Macrophages play critical, but opposite roles, in acute and chronic inflammation and cancer1–5. In response to pathogens or injury, inflammatory macrophages express cytokines that stimulate cytotoxic T cells, while highly abundant macrophages in neoplastic and parasitic diseases express anti-inflammatory cytokines that induce immune suppression and may promote resistance to T cell checkpoint inhibitors1–7. Here we show that macrophage PI(3)Kinase γ controls a critical switch between immune stimulation and suppression during inflammation and cancer. PI3Kγ signaling through Akt and mTor inhibits NFκB activation while stimulating C/EBPβ activation, thereby inducing a transcriptional program that promotes immune suppression during inflammation and tumor growth. In contrast, selective inactivation of macrophage PI3Kγ stimulates and prolongs NFκB activation and inhibits C/EBPβ activation, thus promoting an immunostimulatory transcriptional program that restores CD8+ T cell activation and cytotoxicity and synergizes with checkpoint inhibitor therapy to promote tumor regression and extend survival in mouse models of cancer. As PI3Kγ-directed, anti-inflammatory gene expression predicted survival probability in cancer patients, our findings demonstrate that therapeutic targeting of intracellular signaling pathways that regulate the switch between macrophage polarization states can control immune suppression in cancer and other disorders.
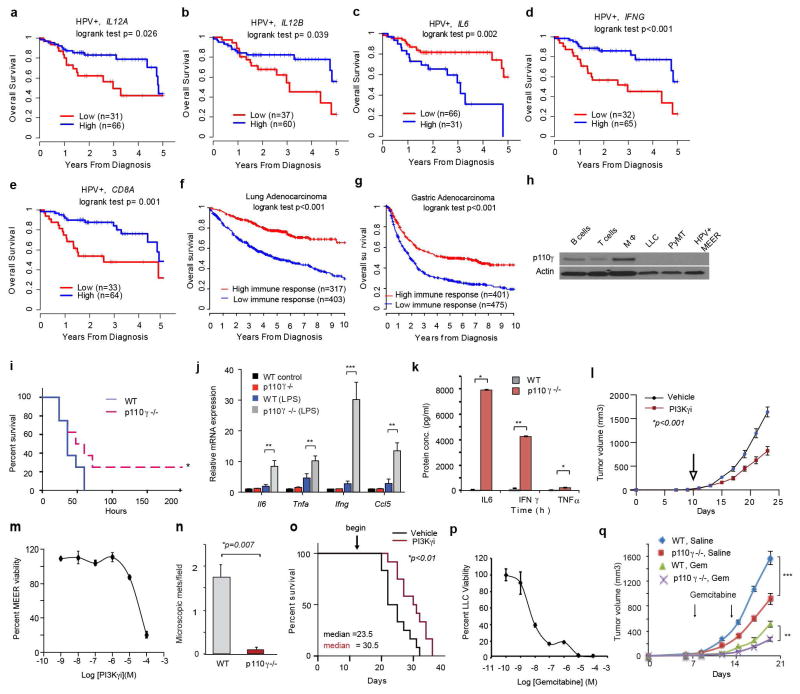
We investigated the association between immune responses and survival in primary tumors from HPV+ (n=97) and HPV− (n=423) head and neck squamous cell carcinoma (HNSCC) cohorts from TCGA.8 High expression levels of pro-inflammatory mRNAs IL12A, IL12B, IFNG, and CD8A were associated with increased survival in HPV+ but not HPV− cohorts, while high expression of IL6 was negatively associated with survival (Extended Data Fig. 1a–e). HPV+ patients with this favorable immune expression profile (n=35) had 97% survival at 3 years compared with 57% survival for patients without this profile (n=62) (Fig. 1a). Similar associations were observed in lung adenocarcinoma and gastric carcinoma patients (Extended Data Fig. 1f–g). These results suggested that therapeutic approaches that stimulate pro-inflammatory gene expression might enhance cancer patient survival.
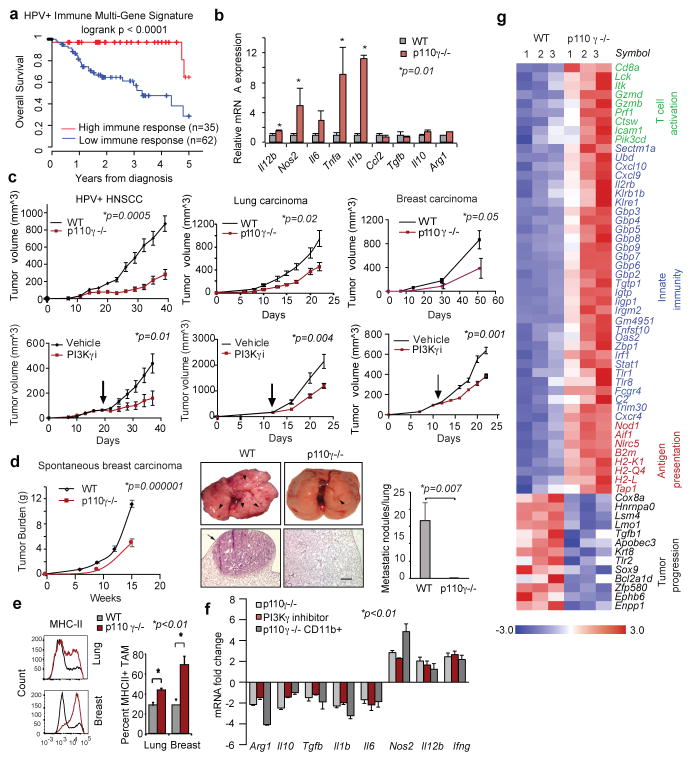
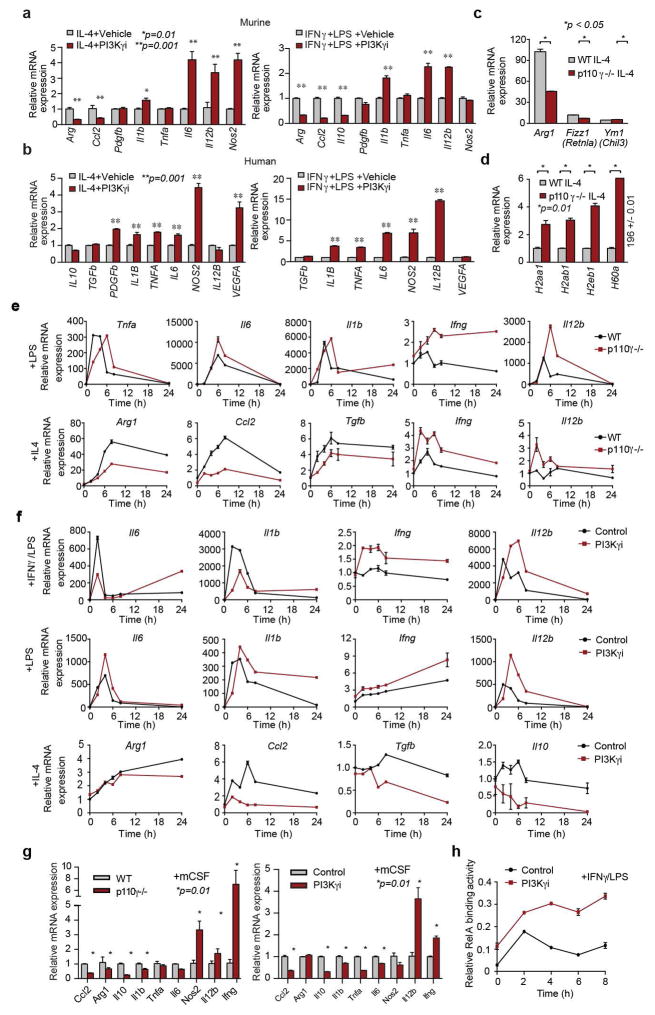
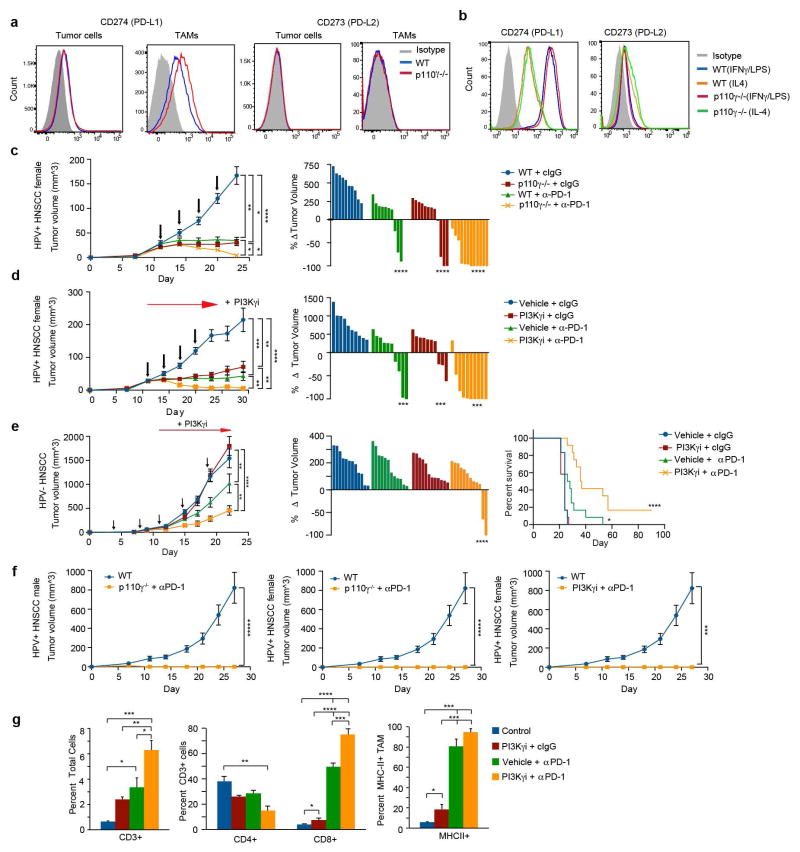
Figure 1. PI3Kγ promotes immune suppression.
(a) Multivariate immune response mRNA signature in HPV+ HNSCC patients (n=97). (b) Immune response mRNA expression in p110γ−/− or WT peritoneal macrophages (n=3). (c) Mean +/− sem tumor growth in WT, p110γ−/− and PI3Kγ-inhibitor-treated mice (n=15). Arrow, daily treatment initiation. (d–f) Mean +/− sem (d) spontaneous breast carcinoma growth and metastasis (bar, 200μm) in WT (n=21) and p110γ−/− (n=8) animals; (e) MHCII expression on WT vs p110γ−/− TAMs (n=3) and (f) fold change mRNA expression in tumors and tumor-derived CD11b+ cells from p110γ−/− and PI3Kγ inhibitor-treated mice (n=5). (g) Heatmap of immune response mRNA expression in tumors from WT and p110γ−/− mice (n=3). n=biological replicates.
