3 mM EDTA, and 0.05% P-20 (pH 7.4)] was then bound to the AHC tips for 4 min and washed in buffer for 1 min to remove any unbound antibody and reach a stable baseline. The antibody–AHC tips were then dipped into gp120 samples, which were prepared independently with a 2-fold dilution series. After association for several minutes, the tips were then placed back into the baseline buffer to allow dissociation to occur. The time ranges for antibody loading, association, and dissociation were optimized according to different antibodies. FortéBio’s Data Analysis 7.0 was used to analyze changes in refractive index. The averaged reference measurements were subtracted prior to data processing. The association and dissociation curves were fit using a 1:1 binding model. The final association (ka) and dissociation (kd) rate constants were used to calculate the ensemble dissociation constants (KD = kd/ka). Standard deviations were calculated from the results of two independent measurements.
RESULTS
Deletion of Variable Loops Does Not Alter the Large-Scale Morphology of the gp120 Core
Small angle X-ray scattering (SAXS) was used to study the overall structural features of unliganded full-length YU2 gp120 monomer (FL) and its truncated variants (ΔV3, ΔV1/V2, and Coree). Size exclusion chromatography in-line with the sample cell was used to gather SAXS data from monodisperse gp120 constructs (Figure 1). The Guinier regions showed good linearity for all data sets, indicating all the specimens were well-behaved in solution. Radii of gyration (Rg) obtained from Guinier analysis agreed well with those derived from P(r) pairwise distance distribution plots.50 Differences in SAXS measurements were consistent with values obtained by static light scattering and with the size of the different YU2 gp120 proteins expected on the basis of their composition (FL > ΔV3 > ΔV1/V2 > Coree) (Table 1).
Figure 1.
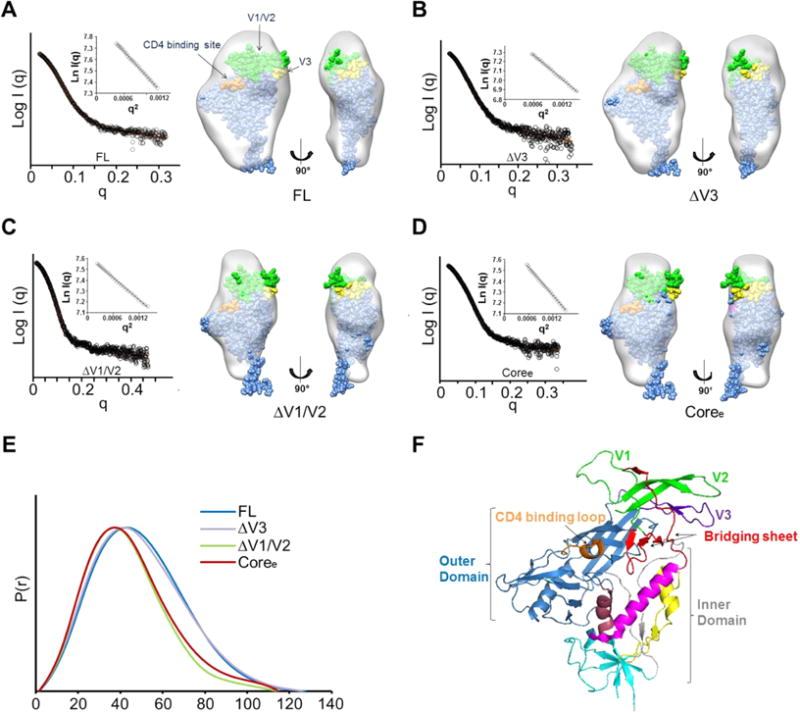
Global gp120 subunit structures determined by small angle X-ray scattering. (A) SAXS pattern (black) for unliganded YU2 FL. The orange line represents the SAXS pattern from a DAMMIN ab initio SAXS reconstruction. Insets show the linear Guinier fit. DAMMIN reconstruction for FL. The structure of the gp120 monomer from the BG505 SOSIP trimer structure (PDB entry 4TVP)93 was fitted into the SAXS density. The V1/V2 residues are colored green. The V3 residues are colored yellow. The contact residues at the CD4-binding site are colored orange. (B) SAXS pattern (black) for unliganded ΔV3 and DAMMIN model for ΔV3. (C) SAXS pattern (black) for unliganded ΔV1/V2 and DAMMIN model for ΔV1/V2. (D) SAXS pattern (black) for unliganded Coree and DAMMIN model for Coree. ΔV3, ΔV1/V2, and Coree SAXS models show a loss of crown density leading to a poorer fit of the V1/V2 and V3 regions in the gp120 subunit as organized in the SOSIP trimer crystal structure. (E) Pairwise distance distributions [P(r)] for FL (blue), ΔV3 (purple), ΔV1/V2 (green), and Coree (red) show changes in size consistent with variable loop truncation. (F) HIV gp120 architecture (PDB entry 4NCO). Different regions in gp120 are highlighted by color: outer domain (blue); inner domain (gray), including layer 1 (yellow), layer 2 (pink), layer 3 (dark red), and seven-stranded β-sandwich (cyan); bridging sheets (red); CD4-binding loop (orange); V1/V2 loop (green); and V3 loop (purple).
Table 1.
Dimensions of Molecules Derived from SAXS and SLS
| sample |
Rg (Å)
|
Dmaxa | Porod volume (Å3)b | mass (kDa)
|
||
|---|---|---|---|---|---|---|
| Guinier | GNOMa | SLSc | expectedd | |||
| FL | 36.5 ± 0.09 | 37.1 ± 0.04 | 127.9 | 222581 | 93.735 | 96.046 |
| ΔV3 | 35.9 ± 0.03 | 37.1 ± 0.06 | 125.7 | 201932 | 89.969 | 88.485 |
| ΔV1/V2 | 32.6 ± 0.03 | 32.0 ± 0.04 | 113.9 | 157757 | 70.995 | 72.563 |
| Coree | 32.4 ± 0.07 | 33.5 ± 0.05 | 113.7 | 151814 | 69.528 | 68.709 |
Real space Rg and Dmax calculated from GNOM.50
Porod volumes determined by Autoporod.57
Molecular mass determined by static light scattering (SLS). Uncertainty is largely dependent on the accuracy of the concentration determination, typically ±10%.
The expected molecular mass was based on the predominant glycoforms observed during H/D-exchange MS analysis.
To determine whether the variable loop truncations led to large-scale conformational changes, ab initio shape reconstruction was applied to SAXS data for the gp120 constructs (Figure 1). As shown in Figure 1A, the FL showed the same features of a ventral stalk, central core, and a crest as observed in previous studies.30,31 SAXS models of the gp120 truncation mutants, including ΔV3, ΔV1/V2, and Coree, lost density in the crest region (Figure 1B–D). Overall, the raw SAXS data and the reconstructed models confirmed the reduced size due to truncations of different variable loops. However, no obvious conformational changes of the core domain were detected from the low-resolution SAXS measurements.
H/D-Exchange Mass Spectrometry Profiles of Structural Dynamics in gp120 Constructs
To detect more detailed local structural changes resulting from variable loop truncations, HDX-MS was used to examine structural ordering in FL, ΔV3, ΔV1/V2, Coree, and FL with sCD4 bound (FL +sCD4). Deuterium exchange could be monitored for peptides covering 73% of the YU2 full-length sequence (Figure S2). The loss of coverage for certain segments, including most of V1/V2, part of V3, part of the CD4-binding loop, and a portion of V4, is most likely due to the high level of heterogeneous glycosylation in these regions. Here we primarily compared deuterium uptake of identical peptides in the conserved core domain that are shared by the truncation mutants.
To establish a baseline for the changes in dynamics we might expect to see in the set of gp120 constructs, we first compared FL in unliganded and sCD4-bound forms. Consistent with previous studies of full-length gp120 monomers,30,31 binding of CD4 to FL induces a dramatic decrease in the rate of deuterium uptake in the gp120 subunit, reflecting an increased level of structural order in the three layers of the inner domain, bridging sheets, and CD4-binding site (Figures 1F and 3 and Figures S3–S5). Interestingly, a small V2 loop peptide (residues 173–176, sequence YALF) showed some exchange protection in unliganded gp120 but rapid deuterium exchange at the earliest time point in CD4-bound gp120 (Figure 3A,J). This hints that a portion of the V2 loop may undergo a transition from being partially ordered in unliganded gp120 to a disordered state upon CD4 binding, echoing changes observed in trimeric forms of Env.25 The high degree of glycosylation of V1/V2 prevented additional portions of the loops from being monitored by HDX-MS.
Figure 3.
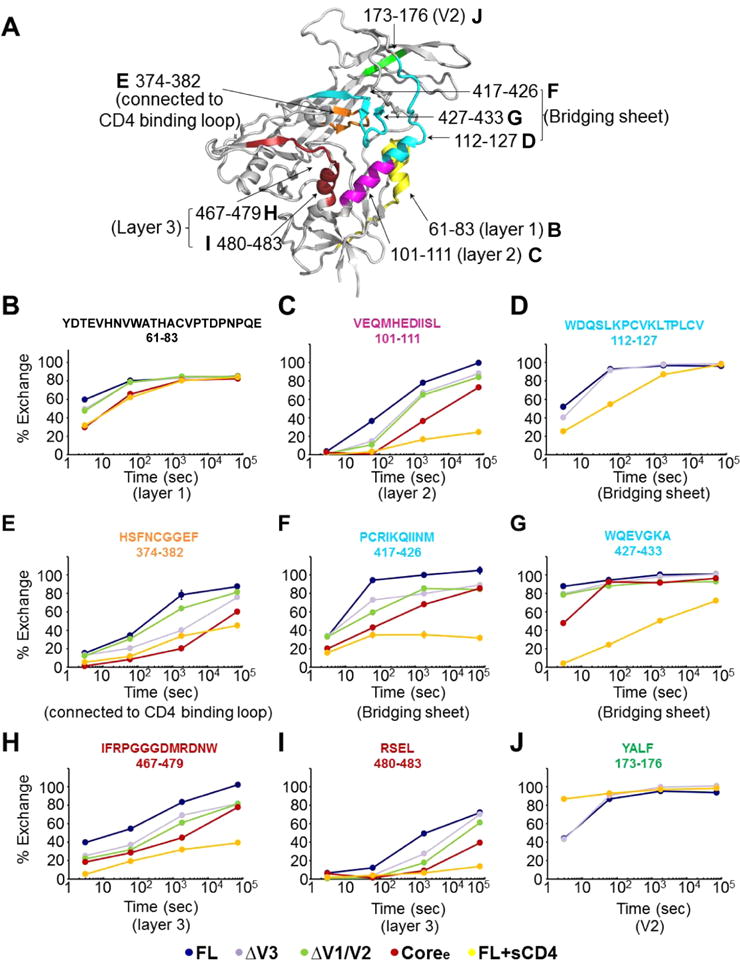
Deuterium-exchange profiles for individual peptides show significant differences in local structural order in variable loop truncation variants. (A) Mapping of selected peptides showing differences in deuterium exchange between FL and other proteins (ΔV3, ΔV1/V2, Coree, and FL+sCD4) on the structure of the gp120 monomer (PDB entry 4NCO). (B–J) Individual deuterium uptake profiles for different peptides from layer 1 (B), layer 2 (C), and layer 3 (H and I) of the inner domain; the bridging sheets (D, F, and G); the CD4-binding loop (E); and the V2 loop (J). HXB2 numbering is applied for the primary sequence.
