Abstract
Recent publications have shown that active RNA polymerase (RNAP) from Mycobacterium tuberculosis (MtbRNAP) can be produced by expressing all four subunits in a single recombinant Escherichia coli strain [1–3]. By reducing the number of plasmids and changing the codon usage of the Mtb genes in the co-expression system published by Banerjee et. al [1], we present a simplified, detailed and reproducible protocol for the purification of recombinant MtbRNAP containing the ω subunit. Moreover, we describe the formation of ternary elongation complexes (TECs) with a short fluorescence-labeled RNA primer and DNA oligonucleotides, suitable for transcription elongation studies. The purification of milligram quantities of the pure and highly active holoenzyme omits ammonium sulfate or polyethylene imine precipitation steps [4] and requires only 5 g of wet cells. Our results indicate that subunit assemblies other than α2ββ′ω·σA can be separated by ion-exchange chromatography on Mono Q column and that assemblies with the wrong RNAP subunit stoichiometry lack transcriptional activity. We show that MtbRNAP TECs can be stalled by NTP substrate deprivation and chased upon the addition of missing NTP(s) without the need of any accessory proteins. Finally, we demonstrate the ability of the purified MtbRNAP to initiate transcription from a promoter and establish that its open promoter complexes are stabilized by the M. tuberculosis protein CarD.
Keywords: Promoter initiation, open complex, elongation complex assembly
INTRODUCTION
Bacterial RNA polymerase (RNAP) is a complex multimeric enzyme of high interest for fundamental research because of its essential role in gene expression, and for biomedical research as a target of antibiotics [5,6]. In particular, rifampin is a potent inhibitor of RNAP activity [3,7] that has allowed the development of successful short course antibiotic treatments against tuberculosis [8,9]. Thus, convenient protocols for the production and activity assays of Mycobacterium tuberculosis RNAP (MtbRNAP) will be essential for the development of new inhibitors and for the characterization of its polymorphic variants, particularly those linked to drug resistance [10].
A number of methods have been used for the purification of prokaryotic RNA polymerases [11]. Difficulties arise from its tendency to misfold, relatively low stability, and a tendency to form oligomers [12] which are likely to lose activity during storage (Kashlev, unpublished observation). The vast majority of biochemical studies on prokaryotic RNAP have been performed on the E. coli enzyme. However, RNAP’s activity is finely tuned by regulatory DNA sequences, GC content and protein transcription factors [13,14], all of which vary greatly among bacterial groups, implying that the E. coli enzyme might be a poor model to investigate the medically relevant aspects of gene expression in M. tuberculosis [15] as well as its interaction with drugs.
Purification of MtbRNAP from M. tuberculosis cultures [16] involves severe risks and costs, therefore some research groups have developed alternative sources for mycobacterial RNAP, such as M. bovis or M. smegmatis [17] which offer the advantage of growing faster than M. tuberculosis and of posing no significant risk for humans. However, despite the sequence similarity of these enzymes to MtbRNAP, it is not known to what extent their biochemical properties and in vivo regulatory mechanisms differ from the MtbRNAP enzyme.
The heterologous expression of each of the MtbRNAP subunits (α, β, β′, ω, and σA) in separate E. coli clones allowed a high yield of protein, but with low specific activity, probably due to misfolding during the reconstitution in vitro [18]; moreover, our own trials with similar protocols showed the presence of E. coli β subunit in the final product, raising concerns about the formation of inter-species chimeras (data not shown). More recently, it has been shown that co-expression of all MtbRNAP subunits in a single E. coli clone can produce an active enzyme [3,19] that can be expressed also in high yield without the presence of chimeras [1]. This strategy has also been used in the co-expression of RNAP of M. bovis with the β and β′ subunits linked as a single peptide [2]. In general, however, until now there has been no detailed protocol to separate out the main undesirable sub-assemblies formed during the co-expression of the MtbRNAP in E. coli.
In this work, we use two plasmids to co-express all the subunits of MtbRNAP holoenzyme in E. coli, having optimized the codon usage of the genes for their expression in this host. Using a two-step purification through a Ni2+ column and a Mono Q ion exchange column, we identify the ω subunit-containing MtbRNAP holoenzyme in Coomassie blue-stained gels. The purified enzyme was used to assembly ternary elongation complex (TEC) [20] carrying a fluorescein-labeled RNA [21,22]. Moreover, we show that the purified MtbRNAP holoenzyme is able to form initiation complexes with promoter DNA and that these are stabilized by addition of M. tuberculosis CarD.
MATERIALS AND METHODS
Strains and bacterial growth
E. coli BL21 (DE3) strain (New England Biolabs) was used for expression of the recombinant RNAP of M. tuberculosis and the transcription factor CarD. All MtbRNAP preparations produced in this work originated from the simultaneous expression of α, β, β′, and ω core subunits along with σA. Therefore, all MtbRNAP entries below refer to the holoenzyme containing the σA subunit.
Plasmids
The sequences of M. tuberculosis genes rpoA, rpoB, rpoC, rpoD and rpoZ, were modified in order to optimize their codon usage for expression in E. coli and a decahistidine tag was added to the N-terminus of the α subunit. The corresponding DNA constructs were synthesized by GenScript and inserted in the plasmids pET Duet-1 and pACYC Duet-1 (Merck Millipore) to produce the plasmids pET-Duet-BC (carrying rpoB and rpoC genes) and pACYC-Duet-AZD (carrying rpoA, rpoZ and rpoD genes). Similarly, the carD gene of M. tuberculosis with the E. coli-optimized codon usage was synthesized and inserted in pET28a plasmid using the sites NdeI and HindIII to produce plasmid pET28-His-CarD.
Protein expression
E. coli cells transformed with the plasmids pET-Duet-BC and pACYC-Duet-AZD were cultured in Luria-Bertani (LB) medium supplemented with 100 μg/ml ampicillin (Amp) and 30 μg/ml chloramphenicol (Cm) overnight at 37°C with 220 rpm agitation. 5 ml of this starter culture was used to inoculate 1 L of LB + Amp + Cm, which was then incubated for 3–4 hours at 37°C to reach an optical density (OD600) of ~0.7. The culture was then allowed to cool down for 30 min at room temperature (~25°C) and IPTG was added to a final concentration of 0.3 mM. The induced culture was kept at RT overnight while agitated by a magnetic stir bar (700 – 800 rpm). The cells were centrifuged (10,000 × g for 15 min) at 4 °C and washed once with cold phosphate buffer saline (PBS) pH 7.4 (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4). The pellets from three separate one liter cultures were collected and frozen at −80°C.
Purification of MtbRNAP
All purification procedures were carried out at 4°C. Frozen cell pellet (5 g) was re-suspended in 30 ml of lysis buffer (all buffer solutions are listed in Table 1), and sonicated for 4.5 minutes on ice with the power set at 45% and ON/OFF cycles of 9 s (Sonicator 3000®). The lysate was clarified by centrifugation at 20,000 × g for 30 minutes. The soluble fraction was mixed with buffer A-1000 to adjust the volume to 200 ml, and loaded at 1.0 ml/min flow rate onto a 5 ml HisTrap™ pre-packed column (GE Healthcare), pre-equilibrated with buffer A-1000 using ÄKTA system (GE Healthcare). The column was washed with approximately 75 ml (15 column volumes) of buffer A-1000, until a constant low absorbance at 280 nm (A280) was reached, and with 25 ml of buffer A-200. The flow rate was increased to 2 ml/min, and a 40 ml gradient of imidazole from 0 to 120 mM in buffer A-200 was applied to the column to elute and discard the major contaminants. After the A280 plateau was reached, MtbRNAP was eluted with buffer A-200 supplemented with 200 mM imidazole. Finally, the column was washed with buffer A-1000 supplemented with 400 mM imidazole.