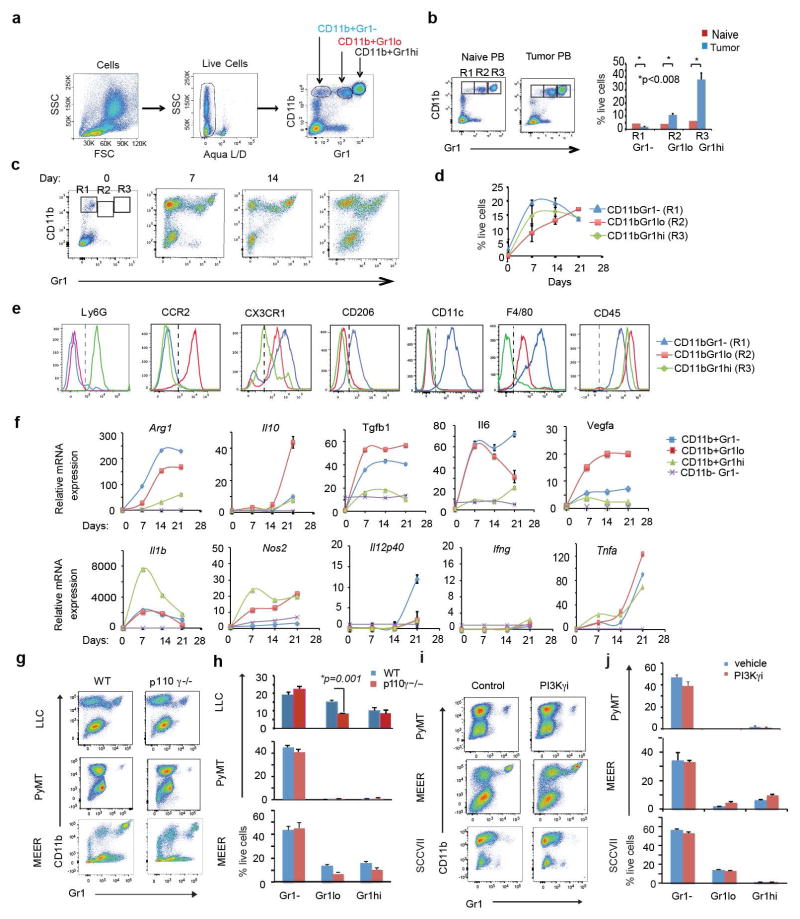
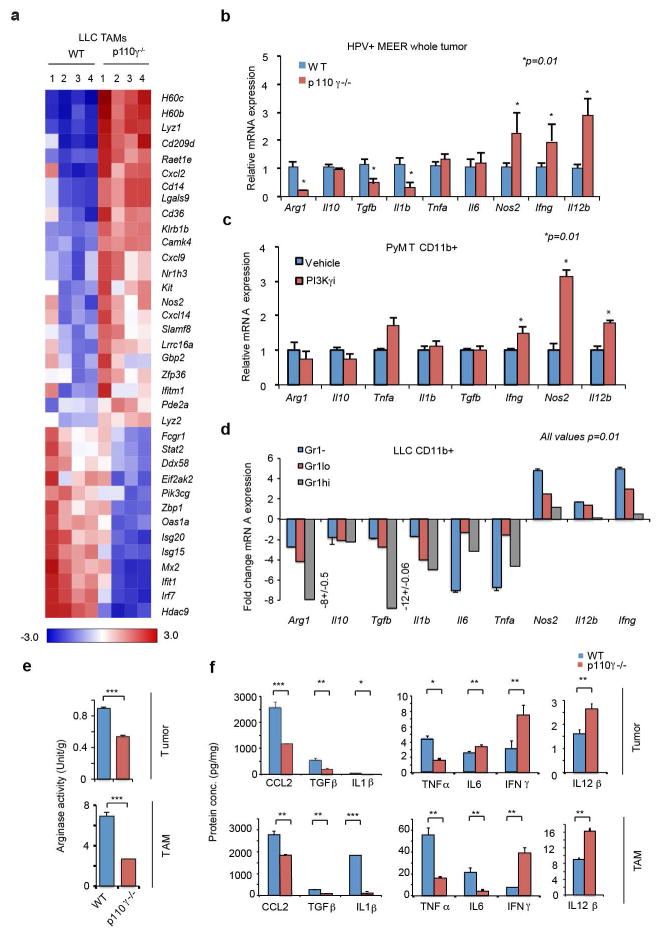
We suspected that macrophage signaling pathways, such as those regulated by Class IB isoform PI3Kγ, might control the switch between immune stimulation and suppression in inflammation and cancer. PI3Kγ is abundantly expressed in myeloid but not cancer cells (Extended Data Fig. 1h)9–12 and promotes myeloid cell trafficking during inflammation and cancer11–15. Mice lacking PI3Kγ (p110γ−/−) mounted exaggerated, macrophage-mediated pro-inflammatory responses upon exposure to pathogenic stimuli (Fig. 1b; Extended Data Fig. 1i–k), suggesting that PI3Kγ inhibits macrophage inflammatory responses and might do so in the tumor microenvironment. Mice lacking PI3Kγ and mice that were treated with PI3Kγ antagonists (TG100–11512 or IPI-54916) exhibited significantly suppressed growth of implanted HPV+ (MEER) and HPV− (SCCVII) HNSCC, lung carcinoma (LLC), and breast carcinoma (PyMT) tumors (Fig. 1c). PI3Kγ inhibition did not directly affect the growth or survival of tumor cells, which do not express the kinase (Extended Data Fig. 1h, 1l–m)12–15. PI3Kγ inhibition suppressed long-term growth and metastases of spontaneous breast tumors, extended survival of mice with orthotopic breast tumors and enhanced the sensitivity of tumors to the nucleoside analogue gemcitabine (Fig. 1d; Extended Data Fig. 1n–q). While PI3Kγ inhibition did not affect the accumulation of CD11b+Gr1-F4/80+ tumor-associated macrophages (TAMs) in tumors (Extended Data Fig. 2a–f), it enhanced expression of MHCII and pro-inflammatory cytokines and inhibited expression of immune suppressive factors in tumors and TAMs, indicating that PI3Kγ controls the TAM switch between immune suppression and immune stimulation (Fig.1e–g Extended Data Fig. 2g–j, 3a–f).
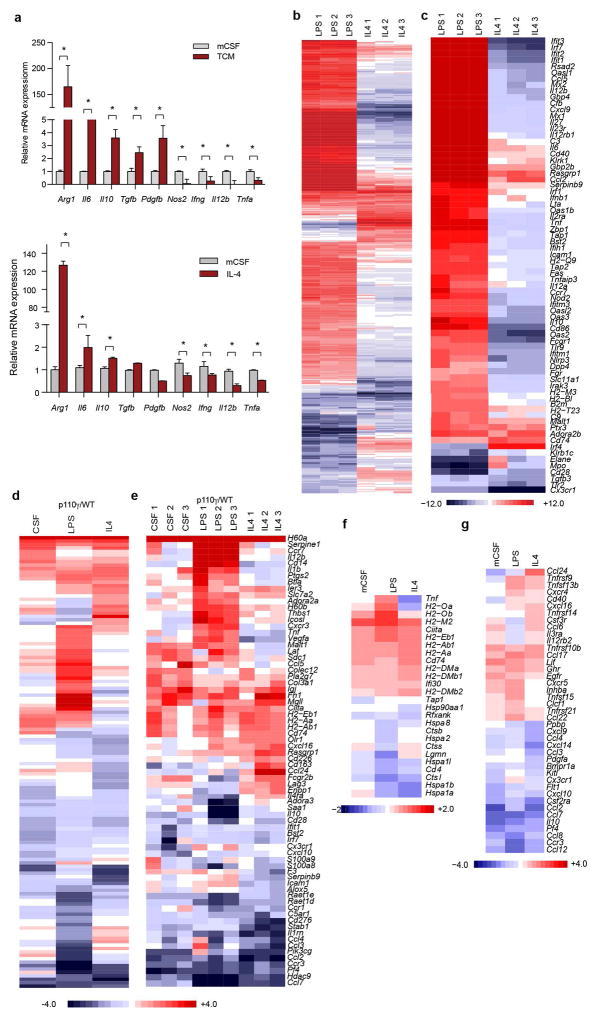
To determine whether PI3Kγ directly regulates macrophage polarization, we analyzed mRNA and protein expression in primary murine macrophages stimulated in vitro with basal (CSF-1), pro-inflammatory (IFNγ/LPS + CSF-1) or anti-inflammatory (IL-4 + CSF-1) conditions. Pro-inflammatory stimuli upregulate macrophage expression of innate immune proteins, cytokines and cell surface receptors, while anti-inflammatory stimuli induces expression of immunosuppressive factors similar to those expressed in TAMs (Extended Data Fig. 4a–c)1. Genes and proteins associated with immune activation, antigen-presentation and T cell activation were upregulated In p110γ−/− and PI3Kγ inhibitor-treated macrophages (Fig. 2a–c, Extended Data Fig. 4c–f, 5a–g). In contrast, genes associated with immune suppression and chemoattraction were inhibited (Fig. 2a–c, Extended Data Fig. 4c–f, 5a–g). These results confirm that PI3Kγ controls a macrophage switch between immune stimulation and suppression.
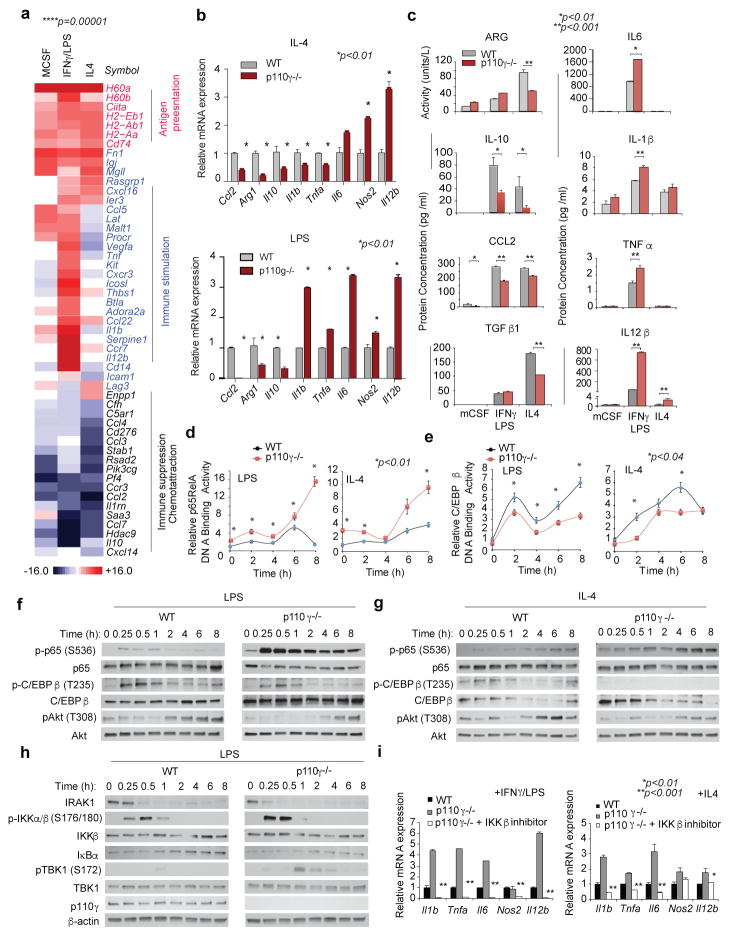
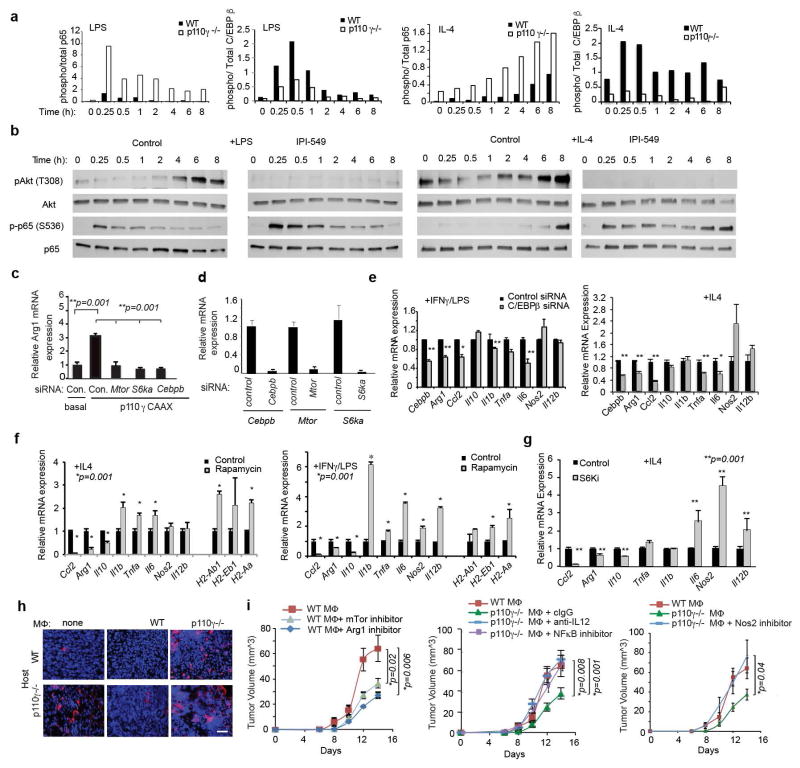
Figure 2. PI3Kγ regulates NFκB and C/EBPβ during macrophage polarization.
