Abstract
The heterocyclic amine 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) targets multiple organs for tumorigenesis in the rat, including the colon and the skin. PhIP-induced skin tumors were subjected to mutation screening, which identified genetic changes in Hras (7/40, 17.5%) and Tp53 (2/40, 5%), but not in Ctnnb1, a commonly mutated gene in PhIP-induced colon tumors. Despite the absence of Ctnnb1 mutations, β-catenin was overexpressed in nuclear and plasma membrane fractions from PhIP-induced skin tumors, coinciding with loss of p120-catenin from the plasma membrane, and the appearance of multiple p120-catenin-associated bands in the nuclear extracts. Real-time RT-PCR revealed that p120-catenin isoforms 1 and 4 were upregulated in PhIP-induced skin tumors, whereas p120-catenin isoform 3 was expressed uniformly, compared with adjacent normal-looking tissue. In human epidermoid carcinoma and colon cancer cells, transient transfection of p120-catenin isoform 1A enhanced the viability and cell invasion index, whereas transient transfection of p120-catenin isoform 4A increased cell viability and cell proliferation. Knockdown of p120-catenin revealed a corresponding reduction in the expression of β-catenin and a transcriptionally regulated target, Ccnd1/Cyclin D1. Co-immunoprecipitation experiments identified associations of β-catenin with p120-catenin isoforms in PhIP-induced skin tumors and human cancer cell lines. The results are discussed in the context of therapeutic strategies that might target different p120-catenin isoforms, providing an avenue to circumvent constitutively active β-catenin arising via distinct mechanisms in skin and colon cancer.
Keywords: Adherens junction, β-catenin, E-cadherin, CTNND1, CTNNB1
INTRODUCTION
There is growing interest in p120-catenin and its role in cancer etiology [1–6]. As a member of the Armadillo family, p120-catenin is a key component of the adherens junction (AJ), with links to cell-cell interactions and signaling within the nucleus [1]. Cell adhesion and signaling functions have been defined for other AJ members, including β-catenin and E-cadherin [6–9]. According to subcellular localization, p120-catenin can regulate the turnover of plasma membrane-associated cadherins and their protein partners, the activation of cytoplasmic small RhoGTPases, or transcriptional mechanisms within the nucleus [1]. Post-translational modifications and alternative splicing leads to the formation of multiple p120-catenin isoforms, with poorly characterized roles in physiology and pathophysiology [1,6].
Loss of p120-catenin has been linked to chronic inflammation in the skin, resulting in atopic dermatitis [10]. Conditional targeting in mice showed that p120 null neonatal epidermis had reduced AJ components, and that as mice age, they displayed epidermal hyperplasia with chronic inflammation and skin tumor formation linked to NFκB activation [11,12]. In humans, squamous cell carcinoma of the skin was associated with aberrant localization of p120-catenin [13]. Specifically, normal and benign tissues had p120-catenin in the cell-cell boundaries, whereas squamous cell carcinomas had p120-catenin localized away from the cell periphery, including p120-catenin isoforms 3A and 4 [13]. p120-catenin and other members of the catenin family were commonly deregulated in human cutaneous melanoma [14].
In the heterocyclic amine-induced multi-organ rat carcinogenesis model, constitutive activation of β-catenin has been identified as a key driver of tumor formation in the colon [15–21]. The current investigation focused, for the first time, on the corresponding skin tumors induced in the rat by 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP). In the absence of genetic changes in Ctnnb1, PhIP-induced skin tumors nonetheless exhibited high levels of β-catenin protein expression in the nucleus and plasma membrane. Subsequent work implicated a role for p120-catenin relocalization from the plasma membrane to the nucleus, and for specific p120-catenin isoforms influencing cell viability, proliferation, and invasiveness.
MATERIALS AND METHODS
Source of tumors
Forty skin tumors and adjacent normal-looking tissues were obtained from a 1-year carcinogenicity study in the rat [17]. A portion of each PhIP-induced skin tumor was processed for histopathology, as reported [17], and the remainder was stored at −80°C for molecular analyses.
Mutation screening
Gene targets were examined using the PCR-based single strand conformation polymorphism and direct sequencing methodology reported [22–24].
RNA expression
Frozen tumor samples and their matched normal tissues were extracted via the RNeasy kit (Qiagen, Valencia, CA), followed by cDNA synthesis using Superscript III (Invitrogen, Carlsbad, CA). After 40 cycles of qPCR, gene targets were normalized to glyceraldehyde-3-phosphate dehydrogenase (Gapdh). The amount of specific mRNA was quantified by determining the point at which the fluorescence accumulation entered the exponential phase (Ct), and the Ct ratio of the target gene to Gapdh was calculated. At least two separate experiments were performed for each sample.
Conventional RT-PCR also was used to investigate different isoforms of p120-catenin, with primer sets designed around alternate translation initiation codons. Primer sequences were as follows: Ctnnd1 1F: 5′-TTC ACC TTG TCA TTG CGG TA-3′; Ctnnd1 3R: 5′-TCC CAT CAT CTG AGG TCT CC-3′; Ctnnd1 4R: 5′-CAT TCC CTT GCG CAG ACT AT-3′. Thirty five cycles of PCR (95°C/30s, 58°C/30s, 72°C/30s) were run in a 20-μl reaction volume containing cDNAs and isoform-specific primer pairs Ctnnd1 1F/Ctnnd1 3R for isoforms 1, 2 and 3, or Ctnnd1 1F/Ctnnd1 4R for isoforms 1, 2, 3 and 4. Reaction products were visualized on a 1.5% agarose gel that included a molecular weight marker.
Immunoblotting
ProteoExtract Native membrane protein extraction kit (EMD Biosciences Inc., San Diego, CA) was used for enrichment of membrane proteins, whereas NE-PER nuclear and cytoplasmic reagents (Pierce Biotechnology, Rockford, IL) were used to separate cytoplasmic and nuclear fractions. The basic immunoblotting methodology was as reported [25–28], with primary antibodies to β-catenin (#9581, Cell Signaling Technology, Danvers, MA), E-cadherin (ab53226, Abcam, Cambridge, MA), p120-catenin (#4989, Cell Signaling; #339600, Zymed Laboratories Inc., San Francisco, CA), p53 (sc-126, Santa Cruz Biotechnology, Dallas, TX), β-actin (AC-74, Sigma-Aldrich, St. Louis, MO), and histone H1 (sc-8030, Santa Cruz).
p120-catenin overexpression and knockdown
Human epidermoid carcinoma cell line A431NS and human colorectal cancer cell line SW480 (American Type Culture Collection, Manassas, VA) were maintained in McCoy’s 5A medium (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (FBS, Hyclone Laboratories Inc., Logan, UT), 100 units/ml penicillin, and 100 μg/ml streptomycin at 37°C in 5% CO2. In knockdown experiments, human shRNA against p120-catenin and non-specific control shRNAs were purchased from OriGene Technologies (Rockville, MD) and transfected according to the manufacturer’s instructions. Briefly, after cells (1.5 ×105) were seeded in 6-well plates overnight, the media was aspirated and replaced with antibiotic free-transfection media containing Lipofectmine2000 plus 1 μg shRNA, or Lipofectmine2000 alone (mock controls). For overexpression experiments, human p120-catenin isoforms 1A, 3A and 4A were kindly provided by Dr. P.Z. Anastasiadis (Mayo Clinic, Rochester, MN). After cells (1.5 ×105) were seeded in 6-well plates overnight, the media was aspirated and replaced with antibiotic-free transfection media containing Lipofectmine2000, 2 μg plasmid DNA for p120-catenin isoform 1A, or 3A, or 4A, or pcDNA3.1 empty vector as control. In both the knockdown and overexpression experiments, 5 h after transfection FBS was added to a final concentration of 10%, and at 72 h cells were harvested for molecular analyses, as detailed below.