Deletion of Variable Loops Progressively Stabilizes Unliganded gp120 Constructs
Truncation of the V3 loop led to detectable increases in the level of ordering at sites distal to the loop region itself (Figure 2 and Figures S3–S5). Modest ordering was observed in select regions of the inner domain, including in layer 2 (residues 101–111) and layer 3 (residues 467–479 and residues 480–483) (Figure 3C,H,I). A peptide covering layer 1 (residues 66–83) showed exchange protection at the 3 s time point (Figure 3B). The β2 peptide (residues 112–127), which forms part of the bridging sheet, remained highly dynamic with only a slight measurable increase in protection from solvent exchange in ΔV3 gp120 at 3 s (Figure 3D). β20 and β21 peptides (residues 417–426 and 427–433, respectively) by contrast showed significant protection in ΔV3 gp120, indicating that loop truncation led to partial ordering of the bridging sheet (Figure 3F,G). The CD4-binding loop also experienced an increase in its level of order compared to that in unliganded FL (Figure 3E).
Figure 2.
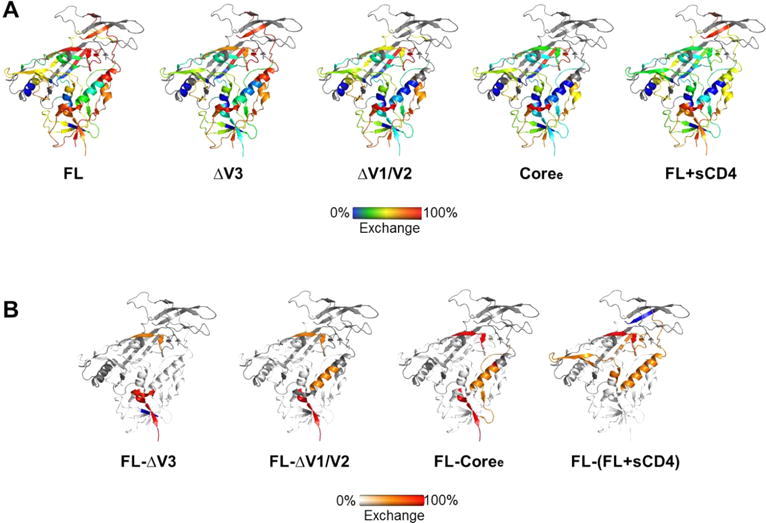
Stabilization of gp120 monomers caused by deletions of variable loops as detected by H/D exchange. (A) Deuterium exchange for FL, ΔV3, ΔV1/V2, Coree, and FL+sCD4 at 1 min plotted on the heat map (from left to right). (B) Differences in deuterium exchange between FL and ΔV3, FL and ΔV1/V2, FL and Coree, and FL and FL+sCD4 at 1 min plotted on the heat map. The degrees of change between the two proteins are colored from white (<20% difference) to orange (20–50% difference) to red (>50% difference).
The ΔV1/V2 truncation mutant, like the V3 truncation mutant, exhibited an increase in the level of structural order of the conserved core (Figure 2 and Figures S3–S6). The inner domain in ΔV1/V2, including layers 1–3, showed exchange protection measurably greater than that of the ΔV3 variant (Figures 2 and 3 and Figure S6C). The same trend was observed in the β20 and β21 peptides of the bridging sheet subdomain in the ΔV1/V2 variant (Figure 3F,G and Figure S6C). By contrast, in ΔV1/V2, the CD4-binding loop exhibited local dynamics similar to that in unliganded full-length gp120 and did not exhibit the same protection as in ΔV3 (Figure 3E and Figure S6C).
The gp120 Coree construct with V1/V2 and V3 loops truncated showed a high degree of protection from solvent exchange and exhibited an H/D-exchange profile more similar to that of sCD4-bound than unliganded full-length gp120 (Figure 2 and Figures S3–S5). All of the available inner domain peptides (Figure 3B,C,H,I), bridging sheet peptides (Figure 3D,F,G), and the CD4-binding loop peptide (Figure 3E) showed increases in protection from deuterium uptake, indicating that deletion of V1/V2 and V3 loops led to substantially increased levels of structural ordering of the inner domain, formation of the bridging sheet, and stabilization of the CD4-binding site. These findings mirror the observation that unliganded Coree adopts a structure nearly identical to that of the sCD4-bound core when crystallized.33 Though we note that while some structural elements such as layer 1 of the core exhibited the same protection from solvent exchange as that of FL+sCD4, overall the unliganded core retained a greater level of deuterium exchange and conformational flexibility in solution than sCD4-bound gp120 (Figures 2 and 3 and Figures S3–S5). Thus, it is likely that crystallization helped to lock in the receptor-bound conformation of the core, while in solution it is more conformationally dynamic than suggested by the crystal structure.
Taken together, these data suggest that the greater the extent of variable loop V1/V2 and V3 truncation, the greater the level of ordering of the gp120 core domain elements toward a conformation and ordered profile resembling the receptor-bound gp120. However, only sCD4 binding induces the full extent of structural ordering consistent with the reported crystal structures.
Deletion of Variable Loops Stabilizes CD4-Induced Epitopes and Improves Recognition by CD4-Induced Antibodies
To determine whether the differences in the structural ordering of FL, ΔV3, ΔV1/V2, and Coree detected by HDX-MS correlated with changes in antigenicity of the glycoproteins, we used ELISAs to assess binding of the antibody to gp120s and Fortébio Octet biolayer interferometry (BLI) to measure the kinetics of antibody–gp120 binding.
Bridging sheet-specific antibodies 17b and 48d and inner domain-specific antibodies A32 and N5i5 recognize CD4-induced (CD4i) epitopes36,42,60–62 (Figure 4A). On the basis of the HDX-MS analysis, the bridging sheet (recognized by 17b and 48d) and gp120 inner domain (recognized by A32 and N5i5) become more ordered with more extensive variable loop truncation (Figure 4B–F); hence, we anticipated these antibodies might exhibit enhanced binding to the truncation mutants. First, monoclonal antibody B18 that recognizes the linear epitope VEQMHEDIIS in helix 1, shared by all the gp120 constructs, was used as a control in ELISA experiments.43,60,63 Indeed, differences in binding of antibody B18 to the four gp120s were negligible as determined by an ELISA (Figure 5A). The gp120 FL displayed the weakest binding to 17b among the four gp120s and Coree showed the strongest binding. In an ELISA, the level of binding of 17b increased in the following order: FL < ΔV3 < ΔV1/V2 < Coree (Figure 5B). Similarly, Octet BLI kinetic analysis showed that gp120 FL had the lowest association rate while Coree had the highest on-rate with CD4i-binding antibodies. The association rates of the four gp120s with CD4i antibodies 17b, 48d, A32, and N5i5 increased in the following order: FL < ΔV3 < ΔV1/V2 < Coree (Figure 5C and Table 2).
Figure 4.
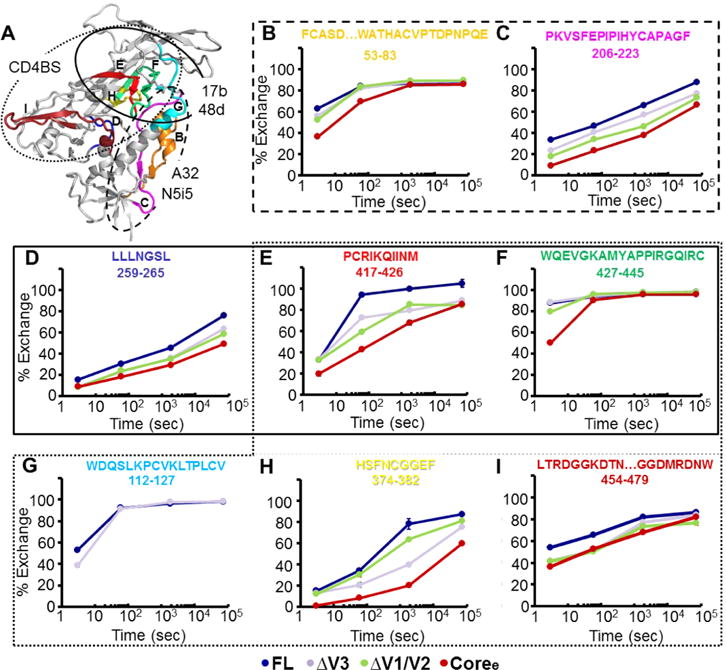
Deuterium-exchange profiles for conformation-dependent antibody epitopes. (A) Indication of 17b or 48d,94 A32, or N5i542,62,95 epitopes on the gp120 monomer (PDB entry 4NCO). The CD4-binding site21,52 is targeted by CD4-binding antibodies, including VRC01,12 VRC03,67 F105,68 and b12.73 The peptides detected in H/D exchange, which cover the epitopes, are colored. (B–I) Individual deuterium uptake profiles for selected peptides positioned at conformation-dependent antibody epitopes. HXB2 numbering is applied for the primary sequence.
Figure 5.