(a) Heatmap of mRNA expression in p110γ−/− vs WT macrophages (n=3). (b–c) Mean +/− sem (b) mRNA (n=3) and (c) protein (n=4) expression. (d–e) Mean +/− sem (d) p65 RelA and (e) C/EBPβ DNA-binding activity in WT and p110γ−/− macrophages (n=4). (f–g) Immunoblotting of pRelA/RelA, pC/EBPβ/C/EBPβ, and pAkt/Akt in (f) LPS and (g) IL-4 stimulated WT and p110γ−/− macrophages. (h) Immunoblotting of IRAK1, pIKKβ/IKKβ, IκBα, pTBK1/TBK1, p110γ and actin in WT and p110γ−/− macrophages. (i) Mean +/− sem mRNA expression in IKKβ inhibitor-treated macrophages (n=3). n=biological replicates.
To identify how PI3Kγ regulates macrophage immune responses, we evaluated DNA binding activities of NFκB p65 RelA and the C/CAAT enhancer binding protein C/EBPβ in WT and p110γ null macrophages, as NFκB promotes expression of inflammatory cytokines17, while C/EBPβ promotes expression of the immunosuppressive factor Arg1.18–19 PI3Kγ ablation rapidly and sustainably stimulated RelA DNA binding activity in macrophages (Fig. 2d, Extended Data Fig. 5h). In contrast, PI3Kγ ablation suppressed DNA binding activity of C/EBPβ (Fig. 2e). Consistent with these findings, PI3Kγ inhibition stimulated and sustained p65 RelA phosphorylation and simultaneously inhibited C/EBPβ and Akt phosphorylation (Fig. 2f–g, Extended Data Fig. 6a–b).
We next examined the effect of PI3Kγ inhibition on the stability and phosphorylation of proteins that activate NFκB, including the TLR4-associated adaptor protein IRAK-1 and inhibitory kappa B kinase β, IKKβ, which promotes degradation of IκBα, with subsequent release of NFκB from an inhibitory IκB-NFκB complex17. PI3Kγ deletion enhanced phosphorylation of IKKβ and TBK1 and degradation of IRAK-1 and IκBα in LPS-stimulated p110γ−/− macrophages (Fig. 2h). As an IKKβ inhibitor suppressed the inflammatory phenotype observed in p110γ−/− macrophages (Fig. 2i), these results indicate PI3Kγ is both a feedback inhibitor of the TLR4-NFκB activation pathway and a promoter of IL-4 and C/EBPβ signaling.
C/EBPβ has been previously linked with tumor immune suppression through its control of Arg-1 expression18–19. Expression of constitutively activated PI3Kγ (p110γCAAX)11 was sufficient to induce Arg-1 expression, in a manner that was inhibited by Cebpb, Mtor, or S6ka knockdown (Extended Data Fig. 6c–d). Cebpb knockdown as well as inhibitors of S6K and mTOR suppressed expression of immune suppressive factors and stimulated expression of pro-inflammatory cytokines (Extended Data Fig. 6e–g). These results show that PI3Kγ promotes immune suppression by activating mTor-S6Kα-C/EBPβ and inhibiting NFκB, thereby controlling a switch that regulates the balance between immune suppression and stimulation.
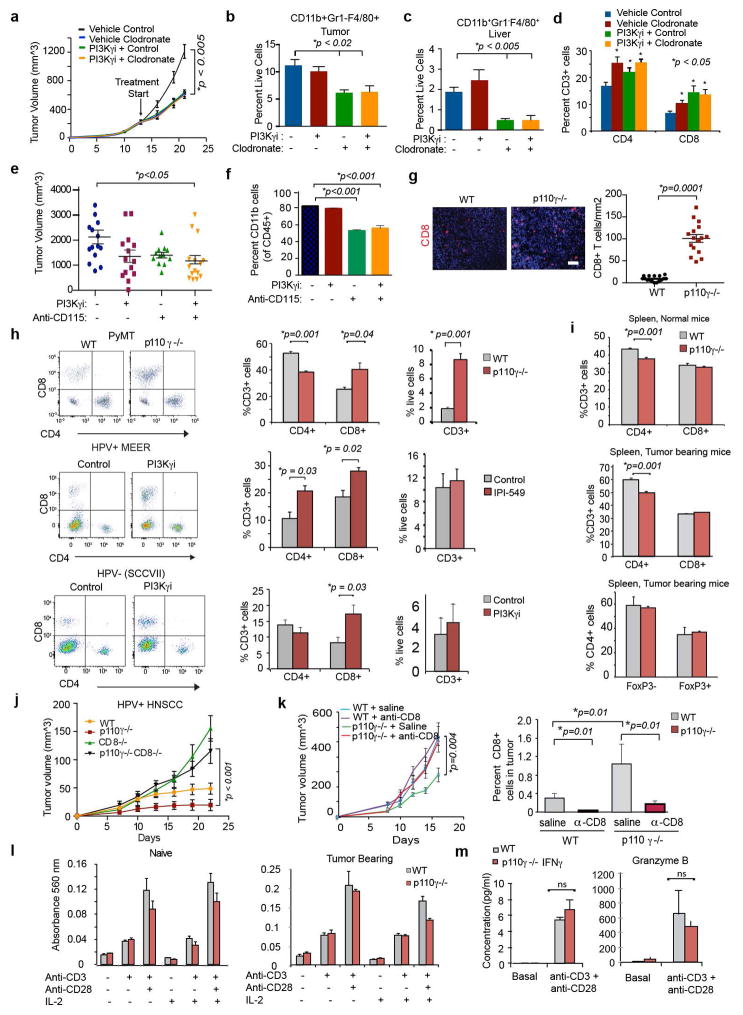
Since PI3Kγ blockade stimulates pro-inflammatory responses in macrophages, we asked whether macrophage PI3Kγ blockade promotes adaptive immunity. TAMs were isolated from WT and p110γ−/− tumors, mixed with tumor cells and adoptively transferred into new WT or p110γ−/− recipient mice (Fig. 3a). Tumor growth was significantly inhibited in tumors containing p110γ−/− TAMs but not WT TAMs (Fig. 3b). CD8+ T cells were significantly increased in tumors with p110γ−/− but not WT macrophages (Fig. 3c, Extended Data Fig. 6h), indicating that PI3Kγ signaling in TAMs inhibits CD8+ T cell recruitment to tumors. To determine whether macrophage-derived cytokines control tumor growth, we implanted tumor cells mixed with in vitro cultured macrophages or conditioned medium (CM) into WT mice. Tumor growth was enhanced by IL-4 stimulated WT macrophages and CM but inhibited by IL-4 stimulated p110γ−/− macrophages and CM from p110γ−/− or PI3Kγ inhibitor-treated macrophages and by all LPS-stimulated macrophages and CM (Fig. 3d–e). To determine which macrophage-derived immune factors affect tumor growth in vivo, we treated WT and p110γ−/− TAMs ex vivo with inhibitors of mTor, Arginase, IKKβ, IL-12 or NOS2 prior to mixing with tumor cells and implanting in mice (Extended Data Fig. 6i). Blockade of mTOR or Arginase in WT macrophages suppressed tumor growth, while inhibition of NOS2, IL-12 or IKKβ in p110γ−/− macrophages stimulated tumor growth. These results indicate that PI3Kγ-mTOR mediated immune suppression promotes tumor growth and that PI3Kγ inhibition reverses these effects by shifting macrophages toward NFκβ-dependent pro-inflammatory polarization.
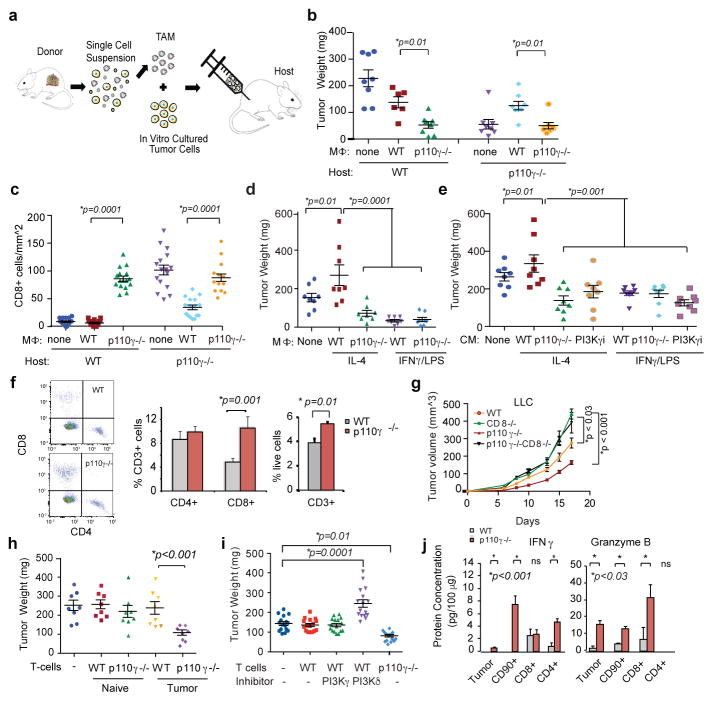
Figure 3. Macrophage PI3Kγ suppresses T cell activation.
(a) Adoptive transfer method. (b) Weights of tumors implanted with p110γ−/−, WT or no TAMs (n=8). (c) CD8+ T cells from b (n=16). (d, e) Weights of tumors implanted with (d) in vitro cultured macrophages (n=8) or (e) macrophage-conditioned medium (CM) (n=8). (f) Percent T cells in WT and p110γ−/− tumors (n=3). (g) Tumor volumes in p110γ and/or CD8−/− mice (n=6). (h–i) Weights of tumors implanted with (h) naïve or tumor-derived T cells (n=8) or (i) inhibitor-treated T cells (n=16). (j) IFNγ and Granzyme B expression in tumors and T cells from WT and p110γ−/− animals (n=3). All graphs show mean +/− sem of biological replicates.
To confirm that macrophage PI3Kγ controls tumor growth, mice bearing pre-established tumors were treated with PI3Kγ inhibitors in combination with clodronate liposomes, which deplete macrophages from tissues.20 PI3Kγ inhibitor and clodronate liposome treatment each partially inhibited tumor growth and stimulated T cell recruitment, but the combination had no additive effects, confirming that PI3Kγ in macrophages, rather than other cell types, promotes tumor growth (Extended Data Fig. 7a–d). Similar results were noted when CSF1R inhibition21 and PI3Kγ inhibition were combined (Extended Data Fig. 7e–f).