Co-immunoprecipitation
Whole cell lysates from cell-based assays and from PhIP-induced skin tumors were subjected to co-immunoprecipitation (co-IP) assays, following the protocols reported elsewhere [25–27]. The antibodies listed above to p120-catenin and β-catenin were used to immunoprecipitate (IP) the corresponding endogenous protein, or transiently transfected p120-catenin isoforms, followed by immunoblotting (IB) of the reverse molecular target. In other experiments, whole cell lysates from PhIP-induced skin tumors and adjacent normal tissue were subjected to IP/IB as in the cell-based assays. While optimizing the co-IP conditions, lysates from cell-based assays in which p120-catenin isoforms 1A, 3A and 4A had been transiently transfected were used as reference for the position of each p120-catenin isoform in the gel (data not shown). Results presented here were representative of the findings from two or more independent experiments.
Cell viability
Cells (1×103) in 100 μl media were seeded in 96-well plates overnight and transfected with p120-catenin isoform 1A, 3A, 4A, or empty vector (as stated above). At selected times, 3-(4,5-dimethythiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) was added and formazan dye was assessed at 570 nm [25].
Cell invasion
Real-time invasion was monitored using the xCELLigence System (ACEA Biosciences Inc., San Diego, CA). Cells were transiently transfected with p120-catenin isoform 1A, 3A, 4A, or empty vector, and harvested at 72 h. Cells (105) in 100 μl serum-free media were loaded onto the upper chamber of CIM-Plate 16 pre-coated with 5% Matrigel (BD Biosciences, San Jose, CA), and the lower chamber was filled with McCoy’s 5A medium containing 10% FBS. Measurements were recorded at 10 min intervals for up to 16 h [29]. For each cell line and treatment condition, experiments were repeated at least three times.
Cell proliferation
Cells were transfected with p120-catenin isoform 1A, 3A, 4A, or empty vector, and harvested at 72 h as described above. Before harvesting, cells were treated for 3 h with 10 μM bromodeoxyuridine (BrdU, Sigma-Aldrich). BrdU incorporation was assessed using the BrdU in situ detection kit (BD Biosciences), according to the manufacturer’s instructions.
Statistics
Data were expressed as mean±SD and compared using the Student’s t-test for paired samples, and analysis of variance (ANOVA) for group comparisons. Significant results were indicated in the figures as follows: *P<0.05, **P<0.01, ***P<0.001. Results shown in the figures were from replicate experiments that gave similar results, each repeated at least three times, unless indicated otherwise.
RESULTS
PhIP-induced skin tumors lack Ctnnb1 mutations but overexpress β-catenin
Ctnnb1 mutations were reported in heterocyclic amine-induced rat colon tumors [15], and the same methodology was used to examine genetic changes in the corresponding PhIP-induced skin tumors. Shifted bands were detected for Hras and Tp53, but not for Ctnnb1 (Figure 1) or Ctnnd1 (data not shown). Sequencing confirmed the presence of a codon 174 mutation in Tp53, and codon 12 and codon 61 mutations in Hras (Table 1). Final results indicated that the skin tumors had mutation frequencies of 7/40 (15.5%) in Hras, 2/40 (5%) in Tp53, and 0/40 (0%) in Ctnnb1 and Ctnnd1 (Table 2).
Figure 1.
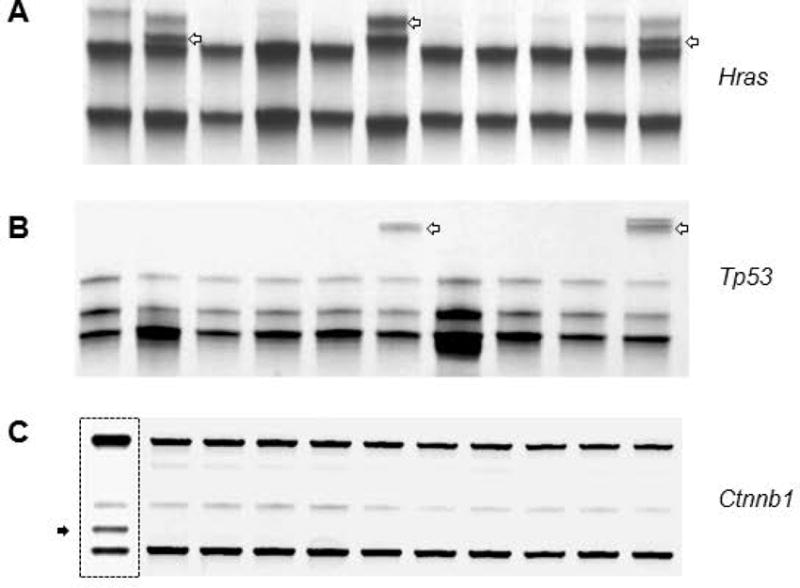
Mutation screening of PhIP-induced skin tumors in the rat. Representative data from PCR-based single-strand conformation polymorphism analysis. (A-B) open arrows indicate bands that were sequenced and confirmed to harbor Hras or Tp53 mutations (see Table 1). (C) No shifted bands were detected for Ctnnb1 in skin tumors. Hatched box, positive control from a PhIP-induced rat colon tumor, in which the shifted band (solid arrow) was sequenced and confirmed to harbor a β-catenin mutation [17]. A random screening of non-shifted bands confirmed the wild-type status of Hras, Tp53, and Ctnnb1 in other skin tumor samples (data not shown).
Table 1.
Summary of gene mutations in PhIP-induced skin tumors
| Gene | Codon | Mutation | Frequency | Substitution | Histopathology |
|---|---|---|---|---|---|
|
| |||||
| Hras | 12 | GGA→GAA | 2 | G12E | Squamous cell carcinoma |
| GGA→GTA | 1 | G12V | Sebaceous epithelioma | ||
| 61 | CAA→CAT | 2 | G61H | Squamous papilloma, basal cell carcinoma | |
| CAA→AAA | 2 | Q61K | Squamous cell carcinoma, sebaceous epithelioma | ||
| Tp53 | 174 | GGC→GCC | 2 | G174A | Sebaceous adenoma, sebaceous squamous cell carcinoma |
Table 2.
Mutation frequency in PhIP-induced skin tumors
| Hras | Tp53 | Ctnnb1 (β-catenin) | Ctnnd1 (p120ctn) |
|---|---|---|---|
| 7/40(17.5 %) | 2/40 (5%) | 0/40 | 0/40 |
Nuclear extracts from PhIP-induced skin tumors and adjacent normal tissues were subjected to immunoblotting (Figure 2A), revealing a decrease in p53 levels and an increase in β-catenin expression in the tumor samples. Real-time qRT-PCR assays confirmed that β-catenin-dependent transcriptional targets c-myc, c-jun, and Ccnd1 were upregulated significantly in the skin tumors, as was Ctnnb1 (Figure 2B). When the corresponding plasma membrane-enriched fractions were immunoblotted (Figure 3A), increased expression of β-catenin and E-cadherin was detected in the skin tumors. In marked contrast, p120-catenin levels were reduced dramatically in the plasma membrane (Figure 3A), coinciding with multiple bands appearing in the nuclear extracts (Figure 3B). These findings suggested that, in the absence of Ctnnb1 mutation providing a driver for β-catenin overexpression, remodeling of AJ might redistribute β-catenin and p120-catenin to the nuclear compartment to affect transcriptional activity in the skin tumors.