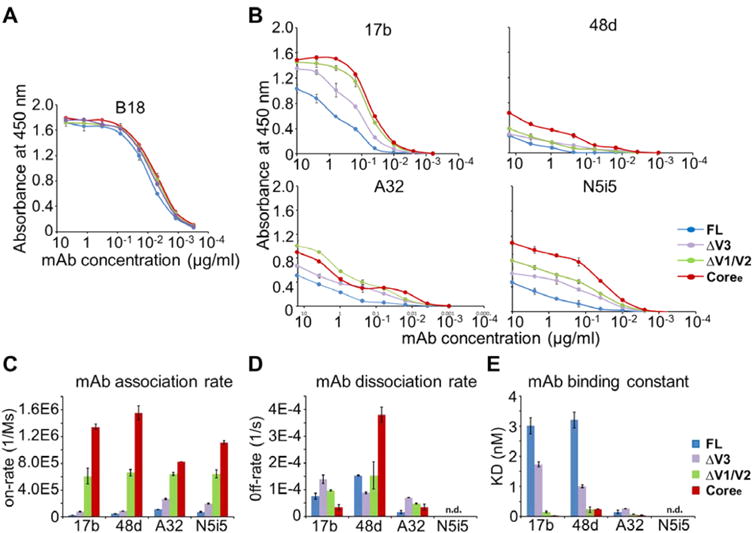
gp120 loop truncation variants show dramatic differences in the binding of the antibody to conserved CD4i conformational epitopes. (A) Binding of FL, ΔV3, ΔV1/V2, and Coree to B18 antibody as determined by an ELISA. B18 binds a linear epitope in the inner domain. (B) Binding of FL, ΔV3, ΔV1/V2, and Coree to 17b, 48d, A32, and N5i5 as determined by an ELISA. The binding curves are colored by different gp120s. Antibody concentrations are shown on the x-axis. The absorbance at 450 nm is shown on the y-axis, representing antibody binding levels. The error bars represent standard deviations from duplicate independent measurements. (C–E) Binding to CD4i-binding antibodies measured by a Fortebio Octet biolayer interferometry platform. (C) Association rates (on-rate) for binding of FL, ΔV3, ΔV1/V2, and Coree to 17b, 48d, A32, and N5i5. (D) Dissociation rates (off-rate) for binding of the four gp120s to 17b, 48d, A32, and N5i5. (E) KD values for binding of the four gp120s to 17b, 48d, A32, and N5i5. Raw data and fitted curves are shown in Figure S6. Error bars represent standard deviations of two independent measurements.
Table 2.
Octet Biolayer Interferometry Binding Parametersa for gp120–Antibody Interactions with CD4i-Binding Antibodiesb
| antibody | parameter | FL | ΔV3 | ΔV1/V2 | Coree |
|---|---|---|---|---|---|
| 17b | ka (M−1 s−1) | 2.5(1) × 104 | 8.1(5) × 104 | 6(1) × 105 | 1.34(5) × 106 |
| kd (s−1) | 8(1) × 10−5 | 1.4(2) × 10−4 | 9.7(2) × 10−5 | 3.5(9) × 10−5 | |
| KD (nM) | 3.0(3) | 1.73(8) | 1.6(3) × 10−1 | 2.6(8) × 10−2 | |
| 48d | ka (M−1 s−1) | 4.8(4) × 104 | 8.9(1) × 104 | 6.6(5) × 105 | 1.6(1) × 106 |
| kd (s−1) | 1.54(1) × 10−4 | 8.9(3) × 10−5 | 1.5(5) × 10−4 | 3.8(3) × 10−4 | |
| KD (nM) | 3.2(3) | 1.01(5) | 2.4(9) × 10−1 | 2.42(7) × 10−1 | |
| A32 | ka (M−1 s−1) | 1.12(1) × 105 | 2.69(8) × 105 | 6.4(2) × 105 | 8.220(3) × 105 |
| kd (s−1) | 1.72(6) × 10−5 | 7.04(3) × 10−5 | 4.8(3) × 10−5 | 4(1) × 10−5 | |
| KD (nM) | 1.54(5) × 10−1 | 2.62(7) × 10−1 | 7.5(7) × 10−2 | 4.1(9) × 10−2 | |
| N5i5 | ka (M−1 s−1) | 7.7(4) × 104 | 1.97(9) × 105 | 6.4(6) × 105 | 1.11(3) × 106 |
| kd (s−1) | ndc | ndc | ndc | ndc | |
| KD (nM) | ndc | ndc | ndc | ndc |
ka, rate of association; kd, rate of dissociation; KD, equilibrium dissociation constant.
Values are means ± standard deviations. Standard errors are for two independent measurements, presented in parentheses with respect to the smallest number place value: 2.4(1) × 104 is equivalent to 24000 ± 1000.
Because of slow dissociation, dissociation rates or binding constants could not be reliably fit.
To examine the correlation of structural dynamics and binding of the antibody to the CD4i epitopes, we plotted the equilibrium dissociation constant (KD) and on-rate versus deuterium exchange of a set of peptides located in the epitopes (% exchange). As shown in Figure 6, KD increased in concert with increased levels of local dynamics measured by HDX-MS, suggesting that enhanced structural flexibility within antibody epitopes correlates with weaker binding affinities. This appeared to be driven primarily by differences in on-rates, as the rate of antibody association decreased as structural dynamics of the peptides increased. A significant linear regression negative correlation (R2 > 0.7) was observed between the on-rate of binding to CD4i antibodies and deuterium exchange (Figure 6). The dissociation rates (Figure 5D) by contrast did not show a detectable correlation with local structural dynamics.
Figure 6.
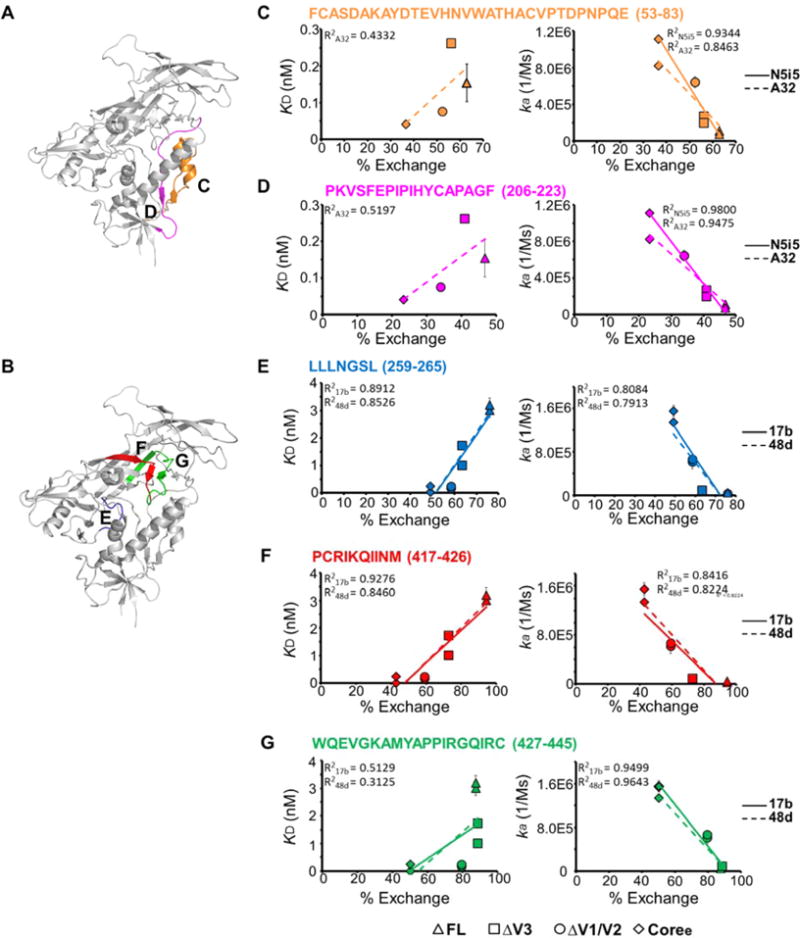
Correlation of binding of antibodies to CD4-induced epitopes and local structural dynamics. (A) Peptides detected via H/D exchange, which cover the critical interface residues on the gp120 monomer (PDB entry 4NCO) of A32 or N5i5, are colored.42 (B) Peptides detected in H/D exchange, which cover the interface residues on the gp120 monomer (PDB entry 4NCO) of 17b or 48d (crayon blue), are colored.94 (C–G) Plots of deuterium exchange at a single time point, where the largest differences were observed for the four gp120s, of different peptides covering the binding interface of 17b or 48d vs KD (left) or association rates (on-rate) (right) with linear regression. Data for FL, ΔV3, ΔV1/V2, and Coree are shown as triangles, squares, circles, and diamonds, respectively. Error bars represent standard deviations for two independent measurements.
Taken together, the conformational changes resulting from variable loop truncation led to greater ordering of the gp120 core and, as a result, more effective presentation of CD4-induced epitopes, resulting in stronger binding of 17b, 48d, A32, and N5i5 to the more ordered gp120 constructs.
Deletion of Variable Loops Stabilizes CD4-Binding-Site-Specific (CD4bs) Epitopes and Improves Recognition by CD4-Binding-Site Antibodies
The binding interface for CD4 on gp120 includes portions of the inner domain, bridging sheet, and outer domain,21,64 with the majority of critical residues located on the outer domain of gp120 (Figure 4A). Deuterium uptake plots for the CD4-binding interface peptides that could be monitored by HDX-MS are shown in Figure 4. These include bridging sheet strands (Figure 4E–G), the CD4-binding loop (Figure 4H), β23 and β24 of the outer domain, V5, and part of helix 5 (Figure 4I). As with peptide segments in the CD4i epitopes, the CD4bs epitopes follow a rank order of structural flexibility as assessed by deuterium uptake: FL > ΔV3 > ΔV1/V2 > Coree.
As determined by an ELISA, the most highly ordered gp120 construct, Coree, was observed to bind to CD4-IgG265 slightly better than the less truncated constructs (Figure 7A). As determined by Octet BLI, the differences were more prominent. Coree had the highest association rate and lowest dissociation rate upon CD4-IgG2 binding with a KD for Coree reflecting an affinity ~10-fold greater than those for FL and ΔV3 and ∼3-fold greater than that for ΔV1/V2 (Figure 7B–D and Table 3). To probe for correlations between structural order in the gp120 constructs and CD4-binding site antibody recognition of gp120, we also examined binding of a panel of CD4-binding site-directed (CD4bs) antibodies to the gp120 truncation variants by an ELISA and Octet BLI. Antibodies VRC01, NIH45–46, VRC03, b12, and F105 share critical interface residues on gp120 such as those on the CD4-binding loop and bridging sheets but differ in some specific contact residues and angles of approach.21,36,66–69 As shown in Figure 4A, the important interface residues for the CD4bs antibodies overlap substantially with the binding interface of CD4.12,67,68,70 As with CD4-IgG2, Coree displayed the strongest binding to VRC01 as monitored by an ELISA, while FL displayed the weakest binding (Figure 7A). The same trend was reported by Octet BLI with association rates and binding affinities: FL < ΔV3 < ΔV1/V2 < Coree (Figure 7B,D). The dissociation rates of FL and ΔV3 were higher than that of ΔV1/V2 or Coree (Figure 7C). A similar trend was observed for NIH45–46, a more potent variant of VRC01 with greater breadth1 (Figure 7).
Figure 7.