Table 1.
Buffer solutions for protein purification and functional assays
| Buffer | Composition |
|---|---|
| Lysis buffer | 150 mM Tris-acetate pH 7.9 at 25°C, 50 mM potassium acetate, 5 mM MgCl2, 0.5 mM EDTA, 10 μM ZnSO4, 2 mM β-mercaptoethanol, 0.3 g/ml lysozyme, 1X protease inhibitors |
| 100X protease inhibitors | 8.5 mg/ml PMSF, 33 mg/ml benzamidine, 0.03 mg/ml leupeptin, 0.14 mg/ml pepstatin, 0.2 mg/ml chymostatin in ethanol |
| A-200 | 20 mM Tris-HCl pH 7.9 at 25°C, 10 mM MgCl2, 200 mM KCl, 1.4 mM βME, 0.1 mM PMSF |
| A-1000 | 20 mM Tris-HCl pH 7.9 at 25°C, 10 mM MgCl2, 1000 mM KCl, 1.4 mM βME, 0.1 mM PMSF |
| TGEB | 40 mM Tris-HCl pH 8.0 at 25°C, 0.5 mM EDTA, 5% glycerol, 0.1 mM PMSF, 1.4 mM βME |
| Storage buffer | 40 mM Tris-HCl pH 8.0 at 25°C, 360 mM KCl, 20 μM ZnSO4, 2X protease inhibitors, 2 mM DTT |
| Transcription buffer (TB40) | 20 mM Tris-HCl pH 7.9 at 25°C, 40 mM KCl, 5 mM MgCl2 |
| Chase buffer 10x | 1 mM GTP, 1 mM ATP, 1 mM CTP, 1 mM UTP, 0.1 μCi/μl [0.3 μM] α-[P]32 GTP |
| Gel loading buffer | 10 M urea, 0.025% bromophenol blue, 0.025% xylene cyanol, 100 mM EDTA |
Four fractions containing MtbRNAP subunits were pooled and diluted with buffer TGEB to 50 ml and loaded to a 1 ml Mono Q 5/5 column (GE Healthcare) equilibrated with buffer TGEB at a 1 ml/min flow rate. The Mono Q column was washed with 15 ml TGEB supplemented with 200 mM NaCl and 15 ml TGEB with 300 mM NaCl. Then a 22 ml linear gradient from 300 to 370 mM NaCl, followed by an 18 ml gradient from 370 to 490 mM NaCl, and a 20 ml gradient from 490 to 1000 mM NaCl were consecutively applied. The eluate was collected in 1 – 1.3 ml fractions.
The fractions containing the active MtbRNAP holoenzyme were pooled and concentrated in a 15 ml Amicon 3 kDa MWCO co (Millipore) at 3,000 g for 20–40 min at 4°C. The retentate solution was diluted two-fold with a 2X storage buffer, concentrated again, and mixed with an equal volume of sterile glycerol. The sample was divided in 100 μl and 10 μl aliquots that were flash frozen by immersion in liquid nitrogen, and stored at −80°C.
Expression and purification of CarD from M. tuberculosis
Two one liter cultures of E. coli carrying pET28a-His6-CarD plasmid were grown in LB medium supplemented with 50 μg/ml kanamycin until OD600 of 0.4–0.6 was reached, and were then induced with 0.3 mM IPTG at 25°C for 3 hours. The bacterial pellet was lysed in 50 mM Tris-HCl, pH 8.0, 250 mM NaCl, 1 mM DTT and the clarified soluble fraction was loaded onto a 5 ml HisTrap™ pre-packed column (GE Healthcare) previously equilibrated with the same buffer. The column was washed with 40mM imidazole in the same buffer and the protein elution was performed with 50 mM Tris-HCl, pH 7.6, 150 mM NaCl containing 500 mM imidazole. The eluted protein was additionally purified on a Superdex 200 10/300 GL column (GE Healthcare). The main peak that contained the CarD dimer was collected and dialyzed against lysis buffer containing 50% of glycerol.
Protein analyses
The cells lysates, chromatographic fractions, and purified protein preparations were analyzed in NuPAGE 4–20% SDS protein gels resolved in MES buffer (Invitrogen). The gels were stained with Simply Blue Safe Stain (Invitrogen). Protein concentration was determined with Bradford reagent (Bio-Rad) using BSA as a standard.
Formation and walking of the TECs
The TEC assembly [20,22,23] was initiated by annealing of 15 μM each of 5′-fluorescein-labeled RNA to a template DNA oligonucleotide to form an RNA-DNA hybrid. 0.03–0.3 μM MtbRNAP was added to 0.09 – 3 μM of the RNA-DNA hybrid and incubated in TB40 at 25°C for 10 min. In most experiments a DNA oligonucleotide complementary to the template DNA strand was added to this pre-formed single-strand DNA complex at a 2-fold molar excess over the RNA-DNA hybrid, and incubated for 10 min. The TECs stalled downstream from the assembly site were obtained by addition of a 20 μM NTP subset containing only three of the four NTPs for 10 min. RNA elongation to form the run-off product was re-started from the stall site by adding all four NTPs. Transcription was stopped by addition of the EDTA- and urea-containing gel loading buffer. The RNA was denatured by heating at 95°C for 2 min. The transcription products were resolved in a 20% PAGE (19:1 acrylamide:bis-acrylamide) in the presence of 7 M urea and 1x TBE at constant 50 W for 50–60 min. Gels were scanned with the 480 nm (blue) laser, and the RNA bands were detected at 526 nm emission using Typhoon 9400 Trio scanner (GE Healthcare).
Blue native PAGE analysis [24] of the TECs
The native gel was prepared in a Mini-PROTEAN tetra cell casting module (BioRad) using glass plates separated by 1.5 mm spacers. The casting module was filled to 80% capacity with a 13% polyacrylamide native gel solution (50 mM Tris-HCl, pH 7.0 at 4°C, 13% acrylamide:bisacrylamide 32:1, 20% glycerol, 0.03% ammonium persulfate and 0.03% TEMED), and filled with the 4% polyacrylamide stacking gel solution (50 mM Bis-Tris-HCl, pH 7.0 at 4°C, 4% acrylamide:bisacrylamide 32:1, 0.06% ammonium persulfate and 0.06% TEMED). After polymerization at 25°C, the gels were stored at 4°C. The run was performed at 4°C in the cold room. During the run set up, the bottom tank was filled with the anode buffer (50 mM Bis-Tris-HCl, pH 7.0 at 4°C), and the gel wells were washed with the cathode buffer A (50 mM Tricine, 15 mM Bis-Tris-HCl, pH 7.0 at 4°C). The internal tank was carefully filled with the cathode buffer B (0.02% Coomassie Blue G-250 in the cathode buffer A). The samples were diluted 1:1 with 50 mM Bis-Tris-HCl, pH 7.0 at 4°C, 0.5 mM EDTA, 5% Coomassie Blue G-250, and loaded to the gel wells. Electrophoresis was performed at 30 V constant for 30 minutes. The voltage was then increased to 80 V, and kept constant for 3 h or until the free Coomassie Blue dye reached half of the gel’s length. After that the cathode buffer B in the internal tank was changed to the cathode buffer A, and electrophoresis was resumed for 3 more hours, or until the dye reached the bottom of the gel. After visual inspection and white-light digital scanning, the RNA in the gel was detected at 526 nm emission using Typhoon 9400 Trio scanner (GE Healthcare) as described above.