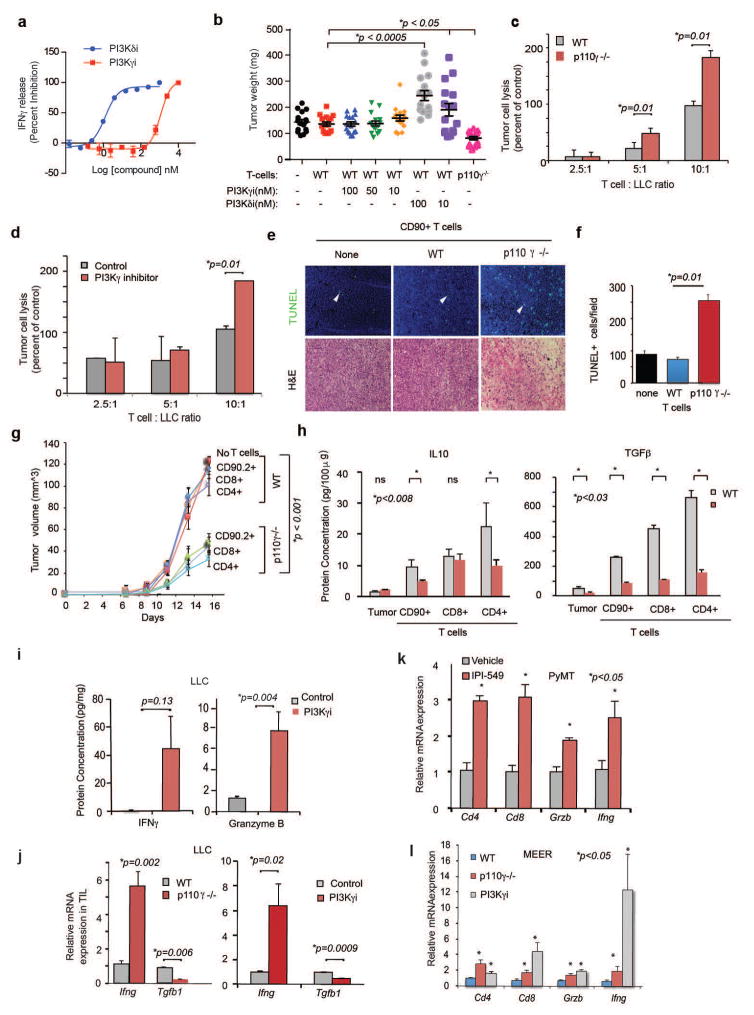
PI3Kγ blockade stimulated T cell recruitment into tumors, as total and CD8+ T cell content increased in tumors from p110γ−/− mice without significantly altering systemic T cell content (Fig. 3f; Extended Data Fig. 7g–i). PI3Kγ inhibition did not suppress tumor growth in CD8 null or antibody-depleted mice, suggesting PI3Kγ inhibition blocks tumor growth by recruiting and/or activating CD8+ T cells (Fig. 3g, Extended Data Fig. 7j–k). When T cells were isolated from tumor-bearing or naive animals, mixed with tumor cells and implanted in mice, only T cells from p110γ−/− tumor-bearing animals suppressed tumor growth (Fig. 3h). However, PI3Kγ inhibition did not directly activate T cells, as neither PI3Kγ deletion nor treatment of T cells with PI3Kγ inhibitors ex vivo affected T cell proliferation or activation; in contrast, PI3Kδ inhibition suppressed T cell activation in vitro and promoted tumor growth in vivo (Fig. 3i; Extended Data Fig. 7l–m, 8a–b). PI3Kγ inhibition promoted T cell mediated cytotoxicity, as T cells isolated from p110γ−/− or PI3Kγ inhibitor-treated tumors stimulated tumor cell cytotoxicity (Extended Data Fig. 8c–g). T cells from p110γ−/− or PI3Kγ inhibitor-treated animals expressed significantly more IFNγ and Granzyme B and significantly less TGFβ1 and IL10 protein and mRNA than T cells from WT animals (Fig. 3j; Extended Data Fig. 8h–l). Together, these results indicate that macrophage PI3Kγ inhibition indirectly promotes both Th1 and cytotoxic adaptive immune responses.
To determine whether PI3Kγ inhibition interacts with other immune therapies, we combined PI3Kγ and the checkpoint inhibitor anti-PD-1 in mouse tumor models. PD-L1, but not PD-L2, was expressed on macrophages in vitro and in vivo (Extended Data Fig. 9a–b). PI3Kγ inhibition synergized with anti-PD-1 to suppress the growth of HPV+ HNSCC tumors in p110γ−/− or inhibitor-treated male or female animals, inducing tumor regression in 86% of male and 90–100% of female animals, as well as continuous survival to date in 60% of male mice and 90–100% of female mice (Fig. 4a–c, Extended Data Fig. 9c–d). Importantly, PI3Kγ inhibition also synergized with anti-PD-1 to reduce tumor growth, extending survival and inducing tumor regression in 30% of mice bearing HPV− HNSCC (SCCVII) tumors (Fig. 4d–f, Extended Data Fig. 9e). The combination of PI3Kγ and anti-PD-1 inhibitors activated T cell memory, as 100% of mice that had previously cleared HPV+ tumors efficiently suppressed re-challenge with HPV+ tumor cells and remained cancer free (Extended Data Fig. 9f). PI3Kγ and PD-1 inhibitors each stimulated immune response gene expression and inhibited immune suppressive gene expression, MHCII expression in TAMs, and CD8+ T cell recruitment to tumors; combination therapy further elevated these parameters (Fig. 4g–i, Extended Data Fig. 9g). These studies show that PI3Kγ inhibition can synergize with T cell targeted therapy to promote anti-tumor immune responses that induce sustained tumor regression in murine models of cancer.
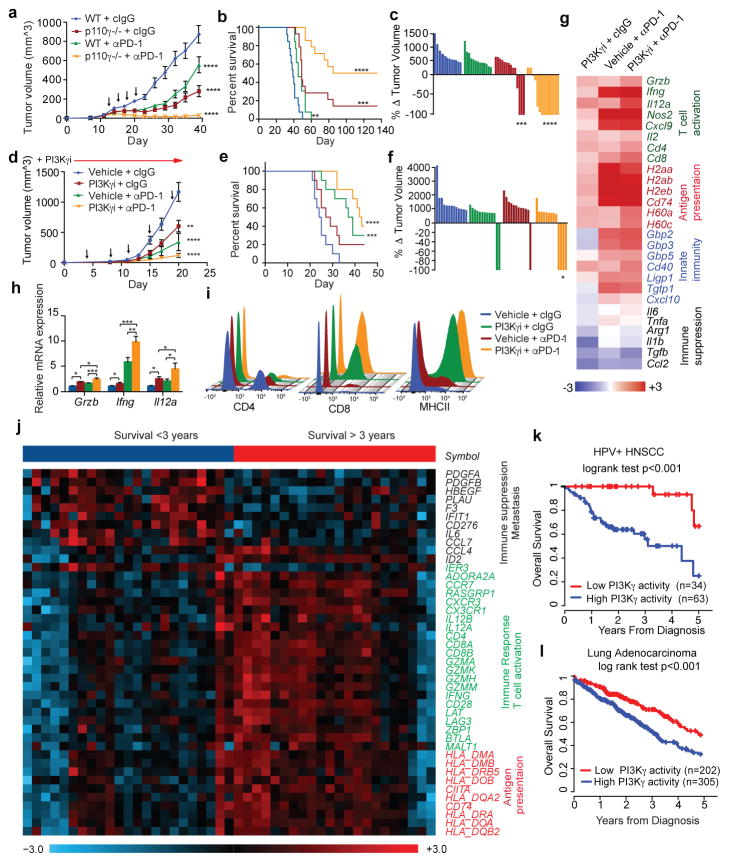
Figure 4. PI3Kγ inhibition synergizes with anti-PD-1.
(a, d) Mean +/− sem tumor volumes in anti-PD-1 (black arrows) treated (a) WT or p110γ−/− mice with HNSCC HPV+ tumors (n=13) and (d) PI3Kγ inhibitor-treated mice with HPV− HNSCC tumors (n=13). (b, e) Percent survival of mice in a, d. (c, f) Change in tumor volumes in a, d. (g–h) Heatmap (g) and graph (h) of mRNA expression from d (n=3). (i) Immune cell profiles from d. (j) Heatmap of PI3Kγ-regulated mRNA expression in HPV+HNSCC patients (n=45). (k–l) Multivariate PI3Kγ-regulated immune signature in (k) HPV+HNSCC patients (n=97) and (l) lung adenocarcinoma patients (n=507). n=biological replicates.
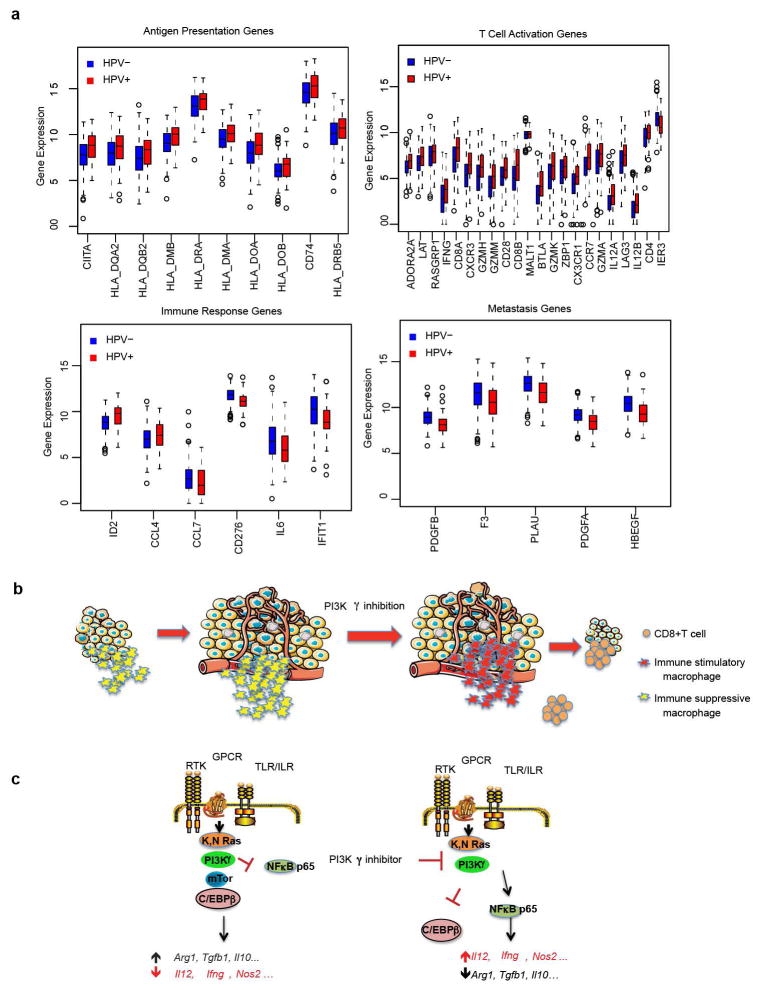
PI3Kγ-regulated immune responses might also affect outcome in cancer patients. We identified 43 PI3Kγ-regulated genes that significantly associated with survival in TCGA HPV+ HNSCC patients (Fig. 5j). HPV+ HNSCC patients with a low PI3Kγ activity profile experienced 100% survival at 3 years, compared with 56% survival for the remaining 63 patients (Fig. 5k). In HPV− HNSCC patients, 39 of these genes were significantly shifted in the direction of high PI3Kγ activity, consistent with a pattern of pervasive immune suppression and reduced survival in HPV− disease (Extended Data Fig. 10a). In lung adenocarcinoma patients, 18 genes predicted survival; patients with a low PI3Kγ activity profile had 73% survival at 3 years, compared with 55% survival for others (Figure 5l). These results suggest that a PI3Kγ-regulated immune suppression signature is associated with survival in cancer patients and that PI3Kγ inhibitors might provide clinical benefit in cancer patients.
Here we have shown that PI3Kγ regulates innate immunity during inflammation and cancer (Extended Data Fig.10b–c). Prior studies have implicated PI3Ks in the regulation of pro-inflammatory immune responses in macrophages, as pan-PI3K inhibitors and null mutations in the PI3Kγ effectors PDK1, Akt1 and TSC enhanced pro-inflammatory NFκB dependent transcription in macrophages,22–24 while inhibition of PTEN and SHIP, which oppose PI3Kinase function, promotes immune suppression.25–26 As macrophage reprogramming can enhance the activity of checkpoint inhibitors in cancer5,13,21,27, our studies indicate that inhibitory targeting of macrophage signaling pathways may provide novel approaches to improve the long-term survival of cancer patients.