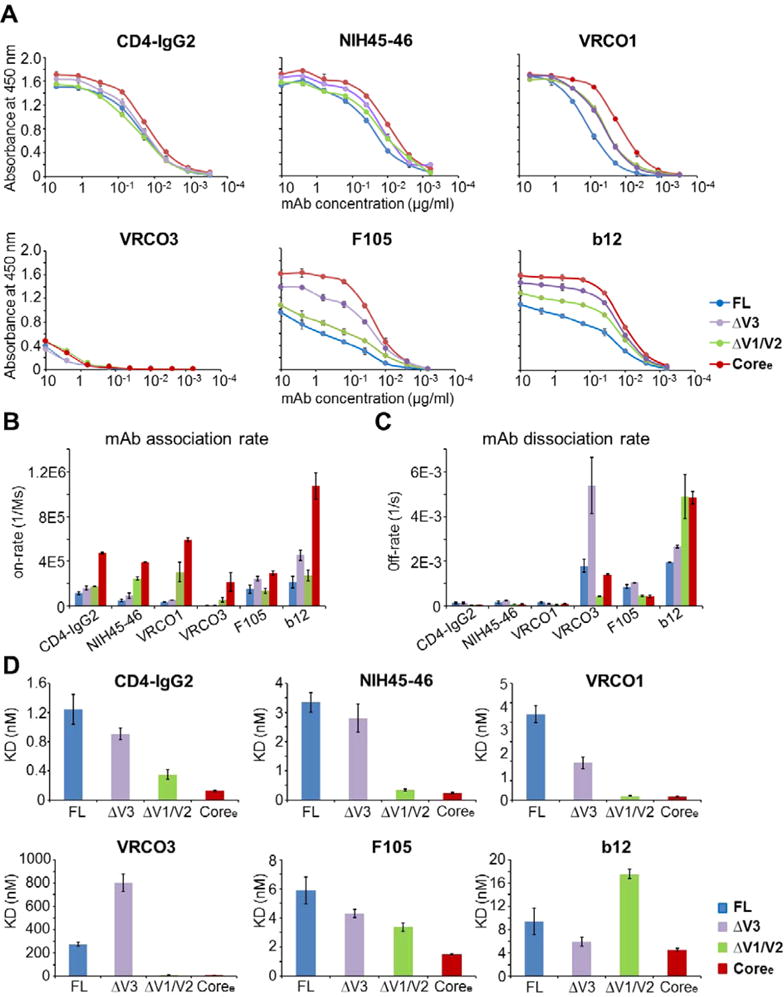
gp120 loop truncation variants show dramatic differences in binding of the antibody to the CD4-binding site epitope. (A) Binding of FL, ΔV3, ΔV1/V2, and Coree to CD4-IgG2, NIH45–46, VRC01, VRC03, F105, and b12 (from left to right, respectively) as determined by an ELISA. The binding curves are colored by different gp120s. Antibody concentrations are shown on the x-axis. The absorbance at 450 nm is shown on the y-axis, representing antibody binding levels. The error bars represent standard deviations of duplicate independent measurements. (B–D) Binding to CD4bs antibodies measured by a Fortebio Octet biolayer interferometry platform. (B) Association rates (on-rate) for binding of FL, ΔV3, ΔV1/V2, and Coree to CD4-IgG2, NIH45–46, VRC01, VRC03, F105, and b12. (B) Dissociation rates (off-rate) for binding of the four gp120s to CD4-IgG2, NIH45–46, VRC01, VRC03, F105, and b12. (C) KD values for the four gp120s with CD4-IgG2, NIH45–46, VRC01, VRC03, F105, and b12. Raw data and fitted curves are presented in Figure S7. Error bars represent standard deviations of two independent measurements.
Table 3.
Octet Biolayer Interferometry Binding Parametersa for gp120–Antibody Interactions with CD4-IgG2- and CD4bs-Binding Antibodiesb
| antibody | parameter | FL | ΔV3 | ΔV1/V2 | Coree |
|---|---|---|---|---|---|
| CD4-IgG2 | ka (M−1 s−1) | 1.2(1) × 105 | 1.6(2) × 105 | 1.780(7) × 105 | 4.74(6) × 105 |
| kd (s−1) | 1.5(4) × 10−4 | 1.5(3) × 10−4 | 5.4(3) × 10−5 | 5.9(8) × 10−5 | |
| KD (nM) | 1.3(2) | 9.1(8) × 10−1 | 3.4(7) × 10−1 | 1.2(1) × 10−1 | |
| NIH45−46 | ka (M−1 s−1) | 5(1) × 104 | 9(2) × 104 | 2.5(1) × 105 | 3.94(2) × 105 |
| kd (s−1) | 1.8(6) × 10−4 | 2.6(3) × 10−4 | 8.2(4) × 10−5 | 9(1) × 10−5 | |
| KD (nM) | 3.4(3) | 2.8(4) | 3.3(4) × 10−1 | 2.4(3) × 10−1 | |
| VRC01 | ka (M−1 s−1) | 3.8(3) × 104 | 5.4(1) × 104 | 3.0(9) × 105 | 5.9(2) × 105 |
| kd (s−1) | 1.7(3) × 10−4 | 1.1(2) × 10−4 | 6.4(3) × 10−5 | 10(2) × 10−5 | |
| KD (nM) | 4.4(4) | 1.9(3) | 2.1(3) × 10−1 | 1.59(4) × 10−1 | |
| VRC03 | ka (M−1 s−1) | 6.5(6) × 103 | 6.7(9) × 103 | 6(2) × 104 | 2.2(8) × 105 |
| kd (s−1) | 1.8(3) × 10−3 | 5(1) × 10−3 | 4.5(3) × 10−4 | 1.41(3) × 10−3 | |
| KD (nM) | 275(16) | 803(75) | 8(3) | 7(2) | |
| F105 | ka (M−1 s−1) | 1.5(4) × 105 | 2.5(2) × 105 | 1.4(2) × 105 | 2.9(2) × 105 |
| kd (s−1) | 8.7(8) × 10−4 | 1.05(1) × 10−3 | 4.6(4) × 10−4 | 4.4(4) × 10−4 | |
| KD (nM) | 5.9(9) | 4.3(3) | 3.4(3) | 1.50(4) | |
| b12 | ka (M−1 s−1) | 2.2(5) × 105 | 4.5(5) × 105 | 2.7(5) × 105 | 1.1(1) × 106 |
| kd (s−1) | 1.960(7) × 10−3 | 2.66(5) × 10−3 | 4.9(9) × 10−3 | 4.8(3) × 10−3 | |
| KD (nM) | 9(2) | 5.9(7) | 17.6(8) | 4.5(2) |
ka, rate of association; kd, rate of dissociation; KD, equilibrium dissociation constant.
Values are means ± standard deviations. Standard errors are for two independent measurements, presented in parentheses with respect to the smallest number place value.
Although the VRC03 gp120-binding interface is similar to that of VRC01 (Figure 4A), VRC03 behaves differently; for example, binding of VRC03 to gp120 does not increase the level of binding of 17b.38,71 It has also been reported that the binding affinity of VRC03 for gp120 is lower than that of VRC01.38 As shown in Figure 8D, the KD for binding of ΔV3 to VRC03 was much higher than those of the other truncations and even higher than that of the unliganded full-length gp120. The association rates of the four gp120s with VRC03 were ranked like those for binding to CD4-IgG2 or VRC01 (FL < ΔV3 < ΔV1/V2 < Coree) (Figure 7B). This is consistent with the dynamic differences among the four gp120s (Figure 4). However, the dissociation rate of ΔV3 upon binding to VRC03 was the highest among those of the four gp120s (Figure 7C). This indicates that the ΔV3–VRC03 complex is relatively unstable. Two possible factors may contribute to destabilize the ΔV3–VRC03 complex. First, the truncation of V3 may remove a constraint upon the large V1/V2 loop region and increase the spatial occupancy of V1/V2 loops, which possibly increases the number of steric clashes with the VRC03 heavy chain and destabilizes the ΔV3–VRC03 complex. Second, at the binding interface between VRC03 and the gp120 core, a β-sheet involving an extra residue inserted into the heavy chain of VRC03 that is not present in the VRC01 complex is formed.38 This may increase the number of steric clashes between ΔV3 and VRC03 and lead to destabilization of the ΔV3–VRC03 complex.
Figure 8.
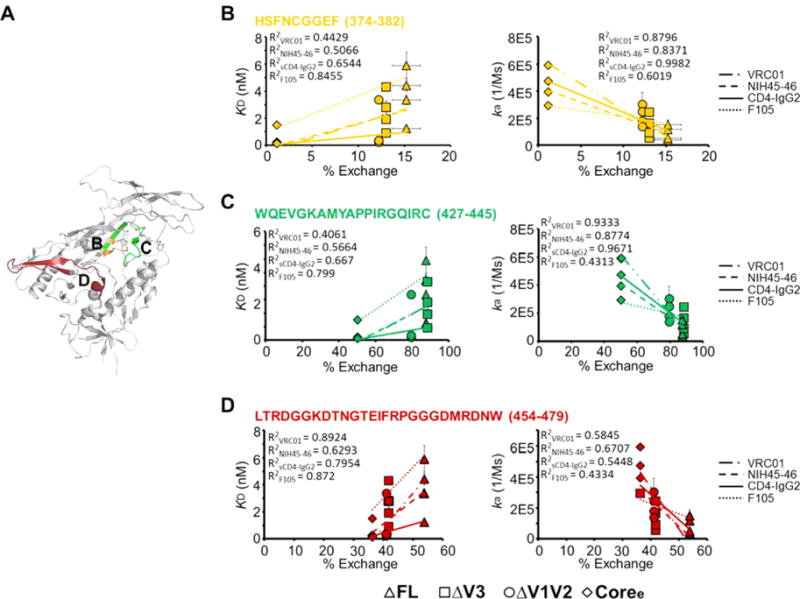
Correlation of CD4-binding site antibody binding and local structural dynamics. (A) Peptides detected in H/D exchange, which cover the critical interface residues on the gp120 monomer (PDB entry 4NCO) upon binding of CD4bs antibodies, are colored. (B–D) Plots of deuterium exchanges at a single time point, where the largest differences were observed for the four gp120s, of different peptides covering the binding interface of CD4bs-binding antibodies vs KD (left) or on-rate (right) with linear regression. Data for FL, ΔV3, ΔV1/V2, and Coree are shown as triangles, squares, circles, and diamonds, respectively. Error bars represent standard deviations for two independent measurements.