Figure 2.
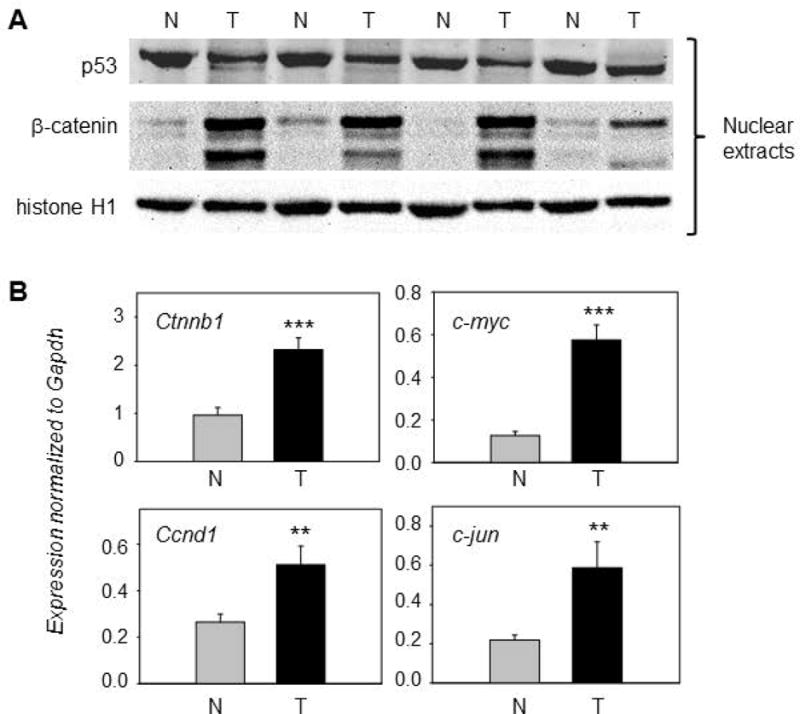
Overexpression of β-catenin in PhIP-induced skin tumors. (A) Representative immunoblots of β-catenin and p53 in nuclear fractions; histone H1, loading control; T, tumor; N, normal-looking tissue adjacent to tumor. An additional set of five T/N pairs generated similar data for the proteins indicated (immunoblot data not presented). (B) qRT-PCR analyses of selected β-catenin target genes normalized to glyceraldehyde-3-phosphate dehydrogenase (Gapdh); mean±SD, n=12, **P<0.01, ***P<0.001. Each panel is representative of an experiment that was repeated at least three times, for each gene mentioned, using twelve matched pairs of T/N samples.
Figure 3.
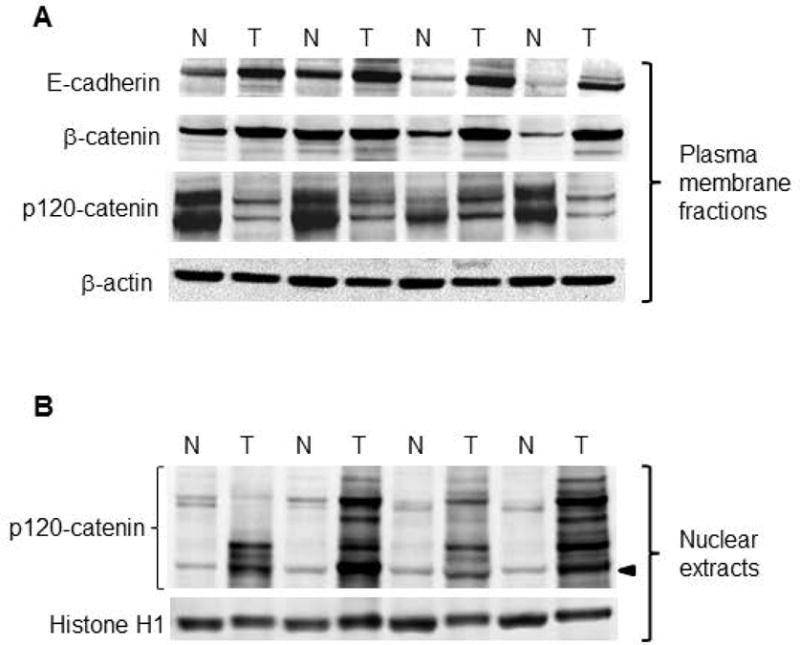
Subcellular redistribution of p120-catenin in PhIP-induced skin tumors. (A) Representative immunoblots of Adherens junction proteins, with β-actin as loading control. Two high molecular weight bands were detected at 120 kD and 100 kD for p120-catenin in the plasma membrane. (B) Nuclear fractions of the corresponding skin tumors, normalized to histone H1. Multiple p120-catenin-associated bands were detected in the nucleus; arrowhead designates 65 kD (the position of p120-catenin isoform 4A, see below). An additional set of five T/N pairs generated similar data for the proteins shown in the figure, in experiments that were repeated at least twice (immunoblot data not presented).
p120-catenin is overexpressed in different human skin cancer subtypes
Human tissue microarrays immunostained for p120-catenin protein revealed a general pattern of overexpression in squamous cell carcinoma, basal cell carcinoma, and malignant melanoma (Figure 4). Focal areas of intense p120-catenin expression were detected in squamous cell carcinoma of the skin, whereas adjacent normal-looking tissue had moderate p120-catenin levels, largely restricted to the cell periphery (Figure 4A). Overexpression of p120-catenin in basal cell carcinoma, malignant melanoma, and lymph node metastases (Figures 4B–D) was recapitulated in human keratinocyte, melanoma, and epidermoid carcinoma cell lines (Figures 4E–G). Immunofluorescence staining detected p120-catenin not only in the cell periphery, but also in the cytoplasm and nucleus (Figures 4H–K). Interestingly, cells that stained strongly for p120-catenin in the cell periphery had less nuclear/cytoplasmic p120-catenin (Figure 4H), whereas cells with p120-catenin in the cytoplasm and nucleus tended to exhibit more restricted immunostaining of p120-catenin at the cell periphery. These observations are in accordance with findings from PhIP-induced skin tumors, implicating p120-catenin relocalization from AJ to the nucleus.
Figure 4.