METHODS
Immune-related gene expression signature analysis in TCGA data
We analyzed TCGA data for association between mRNA expression level of sixteen candidate immune-related genes (ARG1, IL10, FOXP3, CD68, IL12A, IL12B, IFNG, CD8A, CD4, CD11B, CD14, TNFA, IL1A, IL1B, IL6 and CCL5) and 5-year overall survival. Illumina HiSeq RNAseqV2 mRNA expression and clinical data for 520 Head and Neck squamous cell carcinoma samples were downloaded from the TCGA data portal. Median follow-up from diagnosis was 1.8 years with range 0.01 years to 17.6 years. Follow-up time was truncated at 5-years for analysis and 200 deaths occurred in this period. For each of the 16 candidate immune response genes, we scored subjects as above (high) or below (low) the median expression and compared survival using a log rank test at 5% significance. HPV+ patients were stratified into a favorable immune profile if they had expression above the median for the significant genes IL12A, IL12B IFNG, CD8A and below the median for IL6. Kaplan-Meier curves were plotted for these two groups. Similar methods were used to examine association of these 16 genes with 720 lung adenocarcinoma, and 876 gastric carcinoma samples using the publically available data from KM Plotter28. In lung adenocarcinoma, 12 genes were significantly associated with survival; patients were scored as having a favorable immune profile if 7 or more of the 12 significant genes had expression in the favorable direction. In 876 gastric cancer samples, 8 genes were significantly associated with survival. patients were scored as having a favorable immune profile if 5 out of the 8 genes had expression in the favorable direction.
PI3Kγ−regulated gene expression signature analysis in TCGA data
We investigated 66 immune-related genes in four functional classes (17 genes related to antigen presentation (HLA class I and II molecules), 24 genes surveying T cell activation, 20 innate immune response genes (IL6, CCL7 and others), and 5 genes related to cancer cell signaling) that changed expression in response to PI3Kγ inhibition for association with survival in HPV+ and HPV− TCGA HNSCC and lung adenocarcinoma cohorts. Within each cancer type, we scored subjects as above or below the median expression for each gene and compared survival using a log rank test, using 10% FDR within each class as the significance threshold. HPV+ and HPV− HNSCC survival were investigated separately, as HPV− HNSCC has generally worse prognosis. Within each cohort, patients were classified as having a favorable PI3Kγ immune response profile if they had expression levels above or below the median in the direction of low PI3Kγ activity for the genes identified as significant. We compared the survival experience of favorable vs less-favorable patients using Kaplan-Meier curves.
Out of the 66 experimentally identified PI3Kγ regulated genes 43 showed significant association with overall survival in the HPV+ cohort (FDR<10% within each functional class). Comparison of these genes between HPV+ and HPV− cohort showed that HPV− samples generally had significantly (p<0.05) lower expression of 42 genes in the antigen presentation and T cell activation classes, consistent with a pattern of adaptive immune suppression, and higher expression of genes in the innate immune response and cancer cell signaling class, which were negatively associated with survival. Only Malt1 was not differentially expressed between the two groups (p=0.7).
Mice
Pik3cg−/− (p110γ−/−) and Pik3cg−/−; PyMT animals were generated as previously described12. Cd8−/− and Cd4−/− animals in the C57Bl6 background were purchased from the Jackson Laboratory, Bar Harbor, ME and crossed with syngeneic Pik3cg−/− animals. All animal experiments were performed with approval from the Institutional Animal Care and Use Committee of the University of California, San Diego, La Jolla, CA. Animal were euthanized before the maximum IACUC allowable tumor burden of 2 cm3/mouse was exceeded.
Tumor studies
Wildtype or p110γ−/− 6–8 week-old female or male syngeneic C57Bl/6J (LLC lung, PyMT breast, and MEER HPV+ HNSCC) or C3He/J (SSCVII HPV− HNSCC) mice were implanted with 1×106 tumor cells by subcutaneous injection (LLC, MEER, SCCVII) or by orthotopic injection (PyMT) (n=10–15) and tumor growth was monitored for up to 30 days. Tumor dimensions were measured once when tumors were palpable. Tumor volumes were calculated using the equation (l2 x w)/2. In some studies, WT and p110γ−/− animals with LLC tumors were treated with gemcitabine (150 mg/kg) or saline by i.p injection on d7 and d14 (n=10). LLC were acquired from ATCC, PyMT were from Lesley Ellies (University of California, San Diego), HPV+ MEER were from John Lee (Cancer Biology Research Center, Sanford Research/USD, Sioux Falls, SD) and SCCVII squamous carcinoma cells were from Stephen Schoenberger (La Jolla Institute for Allergy and Immunology). All cell lines were tested for mycoplasma and mouse pathogens and checked for authenticity against the International Cell Line Authentication Committee (ICLAC; http://iclac.org/databases/cross-contaminations/) list.
In some studies, mice bearing LLC, PyMT, HPV+ MEER or HPV− HNSCC tumor cells were treated once daily by oral gavage with vehicle (5%NMP/95% PEG 400), 15mg/kg/day of the PI3Kγ inhibitor IPI-549 or by i.p. injection with 2.5 mg/kg b.i.d of TG100–1159,43 beginning on day 8 post-tumor injection and continuing daily until sacrifice. IPI-549 is an orally bioavailable PI3Kγ inhibitor with a long plasma half-life and a KD value of 0.29 nM for PI3Kγ with >58-fold weaker binding affinity for the other Class I PI3K isoforms26. Enzymatic and cellular assays confirmed the selectivity of IPI-549 for PI3Kγ (>200-fold in enzymatic assays and >140-fold in cellular assays over other Class I PI3K isoforms26. To study the effect of IPI-549 on lung tumor growth, LLC tumor cells were passaged three times in C57/BL6 Albino male mice. When tumor volume reached 1500 mm3, tumors were harvested and single-cell suspensions were prepared. This tumor brei inoculum was implanted subcutaneously in the hind flank of C57/BL6 Albino male mice at 1×106 cells/ mouse. Prior to initiating treatment with once daily IPI-549 (15 mg/kg p.o.), groups were normalized on the basis of tumor volume. In some studies, WT and p110γ−/− tumor-bearing mice were treated with 100μg of anti-CD8 (clone YTS 169.4) or isotype control clone (LTF-2) from BioXCell administered by i.p. injections on day 7, 10 and 13 of tumor growth. For all tumor experiments, tumor volumes and weights were recorded at sacrifice.
Anti-PD-1 tumor studies
C57Bl/6J (wildtype) or p110γ−/− 6–8 week-old male or female mice (MEER HPV+ HNSCC) or C3He/J (SCCVII HPV− HNSCC) were implanted with tumor cells by subcutaneous injection (1 × 106 MEER or 1 × 105 SCCVII). In HPV+ MEER studies, wildtype and p110γ−/− animals were treated with 4 doses of 250 μg of anti-PD-1 antibody (Clone RMP-14, Bioxcell) or Rat IgG2a isotype control (Clone2A3, Bioxcell) every 3 days starting when tumors became palpable on day 11 (n = 12–14 mice per group). Wildtype mice bearing HPV+ tumors were also treated with the p110γ inhibitor TG100–11543 b.i.d. by i.p. injection, beginning on day 11. Tumor regressions were calculated as a percentage of the difference in tumor volume between the date treatment was initiated and the first date of sacrifice of the control group. For HPV-SCCVII studies, C3He/J mice were treated with PI3Kγ inhibitor (2.5mg/kg TG100–115 i.p.) beginning on day 6 post-tumor inoculation and with 6 doses of anti-PD-1 antibody (250μg Clone RMP-14, Bioexcell) or Rat IgG2a isotype control Clone 2A3, Bioxcell) every 3 days beginning on day 3 (n=12 mice per group) or with a combination of the two. Alternatively, mice were treated with 5mg/kg TG100–115 b.i.d. +/− anti-PD-1 (250μg every 3 days) beginning on day 1 (Figure 4). Mice that completely cleared HPV+ MEER tumors were re-injected with HPV+ tumor cells contralateral to the initial tumor injection and tumor growth was monitored.
PyMT models of mammary carcinoma
The growth and metastasis of spontaneous mammary tumors in female PyMT+ (n=13) and p110γ−/− PyMT+ (n=8) animals was evaluated over the course of 0–15 weeks. Total tumor burden was determined by subtracting the total mammary gland mass in PyMT− animals from the total mammary gland mass in PyMT+ animals. Lung metastases were quantified macroscopically and microscopically in H&E tissue sections at week 15.
LPS induced septic shock
Septic shock was induced in WT and p110γ−/− mice via intraperitoneal injection of 25 mg/kg LPS (Sigma, B5:005). Survival was monitored every 12h and liver, bone marrow and serum were collected 24h post LPS injection.
Macrophage depletion studies
C57Bl/6J female mice were implanted with 1 × 106 LLC tumor cells by subcutaneous injection. When the average tumor size was 250 mm3, mice were treated by i.p. injection with 1 mg/mouse clodronate or control liposomes (Clodronateliposomes.com, Amsterdam, The Netherlands) every 4 days for 2 weeks in combination with daily administration of vehicle or IPI-549 (15 mg/kg/day p.o.). In other studies, six week old female BALB/c mice were injected subcutaneously with 2.5×105 CT26 murine colon carcinoma cells in 100μl PBS into the right flank. Eight days later, tumor-bearing mice were arranged into four groups (n=15) with an average tumor volume of 70mm3. Oral administration of IPI-549 (15mg/kg) or Vehicle (5%NMP/95% PEG 400) and anti-CSF-1R antibody (50mg/kg i.p. 3× per week, Clone AFS98, Bioexcell) began on day 8 post-tumor injection via gavage at a 5 mL/kg dose volume and continued daily for a total of 18 doses.
Tumor infiltrating myeloid cell analysis
Six week old female BALC/c mice were injected subcutaneously with 2.5×105 CT26 murine colon carcinoma cells in 100μl PBS into the right flank. On day 8 post tumor injection, tumor-bearing animals were grouped and treated with IPI-549 (15 mg/kg, QD, PO) or Vehicle (5%NMP/95% PEG 400). In addition, mice were injected i.p. with 50mg/kg anti-CD115 (BioXcell Clone: AFS98 Cat: BE0213) or 50mg/kg rat IgG2a isotype control (BioXcell Clone: 2A3 Cat: BE0089-R005) antibodies as described above for a total of three injections. Two days after the final injection animals were euthanized, tumors were digested in a mixture of 0.5 mg/ml Collagenase IV and 150 U/ml DNAse I in RPMI-1640 for 30 minutes at 37°C, and tumor-infiltrating myeloid cells were analyzed by flow cytometry.
In vivo macrophage adoptive transfer experiments
CD11b+Gr1− cells were isolated from single cell suspensions of LLC tumors from donor mice by FACS sorting or serial magnetic bead isolation. Additionally, for some experiments, primary bone marrow derived macrophages were polarized and harvested into a single cell suspension. Purified cells were admixed 1:1 with LLC tumor cells and 5 × 105 total cells were injected subcutaneously into new host mice. Tumor dimensions were measured 3 times per week beginning on day 7. In antibody blocking studies, CD11b+Gr1− cells were incubated with 5 μg of anti-IL-12 clone RD1-5D9 or isotype clone LTF-2 (BioXCell) for 30 min prior to the addition of tumor cells. Mice were additionally treated intradermally with 5μg of antibody 3 and 6 days after tumor cell inoculation. In some studies, CD11b+Gr1− cells were pre-incubated with inhibitors of Arginase (nor-NOHA, 50 μM, Cayman Chemical), iNOS (1400W dihydrocholoride, 100 μM, Tocris), mTOR (rapamycin, 10 μM Calbiochem), or IKKβ (ML120B, 30 μM, Tocris) for 30 min before the addition of tumor cells. Inoculated mice were further treated by intradermal injection with inhibitors at 3 and 6 days post inoculation.
T-cell adoptive transfer
Donor C57Bl/6J (wildtype) or p110γ−/− mice were implanted with 1 × 106 LLC tumor cells by subcutaneous injection. On day 14 after tumor implantation, CD90.2+, CD4+ or CD8+ cells were harvested by magnetic bead isolation (Miltenyi Biotec). T cells were mixed 1:1 with viable LLC tumor cells. Cell mixtures containing 5 × 105 total cells were injected into the flanks of naïve WT or p110γ−/− mice (n=8–10 per group). Tumor growth, intratumoral apoptosis and necrosis were investigated over 0–16 days. In other studies, WT T cells were incubated at 37 °C/5% CO2 for 6h with 10 or 100 nM IPI-549 (Infinity Pharmaceuticals) or Cal-101 (Selleck Chem). After 6h, T cells were washed, admixed 1:1 with LLC tumor cells, and 1 × 106 total cells were injected subcutaneously into recipient mice. Tumor growth, was monitored for 14 days.