Electromobility shift assay (EMSA)
200 nM MtbRNAP was incubated with 2 μM hexahistidine-tagged CarD at 25°C for 15 min. A 150 bp DNA fragment containing the M. tuberculosis ribosomal promoter P1Pcl1 [21] was labeled at both 5′ ends with FAM by PCR amplification with /56FAM/ATCGAACGGGTATGCTGTTAG and /56FAM/TTGAGTTCTCAAACAACACGCT primers. 20 nM of this construct was added to free MtbRNAP or to the MtbRNAP-CarD complex in TB40, and incubated for 5 min at 25°C. 10, 20, 30, 40, or 50 μg/ml heparin were added followed by incubation for additional 5 min at 25°C. At the end of the incubation, loading buffer (50% glycerol, 0.5% bromophenol blue) was added to the samples, and the DNA-protein complexes were immediately loaded on to a 4% polyacrylamide gel and resolved at 80V for 100 min in TBE buffer containing 1% glycerol.
Promoter-dependent initiation on P1Pcl1 promoter
200 nM MtbRNAP and 200 nM DNA template containing the M. tuberculosis ribosomal promoter P1Pcl1 in TB were diluted in TB40 and incubated with 2 μM CarD or with CarD storage buffer for 5 min at 37°C. The reaction volume was 24 μl. The resulting complex was split in half and 50 μg/ml heparin (final concentration) was added to one of the two aliquots for 5 min at 37°C. 100 μM each CTP, UTP, and GTP (final concentration) was added to each of the four parallels for 2 min. The resulting TECs were immobilized by a 10 min incubation with 15 μl 50% Ni-NTA agarose suspension pre-washed with TB40 (no heparin parallels) or TB40 containing 50 μg/ml heparin at 25°C. The TECs were washed four times with 1 ml TB40, and the transcript was labeled by incubation with 0.3 μM α-[32P] ATP (3000 μCi/mmol) for 5 min at 25°C. The labeled TECs were washed four times with 1 ml TB40, and chased with 1 mM of all four NTPs for 1 min at 25°C. The reaction was stopped by addition of an equal volume of a 2X gel loading buffer (10 M urea, 0.025% bromophenol blue, 0.025% xylene cyanol, 100 mM EDTA). The samples were resolved in 20% PAGE (19:1 acrylamide: bisacrylamide) containing 7 M urea and 1X TBE at 50 W for 50 min.
Promoter-dependent initiation on λ PR promoter
A DNA fragment containing bacteriophage λ pR and pRM promoters was amplified from plasmid pPIA2-6 [25] with the following primers: 5′-GCGGATACATATTTGAATGT-3′ and 5′-ATTTCAACAAAACGCTGGTCCTTTCCGG CAATCAGGCG-3′. The resulting 5507 bp DNA template was used for in vitro transcription assay. Approximately 4 nM of DNA was incubated with 100 nM MtbRNAP at 25°C for 15 min. Subsequently, TB40 with or without 50 μg/ml heparin was added, and the mixture was incubated for 1 min. Transcription elongation was started by addition of 100 μM each GTP, ATP, CTP, and UTP spiked with 0.1 μCi/μl [0.3 μM] α-[32P] GTP at 25°C. The reaction was stopped by addition of an equal volume of a 2X gel loading buffer (10 M urea, 0.025% bromophenol blue, 0.025% xylene cyanol, 100 mM EDTA). The samples were run in 4% PAGE (29:1 acrylamide: bisacrylamide) containing 7 M urea and 1X TBE starting at 1300 V and kept at constant power of 18 W.
RESULTS AND DISCUSSION
Ni2+ affinity chromatography
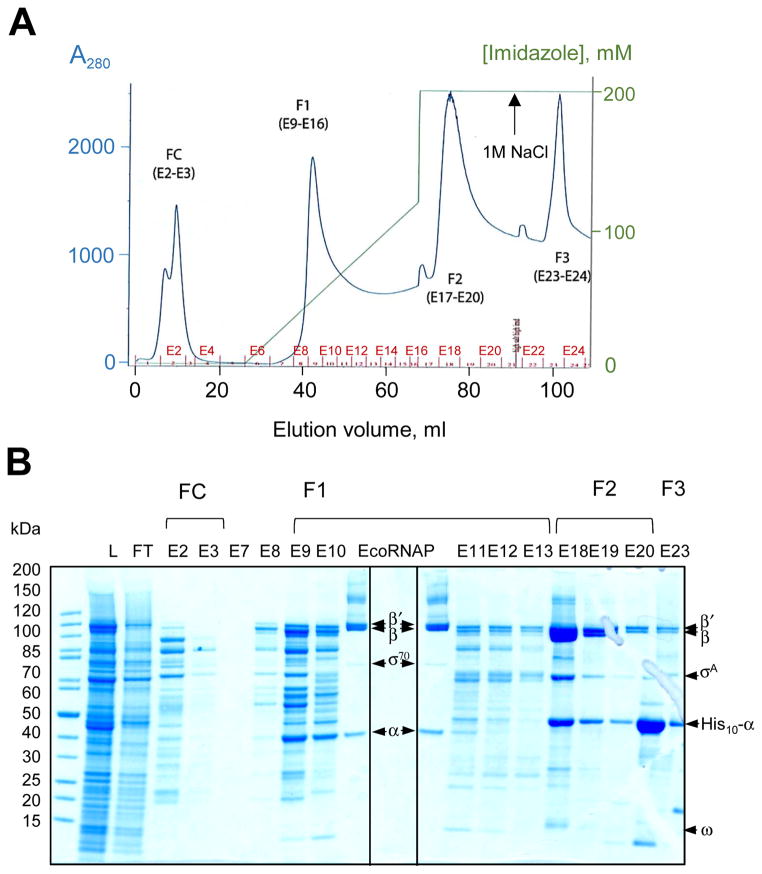
Recombinant MtbRNAP was separated from the endogenous EcoRNAP by Ni2+ affinity chromatography using the N-terminal decahistidine tag on the Mtb α subunits. A combination of stepwise salt increments and a continuous imidazole gradient eliminated most of E. coli lysate proteins through this step (Fig. 1). The protein composition of the chromatography fractions indicated that MtbRNAP was eluted with 200 mM imidazole (Fig. 1B, fractions E18–E20), in agreement with the previous report [1]. The amounts of β and β′ subunits were similar, and their ratio was comparable to the stoichiometric ratio of β and β′ in the E. coli holoenzyme run along as a control (In Fig. 1B, compare fraction E19 with the lanes showing EcoRNAP). Increase of NaCl concentration in the elution buffer containing 200 mM imidazole resulted in the appearance of a new peak containing a large excess of α subunit (Fig. 1A, F3 and Fig. 1B, fraction E23).
Figure 1. Co-expression of MtbRNAP Subunits in E. coli and Ni2+ affinity chromatography.
(A) Elution profile of the E. coli whole cell lysate containing recombinant MtbRNAP subunits from Ni2+ affinity column with imidazole gradient and a stepwise increase in [NaCl]. The fraction numbers are shown on top of the horizontal axis in red. (B) SDS-PAGE analysis of the eluted fractions.