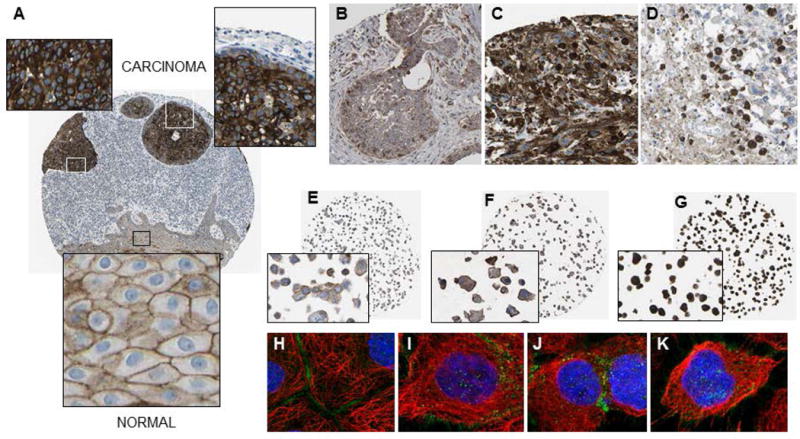
p120-catenin expression in human skin cancer. (A) Squamous cell carcinoma (male, age 93, ID: 2809); (B) basal cell carcinoma (female, age 58, ID: 2079); (C) malignant melanoma (female, age 104, ID: 777); (D) malignant melanoma in a lymph node metastatic site (male, age 59, ID: 1414); (E) HaCaT keratinocytes; (F) SK-MEL-30 melanoma cells; (G) A431 epidermoid carcinoma cells; (H-K) immunofluorescence staining of A431 cells showing p120-catenin positivity along the plasma membrane, as well as in the cytoplasm and nucleus (green dots); blue, nucleus; red, microtubules (http://www.proteinatlas.org/).
p120-catenin isoforms 1 and 4 are upregulated in PhIP-induced skin tumors
No significant differences were observed between skin tumors and the adjacent normal-looking tissue in the rat when Ctnnd1 mRNA levels were quantified under experimental conditions that did not discriminate between the different p120-catenin isoforms (data not shown). Subsequently, primers were designed to examine different isoforms of p120-catenin, arising from alternative start codons (Figure 5A). By conventional RT-PCR, isoform 3 was identified as the dominant p120-catenin transcript, being equally abundant in skin tumors and adjacent normal-looking tissues. On the other hand, in >90% of the PhIP-induced skin tumors examined, p120-catenin isoforms 1 and 4 were upregulated, compared to adjacent normal-looking tissue (Figure 5B, boxes).
Figure 5.
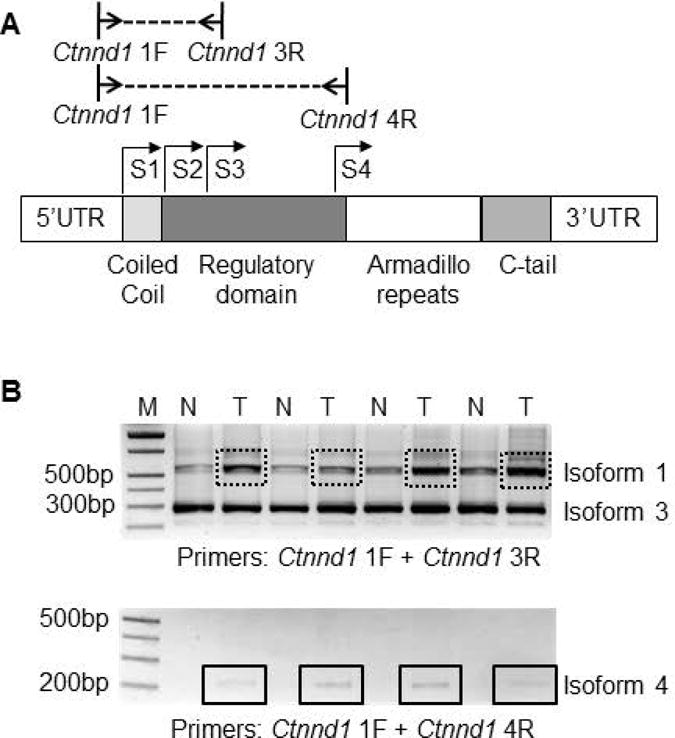
(A) Schematic diagram of rat Ctnnd1 and primer locations used to examine different p120-catenin isoforms; S1–S4, alternative start codons. (B) Representative RT-PCR data for Ctnnd1 isoforms 1, 3 and 4; T, tumor; N, normal-looking tissue; M, molecular marker (DNA ladder). Boxes, increased expression of p120-catenin isoform 1 and isoform 4 in tumor vs. normal.
p120-catenin isoforms differentially regulate the viability, proliferation, and invasiveness of cancer cells
Transient transfection studies were conducted with p120-catenin isoforms 1A, 3A, and 4A in human epidermoid carcinoma cells. At 72 h post-transfection, immunoblotting confirmed the increased levels of p120-catenin isoform 1A and 4A, relative to the vector control (Figure 6A, boxes). In A431NS cells, high constitutive levels of isoform 3A masked any changes from the corresponding transient transfection (Figure 6A, oval), although there was a noticeable reduction in cell viability due to exogenous isoform 3A (Figure 6B). Interestingly, the opposite effect on cell viability was observed for p120-catenin isoform 1A and, in particular, isoform 4A (Figure 6B). A significant increase was detected in the number of cells staining positive for BrdU incorporation following transient transfection of p120-catenin isoform 4A, as compared with the other treatments (Figure 6C). In real-time cell invasion assays, p120-catenin isoform 1A increased the slope significantly (Figure 6D, dotted red line), indicating enhanced invasiveness, in contrast to isoforms 3A and 4A that had the same slope as the vector controls.
Figure 6.
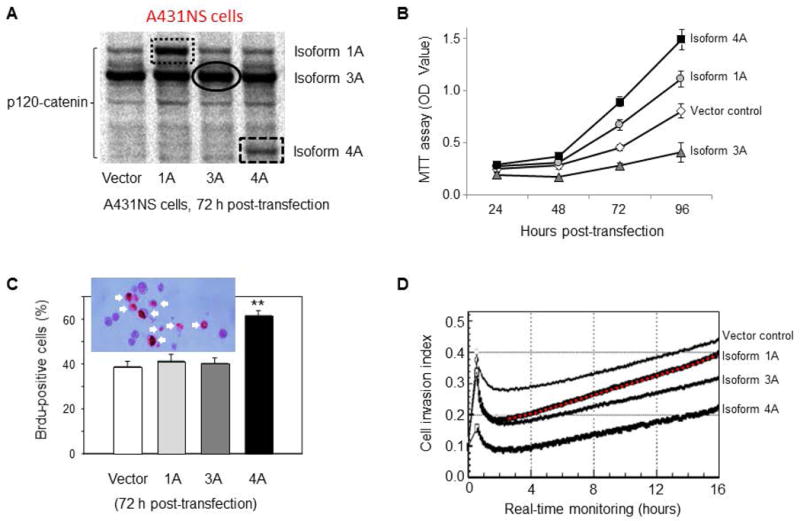
Effects of p120-catenin isoforms in human epidermoid carcinoma cells. (A) Transient transfection increased the levels of p120-catenin isoforms 1A and 4A (hatched boxes). Immunoblotting did not reveal a corresponding increase in p120-catenin isoform 3A (oval), possibly due to the high endogenous levels of this isoform in A431NS cells. Effects of p120-catenin isoforms were examined on (B) cell viability; at the 96-h time-point, p120-catenin isoforms 1A, 3A, and 4A were significantly different from the vector control (p<0.05). (C) Bromodeoxyuridine (BrdU) incorporation, mean±SD, n=3; insert shows BrdU-positive cells, white arrows; (D) cell invasion assays, using real-time monitoring as reported by Parasramka et al. [29]; representative findings from at least three replicate experiments, each giving similar outcomes. The slope of the line for p120-catenin isoform 1A was significantly different from the other slopes shown, including the vector control, p<0.05.