Isolation of single cells from murine tumors
Tumors were isolated, minced in a petri dish on ice and then enzymatically dissociated in Hanks Balanced Salt Solution containing 0.5 mg/ml Collagenase IV (Sigma), 0.1 mg/ml Hyaluronidase V (Sigma), 0.6 U/ml Dispase II (Roche) and 0.005 MU/ml DNAse I (Sigma) at 37°C for 5–30 min. The duration of enzymatic treatment was optimized for greatest yield of live CD11b+ cells per tumor type. Cell suspensions were filtered through a 70μm cell strainer. Red blood cells were solubilized with red cell lysis buffer (Pharm Lyse, BD Biosciences, San Jose, CA), and the resulting suspension was filtered through a cell strainer to produce a single cell suspension. Cells were washed one time with PBS prior to use in flow cytometry analysis or magnetic bead purification.
Peritoneal Macrophage Isolation
Thioglycollate elicited peritoneal macrophages were collected 96h after i.p. injection of a 3% thioglycollate solution. Cells were harvested from the peritoneal cavity in 10ml of PBS and macrophage enrichment was performed by plating cells in RPMI with 10% FBS and 1% pen/strep for 2h at 37C 5% CO2. After 2h, nonadherent cells were removed with three PBS washes, and cells were analyzed via flow cytometry and qPCR analysis.
Flow cytometry staining and analysis
Single cell suspensions (10^6 cells in 100 μL total volume) were incubated with Aqua Live Dead fixable stain (Life Technologies, Carlsbad, CA), FcR-blocking reagent (BD Biosciences, San Jose, CA) and fluorescently labeled antibodies and incubated at 4°C for 1h. Primary antibodies to cell surface markers directed against F4/80 (BM8), CD45 (30-F11), CD11b (M1/70), Gr1 (RB6-8C5), CD3 (145-2C11), CD4 (GK1.5), CD8 (53–6.7), CD273 (B7-DC), CD274 (B7-H1) were from eBioscience; Ly6C (AL-21), Ly6G (1A8), CD11c (HL3), and MHC-II (AF6-120.1) from BD Pharmingen, CCR2 (475301) from R&D Systems and CD206 (MR5D3) from AbD Serotech. For intracellular staining, cells were fixed, permeabilized using Transcription Factor Staining Buffer Set (eBioscience) and then incubated with fluorescently labeled antibodies to FoxP3 (FJK-16s) from eBioscience. Multicolor FACS Analysis was performed on a BD Canto RUO 11 Color Analyzer. All data analysis was performed using the flow cytometry analysis program FloJo (Treestar).
Magnetic bead purification of myeloid cells
Single cell preparations from bone marrow or tumors were incubated with FcR-blocking reagent (BD Biosciences) and then with 20μl magnetic microbeads conjugated to antibodies against CD11b, Gr1, CD90.2, CD4 and CD8 (Miltenyi Biotech MACS Microbeads)/1xE7 cells for 20 min at 4°C. Cells bound to magnetic beads were then removed from the cell suspension according to manufacturers instructions.
Flow cytometric sorting of cells from tumors and bone marrow
For cell sorting, single cell suspensions were stained with Aqua Live Dead fixable stain (Life Technologies) to exclude dead cells and anti-CD11b-APC (M1/70, eBioscience) and anti-Gr1-FITC (RB6-8C5, eBioscience) antibodies. FACS sorting was performed on a FACS Aria 11 color high speed sorter at the Flow Cytometry Core at the UC San Diego Center for AIDS Research. Live cells were sorted into the following populations: CD11b+Gr1−, CD11b+Gr1lo, CD11b+Gr1hi and CD11b−Gr1− cells. CD11b positive cells were defined by increased staining over the isotype control, and Gr1 levels were defined both by comparison to the isotype control and relative staining to other populations.
Murine macrophage differentiation and culture
Bone marrow derived cells (BMDC) were aseptically harvested from 6–8 week-old female mice by flushing leg bones of euthanized mice with phosphate buffered saline (PBS), 0.5% BSA, 2mM EDTA, incubating in red cell lysis buffer (155 mM NH4Cl, 10 mM NaHCO3 and 0.1 mM EDTA) and centrifuging over Histopaque 1083 to purify the mononuclear cells. Approximately 5X107 BMDC were purified by gradient centrifugation from the femurs and tibias of a single mouse. Purified mononuclear cells were cultured in RPMI + 20% serum + 50ng/ml M-CSF (PeproTech).
Human macrophage differentiation and culture
Human leukocytes concentrated by from apheresis were obtained from the San Diego Blood Bank. Cells were diluted in phosphate buffered saline (PBS), 0.5% BSA, 2mM EDTA, incubated in red cell lysis buffer (155 mM NH4Cl, 10 mM NaHCO3 and 0.1 mM EDTA) and centrifuged over Histopaque 1077 to purify mononuclear cells. Approximately 109 BMDC were purified by gradient centrifugation from one apheresis sample. Purified mononuclear cells were cultured in RPMI + 20% serum + 50ng/ml Human M-CSF (PeproTech). Non-adherent cells were removed after 2h by washing, and adherent cells were cultured for 6 days to differentiate macrophages fully.
Macrophage polarization
Bone marrow derived macrophages were polarized with either IFNγ (20 ng/ml, Peprotech) plus LPS (100 ng/ml, Sigma) or LPS alone for 24h or IL-4 (20 ng/ml, Peprotech) for 24–48h. For inhibitor studies, PI3Kγ inhibitors (1 μM) (IPI-549, Infinity Pharmaceuticals and TG100–115, Targegen/Sanofi-Aventis), rapamycin (10 μM) (Selleck), or ML120B (30 μM) were incubated with macrophages 1h prior to the addition of polarizing stimuli. Total RNA was harvested from macrophages using the RNeasy Mini Kit (Qiagen) according to the manufacture’s instructions.
RNA sequencing
Freshly isolated mouse bone marrow cells from 9 WT and 9 p110γ−/− mice were pooled into 3 replicates sets of WT or p110g−/− cells and differentiated into macrophages for six days in RPMI + 20% FBS+ 1%Pen/Strep+ 50 ng/ml M-CSF. Each replicate set of macrophages was then treated with mCSF, IL-4 or IFNg/LPS. Macrophages were removed from dishes, and RNA was harvested using Qiagen Allprep kit. In addition, RNA was harvested from day 14 (500mm3) LLC tumors or purified CD11b+Gr1-F480+ TAMs from WT (C57BL/6) and p110γ−/− null mice. RNA was harvested using Qiagen Allprep kit. RNA libraries prepared from 1 μg RNA per sample were prepared for sequencing using standard Illumina protocols. RNA sequencing was performed by the University of California, San Diego Institute for Genomic Medicine. mRNA profiles were generated by single read deep sequencing, in triplicate, using Illumina HiSeq2000.
Sequence analysis
Sequence analysis was performed as previously described.15 Sequence files from Illumina HiSeq that passed quality filters were aligned to the mouse transcriptome (mm9 genome build) using the Bowtie2 aligner4. Gene-level count summaries were analyzed for statistically significant changes using DESeq. Individual p-values were adjusted for multiple testing by calculating Storey's q-values using fdrtooltrimmer. For each gene, the q-value is the smallest false discovery rate at which the gene is found significant. We analyzed biological processes as defined by the Gene Ontology Consortium. Each gene ontology term defines a set of genes. The entire list of genes, sorted by the q-value in ascending order, is subjected to a non-parametric variant of the Gene Set Enrichment Analysis (GSEA), in which the parametric Kolmogorov-Smirnov p-value is replaced with the exact rank-order p-value. We perform a Bonferroni adjustment of gene set p-values for the number of gene sets tested. Heatmaps of expression levels were created using in-house hierarchical clustering software that implements Ward clustering. The colors qualitatively correspond to fold changes. Complete sequence data can be observed at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=yfwtskeirrefbsp&acc=GSE58318 (in vitro macrophage samples) and http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=kpqnqyqalfgnlgd&acc=GSE84535 (in vivo tumor and tumor associated macrophages samples).
Individual quantitative RT-PCR
cDNA was prepared using 1μg RNA with the qScript cDNA Synthesis Kit (Quanta Biosciences). Sybr green-based qPCR was performed using human and murine primers to Arg1, Ifng, Il10, Il12p40, Il1b, Il6, Ccl2, Vegfa, Gapdh, Nos2, Tgfb1, Tnfa and murine H2-Aa, H2-Ab1, H2-Eb1, and H60a (Qiagen QuantiTect Primer Assay). mRNA levels were normalized to Gapdh (dCt = Ct gene of interest – Ct Gapdh) and reported as relative mRNA expression (ddCt = 2^-(dCt sample – dCt control)) or fold change.
siRNA mediated knockdown and gene transfection
Freshly isolated bone marrow derived CD11b+ myeloid cells or differentiated macrophages were transfected by electroporation using an AMAXA Mouse Macrophage Nucleofection Kit with 100nM of siRNA or 2μg p110gCAAX or pcDNA control plasmid. Non-silencing (Ctrl_AllStars_1), Cebpb (MmCebpb_4 and MmCebpb_6), Mtor (Mm_Frap1_1 and Mm_Frap1_2) were purchased from Qiagen. After transfection, cells were cultured for 36–48 h in RPMI containing 10% serum and 10ng/ml M-CSF (PeproTech) or polarized as described above.
ELISA assays
Whole tumors, CD11b+Gr1− cells, CD90.2+ cells, CD4+ cells and CD8+ cells isolated from LLC tumors were lysed in RIPA buffer and total protein concentration was determined using a BCA Protein Assay (Pierce). Macrophage supernatants (100 μl) or 500 μg of total protein lysate from tumors were used in ELISAs to detect CCL2, TGFβ, IL-1β, TNFα, IL-6, IFNγ, IL-10, IL-12 and Granzyme B (Ready Set Go ELISA, eBioscience). Protein expression was normalized to total volume (supernatants) or mg total protein (tumor lysates).
Quantitative Colorimetric Arginase Determination
The QuantiChrom Arginase Assay Kit (DARG-200, BioAssay Systems) was used to measure Arginase activity in primary murine bone marrow derived macrophages from wildtype and p110γ−/− mice according to manufacturer’s instructions. For all conditions, cells were harvested and lysed in 10 mM Tris (pH7.4) containing 1 μM pepstatin A, 1 μM leupeptin, and 0.4% (w/v) TritonX-100. Samples were centrifuged at 20,000g at 4°C for 10 min.
Transcription Factor Assays
To measure NFκB and C/EBPβ activation, TransAM NFκB Family and C/EBPa/b Transcription Factor Assay Kits (43296 and 44196, Active Motif, Carlsbad, CA) were used according to manufacturer’s protocol. Briefly, wild type and p110γ−/− bone marrow derived macrophages were stimulated with LPS (100 ng/ml) or IL-4 (20 ng/ml) and nuclear extracts were prepared in lysis buffer AM2. Nuclear extracts were incubated with the immobilized consensus sequences and RelA, cRel or C/EBPβ were detected using specific primary antibodies. Quantification was performed via colorimetric readout of absorbance at 450 nm.
Immunoblotting
IL-4 and LPS macrophage cultures were solubilized in RIPA buffer containing protease and phosphatase inhibitors. Thirty μg protein was electrophoresed on Biorad precast gradient gels and electroblotted onto PVDF membranes. Proteins were detected by incubation with 1:1000 dilutions of primary antibodies, washed and incubated with Goat anti-rabbit-HRP antibodies and detected after incubation with a chemiluminescent substrate. Primary antibodies directed against Akt (11E7), p-Akt (244F9), IkBa (L35A5), IKKb (D30C6), p-IKKa/b (16A6), NFkBp65 (D14E12), pNFkBp65 (93H1), C/EBPb (#3087), p-CEBPb (#3082), IRAK1 (D51G7), TBK1 (D1B4), and p110γ (#4252) were from Cell Signaling Technology and pTBK1 (EPR2867(2)) was from Abcam.