Mono Q column
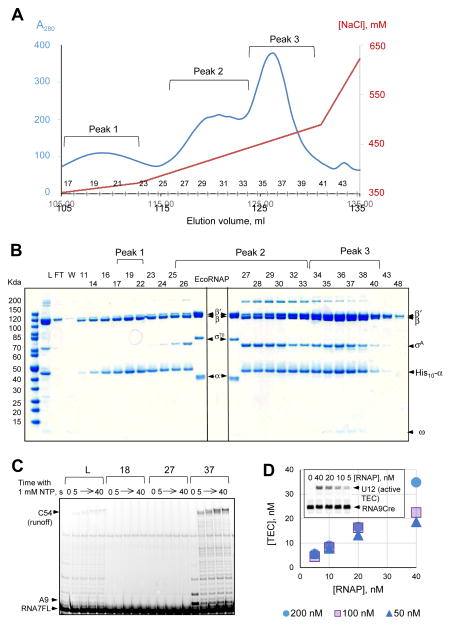
In order to obtain highly pure MtbRNAP holoenzyme, and to discriminate against the subunit assemblies with the stoichiometries distinct from the correct α2ββ′ωσA, we used a procedure similar to one developed by Hager and co-authors [26] for the EcoRNAP holoenzyme purification. The pooled E18–E20 fractions (10.5 mg) were diluted and loaded to the Mono Q column. The proteins were eluted by a NaCl gradient (Fig. 2A), and the protein content of the eluted fractions was analyzed by SDS PAGE (Fig. 2B). Notably, the material eluted at [NaCl] below 390 mM (fractions 11–27) contained significant excess of β subunit over the β′ subunit. Application of the [NaCl] gradient from 390 to 480 mM eluted two distinct peaks (Figure 2A and B, fractions 28–34 and 35–40, respectively). Only the last protein-containing peak showed the correct subunit stoichiometry and contained a detectable amount of ω subunit (Fig. 2B, peak 3). These fractions (35–40) were pooled, concentrated, and diluted with a 2X storage buffer as described in Materials and Methods. The protein concentration in the final preparation was 1 mg/ml (~ 2 μM), and the total MtbRNAP yield was 2.5 mg. The presence of the subunit sub-assemblies and their separation from the holoenzyme on the MonoQ column was reproducible in different purification runs.
Figure 2. MonoQ Purification of the MtbRNAP.
(A) Elution profile of Ni2+-affinity purified MtbRNAP from MonoQ column with a NaCl gradient. The odd fraction numbers are indicated on top of the horizontal axis. (B) SDS-PAGE analysis of the fractions eluted from MonoQ column. L (for Loading) is the material eluted from the Ni2+ affinity column (F2); FT and Wash are the fractions collected before application of the salt gradient; Peak1, Peak2, and Peak3 corresponding to the peaks in the A280 profile in (A); E. coli RNAP holoenzyme was used as reference. (C) Evaluation of the transcriptional activity of the fractions from MonoQ column. 12 pmol of RNA7FL-TDS76 hybrid (Table 2) in 8 μl TB40 containing 20 μM each ATP and GTP were used for the TEC assembly with 3 μl of the F2 fraction of the imidazole eluate (L) or fractions 17, 27, and 37 from MonoQ; 15 pmol NDS79 were added to each reaction. The TEC was chased with 1 mM NTPs for up to 40s. (D) Quantification of MtbRNAP activity. The TECs were assembled with 0.5, 1, or 2 pmol of RNA9Cre-TDS57 hybrid, 0.4, 0.2, 0.1, and 0.05 pmol MtbRNAP, and a 2-fold molar excess of NDS57 in 10 μl TB. The fraction of the active TEC was quantified as the % of the RNA extended after 5 min incubation with 10 μM each GTP + UTP and the [TEC] was quantified based on the total [RNA] and plotted vs [RNAP]. Each series represents the results obtained with a certain concentration of the RNA-DNA hybrid. The inset shows a gel for the “100 nM” (1 pmol of RNA9Cre-TDS57 hybrid) series.
Identification of fractions containing the active MtbRNAP
A convenient promoter-independent activity test was adapted from [22] to evaluate RNAP activity in the representative fractions eluted from Mono Q column. The material loaded on the column, along with the fractions from 18, 27, and 37, were used for the TEC assembly. The fractions assayed for RNAP activity from peaks 1 and 2 were chosen from the left shoulder of each peak to ensure that they do not contain any material from the subsequent peaks. Fraction 37 from peak 3 was chosen for the activity analysis because it contained an apparently stoichiometric amount of ω subunit (Fig. 2B). The assembled TECs were chased with all four NTPs, and the extension of the RNA, as well as the accumulation of the runoff product were used as indicators of the presence of the catalytically active RNAP. Fig. 2C illustrates that only the imidazole eluate and a representative fraction from peak 3 contained any detectable RNA polymerase activity. We conclude that the other peaks resolved by the anion exchange column correspond to inactive sub-assemblies of the recombinant MtbRNAP subunits. This conclusion is consistent with the wrong stoichiometry of α, β and β′ subunits and the absence of ω subunit observed in peaks 1 and 2.
MtbRNAP activity quantification
To obtain a quantitative estimate of the purified MtbRNAP activity, we looked for the conditions, at which the amount of the assembled TEC is proportional to the amount of MtbRNAP added. The plot in Figure 2D shows that while the amount of the TEC was almost precisely proportional to the amount of RNAP added when the assembly was performed with 200 nM RNA-DNA hybrid, the amount of the resulting TEC plateaued when 100 nM and 50 nM concentrations of the hybrid were used. It appears that the RNA-DNA hybrid should be added in at least 10-fold molar excess for the accurate MtbRNAP activity measurement. A similar result was obtained for EcoRNAP (Supplemental Fig. 1). Averaging of the six data points obtained with a 10-fold or higher molar excess of the RNA-DNA hybrid, nearly all (95% with a 6% standard error) purified MtbRNAP molecules formed an active TEC (Fig. 2D). We conclude that the removal of the inactive sub-assemblies by chromatography on MonoQ column is essential for obtaining highly active and homogeneous holoenzyme. Next, we subjected this preparation of MtbRNAP to additional functional tests.
Stability of the TECs
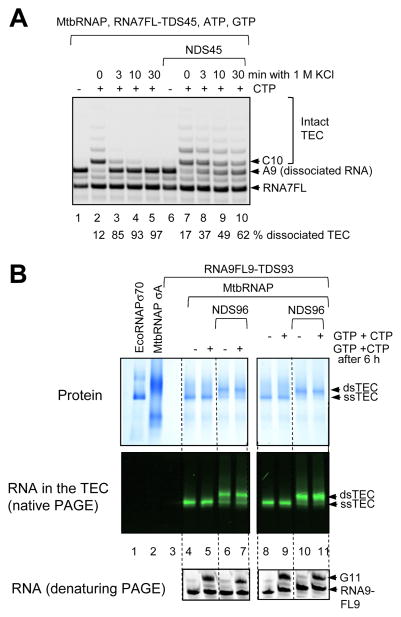
EcoRNAP requires the 9 bp RNA-DNA hybrid and at least 10 bp dsDNA downstream its active site to form stable TECs [20]. To test if the downstream dsDNA is required for MtbRNAP TEC, the TECs were assembled on a short template with only 13 nt downstream from the RNA 3′ end to rule out formation of a hairpin by the downstream ssDNA (Fig. 3A and Table 2, TDS45 and RNA7FL extended to RNA A9 by incorporation of GMP and AMP). The TEC formed in the absence of the non-template NDS45 DNA strand dissociated after exposure to 1 M KCl (Fig. 3A, compare lane 2 and lanes 3 – 5), while about ½ of the dsTEC retained catalytic activity even after 30 min incubation with 1 M KCl (lane 10). Therefore, the requirement for the downstream dsDNA is conserved between MtbRNAP and EcoRNAP.
Figure 3. Stability of MtbRNAP TECs.