The latter experiments were repeated in SW480 colon cancer cells, which harbor high endogenous β-catenin due to a mutation in APC [30]. Forced expression of p120-catenin isoform 4A (Figure 7A) resulted in a marked increase in cell viability compared with the vector controls (Figure 7B). Under these conditions, p120-catenin isoforms 1A and 3A had a less dramatic effect on cell viability. In SW480 cells, a significant increase in the number of cells staining positive for BrdU incorporation was detected after transient transfection of p120-catenin isoform 4A (Figure 7C). In real-time invasion assays, p120-catenin isoform 1A increased the slope compared with the vector control, whereas p120-catenin isoforms 3A and 4A had the opposite effect (Figure 7D). The latter findings suggested reduced invasiveness of SW480 cells, which was not observed in A431NS cells transfected with p120-catenin isoforms 3A and 4A (Figure 6D). Differences in phenotypic response among cancer cell lines might be related, in part, to the endogenous levels of each p120-catenin isoform [31]. Nonetheless, the findings reaffirmed the opposing activities of some p120-catenin isoforms towards cell viability, proliferation, and invasiveness.
Figure 7.
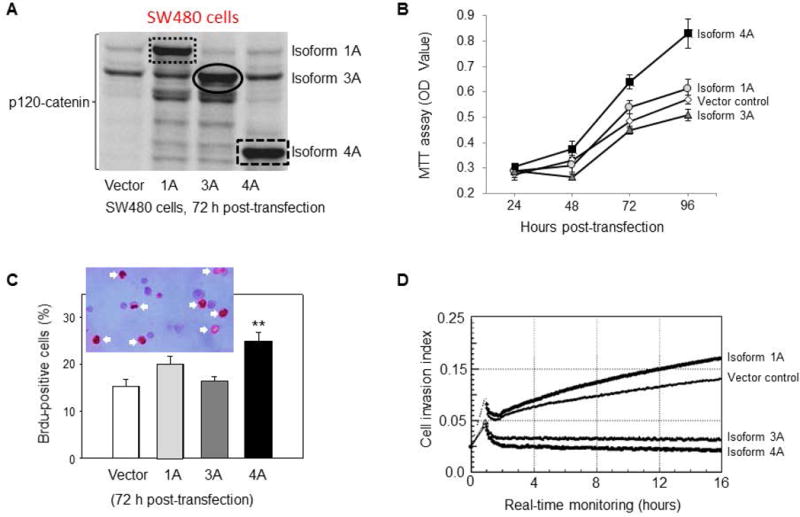
Effects of p120-catenin isoforms in human colon cancer cells. (A) Transient transfection increased the levels of p120-catenin isoforms 1A, 3A and 4A in SW480 cells. Effects of p120-catenin isoforms on (B) cell viability, at the 96-h time-point, p120-catenin isoform 4A was significantly different from the vector control (p<0.05). (C) BrdU incorporation, and (D) cell invasion were as detailed in Figure 6, legend. Slopes of the p120-isoform treatments were each significantly different from the vector control, p<0.05.
p120-catenin/β-catenin interactions in cancer cells
Next, shRNA experiments were designed to knockdown p120-catenin in A431NS and SW480 cells. Immunoblotting confirmed the reduced expression of p120-catenin, and a corresponding lowering of β-catenin and Cyclin D1 protein levels (hatched boxes, Figure 8A), relative to mock and non-target shRNA controls. Under the same experimental conditions, qRT-PCR assays revealed a significant loss of Ccnd1 mRNA expression (Figure 8B), without changes in Ctnnb1 mRNA levels (data not shown). Given that Ccnd1 is a direct transcriptional target of β-catenin [32], we postulated that reduced p120-catenin levels lowered the expression of β-catenin protein, and the loss of β-catenin transcriptionally downregulated Cyclin D1.
Figure 8.
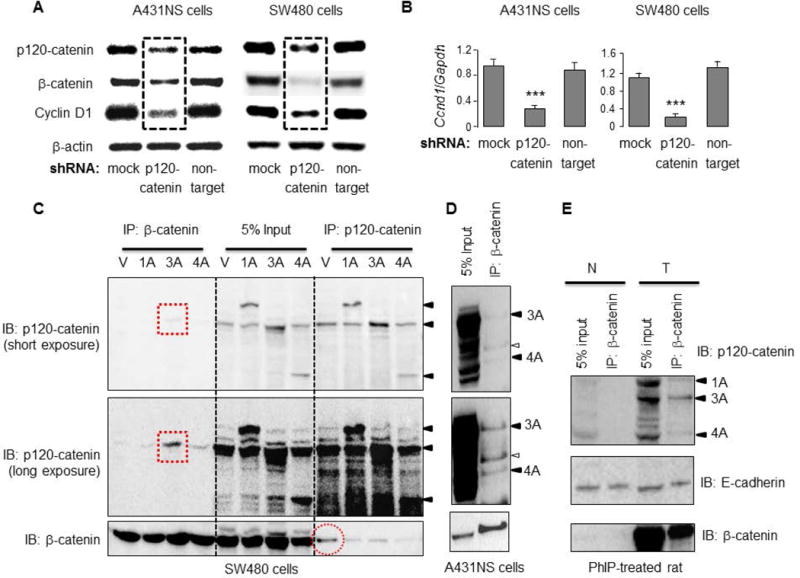
p120-catenin/β-catenin interactions in human cancer cells and PhIP-induced skin tumors. (A) Target-specific shRNA was used to knockdown p120-catenin, and was compared with vector and non-target shRNA controls. Immunoblotting in A431NS and SW480 cells confirmed the reduced expression of p120-catenin protein, as well as β-catenin and Cyclin D1 (hatched boxes); (B) shRNA knockdown of p120-catenin lowered Ccnd1 (Cyclin D1) mRNA levels significantly, relative to non-target and mock controls; n=3, p<0.001. No changes were detected in Ctnnb1 (β-catenin) mRNA levels under these conditions, data not shown; (C) Co-immunoprecipitation (co-IP) experiments in SW480 cells transiently transfected with p120-catenin isoforms 1A, 3A, 4A, or the vector control, as in Figure 7; solid arrowheads indicate the positions of the three p120-catenin isoforms transfected. (D) Endogenous interactions of β-catenin with p120-isoforms in A431NS cells; open arrowhead designates a putative p120-catenin isoform (or splice variant) of intermediate size between isoforms 3A and 4A, shown with solid arrowheads; (E) co-IP experiments were performed on whole cell lysates from PhIP-induced skin tumors (T) and adjacent normal-looking tissue (N). Data shown are from a single T/N pair run in duplicate, and are representative of the findings from a least three separate rat tumor samples.
To examine protein-protein interactions involving p120-catenin and β-catenin, we took cell lysates from SW480 cells transiently transfected with different p120-catenin isoforms and performed co-IP assays (Figure 8C). Pulling-down with β-catenin in the IP step, followed by IB with p120-catenin, revealed a band corresponding to p120-catenin isoform 3A (Figure 8C, red hatched boxes). A faint band corresponding to p120-catenin isoform 3A was detected in all of the lanes, including the vector control, hinting at interaction of endogenous p120-catenin isoform 3A with β-catenin in SW480 cells. In the reverse pull-down, IP with p120-catenin antibody revealed an interaction with β-catenin, including in vector controls representing the endogenous proteins (Figure 8C, red dotted circle).