In vitro cytotoxicity assay
CD90.2+ tumor derived T cells were purified from LLC tumor-bearing WT and p110γ−/− or TG100–115 and control treated mice and then co-incubated with LLC tumor cells (target cells) at 2.5:1, 5:1 and 10:1 ratios of T cells to tumor cells (2×103 LLC tumor cells per well) for 6 hours. Target cell killing was assayed by collecting the supernatants from each well for measurement of the lactate dehydrogenase release (Cytotox96 Non Radioactive Cytotoxicity Assay kit, Promega).
Immunohistochemistry
Tumor samples were collected and cryopreserved in O.C.T. Sections (5μm) were fixed in 100% cold acetone, blocked with 8% normal goat serum for 2 hours, and incubated anti-CD8 (53–6.7, 1:50 BD Biosciences) for 2 hours at room temperature. Sections were washed 3 times with PBS and incubated with Alexa594-conjugated secondary antibodies. Slides were counterstained with 4',6-diamidino-2-phenylindole (DAPI) to identify nuclei. Immunofluorescence images were collected on a Nikon microscope (Eclipse TE2000-U) and analyzed using Metamorph image capture and analysis software (Version 6.3r5, Molecular Devices). The detection of apoptotic cells was performed using a TUNEL-assay (ApopTag Fluorescein In Situ Apoptosis Detection Kit, Promega) according to manufacturer’s instructions. Slides were washed and mounted in DAKO fluorescent mounting medium. Immunofluorescence images were collected on a Nikon microscope (Eclipse TE2000-U) and analyzed with MetaMorph Software (version 6.3r5) or SPOT software (version 4.6). Pixels/field or cell number/field were quantified in five 100x fields from 10 biological replicates.
Statistics
Primary tumor samples with mRNA expression data were scored as above or below the median expression level, and tested for association with patient survival using a logrank test at 5% significance. For studies evaluating the effect of drugs on tumor size, tumor dimensions were measured directly before the start of treatment, tumor volumes were computed and mice were randomly assigned to groups so that the mean volume +/− s.e.m. of each group was identical. A sample size of 10 mice/group provided 80% power to detect mean difference of 2.25 standard deviation (SD) between two groups (based on a two-sample t-test with 2-sided 5% significance level). Sample sizes of 15 mice/group provided 80% power to detect one SD difference between two groups. Data were normalized to the standard (control). Significance testing was performed by one-way Anova with Tukey's posthoc testing for multiple pairwise testing with more than two groups and by parametric or nonparametric Student’s t test when only two groups were compared. We used a two-sample t-test (two groups) and ANOVA (multiple groups) when data were normally distributed and a Wilcoxon rank sum test (two groups) when data were not normally distributed. All mouse studies were randomized and blinded; assignment of mice to treatment groups, tumor measurement and tumor analysis was performed by coding mice with randomly assigned mouse number, with the key unknown to operators until experiments were completed. In tumor studies for which tumor size was the outcome, animals removed from the study due to health concerns were not included in endpoint analyses. All experiments were performed at least twice; n refers to biological replicates.
Extended Data
Extended Data Figure 1. Pro-inflammatory gene expression signatures predict survival in cancer patients.
(a–e) Expression levels of IL12A, IL12B, IFNG, and CD8A, and IL6 associated with survival in HPV+ HNSCC patients. (f) Multivariate immune signature for 720 lung adenocarcinoma patients from KM plotter cohorts. (g) Multivariate immune signature in 876 gastric cancer samples from KM plotter cohorts. (h) Western blotting to detect PI3Kγ (p110γ) in B cells, T cells, macrophages (MΦ) and LLC, PyMT and MEER tumor cells. (i) Kaplan Meier survival plot of WT and p110γ−/− mice inoculated with LPS (endotoxin). (j) Pro-inflammatory cytokine mRNA expression in bone marrow from WT and p110γ/ − LPS injected animals (n=4, **p<0.001, **p<0.0001). (k) Circulating inflammatory cytokine levels in p110γ−/− and WT mice 24h after endotoxin administration (n=4, *p<0.01, **p<0.001). (l) Tumor volume of HPV− (SCCVII) head and neck (n=15) carcinomas from vehicle or PI3Kγ inhibitor-treated mice. Arrow, start of drug treatment. (m) Dose response of the effect of PI3Kγ inhibitor IPI549 on in vitro MEER cell viability. (n) Spontaneous PyMT lung metastases per high power field (200X) in WT and p110γ−/− animals (n=8). (o) Kaplan Meier survival plot of mice bearing orthotopic PyMT tumors treated with vehicle or PI3Kγ inhibitor IPI549 initiated as indicated by arrow (n=10). (p) In vitro LLC tumor cell survival in the presence of gemcitabine. (q) Volume of LLC tumors implanted in WT and p110γ−/− animals treated with saline or gemcitabine (n=10, **p<0.001, **p<0.01).
Extended Data Figure 2. Effect of PI3Kγ inhibition on tumor inflammation.
(a) Gating strategy for flow cytometric analysis of myeloid cell populations in peripheral blood leukocytes. (b) Representative flow cytometric analysis and quantification of myeloid cell populations in peripheral blood (PB) of naïve and LLC tumor bearing mice (n = 3). (c) Flow cytometric analysis of myeloid cell populations on days 0, 7, 14 and 21 after subcutaneous inoculation with Lewis lung carcinoma cells (n= 3). (d) Quantification of populations from c. (e) Flow cytometric analysis of Ly6G, CCR2, CX3CR1, CD206, CD11c, F4/80 and CD45 expression on myeloid cell populations from c (n=3). (f) Relative immune response transcript levels in tumor-derived myeloid cells and tumor cells (CD11b-Gr1− cells) isolated at day 0 (n=3), d7 (n=5), d14 (n=3) or d21 (n=4) after LLC cell inoculation (p < 0.002, d21 vs d0). (g) Flow cytometric analysis of CD11b+ myeloid cell populations in WT and p110γ−/− LLC, PyMT and MEER tumors (n=3). (h) Quantification of CD11b+ myeloid cell populations from (g). (i) Flow cytometric analysis of CD11b+ myeloid cell populations in vehicle and PI3Kγ inhibitor treated PyMT, MEER and SCCVII tumors (n=3). (j). Quantification of CD11b+ myeloid cell populations from (i).
Extended Data Figure 3. Effect of PI3Kγ inhibition on TAM expression profile.
(a) Heatmap of differentially expressed immune response genes in TAMs isolated from LLC tumors from WT and p110γ−/− mice (n=3, *p<0.01, lfdr <0.1) obtained by RNA sequencing. (b) Relative mRNA expression of immune response factors in HPV+ HNSCC MEER tumors from p110γ−/− and WT mice (n=4), *p=0.01. (c) Relative mRNA expression of immune response factors in CD11b+ myeloid cells isolated from PyMT tumors grown in vehicle or PI3Kγ inhibitor-treated mice (n=4), *p=0.01. (d) Fold change in mRNA expression in CD11b+Gr1− (macrophage), CD11b+Gr1lo (monocytic) and CD11b+Gr1hi (granulocytic) myeloid cells isolated from LLC tumors grown in p110γ−/− mice (n=5) and normalized to WT control (n=5), p=0.001. (e) Arginase activity in tumors and TAMs isolated from LLC tumors grown in WT and p110γ−/− mice (n=4, ***p<0.0003). (f) Protein expression of cytokines in LLC tumors and TAMs from WT and p110γ−/− mice (n=4, *p<0.01, **p<0.001, ***p<0.0001).
Extended Data Figure 4. Effect of PI3Kγ deletion on in vitro macrophage mRNA expression.
(a) Relative immune response mRNA expression in p110γ−/− and WT murine macrophages stimulated by IL-4 or LLC tumor cell conditioned medium (TCM) as determined by RT-PCR (n=3, *p=0.01). (b) Heat map of differentially expressed immune response transcripts in IL-4 and IFNγ/LPS polarized murine macrophages obtained by RNA sequencing (n=3, p=0.00001). (c) Heat map of select differentially expressed immune response transcripts in in vitro polarized murine macrophages (n=3, p=0.00001). (d) Heat map of immune response transcripts in mCSF, IL-4 and IFNγ/LPS stimulated p110γ−/− murine macrophages obtained by RNA sequencing and normalized to WT (n=3, p=0.00001). (e) Heat map of select differentially expressed immune response transcripts in polarized p110γ−/− murine macrophages normalized to WT (n=3, p=0.00001). (f) Heat map of differentially expressed antigen presentation and processing mRNAs in mCSF, IL-4 and IFNγ/LPS polarized p110γ−/− murine macrophages (n=3). (g) Heat map of differentially expressed chemokine and chemokine receptor mRNAs in polarized p110γ−/− murine macrophages (n=3).
Extended Data Figure 5. Effect of PI3Kγ inhibition on murine and human macrophage polarization.
(a) Relative mRNA expression of immune response transcripts in IL-4 and IFNγ/LPS stimulated vehicle and PI3Kγ inhibitor (IPI-549) treated (a) murine and (b) human macrophages (n=3, p=0.001). (c) Relative mRNA expression of M2 macrophage markers (Arg1, Fizz1 and Ym1) in WT and p110γ−/− IL4-stimulated macrophages (n=3). (d) Relative expression of MHC family members in WT and p110γ−/− IL4-stimulated macrophages (n=3). (e–f) Time course of cytokine mRNA expression in IFNγ/LPS, LPS and IL-4 stimulated (e) WT vs p110γ−/− and (f) vehicle vs. PI3Kγ inhibitor (IPI-549)-treated macrophages (n=3). (g) Relative mRNA expression in mCSF-stimulated WT vs p110γ−/− and IPI-549- vs vehicle-treated macrophages (n=3). (g) Relative nuclear RelA DNA binding activity in IFNγ/LPS stimulated WT and p110γ−/− macrophages (n=3).
Extended Data Figure 6. Mechanism of PI3Kγ mediated gene expression regulation.
(a) Relative levels of phospho/total p65 and phospho/total C/EBPβ in LPS and IL-4 stimulated WT and p110γ−/− macrophages. (b) Immunoblotting to detect pThr308Akt, total Akt, phospho-p65 and total p65 in LPS and IL-4 stimulated, macrophages that were treated with vehicle or the PI3Kγ inhibitor IPI-549. (c) Relative Arg1 mRNA expression in myeloid cells transfected with constitutively active, membrane-targeted PI3Kγ (p110γCAAX) and Mtor, S6ka, Cebpb or control siRNA (n=3). (d) Validation of siRNAs from c. (e) Effect of Cebpb, Mtor or S6ka siRNAs on gene expression in WT macrophages. (f–g) Effect of rapamycin (f) or S6K inhibitor (PF4708671) (g) on macrophage mRNA expression. (h) Immunofluorescence images of CD8+ T cells in 10μm tumor sections from 3c. (i) Mean tumor volumes from tumor cells mixed with WT TAMs pretreated with the mTOR inhibitor Rapamycin or the Arginase inhibitor nor- NOHA and p110γ−/− TAMs pretreated with anti-IL12 or isotype matched control antibody (cIgG), IKKβ inhibitor (MLB120) or NOS2 inhibitor (1400W dihydrochloride) (n=10).
Extended Data Figure 7. No direct effect of PI3Kγ inhibition on T cells.