(A) Stability of MtbRNAP TECs in high salt. The TEC was assembled with 20 pmol MtbRNAP and 45 pmol of RNA7FL-TDS45 hybrid in 40 μl TB40 containing 10 μM each ATP and GTP. 45 pmol NDS45 was added to one half of this ssTEC to form the dsTEC. The TECs were mixed with ½ volume of 3M KCl, and 1 μl aliquots were removed after 3, 10, and 30 min of incubation (lanes 3 – 5 and 8 – 10), mixed with 4 μl of TB40 containing 100 μM CTP, and elongation was stopped by addition of 10 μl of gel loading buffer after 10 s incubation. The control TECs were mixed with ½ volume of water, and 1 μl aliquots were mixed with TB40 with (lanes 2 and 7) or without CTP (lanes 1 and 6), and the gel loading buffer was added after 10 s. The % of dissociated TEC was quantified as fraction of A9 RNA that was not extended after incubation with CTP. (B) Stability of the TECs on the template designed for the single-molecule assays. The TECs were assembled with 6 pmol RNA9-FL9 – TDS93 hybrid and 4 pmol MtbRNAP in 60 μl TB40 to form the ssTEC. 15 pmol NDS96 was added to one half of the ssTEC to form the dsTEC. Each TEC was divided in 4 aliquots, and 20 μM each GTP and CTP were added before or after 6 h incubation on ice. Coomasie Blue dye was added to the MtbRNAP, EcoRNAP, RNA-DNA hybrid and to one half of the TEC aliquots, and the material was analyzed by PAGE in non-denaturing (native) conditions. The Coomassie-stained proteins are shown in the Protein panel. The RNA in the same gel is shown in the middle (RNA in the TEC) panel. 2-nt RNA extension by ssTEC and dsTEC was analyzed in the remaining TEC by PAGE in denaturing conditions. The positions of the ssTEC, dsTEC in the native gel, and of the RNA primer RNA9-FL9 and elongated product G11 in the denaturing PAGE are indicated with arrows.
Table 2.
Oligonucleotides used for TEC assembly
| Name | Sequence, 5′ to 3′ |
|---|---|
| RNA9-FL9* | /56FAM/UUACUUCGACGCCCGA |
| RNA7FL** | /56FAM/UUCAUUCCCGAGAGG |
| RNA9Cre | /56FAM/ CCCCCCCCGUCAUGAAC |
| TDS57 | TGTTTCACTATCCAGGTTACGGATACAGTTCATGACAATATTTACATTGG TCCAGCC |
| NDS57 | GGCTGGACCAATGTAAATATTGTCATGAACTGTATCCGTAACCTGGATAG TGAAAC |
| NDS79** | CCTATAGGATACTTAAGCCATCGAGAGGGACACGGCGAATAGCCATCCC AATCCACACGTCCAACGGGGCAAACCGTA |
| TDS76** | GGTTTGCCCCGTTGGACGTGTGGATTGGGATGGCTATTCGCCGTGTCCCT CTCGATGGCTGTAAGTATCCTATAGG |
| NDS45 | CCTATAGGATACTTAAGCCATCGAGAGGGACACGGCGAATAGCCA |
| TDS45 | ATGGCTATTCGCCGTGTCCCTCTCGATGGCTGTAAGTATCCTATAGG |
| TDS93* | AGCATAATCCTGAATATGGCAAGTTACATAGATAAGTTGGTCGGTTGGG GTTTGTGTGGCTTCGTCGGGCGTCTTCTACATACTACTCCTACC |
| NDS96* | GGTAGGAGTAGTATGTAGAAGACGCCCGACGAAGCCACACAAACCCCA ACCGACCAACTTATCTATGTAACTTGCCATATTCAGGATTATGCTCAT |
To establish whether MtbRNAP TECs retain their integrity upon prolonged storage in TB40, we assembled the TECs using the RNA-DNA scaffold designed for the single-molecule analyses of transcription elongation [23]. The assembly was performed with just an RNA-DNA hybrid (ssTEC) or with the RNA-DNA hybrid followed by the addition of the non-template DNA strand (dsTEC) (Fig. 3B). The TECs were analyzed by native blue PAGE [24]. The protein component and the RNA component were detected in the same gel by Coomassie staining and by fluorescence, respectively, (Fig. 3B, protein and RNA in the TEC panels). The electrophoretic mobility of the TECs is distinct from the mobility of the free MtbRNAP holoenzyme. Note that the latter migrates as two distinct bands. The TECs have an intermediate mobility, suggesting that the slower band in the free MtbRNAP holoenzyme corresponds to a dimer, which dissociates upon formation of the TEC. Fig. 3B demonstrates that the TECs formed both in the absence and in the presence of the non-template DNA strand retain their ability to elongate the RNA primer even after 6 h incubation, suggesting that in the low salt conditions stability of the MtbRNAP TEC on this scaffold is not compromised by omission of the non-template DNA. The incorporation of the non-template DNA strand is efficient, as evidenced by the mobility shift that the majority of the TEC undergoes in the presence of the non-template DNA strand. In this scaffold, 9 nt from the 3′ end of RNA primer is complementary to the template DNA strand. The RNA in the TEC can be extended to 2-nt by enzymatic incorporation of GMP and CMP. In agreement with the non-denaturing gel results, the identical fraction of RNA extended before or after the 6 h incubation of the ssTEC and dsTEC indicates that both variants of the TEC are resistant to dissociation.
Transcription of the single-strand and the double-strand DNA templates and sequence-specific pausing pattern
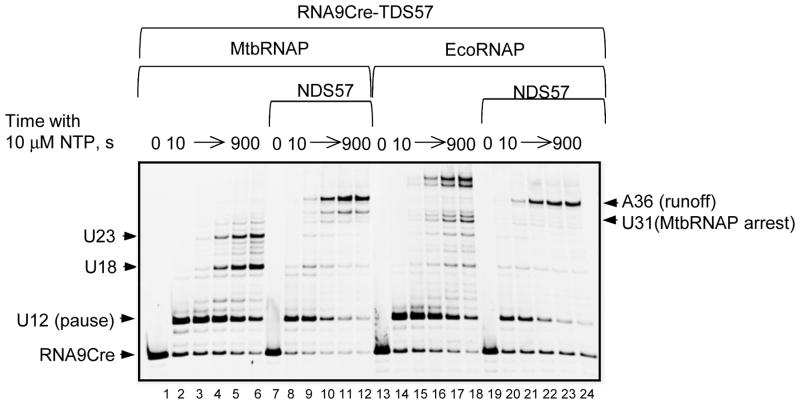
Direct comparison of the transcript elongation patterns within the same sequence context on the single-strand (ss) and the double-strand (ds) DNA, reveal only the minor differences between MtbRNAP and EcoRNAP (Fig. 4). On the dsDNA template, both RNAPs undergo strong sequence-specific pausing at the same position U12, within the sequence context C−10AUGAACUGUA, which differs from the previously reported consensus pause sequence for EcoRNAP [27–29] only by the CMP, not GMP, encoded 10 nt upstream from the pause site. MtbRNAP is more prone to transcription arrest 5 bp from the end of the dsDNA template, at U31 (compare lanes 10–12 and 22–24 in Fig. 4).
Figure 4. Transcription pattern on ssDNA and dsDNA templates.
The TECs were formed as in Fig. 2D with 0.2 pmol MtbRNAP or EcoRNAP and 2 pmol of RNA9Cre-TDS57 hybrid. 2 pmol of NDS57 was added to one half of each TEC. The TECs were purified from the excess of RNA and DNA on centrifugal concentrators with 100 kDa cutoff, and chased with 10 μM NTP at 37 °C. The positions of the RNA primer, main pause and arrest sites on the dsDNA template, and the runoff transcript are indicated at left.