CD4bs Ab F105 is capable of neutralizing only a small subset of relatively neutralization sensitive viruses.43 In the crystal structure of F105 Fab bound to the gp120 core, it was observed that the peptides involved in bridging sheet formation were displaced and the V1/V2 stem tips were shifted relative to the conformation observed in the sCD4-bound conformation.68 In both ELISA and Octet BLI analysis, Coree exhibited the strongest interactions with F105 (Figure 7A,D and Table 3). The unliganded full-length gp120 showed the weakest binding to F105 resulting from a combination of a low association rate and a high dissociation rate (Figure 7). The F105 antibody binding trend of the four gp120 constructs was as follows: FL < ΔV3 < ΔV1/V2 < Coree. This is consistent with the results of previous antigenicity studies, which indicated that F105 bound to ΔV1/V2 and ΔV1/V2/V3 gp120s more strongly than to full-length gp120.72
IgG1-b12 is another CD4bs neutralizing antibody that also recognizes a small portion of V1/V225,73 (Figure 4A). As shown in Figure 7, ΔV1/V2 gp120 exhibited a low association rate and a high dissociation rate in b12 binding. Additionally, the KD for ΔV1/V2 was greater than those of the other three gp120s. This is consistent with previous observations that deletions of V1/V2 lead to a reduction in the level of b12 binding. Additionally, possibly because of the deletions of both V1/V2 and V3 loops, Coree exhibited the highest association rate because of reduced steric hindrance, but also a high dissociation rate because of the deletion of contact residues that contribute to the stability of the complex. This resulted in relatively strong binding of Coree to b12, on par with what was measured for ΔV3 and full-length gp120.
It is perhaps notable that the more poorly neutralizing CD4bs antibodies F105 and b12 in ELISA measurements were observed to exhibit differences in binding to the panel of gp120 constructs greater than those of the antibodies with greater neutralization breadth (Figure 7A). The trend could be inferred to reflect improved focusing of the NAbs with greater breadth on conserved features of the CD4bs, resulting in less sensitivity to variations and dynamic fluctuations in structural features outside of the binding site. Such features influence the interactions of VRC03 (involvement of bridging sheet38) and b12 (involvement of a small element of V225,81) with gp120. However, the CD4bs mAbs showed more similar binding trends in BLI measurements where the glycoprotein is the soluble analyte and presented in a more native state than in the ELISA experiments where gp120 is adsorbed to substrates prior to antibody binding. We generally consider the BLI experiments to yield more definitive measures of antibody–antigen interaction.
To assess whether a correlation exists between gp120 structural dynamics and CD4bs antibody binding, we plotted kinetic parameters of binding versus deuterium uptake of a set of peptides located in the epitopes (% exchange). As shown in Figure 8, a negative correlation (R2 > 0.5) was observed between the on-rate of binding of the gp120 construct to CD4-IgG2, VRC01, and NIH45–46 and the measured deuterium uptake of peptides within the antibodies’ epitopes. The negative correlation between F105 association rate and deuterium exchange was weaker (R2 > 0.4). Overall, increased structural dynamics within the CD4bs was associated with lower on-rates and weaker affinities of gp120s for the CD4bs antibodies, while dissociation rates did not show a detectable correlation with the level of local structural dynamics.
DISCUSSION
The relation of structural flexibility and antigenicity is of fundamental significance and long-standing interest for understanding the structural basis of antibody recognition of target epitopes.74,75 In past studies in which linear epitopes were investigated, conformational flexibility was generally observed to positively correlate with increased antigenicity.64,65 This led to the conclusion that antibodies bind more readily to linear epitopes that displayed high B factors in crystal structures, representing high local flexibility.3,76,77 Other studies, however, indicated that antibodies do not always bind isolated peptides more effectively than the antibodies bind the same peptide in the context of the intact antigen, possibly because of the tertiary context constraining the peptide into a conformation recognized by the antibody.75,78
Relatively few studies have examined the direct correlation of local structural dynamics and antigenicity for complex, discontinuous conformational epitopes such as those targeted by broadly neutralizing antibodies against native antigen on the surface of pathogens. In HIV Env, the conserved epitopes that are of greatest interest for targeting by broadly neutralizing antibodies are these types of complex conformational epitopes, including the CD4-binding site, V1/V2 loops, and around the base of V3.79,80 The coreceptor-binding site, involving bridging sheet and V3 elements, is highly conserved, as well, but in general not highly accessible until CD4 has bound, though in more neutralization sensitive isolates, it appears that conformational sampling and breathing by Env provide some degree of accessibility to the binding site.24,25 Likewise, a conformational epitope on the inner domain is highly conserved and targeted by ADCC-active IgG antibodies, such as N5i5 and A32. All of the sites in the gp120 constructs examined here exhibited measurable differences in structural ordering as probed by HDX-MS.
By connecting kinetic parameters versus deuterium exchange, we observed that inclusion of the variable loops, from ΔV1/V2 and ΔV3 to FL, was associated with increased deuterium exchange, greater conformational flexibility (Figure 9), and slower on-rates of binding. This finding is consistent with the observation that compared to ΔV1/V2, ΔV3, and Coree, full-length gp120 with variable loops showed a significantly reduced level of recognition by antibodies that specifically target the CD4-bound conformation.33 In a previous study of four full-length gp120 subunits from distinct HIV-1 isolates, we observed that one particularly dynamic gp120 was poorly recognized by CD4 and antibodies directed against conformational epitopes.30 A thorough comparison of local epitope structural dynamics and antigenicity was not straightforward, however, in that case, because of differences in sequence among the four variants. In the study presented here, the differences in structural dynamics among gp120 constructs all based upon the YU2 isolate allow us to compare dynamics and antigenicity where the epitope residues and subunit core are identical.
Figure 9.
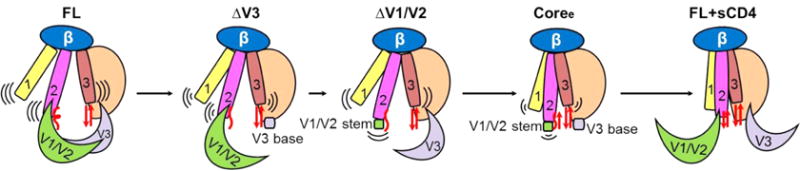
Model of conformational and dynamic changes in the unliganded gp120 monomer upon deletion of V1/V2 and V3 loops. The unliganded wild-type full-length gp120 (FL) (left) had the highest dynamics in three layers of the inner domain with unfolded bridging sheets and masked immature coreceptor-binding site.96 Upon deletion of the V1/V2 and V3 loops, the three layers of the inner domain became less dynamic and more ordered. Coree without all V1/V2 and V3 loops showed more stability toward the CD4-bound state with an ordered inner domain and well-folded bridging sheets. The CD4 engagement (right) induced a fixed, stable conformation with formation of the bridging sheets and movement of V1/V2 loops to expose a mature coreceptor-binding site on the bridging sheet and V3 loop.
The association of increased conformational stability of truncated mutations with faster on-rates was particularly prominent for CD4i and CD4bs antibodies in this study. A similar observation was made in previous studies that indicated a correlation between a faster on-rate and a greater affinity of, for example, CD4i mAb 17b to a series of gp120 cores that were engineered for greater global stability.68,82 We did not, however, observe significant correlation between dissociation rates and local dynamics. Other factors such as the loss of some intermolecular interactions likely influenced the dissociation rates in certain constructs. For example, ΔV1/V2 and Coree showed the fastest dissociation rates from IgG1-b12 due to the loss of important binding interface residues in the V1/V2 loops.25,81 In the case of binding to CD4bs bNAb VRC03, we infer that the rapid dissociation of the antibody from gp120 ΔV3 may be due to the greater degree of freedom afforded to V1/V2 in the absence of interactions with the V3 loop. This may result in greater spatial sampling and steric clashes with the VRC03 heavy chain.
Our results provide evidence of a correlation between local structural dynamics and recognition of those epitopes by a diverse panel of neutralizing antibodies. Env proteins derived from the stabilized gp120 core with surface modifications and masking glycans to focus the immune response on broadly neutralizing epitopes have been proposed as possible immunogens.12,38,67 By constraining the immunogen’s structure and biasing it toward a state that is recognized by desirable antibodies, IgG and B cell receptors have a more focused target and would pay less of an entropic cost to induce the fixed epitope conformation. Support for this is found in studies by Dey et al, which indicated that stabilization of the gp120 core by the introduction of additional disulfide bonds was associated with a significant increase in the on-rate of binding to 17b, CD4, and CD4bs antibodies and increased immunogenicity of the coreceptor-binding site.82 In combination with selection for suitable quaternary structural considerations and angles of approach to epitopes on complex, large antigens such as HIV Env trimers,69 presentation of a conformationally constrained target for antibodies in vaccine immunogens, may help to elicit a better response against the more ordered epitopes.
Indeed, we note that while we have demonstrated the link between structural dynamics and antigenicity in the context of gp120 subunits, the fundamental physical effects linking epitope dynamics and antibody–antigen recognition are general and affect recognition of conformational epitopes on any antigen. Significant attention in HIV vaccines is currently directed toward new constructs that mimic the nativelike trimer structure of the Env ectodomain. These constructs, termed “SOSIP” trimers, have been engineered to maintain the native, prefusion organization of the Env trimer ectodomain through the addition of a gp120–gp41 disulfide bond and mutation of an isoleucine to a proline residue in the HR1 segment.35,83,84 The nativelike SOSIP trimer presents multiple epitopes for broad neutralizing antibodies but few non-neutralizing antibody epitopes.35,85 Although the SOSIP trimer has the ability to induce tier 2 autologous neutralizing antibodies, the induced cross-reactive tier 1 neutralizing antibody response was biased to targeting the V3 epitope.86,87 More recently, two groups have further modified the SOSIP trimer to conformationally fix them in the prefusion, closed state.88,89 In one study, the additional stabilization aimed at reducing conformational sampling and V3 exposure indeed were reported to yield a weaker immune response against V3.89 Similar strategies of stabilizing antigens to more effectively present neutralizing antibody epitopes have been reported for respiratory syncytial virus immunogens8,90 as well as for influenza A virus hemagglutinin-based immunogens.91,92 We thus anticipate that in addition to elucidating a relatively cryptic but important structural property underlying antibody recognition of complex conformational epitopes, an improved understanding of the impact of local structural dynamics on antigenicity and immunogenicity may aid in the further optimization of vaccine immunogens.
Supplementary Material
Acknowledgments
We gratefully acknowledge Drs. Peter D. Kwong and Young Do Kwon of the Vaccine Research Center, NIH, for providing the plasmids used for expression of the YU2 gp120 constructs in this study.
Funding
The work was supported by National Institutes of Health (NIH) Grants R21-AI112389 and R01-GM099989 and by Bill and Melinda Gates Foundation Collaboration for AIDS Vaccine Discovery (CAVD) Grant OPP1033102. Portions of the work were conducted at SSRL and supported by the Structural Molecular Biology Program, Department of Energy’s Office of Biological and Environmental Research, NIH Grants P41-GM103393 and P41-RR001209.