Endogenous protein-protein interactions also were examined in untransfected A431NS cells (Figure 8D). Pulling-down with β-catenin antibody revealed interactions with p120-catenin isoforms 3A and 4A, plus a third putative p120-catenin isoform of intermediate size (open arrowhead, Figure 8D).
Finally, cell lysates from PhIP-induced skin tumors and adjacent normal-looking tissues were subjected to co-IP experiments. Compared with adjacent normal tissue, input controls from tumor lysates had high expression levels of p120-catenin isoforms 1A, 3A and 4A (Figures 8E, arrowheads). Pulling-down with β-catenin antibody in the tumor lysates revealed corresponding bands for p120-catenin isoforms 1A, 3A, and 4A in the IB step (Figure 8E), as well as the positive control, E-cadherin. We conclude that β-catenin can interact not only with E-cadherin, but also with p120-catenin isoforms 1A, 3A and 4A in PhIP-induced skin tumors. Future studies might examine this possibility in more detail via tethering assays such as fluorescence resonance energy transfer.
DISCUSSION
The colon and mammary gland are well-established primary target organs for tumorigenesis in PhIP-treated male and female rats, respectively [33–35], but one-quarter to one-third of the animals also develop skin tumors [17,20]. The skin tumors exhibit a diverse range of histopathology, including squamous adenoma, squamous papilloma, squamous cell carcinoma, basal cell carcinoma, and sebaceous epithelioma (Table 1). These skin tumors, like the corresponding mammary gland and liver tumors [15,36], harbor occasional ras or Tp53 mutations, but lack genetic changes in Ctnnb1 (Table 2). In marked contrast, c-myc, c-jun, and Ccnd1 gene activation is driven by Ctnnb1 mutations that stabilize β-catenin in PhIP-induced colon tumors [15,18,37].
We postulated that, rather than Ctnnb1 mutation leading to β-catenin protein stabilization, a key driver of skin tumorigenesis in the PhIP model might be the deregulation of AJ at the cell periphery, releasing β-catenin and p120-catenin to become active in other subcellular compartments [38–44]. It is known that p120-catenin binds to the juxtamembrane domain of E-cadherin and stabilizes diverse cadherins at the cell membrane [40]. In SW48 colon cancer cells, a genetic deficiency in p120-catenin results in reduced E-cadherin protein expression, and restoring p120-catenin rescues the epithelial phenotype by stabilizing E-cadherin levels [31]. Interestingly, a ‘cadherin switch’ has been described in the transition of normal melanocytes to malignant melanoma, which has features of epithelial-to-mesenchymal transition [45]. No loss of E-cadherin was detected in the current investigation; on the contrary, E-cadherin was consistently upregulated in PhIP-induced skin tumors (Figure 3A), implying that epithelial-to-mesenchymal transition was not a major feature of skin tumorigenesis in this preclinical model.
In the current investigation, we observed that p120-catenin isoform 1 and isoform 4 were upregulated in PhIP-induced skin tumors, compared with adjacent normal tissue in the rat. Cell viability and invasion were enhanced by p120-catenin isoform 1A in A431NS cells, and a similar tendency was observed in SW480 cells, suggesting an oncogenic function. An oncogenic role also was ascribed to the shorter p120-catenin isoform, for the following reasons: (i) p120-catenin isoform 4 was transcriptionally upregulated in most of the PhIP-induced skin tumors (Figure 5B); (ii) a band corresponding in size to isoform 4 appeared in the nuclear extracts of PhIP-induced skin tumors (Figure 3B); (iii) in PhIP-induced skin tumors, co-IP experiments identified p120-catenin isoform 4A in association with β-catenin (Figure 8E); and (iv) transient transfection of isoform 4A resulted in enhanced cell viability and BrdU labeling, indicative of increased cell proliferation, in both human skin and colon cancer cells lines (Figures 6B,C and 7B,C). On the other hand, p120-catenin isoform 3A tended to inhibit cell viability, and reduced the invasion index in SW480 colon cancer cells. Collectively, these findings may be of importance, because they suggest that targeting different p120-catenin isoforms, therapeutically, might provide an avenue to circumvent constitutively active β-catenin, even in the presence of CTNNB1 or APC mutations.
In the present investigation, we were unable to associate specific genetic alterations, or changes in p120-catenin expression, to the corresponding histopathological diagnosis, due to the relatively limited number of forty PhIP-induced skin tumors available. Thus, despite the diverse range of histopathology (Table 1), no firm conclusions could be drawn regarding histological sub-type, origin of cells, aggressiveness, or malignant transformation linked to p120-mediated changes in viability, proliferation, or invasiveness. Data are available for melanoma patients from online sources such as PrognoScan [46]; however, we are cautious not to over-interpret the results as supporting reduced overall survival in cases of high versus low CTNND1 expression, given the small number of samples involved (Figure 9).
Figure 9.
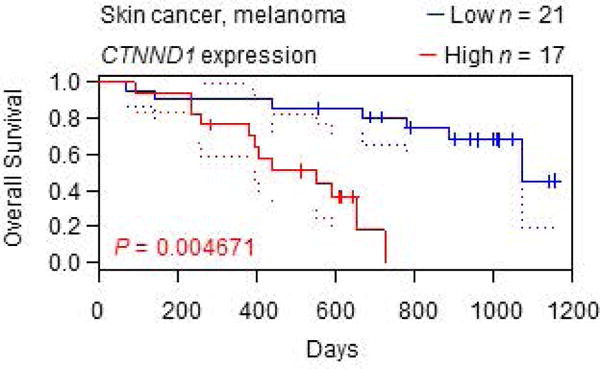
Kaplan-Meier curves for melanoma patients. Data were generated from skin cancer/melanoma dataset GSE19234 in PrognoScan [31], stratified for high vs. low CTNND1 expression.
Conditional targeting of p120-catenin in mice resulted in p120-catenin null neonatal epidermis that had reduced AJ components, and as mice age they displayed epidermal hyperplasia with chronic inflammation and skin tumor formation [11,12]. In another mouse study, conditional knockout of p120-catenin led to the formation of preneoplastic and neoplastic lesions of the oral cavity, esophagus, and squamous forestomach, with tumors containing signficant immune cell infiltration [47]. These reports suggested that total ablation of 120-catenin significantly deregulates cell-cell communication and inflammation, resulting in enhanced tumor formation in the mouse. However, the genetic models provided no specific insights into the opposing roles of different p120-catenin isoforms, and might have obscured crucial ‘stop/go’ checkpoints relevant to human cancer development. In this context, the PhIP-induced skin carcinogenesis model offers certain advantages [48–50], including the ability to examine p120-catenin isoforms at different stages of tumor formation, and the underlying mechanisms impacting cell survival, proliferation, and invasiveness.
Acknowledgments
Grant support: This research was supported in part by NIH grants CA090890, CA122959, ES00210, and ES023512, the John S. Dunn Foundation, and a Chancellor’s Research Initiative.