It was unexpected to observe that EcoRNAP transcribes to the end of the ss template and adds a few more NMPs beyond the end of the template (Fig. 4, lanes 16–18), which is in disagreement with the previously reported low processivity of EcoRNAP in the absence of the non-template DNA strand [30]. It is likely that the non-complementary RNA tail present in the nucleic acid scaffolds used in this work promotes transcript displacement from the RNA-DNA hybrid thus preventing the proposed clash between the EcoRNAP lid domain and the transcript 5′ end [30]. In contrast, MtbRNAP has a lower processivity on the ssDNA than EcoRNAP, and fails to reach the end of the template. MtbRNAP undergoes transcription arrest or dissociation at positions U18 and U23 (Fig. 4, lanes 4–6).
Elongation stalling and escape from the stall
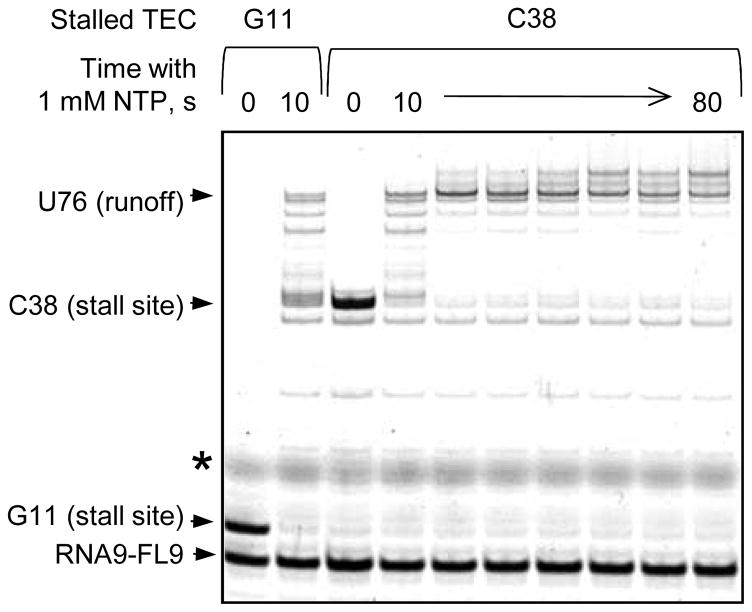
The setup of transcription elongation assays previously developed and optimized for the single-molecule experiments with S. cerevisiae RNAP II [23] requires the assembly of the TEC on the RNA9FL9-TDS93-NDS96 scaffold, and walking of the RNAP away from the assembly site by addition of ATP, CTP, and GTP. Such complex is captured in optical tweezers, and the elongation is resumed by adding all four NTPs. The success of this experimental strategy depends, among other factors, on the efficiency of RNAP escape from the stalled state induced by nucleotide starvation. Figure 5 demonstrates that MtbRNAP assembled on the RNA9FL9-TDS93-NDS96 scaffold efficiently extends the RNA in a template-specific manner. Upon incubation with GTP and CTP, the RNA primer is elongated 2 nt, and MtbRNAP stops because the next cognate ATP is lacking. Upon incubation with ATP, GTP, and CTP, MtbRNAP adds 29 nt to the transcript and is stalled because the next cognate UTP is missing. Importantly, the stalled TECs efficiently resume elongation when 1 mM each of all four NTPs are added to the reaction mix. Apparently, MtbRNAP does not undergo transcription arrest at the stall sites tested and does not depend on any external factors to resume transcript elongation from them.
Figure 5. MtbRNAP escape from the stall on the template designed for the single molecule applications.
MtbRNAP TEC was formed by incubation of 6 pmol RNA9-FLTDS93 hybrid with 4 pmol MtbRNAP and 15 pmol NDS96 in 55 μl TB. The TEC was walked 2 nt or 29 nt downstream by incubation with 20 μM each GTP and CTP to obtain the stalled TEC G11 or with 20 μM each ATP, GTP and CTP to obtain the stalled TEC C38. Elongation was resumed by addition of a 1 mM NTPs for the indicated times. The positions of the RNA primer, stall sites, and the runoff transcript template are indicated at left.
Transcription factor-dependent formation of the initiation complex by MtbRNAP holoenzyme
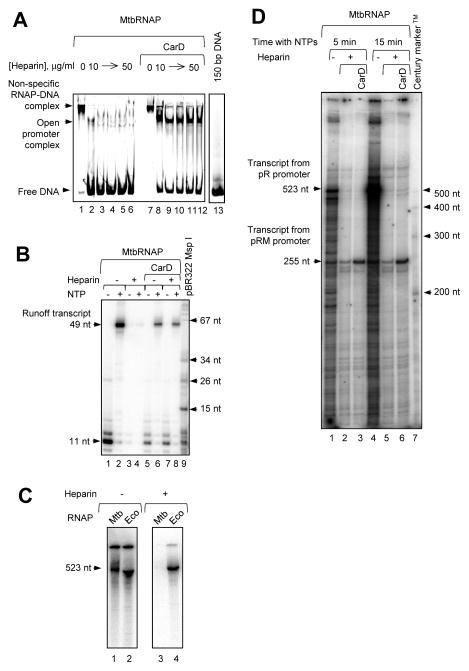
Formation of the initiation complexes at M. tuberculosis ribosomal promoters P1Pcl1 [21] was analyzed by EMSA. The 5 min pre-incubation of promoter DNA with MtbRNAP at 25°C was sufficient for an efficient formation of the RNAP-DNA complex, which is in agreement with the previously reported 24 min half-life of the open complex [31,32]. In the absence of heparin, all the input promoter DNA interacts with MtbRNAP, forming the complexes with very slow mobility (Fig. 6A, lane 1). Later, in order to observe formation of the transcription-competent open promoter complexes, different concentrations of heparin, a negative polyelectrolite able to bind free DNA and actively dissociate non-specific RNAP-DNA complexes and closed promoter complexes [33] were added. Even very low amounts of heparin disrupt the interaction of MtbRNAP with the DNA (Fig. 6A, lanes 2 – 6), suggesting that the DNA shift is caused by weak non-specific binding of MtbRNAP to the DNA, rather than by formation of the stable initiation complex.
Figure 6. Stabilization of MtbRNAP promoter binding by transcription initiation factor CarD.
(A) Initiation complex formation on the Mtb ribosomal promoter. The MtbRNAP alone or MtbRNAP pre-incubated with CarD was added to the DNA fragment bearing Mtb ribosomal promoters P1Pcl1. The DNA-protein complexes were challenged by heparin. The positions of the bands corresponding to the free DNA, heparin-sensitive (non-specific) and heparin-resistant (open promoter) complexes are indicated at left. (B) Transcription initiation on P1Pcl1 promoter and elongation. MtbRNAP with or without CarD was pre-incubated with the same template as in (A), and challenged with 50 μg/ml heparin. Transcription was initiated by addition of CTP, GTP, and UTP, and the resulting TECs were immobilized on Ni-NTA, purified, labeled with α-[32P] ATP and chased with 1 mM of all four NTPs for 1 min. (C) Transcription initiation on a template containing λ pR promoter. MtbRNAP (lanes 1and 3, Mtb) and EcoRNAP holoenzymes (lanes 2 and 4, Eco) were incubated with an 863 bp DNA fragment containing the λ pR and pRM promoters. The complexes were incubated with buffer or challenged with 50 μg/ml heparin before chase in the presence of α-[32P] GTP. The product corresponding to a runoff transcript initiated at λ pR promoter is indicated. (D) CarD effect on transcription by MtbRNAP. The experiment was performed as in (C) with MtbRNAP, which was used alone or pre-incubated with a 20-fold molar excess of CarD. The two main transcription products are indicated at left, and the sizes of the reference bands are shown at right.