ABBREVIATIONS
- HIV
human immunodeficiency virus
- HDX-MS
hydrogen/deuterium-exchange mass spectrometry
- ADCC
antibody-dependent cell-mediated cytotoxicity
- ELISA
enzyme-linked immunosorbent assay
- Env
envelope glycoprotein
- FL
full length
- Coree
extended core
- NMR
nuclear magnetic resonance
- GNA
G. nivalis lectin
- BN-PAGE
Blue-native polyacrylamide gel electrophoresis
- TCEP
tris(2-carboxyethyl)phosphine hydrochloride
- LC–MS
liquid chromatography–mass spectrometry
- UPLC
ultraperformance liquid chromatography
- TFA
trifluoroacetic acid
- ACN
acetonitrile
- SAXS
small angle X-ray scattering
- PBS
phosphate-buffered saline
- BSA
bovine serum albumin
- TBS
Tris-buffered saline
- HRP
horseradish peroxidase
- AHC
Anti-Human IgG Fc Capture
- BLI
biolayer interferometry
- CD4i
CD4-induced
- KD
equilibrium dissociation constant
- ka
association rate
- kd
dissociation rate
- CD4bs
CD4-binding site-specific
- SLS
static light scattering
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.5b01354.
Data for protein purification and the complete hydrogen/deuterium-exchange mass spectrometry data in the form of butterfly plots and deuterium uptake plots for each observable peptide, a peptic digest sequence coverage map, and Octet biolayer interferometry sensor-grams measuring antibody–gp120 binding (PDF)
Notes
The authors declare no competing financial interest.
References
- 1.Diskin R, Scheid JF, Marcovecchio PM, West AP, Jr, Klein F, Gao H, Gnanapragasam PN, Abadir A, Seaman MS, Nussenzweig MC, Bjorkman PJ. Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science. 2011;334:1289–1293. doi: 10.1126/science.1213782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McGuire AT, Hoot S, Dreyer AM, Lippy A, Stuart A, Cohen KW, Jardine J, Menis S, Scheid JF, West AP, Schief WR, Stamatatos L. Engineering HIV envelope protein to activate germline B cell receptors of broadly neutralizing anti-CD4 binding site antibodies. J Exp Med. 2013;210:655–663. doi: 10.1084/jem.20122824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Westhof E, Altschuh D, Moras D, Bloomer AC, Mondragon A, Klug A, Van Regenmortel MH. Correlation between segmental mobility and the location of antigenic determinants in proteins. Nature. 1984;311:123–126. doi: 10.1038/311123a0. [DOI] [PubMed] [Google Scholar]
- 4.Kumar S, Ma B, Tsai CJ, Sinha N, Nussinov R. Folding and binding cascades: dynamic landscapes and population shifts. Protein Sci. 2000;9:10–19. doi: 10.1110/ps.9.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ponomarenko J, Bui HH, Li W, Fusseder N, Bourne PE, Sette A, Peters B. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinf. 2008;9:514. doi: 10.1186/1471-2105-9-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ofek G, Guenaga FJ, Schief WR, Skinner J, Baker D, Wyatt R, Kwong PD. Elicitation of structure-specific antibodies by epitope scaffolds. Proc Natl Acad Sci U S A. 2010;107:17880–17887. doi: 10.1073/pnas.1004728107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van den Elsen J, Vandeputte-Rutten L, Kroon J, Gros P. Bactericidal antibody recognition of meningococcal PorA by induced fit. Comparison of liganded and unliganded Fab structures. J Biol Chem. 1999;274:1495–1501. doi: 10.1074/jbc.274.3.1495. [DOI] [PubMed] [Google Scholar]
- 8.McLellan JS, Chen M, Joyce MG, Sastry M, Stewart-Jones GB, Yang Y, Zhang B, Chen L, Srivatsan S, Zheng A, Zhou T, Graepel KW, Kumar A, Moin S, Boyington JC, Chuang GY, Soto C, Baxa U, Bakker AQ, Spits H, Beaumont T, Zheng Z, Xia N, Ko SY, Todd JP, Rao S, Graham BS, Kwong PD. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science. 2013;342:592–598. doi: 10.1126/science.1243283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.MacRaild CA, Zachrdla M, Andrew D, Krishnarjuna B, Novacek J, Zidek L, Sklenar V, Richards JS, Beeson JG, Anders RF, Norton RS. Conformational dynamics and antigenicity in the disordered malaria antigen merozoite surface protein 2. PLoS One. 2015;10:e0119899. doi: 10.1371/journal.pone.0119899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt SA, Majeed S, Steenbeke TD, Venturi M, Chaiken I, Fung M, Katinger H, Parren PW, Robinson J, Van Ryk D, Wang L, Burton DR, Freire E, Wyatt R, Sodroski J, Hendrickson WA, Arthos J. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420:678–682. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- 11.Myszka DG, Sweet RW, Hensley P, Brigham-Burke M, Kwong PD, Hendrickson WA, Wyatt R, Sodroski J, Doyle ML. Energetics of the HIV gp120-CD4 binding reaction. Proc Natl Acad Sci U S A. 2000;97:9026–9031. doi: 10.1073/pnas.97.16.9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, Yang Y, Zhu J, Nussenzweig MC, Sodroski J, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adhikary R, Yu W, Oda M, Zimmermann J, Romesberg FE. Protein dynamics and the diversity of an antibody response. J Biol Chem. 2012;287:27139–27147. doi: 10.1074/jbc.M112.372698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jimenez R, Salazar G, Yin J, Joo T, Romesberg FE. Protein dynamics and the immunological evolution of molecular recognition. Proc Natl Acad Sci U S A. 2004;101:3803–3808. doi: 10.1073/pnas.0305745101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Addis PW, Hall CJ, Bruton S, Veverka V, Wilkinson IC, Muskett FW, Renshaw PS, Prosser CE, Carrington B, Lawson AD, Griffin R, Taylor RJ, Waters LC, Henry AJ, Carr MD. Conformational heterogeneity in antibody-protein antigen recognition: implications for high affinity protein complex formation. J Biol Chem. 2014;289:7200–7210. doi: 10.1074/jbc.M113.492215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vu TQ, Liu HY. Quantum dot hybrid gel blotting: a technique for identifying quantum dot-protein/protein-protein interactions. Methods Mol Biol. 2009;544:381–391. doi: 10.1007/978-1-59745-483-4_25. [DOI] [PubMed] [Google Scholar]
- 17.Rosen O, Anglister J. Epitope mapping of antibody-antigen complexes by nuclear magnetic resonance spectroscopy. Methods Mol Biol. 2009;524:37–57. doi: 10.1007/978-1-59745-450-6_3. [DOI] [PubMed] [Google Scholar]
- 18.Naik MT, Chang CF, Kuo IC, Kung CC, Yi FC, Chua KY, Huang TH. Roles of structure and structural dynamics in the antibody recognition of the allergen proteins: an NMR study on Blomia tropicalis major allergen. Structure. 2008;16:125–136. doi: 10.1016/j.str.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 19.Julien JP, Lee PS, Wilson IA. Structural insights into key sites of vulnerability on HIV-1 Env and influenza HA. Immunol Rev. 2012;250:180–198. doi: 10.1111/imr.12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Falkowska E, Le KM, Ramos A, Doores KJ, Lee JH, Blattner C, Ramirez A, Derking R, van Gils MJ, Liang CH, McBride R, von Bredow B, Shivatare SS, Wu CY, Chan-Hui PY, Liu Y, Feizi T, Zwick MB, Koff WC, Seaman MS, Swiderek K, Moore JP, Evans D, Paulson JC, Wong CH, Ward AB, Wilson IA, Sanders RW, Poignard P, Burton DR. Broadly neutralizing HIV antibodies define a glycan-dependent epitope on the prefusion conformation of gp41 on cleaved envelope trimers. Immunity. 2014;40:657–668. doi: 10.1016/j.immuni.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, Wilson IA, Ward AB. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, Ward AB, Wilson IA. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munro JB, Gorman J, Ma X, Zhou Z, Arthos J, Burton DR, Koff WC, Courter JR, Smith AB, 3rd, Kwong PD, Blanchard SC, Mothes W. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science. 2014;346:759–763. doi: 10.1126/science.1254426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guttman M, Cupo A, Julien JP, Sanders RW, Wilson IA, Moore JP, Lee KK. Antibody potency relates to the ability to recognize the closed, pre-fusion form of HIV Env. Nat Commun. 2015;6:6144. doi: 10.1038/ncomms7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moody MA, Yates NL, Amos JD, Drinker MS, Eudailey JA, Gurley TC, Marshall DJ, Whitesides JF, Chen X, Foulger A, Yu JS, Zhang R, Meyerhoff RR, Parks R, Scull JC, Wang L, Vandergrift NA, Pickeral J, Pollara J, Kelsoe G, Alam SM, Ferrari G, Montefiori DC, Voss G, Liao HX, Tomaras GD, Haynes BF. HIV-1 gp120 vaccine induces affinity maturation in both new and persistent antibody clonal lineages. Journal of Virology. 2012;86:7496–7507. doi: 10.1128/JVI.00426-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pantophlet R, Burton DR. GP120: target for neutralizing HIV-1 antibodies. Annu Rev Immunol. 2006;24:739–769. doi: 10.1146/annurev.immunol.24.021605.090557. [DOI] [PubMed] [Google Scholar]