Abbreviations
- AJ
adherens junction
- BrdU
bromodeoxyuridine
- CTNNB1/Ctnnb1
human/murine gene coding for β-catenin
- CTNND1/Ctnnd1
human/murine gene coding for p120-catenin
- Ccnd1
murine gene coding for cyclin D1
- Gapdh
murine gene coding for glyceraldehyde-3-phosphate dehydrogenase
- MTT
3-(4,5-dimethythiazol-2-yl)-2,5-diphenyltetrazolium bromide
- PhIP
2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine
Footnotes
Conflict of interest: The authors declare no conflict of interest
References
- 1.Hong JY, Oh IH, McCrea PD. Phosphorylation and isoform use in p120-catenin during development and tumorigenesis. Biochim Biophys Acta. 2016;1863:102–114. doi: 10.1016/j.bbamcr.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang D, Tang N, Liu Y, Wang EH. ARVCF expression is significantly correlated with the malignant phenotype of non-small cell lung cancer. Mol Carcinogen. 2015;54(Suppl 1):E185–E191. doi: 10.1002/mc.22281. [DOI] [PubMed] [Google Scholar]
- 3.Fan C, Jiang G, Zhang V, et al. Zbed3 contributes to malignant phenotype of lung cancer via regulating β-catenin and P120-catenin 1. Mol Carcinogen. 2015;54(Suppl 1):E138–E147. doi: 10.1002/mc.22216. [DOI] [PubMed] [Google Scholar]
- 4.Zhao H, Zhao Y, Jiang G, et al. Dishevelled-3 activates p65 to upregulate p120-catenin transcription via a p38-dependent pathway in non-small cell lung cancer. Mol Carcinogen. 2015;54(Supple 1):E112–E121. doi: 10.1002/mc.22196. [DOI] [PubMed] [Google Scholar]
- 5.Yang ZQ, Zhao Y, Liu Y, et al. Downregulation of HDPR1 is associated with poor prognosis and affects expression levels of p120-catenin and β-catenin in non-small cell lung cancer. Mol Carcinogen. 2010;49:508–519. doi: 10.1002/mc.20622. [DOI] [PubMed] [Google Scholar]
- 6.Pieters T, van Roy F, van Hengel J. Functions of p120ctn isoforms in cell-adhesion and intracellular signaling. Front Biosci. 2012;17:1669–1694. doi: 10.2741/4012. [DOI] [PubMed] [Google Scholar]
- 7.McCrea PD, Maher MT, Gottardi CJ. Nuclear signaling from cadherin adhesion complexes. Curr Top Dev Biol. 2015;112:129–196. doi: 10.1016/bs.ctdb.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCrea PD, Gottardi CJ. Beyond β-catenin: prospects for a larger catenin network in the nucleus. Nat Rev Mol Cell Biol. 2016;17:55–64. doi: 10.1038/nrm.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Du W, Liu X, Fan G, et al. From cell membrane to the nucleus: an emerging role of E-cadherin in gene transcriptional regulation. J Cell Mol Med. 2014;18:1712–1719. doi: 10.1111/jcmm.12340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobielak A, Boddupally K. Junctions and inflammation in the skin. Cell Commun Adhes. 2014;21:141–147. doi: 10.3109/15419061.2014.905930. [DOI] [PubMed] [Google Scholar]
- 11.Perez-Moreno M, Davis MA, Wong E, Pasolli HA, Reynolds AB, Fuchs E. p120-catenin mediates inflammatory responses in the skin. Cell. 2006;124:631–644. doi: 10.1016/j.cell.2005.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perez-Moreno M, Song W, Pasolli HA, Williams SE, Fuchs E. Loss of p120 catenin and links to mitotic alterations, inflammation, and skin cancer. Proc Natl Acad Sci U S A. 2008;105:15399–15404. doi: 10.1073/pnas.0807301105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishizaki Y, Omori Y, Momiyama M, et al. Reduced expression and aberrant localization of p120catenin in human squamous cell carcinoma of the skin. J Dermatol Sci. 2004;34:99–108. doi: 10.1016/j.jdermsci.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 14.Zhang XD, Hersey P. Expression of catenins and p120cas in melanocytic nevi and cutaneous melanoma: deficient alpha-catenin expression is associated with melanoma progression. Pathology. 1999;31:239–246. doi: 10.1080/003130299105052. [DOI] [PubMed] [Google Scholar]
- 15.Dashwood RH, Suzui M, Nakagama H, Sugimura T, Nagao M. High frequency of β-catenin (Ctnnb1) mutations in the colon tumors induced by two heterocyclic amines in the F344 rat. Cancer Res. 1998;58:1127–1129. [PubMed] [Google Scholar]
- 16.Li Q, Dashwood WM, Zhong X, Nakagama H, Dashwood RH. Bcl-2 overexpression in PhIP-induced colon tumors: cloning of the rat Bcl-2 promoter and characterization of a pathway involving β-catenin, c-Myc, and E2F1. Oncogene. 2007;26:6194–6202. doi: 10.1038/sj.onc.1210438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang R, Dashwood WM, Löhr CV, et al. Protective versus promotional effects of white tea and caffeine on PhIP-induced tumorigenesis and β-catenin expression in the rat. Carcinogenesis. 2008;29:834–839. doi: 10.1093/carcin/bgn051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang R, Dashwood WM, Löhr CV, et al. β-Catenin is strongly elevated in rat colonic epithelium following short-term intermittent treatment with 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) and a high fat diet. Cancer Sci. 2008;99:1754–1759. doi: 10.1111/j.1349-7006.2008.00887.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang R, Dashwood WM, Nian H, et al. NADPH oxidase overexpression in human colon cancers and rat colon tumors induced by 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) Int J Cancer. 2011;128:2581–2590. doi: 10.1002/ijc.25610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parasramka MA, Dashwood WM, Wang R, et al. MicroRNA profiling of carcinogen-induced rat colon tumors and the influence of dietary spinach. Mol Nutr Food Res. 2012;56:1259–1269. doi: 10.1002/mnfr.201200117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang R, Löhr CV, Fischer KA, et al. Epigenetic inactivation of endothelin-2 and endothelin-3 in colon cancer. Int J Cancer. 2013;132:1004–1012. doi: 10.1002/ijc.27762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blum CA, Xu M, Orner GA, et al. β-Catenin mutation in rat colon tumors initiated by 1,2-dimethylhdrazine and 2-amino-3-methylimidazo[4,5-f]quinoline, and the effect of post-initiation treatment with chlorophyllin and indole-3-carbinol. Carcinogenesis. 2001;22:315–320. doi: 10.1093/carcin/22.2.315. [DOI] [PubMed] [Google Scholar]