(a) Volumes of LLC tumors treated with vehicle + control liposomes, PI3Kγ inhibitor (IPI-549) + control liposomes, clodronate liposomes + vehicle and PI3Kγ inhibitor + clodronate liposomes (n=10). (b) Quantification of F4/80+ macrophages in tumors from a (n=3). (c) Quantification of F4/80+ macrophages in livers from a (n=3). (d) Quantification of T cells in tumors from a (n=3, *p<0.05). (e) Volumes of CT26 tumors treated with vehicle + cIgG, PI3Kγ inhibitor (IPI-549) + cIgG, anti-CD115 + vehicle and PI3Kγ inhibitor + anti-CD115 (n=15). (f) Quantification of CD11b+ myeloid cells in tumors from e (n=5). (g) Images and quantification of CD8+ T cells in WT and p110g−/− LLC tumors by IHC (n=5). (h) Flow cytometric analysis and quantification of T cell populations in tumors from WT and p110γ−/− or IPI-549 treated animals (n=3). (i) Quantification of T cells in spleens of naïve and LLC tumor-bearing WT and p110γ−/− mice (n=3). (j) Volumes of LLC lung tumors from WT, p110γ−/−, CD8−/− and CD8−/−; p110γ−/− animals (n=12). (k) LLC tumor volume from WT and p110γ−/− animals treated with anti-CD8 antibodies or control (n=10) and percent CD8+ T cells in these tumors (n=3). (l) In vitro proliferation of T cells isolated from naïve and LLC tumor-bearing WT and p110γ−/− mice (n=3). (m) IFNγ and Granzyme B protein expression in T cells from l (n=3).
Extended Data Figure 8. PI3Kγ inhibition relieves T cell exhaustion.
(a) Effect of PI3Kγ and PI3Kδ inhibitors on IFNγ expression by activated human T cells (n=3). (b) Mean weights of tumors derived from a mixture of LLC cells and WT or p110γ−/− tumor derived T cells or WT T cells pre-incubated with 10 or 100 nM PI3Kγ (IPI-549) and PI3Kδ (Cal101) inhibitors prior to implantation (n=16). (c–d) In vitro LLC tumor cell cytotoxicity induced by T cells isolated from LLC tumors from (c) WT and p110γ−/− or (d) control- and PI3Kγ inhibitor-treated mice (n=3, *p<0.001). (e) Images of TUNEL and H&E stained tumors as described in Fig. 5c. (f) Quantification of TUNEL+ cells in tumor sections from e. (g) Mean tumor volumes in WT mice derived from LLC tumor cells mixed 1:1 with CD90.2+, CD4+ and CD8+ T cells or no T cells from WT or p110γ−/− tumor-bearing animals (n=8). (h) IL10 and TGFβ protein expression in lysates from tumor and CD90.2+, CD8+ and CD4+ T cells isolated from LLC tumors grown in WT and p110γ−/− animals (n=3). (i) Interferon gamma and Granzyme B protein expression in PI3Kγ inhibitor or control-treated LLC tumors (n=3). (j) Ifnγ and Tgfb1 mRNA expression in T cells isolated from LLC tumors grown in WT and p110γ−/− or control- and PI3Kγ inhibitor-treated mice (n=3). (k) Relative mRNA expression of Cd4, Cd8, Grzb and Ifng in control and PI3Kγ inhibitor treated PyMT tumors (n=3). (l) Relative mRNA expression of Cd4, Cd8, Grzb, and Ifng in WT and p110γ−/− and PI3Kγ inhibitor treated HPV+ MEER tumors (n=3).
Extended Data Figure 9. PI3Kγ role in the macrophage-mediated tumor immune response.
(a–b) Flow cytometric analysis of PD-L1 and PD-L2 expression on (a) tumor cells and TAMs from WT and p110γ−/− LLC tumors and (b) WT and p110γ−/− in vitro cultured IFNγ/LPS- and IL4-stimulated macrophages (n=3). (c) HPV+ HNSCC tumor growth in female WT or p110γ−/− mice that were treated with anti-PD-1 or isotype matched antibody (cIgG), as indicated by arrows, and percent change in tumor volumes between days 11 and 23. (d) HPV+ HNSCC tumor growth in female WT mice that were treated with PI3Kγ inhibitor (2.5 mg/kg TG100–115 b.i.d). in combination with anti-PD-1 or isotype matched antibody (cIgG), as indicated by arrows, and percent change in tumor volumes 11 and 29. (e) HPV− HNSCC tumor growth in mice that were treated with PI3Kγ inhibitor (2.5 mg/kg TG100–115 b.i.d) in combination with anti-PD-1 cIgG, as indicated by arrows, and percent change in tumor volumes between days 19–26. (f) Tumor rechallenge in HPV+ mice that had cleared previously HPV+ tumors (n=7–12) vs WT mice (n=5). (g) Quantification of percent CD3, CD4+ and CD8+ T cells and MHCII+ macrophages from Figure 5l. (*p<0.05, **p<0.005, ****p<0.00005).
Extended Data Figure 10. PI3Kγ promotes immune suppression.
(a) Comparison of gene expression between HPV+ and HPV− cohorts indicating HPV− samples had significantly (p<0.05) lower expression of adaptive immune genes and higher expression of immune suppressive/pro-metastasis genes (Blue: HPV− samples, Red: HPV+ samples.) (b) Model depicting the effect of PI3Kγ inhibition on tumor immune suppression. PI3Kγ inhibition converts tumor-associated macrophages into pro-inflammatory macrophages that promote a CD8+ T cell response that suppresses tumor growth. (c) Model depicting the PI3Kγ signaling pathway in macrophages. PI3Kγ activation restrains NFκB activation and promotes mTOR-dependent C/EBPβ activation, leading to expression of immune suppressive factors and tumor growth. In contrast, PI3Kγ inhibition inhibits C/EBPβ and stimulates NFκB, leading to altered expression of pro-inflammatory immune response cytokines.
Supplementary Material
Acknowledgments
This work was supported by NIH grants R01CA126820 (JAV), T32HL098062 (MMK), T32CA009523 (SG) and T32CA121938 (SG), the CAPES Foundation and Ministry of Education of Brazil (CF) and by Ralph and Fernanda Whitworth and the Immunotherapy Foundation (JAV and EEC). The authors thank John Lee and Stephen Schoenberger for HPV+MEER HNSCC and SSCVII cells.
Footnotes
The authors declare competing financial interests: details accompany the full-text HTML version of the paper.
Data deposition: RNA sequencing data can be accessing using numbers GSE58318 and GSE84318 at www.ncbi.nlm.nih.gov/geo.
Author contributions
TCGA analysis was performed by HY and KSM, RNA sequencing by KAF, MMK, SG, and RS, flow cytometry by MMK and NR, in vitro studies by MMK, NR, SG, GW, CCF, AVN, MCS, and animal studies by MMK, NR, CL and PF. MP, VJP, JK, KM, MR and DGW provided IPI-549 and Fig.1c, Extended Data Fig. 8a–b. ML120B was contributed by XW and MK. The project was directed by EEWC, KSM and JAV. The manuscript was written by JAV and MMK.
Online content
Supplementary data display items are available in the online version of the paper. For gel source data see Supplementary Figure 1.
References
- 1.Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest. 2012;122:787–795. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schmid MC, Varner JA. Myeloid cells in tumor inflammation. Vasc Cell. 2012;4:14. doi: 10.1186/2045-824X-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature. 2013;496:445–455. doi: 10.1038/nature12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tabas I, Glass CK. Anti-inflammatory therapy in chronic disease: challenges and opportunities. Science. 2013;339:166–172. doi: 10.1126/science.1230720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruffell B, Coussens LM. Macrophages and therapeutic resistance in cancer. Cancer Cell. 2015;27:462–472. doi: 10.1016/j.ccell.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348:56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 7.Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 2015;27:450–461. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. [accessed 12/23/2015]; [accessed 3/14/2016]; https://gdc-portal.nci.nih.gov/projects/TCGA-HNSC. and https://gdc-portal.nci.nih.gov/projects/TCGA-LUAD.
- 9.Martini M, De Santis MC, Braccini L, Gulluni F, Hirsch E. PI3K/AKT signaling pathway and cancer: an updated review. Ann Med. 2014;46:372–383. doi: 10.3109/07853890.2014.912836. [DOI] [PubMed] [Google Scholar]
- 10.Vanhaesebroeck B, Stephens L, Hawkins P. PI3K signalling: the path to discovery and understanding. Nat Rev Mol Cell Biol. 2012;13:195–203. doi: 10.1038/nrm3290. [DOI] [PubMed] [Google Scholar]
- 11.Martin EL, et al. Phosphoinositide-3 kinase gamma activity contributes to sepsis and organ damage by altering neutrophil recruitment. Am J Respir Crit Care Med. 2010;182:762–773. doi: 10.1164/rccm.201001-0088OC. [DOI] [PubMed] [Google Scholar]
- 12.Schmid MC, et al. Receptor tyrosine kinases and TLR/IL1Rs unexpectedly activate myeloid cell PI3kgamma, a single convergent point promoting tumor inflammation and progression. Cancer Cell. 2011;19:715–727. doi: 10.1016/j.ccr.2011.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gunderson AJ, Kaneda MM, et al. Bruton Tyrosine Kinase-Dependent Immune Cell Cross-talk Drives Pancreas Cancer. Cancer Discov. 2015;6:270–285. doi: 10.1158/2159-8290.CD-15-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmid MC, et al. PI3-kinase gamma promotes Rap1a-mediated activation of myeloid cell integrin alpha4beta1, leading to tumor inflammation and growth. PLoS One. 2013;8:e60226. doi: 10.1371/journal.pone.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaneda MM, et al. Macrophage PI3Kγ drives pancreatic ductal adenocarcinoma progression. Cancer Discov. 2016;6:870–875. doi: 10.1158/2159-8290.CD-15-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evans CA, et al. Discovery of a Selective Phosphoinositide-3-Kinase (PI3K) (PI3K)-γ Inhibitor (IPI-549) as an Immuno-Oncology Clinical Candidate. ACS Med Chem Lett. 2016 doi: 10.1021/acsmedchemlett.6b00238. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ben-Neriah Y, Karin M. Inflammation meets cancer, with NF-kappaB as the matchmaker. Nat Immunol. 2011;12:715–723. doi: 10.1038/ni.2060. [DOI] [PubMed] [Google Scholar]
- 18.Poli V. The role of C/EBP isoforms in the control of inflammatory and native immunity functions. J Biol Chem. 1998;273:29279–29282. doi: 10.1074/jbc.273.45.29279. [DOI] [PubMed] [Google Scholar]
- 19.Gray MJ, Poljakovic M, Kepka-Lenhart D, Morris SM., Jr Induction of arginase I transcription by IL-4 requires a composite DNA response element for STAT6 and C/EBPbeta. Gene. 2005;353:98–106. doi: 10.1016/j.gene.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 20.van Rooijen N, Kors N, ter Hart H, Claassen E. In vitro and in vivo elimination of macrophage tumor cells using liposome-encapsulated dichloromethylene diphosphonate. Virchows Arch B Cell Pathol Incl Mol Pathol. 1988;54:241–245. doi: 10.1007/BF02899217. [DOI] [PubMed] [Google Scholar]
- 21.Pyonteck SM, et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat Med. 2013;19:1264–1272. doi: 10.1038/nm.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chaurasia B, et al. Phosphoinositide-dependent kinase 1 provides negative feedback inhibition to Toll-like receptor-mediated NF-kappaB activation in macrophages. Mol Cell Biol. 2010;30:4354–4366. doi: 10.1128/MCB.00069-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arranz A, et al. Akt1 and Akt2 protein kinases differentially contribute to macrophage polarization. Proc Natl Acad Sci U S A. 2012;109:9517–9522. doi: 10.1073/pnas.1119038109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Byles V, et al. The TSC-mTOR pathway regulates macrophage polarization. Nat Commun. 2013;4:2834. doi: 10.1038/ncomms3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yue S, et al. Myeloid PTEN deficiency protects livers from ischemia reperfusion injury by facilitating M2 macrophage differentiation. J Immunol. 2014;192:5343–5353. doi: 10.4049/jimmunol.1400280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rauh MJ, et al. SHIP represses the generation of alternatively activated macrophages. Immunity. 2005;23:361–374. doi: 10.1016/j.immuni.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 27.Baer C, et al. Suppression of microRNA activity amplifies IFN-γ-induced macrophage activation and promotes anti-tumour immunity. Nat Cell Biol. 2016;18:790–802. doi: 10.1038/ncb3371. [DOI] [PubMed] [Google Scholar]
- 28.Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.