- 28.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, Robb ML, Michael NL, Kunasol P, Kim JH. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 29.Fouts TR, Bagley K, Prado IJ, Bobb KL, Schwartz JA, Xu R, Zagursky RJ, Egan MA, Eldridge JH, LaBranche CC, Montefiori DC, Le Buanec H, Zagury D, Pal R, Pavlakis GN, Felber BK, Franchini G, Gordon S, Vaccari M, Lewis GK, DeVico AL, Gallo RC. Balance of cellular and humoral immunity determines the level of protection by HIV vaccines in rhesus macaque models of HIV infection. Proc Natl Acad Sci U S A. 2015;112:E992–999. doi: 10.1073/pnas.1423669112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davenport TM, Guttman M, Guo W, Cleveland B, Kahn M, Hu SL, Lee KK. Isolate-specific differences in the conformational dynamics and antigenicity of HIV-1 gp120. Journal of Virology. 2013;87:10855–10873. doi: 10.1128/JVI.01535-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guttman M, Kahn M, Garcia NK, Hu SL, Lee KK. Solution structure, conformational dynamics, and CD4-induced activation in full-length, glycosylated, monomeric HIV gp120. J Virol. 2012;86:8750–8764. doi: 10.1128/JVI.07224-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kong L, Huang CC, Coales SJ, Molnar KS, Skinner J, Hamuro Y, Kwong PD. Local conformational stability of HIV-1 gp120 in unliganded and CD4-bound states as defined by amide hydrogen/deuterium exchange. Journal of Virology. 2010;84:10311–10321. doi: 10.1128/JVI.00688-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwon YD, Finzi A, Wu X, Dogo-Isonagie C, Lee LK, Moore LR, Schmidt SD, Stuckey J, Yang Y, Zhou T, Zhu J, Vicic DA, Debnath AK, Shapiro L, Bewley CA, Mascola JR, Sodroski JG, Kwong PD. Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops. Proc Natl Acad Sci U S A. 2012;109:5663–5668. doi: 10.1073/pnas.1112391109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang YK, Andjelic S, Binley JM, Crooks ET, Franti M, Iyer SP, Donovan GP, Dey AK, Zhu P, Roux KH, Durso RJ, Parsons TF, Maddon PJ, Moore JP, Olson WC. Structural and immunogenicity studies of a cleaved, stabilized envelope trimer derived from subtype A HIV-1. Vaccine. 2009;27:5120–5132. doi: 10.1016/j.vaccine.2009.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Pena AT, Korzun J, Golabek M, de Los Reyes K, Ketas TJ, van Gils MJ, King CR, Wilson IA, Ward AB, Klasse PJ, Moore JP. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP. 664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013;9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thali M, Moore JP, Furman C, Charles M, Ho DD, Robinson J, Sodroski J. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. Journal of Virology. 1993;67:3978–3988. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moore JP, Thali M, Jameson BA, Vignaux F, Lewis GK, Poon SW, Charles M, Fung MS, Sun B, Durda PJ, et al. Immunochemical analysis of the gp120 surface glycoprotein of human immunodeficiency virus type 1: probing the structure of the C4 and V4 domains and the interaction of the C4 domain with the V3 loop. Journal of Virology. 1993;67:4785–4796. doi: 10.1128/jvi.67.8.4785-4796.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O’;Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Posner MR, Cavacini LA, Emes CL, Power J, Byrn R. Neutralization of HIV-1 by F105, a human monoclonal antibody to the CD4 binding site of gp120. J Acquired Immune Defic Syndr. 1993;6:7–14. [PubMed] [Google Scholar]
- 40.Burton DR, Barbas CF, Persson MA, 3rd, Koenig S, Chanock RM, Lerner RA. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. Proc Natl Acad Sci U S A. 1991;88:10134–10137. doi: 10.1073/pnas.88.22.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garlick RL, Kirschner RJ, Eckenrode FM, Tarpley WG, Tomich CS. Escherichia coli expression, purification, and biological activity of a truncated soluble CD4. AIDS Res Hum Retroviruses. 1990;6:465–479. doi: 10.1089/aid.1990.6.465. [DOI] [PubMed] [Google Scholar]
- 42.Guan Y, Pazgier M, Sajadi MM, Kamin-Lewis R, Al-Darmarki S, Flinko R, Lovo E, Wu X, Robinson JE, Seaman MS, Fouts TR, Gallo RC, DeVico AL, Lewis GK. Diverse specificity and effector function among human antibodies to HIV-1 envelope glycoprotein epitopes exposed by CD4 binding. Proc Natl Acad Sci U S A. 2013;110:E69–78. doi: 10.1073/pnas.1217609110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abacioglu YH, Fouts TR, Laman JD, Claassen E, Pincus SH, Moore JP, Roby CA, Kamin-Lewis R, Lewis GK. Epitope mapping and topology of baculovirus-expressed HIV-1 gp160 determined with a panel of murine monoclonal antibodies. AIDS Res Hum Retroviruses. 1994;10:371–381. doi: 10.1089/aid.1994.10.371. [DOI] [PubMed] [Google Scholar]
- 44.Backliwal G, Hildinger M, Hasija V, Wurm FM. High-density transfection with HEK-293 cells allows doubling of transient titers and removes need for a priori DNA complex formation with PEI. Biotechnol Bioeng. 2008;99:721–727. doi: 10.1002/bit.21596. [DOI] [PubMed] [Google Scholar]
- 45.Weis DD, Engen JR, Kass IJ. Semi-automated data processing of hydrogen exchange mass spectra using HX-Express. J Am Soc Mass Spectrom. 2006;17:1700–1703. doi: 10.1016/j.jasms.2006.07.025. [DOI] [PubMed] [Google Scholar]
- 46.Lisal J, Lam TT, Kainov DE, Emmett MR, Marshall AG, Tuma R. Functional visualization of viral molecular motor by hydrogen-deuterium exchange reveals transient states. Nat Struct Mol Biol. 2005;12:460–466. doi: 10.1038/nsmb927. [DOI] [PubMed] [Google Scholar]
- 47.Guttman M, Weis DD, Engen JR, Lee KK. Analysis of overlapped and noisy hydrogen/deuterium exchange mass spectra. J Am Soc Mass Spectrom. 2013;24:1906–1912. doi: 10.1007/s13361-013-0727-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smolsky IL, Liu P, Niebuhr M, Ito K, Weiss TM, Tsuruta H. Biological small-angle x-ray scattering facility at the Stanford synchrotron radiation laboratory. J Appl Crystallogr. 2007;40:S453–S458. [Google Scholar]
- 49.Svergun DI. Mathematical-Methods in Small-Angle Scattering Data-Analysis. J Appl Crystallogr. 1991;24:485–492. [Google Scholar]
- 50.Svergun DI. Determination of the Regularization Parameter in Indirect-Transform Methods Using Perceptual Criteria. J Appl Crystallogr. 1992;25:495–503. [Google Scholar]
- 51.Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 52.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J Appl Crystallogr. 2003;36:1277–1282. [Google Scholar]
- 53.Petoukhov MV, Svergun DI. Analysis of X-ray and neutron scattering from biomacromolecular solutions. Curr Opin Struct Biol. 2007;17:562–571. doi: 10.1016/j.sbi.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 54.Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing (76pg 2879, 1999) Biophys J. 1999;77:2896. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kozin MB, Svergun DI. Automated matching of high- and low-resolution structural models. J Appl Crystallogr. 2001;34:33–41. [Google Scholar]
- 56.Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J Appl Crystallogr. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Petoukhov MV, Konarev PV, Kikhney AG, Svergun DI. ATSAS 2.1 - towards automated and web-supported small-angle scattering data analysis. J Appl Crystallogr. 2007;40:S223–S228. [Google Scholar]
- 58.Wriggers W, Birmanns S. Using Situs for flexible and rigid-body fitting of multiresolution single-molecule data. J Struct Biol. 2001;133:193–202. doi: 10.1006/jsbi.2000.4350. [DOI] [PubMed] [Google Scholar]
- 59.Fischer H, de Oliveira Neto M, Napolitano HB, Polikarpov I, Craievich AF. Determination of the molecular weight of proteins in solution from a single small-angle X-ray scattering measurement on a relative scale. J Appl Crystallogr. 2010;43:101–109. [Google Scholar]
- 60.Moore JP, McCutchan FE, Poon SW, Mascola J, Liu J, Cao Y, Ho DD. Exploration of antigenic variation in gp120 from clades A through F of human immunodeficiency virus type 1 by using monoclonal antibodies. J Virol. 1994;68:8350–8364. doi: 10.1128/jvi.68.12.8350-8364.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moore JP, Ho DD. Antibodies to discontinuous or conformationally sensitive epitopes on the gp120 glycoprotein of human immunodeficiency virus type 1 are highly prevalent in sera of infected humans. J Virol. 1993;67:863–875. doi: 10.1128/jvi.67.2.863-875.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Acharya P, Tolbert WD, Gohain N, Wu X, Yu L, Liu T, Huang W, Huang CC, Kwon YD, Louder RK, Luongo TS, McLellan JS, Pancera M, Yang Y, Zhang B, Flinko R, Foulke JS, Jr, Sajadi MM, Kamin-Lewis R, Robinson JE, Martin L, Kwong PD, Guan Y, DeVico AL, Lewis GK, Pazgier M. Structural definition of an antibody-dependent cellular cytotoxicity response implicated in reduced risk for HIV-1 infection. J Virol. 2014;88:12895–12906. doi: 10.1128/JVI.02194-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kanduc D, Serpico R, Lucchese A, Shoenfeld Y. Correlating low-similarity peptide sequences and HIV B-cell epitopes. Autoimmun Rev. 2008;7:291–296. doi: 10.1016/j.autrev.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 64.Wyatt R, Kwong PD, Desjardins E, Sweet RW, Robinson J, Hendrickson WA, Sodroski JG. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 65.Allaway GP, Davis-Bruno KL, Beaudry GA, Garcia EB, Wong EL, Ryder AM, Hasel KW, Gauduin MC, Koup RA, McDougal JS, et al. Expression and characterization of CD4-IgG2, a novel heterotetramer that neutralizes primary HIV type 1 isolates. AIDS Res Hum Retroviruses. 1995;11:533–539. doi: 10.1089/aid.1995.11.533. [DOI] [PubMed] [Google Scholar]