- 23.Blum CA, Tanaka T, Zhong X, et al. Mutational analysis of Ctnnb1 and Apc in tumors from rats given 1,2-dimethylhdrazine and 2-amino-3-methylimidazo[4,5-f]quinoline: mutational ‘hotspots’ and the relative expression of β-catenin and c-jun. Mol Carcinogen. 2003;36:195–203. doi: 10.1002/mc.10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzui M, Ushijima T, Dashwood RH, et al. Frequent mutations of the rat β-catenin gene in colon cancers induced by methylazoxymethanol acetate plus 1-hydroxyanthraquinone. Mol Carcinogen. 1999;24:232–237. doi: 10.1002/(sici)1098-2744(199903)24:3<232::aid-mc10>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 25.Rajendran P, Delage B, Dashwood WM, et al. Histone deacetylase turnover and recovery in sulforaphane-treated colon cancer cells: competing actions of 14–3–3 and Pin1 in HDAC3/SMRT corepressor complex dissociation/reassembly. Mol Cancer. 2011;10:68. doi: 10.1186/1476-4598-10-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rajendran P, Kidane AI, Yu TW, et al. HDAC turnover, CtIP acetylation, and dysregulated DNA damage signaling in colon cancer cells treated with sulforaphane and related dietary isothiocyanates. Epigenetics. 2013;8:612–623. doi: 10.4161/epi.24710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang Y, Nian H, Rajendran P, et al. HDAC8 and STAT3 repress BMF gene activity in colon cancer cells. Cell Death Dis. 2014;5:e1476. doi: 10.1038/cddis.2014.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rajendran P, Dashwood WM, Li L, et al. Nrf2 status affects tumor growth, HDAC3 gene promoter associations, and the response to sulforaphane in the colon. Clin Epigenetics. 2015;7:102. doi: 10.1186/s13148-015-0132-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parasramka M, Dashwood WM, Wang R, et al. A role for low-abundance miRNAs in colon cancer: the miR-206/Krüppel-like factor 4 (KLF4) axis. Clin Epigenetics. 2012;4:16. doi: 10.1186/1868-7083-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morin PJ, et al. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 31.Ireton RC, Davis MA, van Hengel J, et al. A novel role for p120 catenin in E-cadherin function. J Cell Biol. 2002;159:465–476. doi: 10.1083/jcb.200205115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shtutman M, Zhurinsky J, Simcha I, Albanese C, D’Amico M, Pestell R, Ben-Ze’ev A. The cyclin D1 gene is a target of the beta-catenin/LEF1 pathway. Proc Natl Acad Sci USA. 1999;96:5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ito N, Hasegawa R, Sano T, et al. A new colon and mammary carcinogen in cooked food, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) Carcinogenesis. 1991;12:1503–1506. doi: 10.1093/carcin/12.8.1503. [DOI] [PubMed] [Google Scholar]
- 34.Yu M, Ryu DY, Snyderwine EG. Genomic imbalance in rat mammary gland carcinomas induced by 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) Mol Carcinogen. 2000;27:76–83. doi: 10.1002/(sici)1098-2744(200002)27:2<76::aid-mc3>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 35.Okochi E, Miyamoto K, Wakazono K, Shima H, Sugimura T, Ushijima T. Reduced Brda1 protein expression in 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine-induced rat mammary carcinomas. Mol Carcinogen. 2002;34:211–218. doi: 10.1002/mc.10065. [DOI] [PubMed] [Google Scholar]
- 36.Li Q, Dixon BM, Al-Fageeh M, Blum CA, Dashwood RH. Sequencing of the rat β-catenin (Ctnnb1) gene and mutational analysis of liver tumors induced by 2-amino-3-methylimidazo[4,5-f]quinoline. Gene. 2002;283:255–262. doi: 10.1016/s0378-1119(01)00839-3. [DOI] [PubMed] [Google Scholar]
- 37.Al-Fageeh M, Li Q, Dashwood WM, Myzak MC, Dashwood RH. Phosphorylation and ubiquitination of oncogenic mutants of β-catenin containing substitutions as Asp32. Oncogene. 2004;23:4839–4846. doi: 10.1038/sj.onc.1207634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peglion F, Etienne-Mannerville S. p120catenin alteration in cancer and its role in tumour invasion. Philos Trans R Soc Lond B Biol Sci. 2013;368:20130015. doi: 10.1098/rstb.2013.0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schackmann RC, Tenhagen M, van de Ven RA, Derksen PW. p120-catenin in cancer – mechanisms, models and opportunities for intervention. J Cell Sci. 2013;126:3515–3525. doi: 10.1242/jcs.134411. [DOI] [PubMed] [Google Scholar]
- 40.Hartsock A, Nelson WJ. Competitive regulation of E-cadherin juxtamembrane domain degradation by p120-catenin binding and Hakai-mediated ubiquitination. PLoS One. 2012;7:e37476. doi: 10.1371/journal.pone.0037476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thoreson MA, Reynolds AB. Altered expression of the catenin p120 in human cancer: implications for tumor progression. Differentiation. 2002;70:583–589. doi: 10.1046/j.1432-0436.2002.700911.x. [DOI] [PubMed] [Google Scholar]
- 42.Kreizenbeck GM, Berger AJ, Subtil A, Rimm DL, Gould Rothberg BE. Prognostic significance of cadherin-based adhesion molecules in cutaneous malignant melanoma. Cancer Epidemiol Biomarkers Prev. 2008;17:949–958. doi: 10.1158/1055-9965.EPI-07-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aho S, Levänsuo L, Montonen O, Kari C, Rodeck U, Uitto J. Specific sequences in p120ctn determine subcellular distribution of its multiple isoforms involved in cellular adhesion of normal and malignant epithelial cells. J Cell Sci. 2002;115:1391–1402. doi: 10.1242/jcs.115.7.1391. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y, Zhao Y, Jiang G, et al. Impact of p120-catenin isoforms 1A and 3A on epithelial-mesenchymal transition of lung cancer cells expressing E-cadherin in different subcellular locations. PLoS One. 2014;9:e88064. doi: 10.1371/journal.pone.0088064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hao L, Ha JR, Kuzel P, Garcia E, Persad S. Cadherin switch from E- to N-cadherin in melanoma progression is regulated by the PI3K/PTEN pathway through Twist and Snail. Br J Dermatol. 2012;166:1184–1197. doi: 10.1111/j.1365-2133.2012.10824.x. [DOI] [PubMed] [Google Scholar]
- 46.Mizuno H, Kitada K, Nakai K, Sarai A. PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med Genomics. 2009;2:18. doi: 10.1186/1755-8794-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stairs DB, Bayne LJ, Rhoades B, et al. Deletion of p120-catenin results in a tumor microenvironment with inflammation and cancer that establishes it as a tumor suppressor gene. Cancer Cell. 2011;19:470–483. doi: 10.1016/j.ccr.2011.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Orner GA, Dashwood WM, Blum CA, et al. Response of Apcmin and A33ΔNβcat mutant mice to treatment with tea, sulindac, and, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) Mutat Res. 2002;506–507:121–127. doi: 10.1016/s0027-5107(02)00158-6. [DOI] [PubMed] [Google Scholar]
- 49.Dashwood RH. Modulation of heterocyclic amine-induced mutagenicity and carcinogenicity: an ‘A-to-Z’ guide to chemopreventive agents, promoters, and transgenic models. Mutat Res. 2002;511:89–112. doi: 10.1016/s1383-5742(02)00005-4. [DOI] [PubMed] [Google Scholar]
- 50.Wang H, Zhou H, Liu A, Guo X, Yang CS. Genetic analysis of colon tumors induced by a dietary carcinogen PhIP in CYP1A humanized mice: identification of mutation of β-catenin/Ctnnb1 as the driver gene for the carcinogenesis. Mol Carcinogen. 2015;54:1264–1274. doi: 10.1002/mc.22199. [DOI] [PMC free article] [PubMed] [Google Scholar]