To increase the specificity and stability of MtbRNAP interaction with the promoter, we purified Mtb transcription factor CarD [34] (Supplemental Fig. 2A and B). The 2 L culture yielded approximately 8 mg of hexahistidine-tagged CarD, which was further purified by a size exclusion column. CarD elution pattern suggested that it forms a dimer (Supplemental Fig. 2C and D). Dimerization of MtbCarD is consistent with the previously reported dimerization of CarD from other mycobacteria [35,36]. When MtbRNAP is pre-incubated with the purified CarD, a heparin-resistant promoter open complex is formed (Fig. 6A, lanes lanes 8 – 12). MtbRNAP was pre-incubated with CarD before the addition of the promoter-containing DNA fragment, because, apparently, this factor interacts exclusively with MtbRNAP holoenzyme, and not with the DNA [31]. The band corresponding to the heparin-resistant open promoter complexes migrates faster than the non-specific heparin-sensitive MtbRNAP-DNA complexes (compare lane 7 in Fig. 6A with lanes 8 – 12).
Next, the CarD effect on transcription initiation was tested in a promoter-specific transcription assay on the same template. The initiation complexes were formed on the same DNA template that was used for the EMSA with or without CarD, challenged with heparin, and then purified CTP, GTP, and UTP were added to promote formation of the TEC with the 9-nt transcript 5′-GUUGCCCCG-3′ stalled because the next cognate ATP was lacking. The TEC was immobilized on Ni-NTA agarose beads, the NTPs were removed by washing the beads with TB40, and the RNA was labeled by incubation with α-[32P] ATP producing the expected 11 nt RNA product (Fig. 6B, lanes 1, 5, and 7). Upon addition of all four NTPs the RNA was extended to the runoff product of the expected length (lanes 2, 6, and 8). Consistently with EMSA results, heparin inhibited transcription initiation in the absence of CarD (lanes 3 and 4).
Despite the predominance of the non-specific RNAP-DNA interactions detected by EMSA in the absence of heparin, MtbRNAP holoenzyme initiated at λ pR promoter with an efficiency similar to EcoRNAP holoenzyme (Fig. 6C, lanes 1 and 2). Heparin did not affect promoter-dependent transcription by EcoRNAP, but completely inhibited MtbRNAP (lanes 3 and 4). A longer exposure of the gels revealed that in the presence of heparin a less prominent, 255-nt transcript is synthesized by MtbRNAP holoenzyme in the presence of heparin (Fig. 6C, lanes 2 and 5). We interpreted this product as a transcript initiated from λ pRM promoter, which is also present in the template DNA, or as a product of premature arrest of elongation from λ pR. Notably, CarD promotes MtbRNAP-dependent transcription from λ pRM in the presence of heparin (Fig. 6D, lanes 3 and 6), probably, by reducing the rate of transcriptional bubble collapse [34]. The latter result indicates that the stabilizing effect of CarD on the open promoter complex is not specific exclusively to Mtb promoters.
CONCLUSIONS
A detailed protocol for the expression and purification of MtbRNAP holoenzyme is presented at a small scale (~5 g of bacterial pellet), based on a strategy of co-expression from two plasmids similar to the one previously described by [1]. The plasmid pACYC Duet-AZD was designed to contain simultaneously the α, ω and σA subunits, thus allowing for the assembly of MtbRNAP holoenzyme. The purified holoenzyme formed stable, heparin-resistant pre-initiation complexes only in the presence of CarD. This transcription initiation factor was also necessary to produce promoter-dependent initiation above the detection limits. In contrast, stable ternary elongation complexes can be assembled from MtbRNAP and synthetic RNA and DNA oligonucleotides without the aid of CarD. These ternary (DNA, RNA and protein) complexes can be used in a convenient activity assay that eliminates the need of radioactively labeled nucleotides.
Supplementary Material
Highlights.
90% active M. tuberculosis RNA polymerase holoenzyme purified to homogeneity
Transcription initiation by this holoenzyme requires initiation factor CarD
Transcription elongation by MtbRNAP occurs without the aid of additional factors
Acknowledgments
This work was supported in part by a special grant contract and a travel award from CONCYTEC, Perú [FONDECYT N° 196-2013 and N° 033-2015-FONDECYT-DEC], by the National Institutes of Health [grant #R01GM032543], the U.S. Department of Energy Office of Basic Energy Sciences Nanomachine Program [contract #DE-AC02-05CH11231], and by the Intramural Research Program of the National Institutes of Health, NCI, Center for Cancer Research.
Footnotes
AUTHOR CONTRIBUTIONS
O.H.-A., D.G.G., C.J.B., M.K., L.L., and M.L.K. designed the protein expression and purification setups and the experiments, O. H.-A. and L.L. performed protein purification, O. H.-A. and M.L.K. performed the experiments, O. H.-A., D.G.G., C.J.B. and M.L.K. wrote the manuscript helped by the input from L.L and M.K.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Banerjee R, Rudra P, Prajapati RK, Sengupta S, Mukhopadhyay J. Optimization of recombinant Mycobacterium tuberculosis RNA polymerase expression and purification. Tuberc Edinb Scotl. 2014;94:397–404. doi: 10.1016/j.tube.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Czyz A, Mooney RA, Iaconi A, Landick R. Mycobacterial RNA polymerase requires a U-tract at intrinsic terminators and is aided by NusG at suboptimal terminators. mBio. 2014;5:e00931. doi: 10.1128/mBio.00931-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gill SK, Garcia GA. Rifamycin inhibition of WT and Rif-resistant Mycobacterium tuberculosis and Escherichia coli RNA polymerases in vitro. Tuberc Edinb Scotl. 2011;91:361–369. doi: 10.1016/j.tube.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Murakami KS. X-ray crystal structure of Escherichia coli RNA polymerase σ70 holoenzyme. J Biol Chem. 2013;288:9126–9134. doi: 10.1074/jbc.M112.430900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ho MX, Hudson BP, Das K, Arnold E, Ebright RH. Structures of RNA polymerase-antibiotic complexes. Curr Opin Struct Biol. 2009;19:715–723. doi: 10.1016/j.sbi.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mariani R, Maffioli SI. Bacterial RNA polymerase inhibitors: an organized overview of their structure, derivatives, biological activity and current clinical development status. Curr Med Chem. 2009;16:430–454. doi: 10.2174/092986709787315559. [DOI] [PubMed] [Google Scholar]
- 7.McClure WR, Cech CL. On the mechanism of rifampicin inhibition of RNA synthesis. J Biol Chem. 1978;253:8949–8956. [PubMed] [Google Scholar]
- 8.Aquinas M. Short-course therapy for tuberculosis. Drugs. 1982;24:118–132. doi: 10.2165/00003495-198224020-00002. [DOI] [PubMed] [Google Scholar]
- 9.Ena J, Valls V. Short-course therapy with rifampin plus isoniazid, compared with standard therapy with isoniazid, for latent tuberculosis infection: a meta-analysis. Clin Infect Dis Off Publ Infect Dis Soc Am. 2005;40:670–676. doi: 10.1086/427802. [DOI] [PubMed] [Google Scholar]