- 66.Moore JP, Sodroski J. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J Virol. 1996;70:1863–1872. doi: 10.1128/jvi.70.3.1863-1872.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, Yang X, Zhang MY, Zwick MB, Arthos J, Burton DR, Dimitrov DS, Sodroski J, Wyatt R, Nabel GJ, Kwong PD. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen L, Kwon YD, Zhou T, Wu X, O’Dell S, Cavacini L, Hessell AJ, Pancera M, Tang M, Xu L, Yang ZY, Zhang MY, Arthos J, Burton DR, Dimitrov DS, Nabel GJ, Posner MR, Sodroski J, Wyatt R, Mascola JR, Kwong PD. Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science. 2009;326:1123–1127. doi: 10.1126/science.1175868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou T, Lynch RM, Chen L, Acharya P, Wu X, Doria-Rose NA, Joyce MG, Lingwood D, Soto C, Bailer RT, Ernandes MJ, Kong R, Longo NS, Louder MK, McKee K, O’;Dell S, Schmidt SD, Tran L, Yang Z, Druz A, Luongo TS, Moquin S, Srivatsan S, Yang Y, Zhang B, Zheng A, Pancera M, Kirys T, Georgiev IS, Gindin T, Peng HP, Yang AS, Mullikin JC, Gray MD, Stamatatos L, Burton DR, Koff WC, Cohen MS, Haynes BF, Casazza JP, Connors M, Corti D, Lanzavecchia A, Sattentau QJ, Weiss RA, West AP, Jr, Bjorkman PJ, Scheid JF, Nussenzweig MC, Shapiro L, Mascola JR, Kwong PD. Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors. Cell. 2015;161:1280–1292. doi: 10.1016/j.cell.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, McKee K, O’Dell S, Perfetto S, Schmidt SD, Shi W, Wu L, Yang Y, Yang ZY, Yang Z, Zhang Z, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Simek M, Burton DR, Koff WC, Doria-Rose NA, Connors M, Mullikin JC, Nabel GJ, Roederer M, Shapiro L, Kwong PD, Mascola JR. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Falkowska E, Ramos A, Feng Y, Zhou T, Moquin S, Walker LM, Wu X, Seaman MS, Wrin T, Kwong PD, Wyatt RT, Mascola JR, Poignard P, Burton DR. PGV04, an HIV-1 gp120 CD4 binding site antibody, is broad and potent in neutralization but does not induce conformational changes characteristic of CD4. J Virol. 2012;86:4394–4403. doi: 10.1128/JVI.06973-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wyatt R, Desjardin E, Olshevsky U, Nixon C, Binley J, Olshevsky V, Sodroski J. Analysis of the interaction of the human immunodeficiency virus type 1 gp120 envelope glycoprotein with the gp41 transmembrane glycoprotein. J Virol. 1997;71:9722–9731. doi: 10.1128/jvi.71.12.9722-9731.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Roben P, Moore JP, Thali M, Sodroski J, Barbas CF, 3rd, Burton DR. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J Virol. 1994;68:4821–4828. doi: 10.1128/jvi.68.8.4821-4828.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wilson IA, Niman HL, Houghten RA, Cherenson AR, Connolly ML, Lerner RA. The structure of an antigenic determinant in a protein. Cell. 1984;37:767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- 75.Fieser TM, Tainer JA, Geysen HM, Houghten RA, Lerner RA. Influence of protein flexibility and peptide conformation on reactivity of monoclonal anti-peptide antibodies with a protein alpha-helix. Proc Natl Acad Sci U S A. 1987;84:8568–8572. doi: 10.1073/pnas.84.23.8568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lerner RA. Tapping the immunological repertoire to produce antibodies of predetermined specificity. Nature. 1982;299:592–596. doi: 10.1038/299592a0. [DOI] [PubMed] [Google Scholar]
- 77.Tainer JA, Getzoff ED, Paterson Y, Olson AJ, Lerner RA. The atomic mobility component of protein antigenicity. Annu Rev Immunol. 1985;3:501–535. doi: 10.1146/annurev.iy.03.040185.002441. [DOI] [PubMed] [Google Scholar]
- 78.Bahraoui EM, Granier C, Van Rietschoten J, Rochat H, el Ayeb M. Specificity and neutralizing capacity of antibodies elicited by a synthetic peptide of scorpion toxin. J Immunol. 1986;136:3371–3377. [PubMed] [Google Scholar]
- 79.Zolla-Pazner S. Identifying epitopes of HIV-1 that induce protective antibodies. Nat Rev Immunol. 2004;4:199–210. doi: 10.1038/nri1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kwong PD, Mascola JR, Nabel GJ. Broadly neutralizing antibodies and the search for an HIV-1 vaccine: the end of the beginning. Nat Rev Immunol. 2013;13:693–701. doi: 10.1038/nri3516. [DOI] [PubMed] [Google Scholar]
- 81.Pantophlet R, Ollmann Saphire E, Poignard P, Parren PW, Wilson IA, Burton DR. Fine mapping of the interaction of neutralizing and nonneutralizing monoclonal antibodies with the CD4 binding site of human immunodeficiency virus type 1 gp120. Journal of Virology. 2003;77:642–658. doi: 10.1128/JVI.77.1.642-658.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dey B, Svehla K, Xu L, Wycuff D, Zhou T, Voss G, Phogat A, Chakrabarti BK, Li Y, Shaw G, Kwong PD, Nabel GJ, Mascola JR, Wyatt RT. Structure-based stabilization of HIV-1 gp120 enhances humoral immune responses to the induced co-receptor binding site. PLoS Pathog. 2009;5:e1000445. doi: 10.1371/journal.ppat.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sanders RW, Vesanen M, Schuelke N, Master A, Schiffner L, Kalyanaraman R, Paluch M, Berkhout B, Maddon PJ, Olson WC, Lu M, Moore JP. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. Journal of Virology. 2002;76:8875–8889. doi: 10.1128/JVI.76.17.8875-8889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Binley JM, Sanders RW, Clas B, Schuelke N, Master A, Guo Y, Kajumo F, Anselma DJ, Maddon PJ, Olson WC, Moore JP. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. Journal of Virology. 2000;74:627–643. doi: 10.1128/jvi.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sliepen K, Medina-Ramirez M, Yasmeen A, Moore JP, Klasse PJ, Sanders RW. Binding of inferred germline precursors of broadly neutralizing HIV-1 antibodies to native-like envelope trimers. Virology. 2015;486:116–120. doi: 10.1016/j.virol.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ringe RP, Yasmeen A, Ozorowski G, Go EP, Pritchard LK, Guttman M, Ketas TA, Cottrell CA, Wilson IA, Sanders RW, Cupo A, Crispin M, Lee KK, Desaire H, Ward AB, Klasse PJ, Moore JP. Influences on the Design and Purification of Soluble, Recombinant Native-Like HIV-1 Envelope Glycoprotein Trimers. J Virol. 2015;89:12189–12210. doi: 10.1128/JVI.01768-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sanders RW, van Gils MJ, Derking R, Sok D, Ketas TJ, Burger JA, Ozorowski G, Cupo A, Simonich C, Goo L, Arendt H, Kim HJ, Lee JH, Pugach P, Williams M, Debnath G, Moldt B, van Breemen MJ, Isik G, Medina-Ramirez M, Back JW, Koff WC, Julien JP, Rakasz EG, Seaman MS, Guttman M, Lee KK, Klasse PJ, LaBranche C, Schief WR, Wilson IA, Overbaugh J, Burton DR, Ward AB, Montefiori DC, Dean H, Moore JP. HIV-1 VACCINES. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science. 2015;349:aac4223. doi: 10.1126/science.aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Do Kwon Y, Pancera M, Acharya P, Georgiev IS, Crooks ET, Gorman J, Joyce MG, Guttman M, Ma X, Narpala S, Soto C, Terry DS, Yang Y, Zhou T, Ahlsen G, Bailer RT, Chambers M, Chuang GY, Doria-Rose NA, Druz A, Hallen MA, Harned A, Kirys T, Louder MK, O’Dell S, Ofek G, Osawa K, Prabhakaran M, Sastry M, Stewart-Jones GB, Stuckey J, Thomas PV, Tittley T, Williams C, Zhang B, Zhao H, Zhou Z, Donald BR, Lee LK, Zolla-Pazner S, Baxa U, Schon A, Freire E, Shapiro L, Lee KK, Arthos J, Munro JB, Blanchard SC, Mothes W, Binley JM, McDermott AB, Mascola JR, Kwong PD. Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat Struct Mol Biol. 2015;22:522–531. doi: 10.1038/nsmb.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.de Taeye SW, Torrents de la Peña A, Julien JP, van den Kerkhof TLGM, Burger JA, Pritchard LK, Pugach P, Yasmeen A, Crampton J, Hu J, Bontjer I, Torres JL, Arendt H, DeStefano J, Koff WC, Schuitemaker H, Eggink D, Berkhout B, Dean H, LaBranche C, Crotty S, Crispin M, Montefiori DC, Klasse PJ, Lee KK, Moore JP, Wilson IA, Ward AB, Sanders RW, Ozorowski G, Guttman M. Immunogenicity of stabilized HIV61 envelope trimers with reduced exposure of non-neutralizing epitopes. Cell. 2015;163:1702–1715. doi: 10.1016/j.cell.2015.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Correia BE, Bates JT, Loomis RJ, Baneyx G, Carrico C, Jardine JG, Rupert P, Correnti C, Kalyuzhniy O, Vittal V, Connell MJ, Stevens E, Schroeter A, Chen M, Macpherson S, Serra AM, Adachi Y, Holmes MA, Li Y, Klevit RE, Graham BS, Wyatt RT, Baker D, Strong RK, Crowe JE, Jr, Johnson PR, Schief WR. Proof of principle for epitope-focused vaccine design. Nature. 2014;507:201–206. doi: 10.1038/nature12966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Impagliazzo A, Milder F, Kuipers H, Wagner M, Zhu X, Hoffman RM, van Meersbergen R, Huizingh J, Wanningen P, Verspuij J, de Man M, Ding Z, Apetri A, Kukrer B, Sneekes-Vriese E, Tomkiewicz D, Laursen NS, Lee PS, Zakrzewska A, Dekking L, Tolboom J, Tettero L, van Meerten S, Yu W, Koudstaal W, Goudsmit J, Ward AB, Meijberg W, Wilson IA, Radosevic K. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science. 2015;349:1301–1306. doi: 10.1126/science.aac7263. [DOI] [PubMed] [Google Scholar]
- 92.Yassine HM, Boyington JC, McTamney PM, Wei CJ, Kanekiyo M, Kong WP, Gallagher JR, Wang L, Zhang Y, Joyce MG, Lingwood D, Moin SM, Andersen H, Okuno Y, Rao SS, Harris AK, Kwong PD, Mascola JR, Nabel GJ, Graham BS. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat Med. 2015;21:1065–1070. doi: 10.1038/nm.3927. [DOI] [PubMed] [Google Scholar]
- 93.Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang GY, Ofek G, Stewart-Jones GB, Stuckey J, Bailer RT, Joyce MG, Louder MK, Tumba N, Yang Y, Zhang B, Cohen MS, Haynes BF, Mascola JR, Morris L, Munro JB, Blanchard SC, Mothes W, Connors M, Kwong PD. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature. 2014;514:455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xiang SH, Doka N, Choudhary RK, Sodroski J, Robinson JE. Characterization of CD4-induced epitopes on the HIV type 1 gp120 envelope glycoprotein recognized by neutralizing human monoclonal antibodies. AIDS Res Hum Retroviruses. 2002;18:1207–1217. doi: 10.1089/08892220260387959. [DOI] [PubMed] [Google Scholar]
- 95.Moore JP, Sattentau QJ, Wyatt R, Sodroski J. Probing the structure of the human immunodeficiency virus surface glycoprotein gp120 with a panel of monoclonal antibodies. J Virol. 1994;68:469–484. doi: 10.1128/jvi.68.1.469-484.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pancera M, Majeed S, Ban YE, Chen L, Huang CC, Kong L, Kwon YD, Stuckey J, Zhou T, Robinson JE, Schief WR, Sodroski J, Wyatt R, Kwong PD. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc Natl Acad Sci U S A. 2010;107:1166–1171. doi: 10.1073/pnas.0911004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