- 10.Goldstein BP. Resistance to rifampicin: a review. J Antibiot (Tokyo) 2014;67:625–630. doi: 10.1038/ja.2014.107. [DOI] [PubMed] [Google Scholar]
- 11.Svetlov V, Artsimovitch I. Purification of bacterial RNA polymerase: tools and protocols. Methods Mol Biol Clifton NJ. 2015;1276:13–29. doi: 10.1007/978-1-4939-2392-2_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kansara SG, Sukhodolets MV. Oligomerization of the E. coli core RNA polymerase: formation of (α2ββ′ω)2-DNA complexes and regulation of the oligomerization by auxiliary subunits. PloS One. 2011;6:e18990. doi: 10.1371/journal.pone.0018990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imashimizu M, Kashlev M. Unveiling translocation intermediates of RNA polymerase. Proc Natl Acad Sci U S A. 2014;111:7507–7508. doi: 10.1073/pnas.1406413111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruff EF, Drennan AC, Capp MW, Poulos MA, Artsimovitch I, Record MT. E. coli RNA Polymerase Determinants of Open Complex Lifetime and Structure. J Mol Biol. 2015;427:2435–2450. doi: 10.1016/j.jmb.2015.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Artsimovitch I, Svetlov V, Anthony L, Burgess RR, Landick R. RNA polymerases from Bacillus subtilis and Escherichia coli differ in recognition of regulatory signals in vitro. J Bacteriol. 2000;182:6027–6035. doi: 10.1128/jb.182.21.6027-6035.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harshey RM, Ramakrishnan T. Purification and properties of DNA-dependent RNA polymerase from Mycobacterium tuberculosis H37RV. Biochim Biophys Acta. 1976;432:49–59. doi: 10.1016/0005-2787(76)90040-x. [DOI] [PubMed] [Google Scholar]
- 17.China A, Nagaraja V. Purification of RNA polymerase from mycobacteria for optimized promoter-polymerase interactions. Protein Expr Purif. 2010;69:235–242. doi: 10.1016/j.pep.2009.09.022. [DOI] [PubMed] [Google Scholar]
- 18.Jacques JF, Rodrigue S, Brzezinski R, Gaudreau L. A recombinant Mycobacterium tuberculosis in vitro transcription system. FEMS Microbiol Lett. 2006;255:140–147. doi: 10.1111/j.1574-6968.2005.00071.x. [DOI] [PubMed] [Google Scholar]
- 19.Hu Y, Morichaud Z, Perumal AS, Roquet-Baneres F, Brodolin K. Mycobacterium RbpA cooperates with the stress-response σB subunit of RNA polymerase in promoter DNA unwinding. Nucleic Acids Res. 2014;42:10399–10408. doi: 10.1093/nar/gku742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sidorenkov I, Komissarova N, Kashlev M. Crucial role of the RNA:DNA hybrid in the processivity of transcription. Mol Cell. 1998;2:55–64. doi: 10.1016/s1097-2765(00)80113-6. [DOI] [PubMed] [Google Scholar]
- 21.Hu Y, Morichaud Z, Chen S, Leonetti JP, Brodolin K. Mycobacterium tuberculosis RbpA protein is a new type of transcriptional activator that stabilizes the σ A-containing RNA polymerase holoenzyme. Nucleic Acids Res. 2012;40:6547–6557. doi: 10.1093/nar/gks346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kireeva ML, Domecq C, Coulombe B, Burton ZF, Kashlev M. Interaction of RNA polymerase II fork loop 2 with downstream non-template DNA regulates transcription elongation. J Biol Chem. 2011;286:30898–30910. doi: 10.1074/jbc.M111.260844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hodges C, Bintu L, Lubkowska L, Kashlev M, Bustamante C. Nucleosomal fluctuations govern the transcription dynamics of RNA polymerase II. Science. 2009;325:626–628. doi: 10.1126/science.1172926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fiala GJ, Schamel WWA, Blumenthal B. Blue native polyacrylamide gel electrophoresis (BN-PAGE) for analysis of multiprotein complexes from cellular lysates. J Vis Exp JoVE. 2011 doi: 10.3791/2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davenport RJ, Wuite GJ, Landick R, Bustamante C. Single-molecule study of transcriptional pausing and arrest by E. coli RNA polymerase. Science. 2000;287:2497–2500. doi: 10.1126/science.287.5462.2497. [DOI] [PubMed] [Google Scholar]
- 26.Hager DA, Jin DJ, Burgess RR. Use of Mono Q high-resolution ion-exchange chromatography to obtain highly pure and active Escherichia coli RNA polymerase. Biochemistry (Mosc) 1990;29:7890–7894. doi: 10.1021/bi00486a016. [DOI] [PubMed] [Google Scholar]
- 27.Imashimizu M, Takahashi H, Oshima T, McIntosh C, Bubunenko M, Court DL, Kashlev M. Visualizing translocation dynamics and nascent transcript errors in paused RNA polymerases in vivo. Genome Biol. 2015;16:98. doi: 10.1186/s13059-015-0666-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larson MH, Mooney RA, Peters JM, Windgassen T, Nayak D, Gross CA, Block SM, Greenleaf WJ, Landick R, Weissman JS. A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science. 2014;344:1042–1047. doi: 10.1126/science.1251871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vvedenskaya IO, Vahedian-Movahed H, Bird JG, Knoblauch JG, Goldman SR, Zhang Y, Ebright RH, Nickels BE. Interactions between RNA polymerase and the “core recognition element” counteract pausing. Science. 2014;344:1285–1289. doi: 10.1126/science.1253458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naryshkina T, Kuznedelov K, Severinov K. The role of the largest RNA polymerase subunit lid element in preventing the formation of extended RNA-DNA hybrid. J Mol Biol. 2006;361:634–643. doi: 10.1016/j.jmb.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 31.Rammohan J, Ruiz Manzano A, Garner AL, Stallings CL, Galburt EA. CarD stabilizes mycobacterial open complexes via a two-tiered kinetic mechanism. Nucleic Acids Res. 2015;43:3272–3285. doi: 10.1093/nar/gkv078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schickor P, Metzger W, Werel W, Lederer H, Heumann H. Topography of intermediates in transcription initiation of E. coli. EMBO J. 1990;9:2215–2220. doi: 10.1002/j.1460-2075.1990.tb07391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dayton CJ, Prosen DE, Parker KL, Cech CL. Kinetic measurements of Escherichia coli RNA polymerase association with bacteriophage T7 early promoters. J Biol Chem. 1984;259:1616–1621. [PubMed] [Google Scholar]
- 34.Davis E, Chen J, Leon K, Darst SA, Campbell EA. Mycobacterial RNA polymerase forms unstable open promoter complexes that are stabilized by CarD. Nucleic Acids Res. 2015;43:433–445. doi: 10.1093/nar/gku1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gangwar SP, Meena SR, Saxena AK. Structure of the carboxy-terminal domain of Mycobacterium tuberculosis CarD protein: an essential rRNA transcriptional regulator. Acta Crystallogr Sect F Struct Biol Commun. 2014;70:160–165. doi: 10.1107/S2053230X13034407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaur G, Dutta D, Thakur KG. Crystal structure of Mycobacterium tuberculosis CarD, an essential RNA polymerase binding protein, reveals a quasidomain-swapped dimeric structural architecture. Proteins. 2014;82:879–884. doi: 10.1002/prot.24419. [DOI] [PubMed] [Google Scholar]
- 37.Kireeva ML, Kashlev M. Mechanism of sequence-specific pausing of bacterial RNA polymerase. Proc Natl Acad Sci U S A. 2009;106:8900–8905. doi: 10.1073/pnas.0900407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.