Abstract
Vernalization is an essential process by which many temperate plant species acquire competence for flowering. Brachypodium distachyon is a model plant for temperate grasses including many cool-season forage and turfgrasses and cereals. Grasses with spring growth habit do not require vernalization for flowering and are typically less winter hardy. Yet the connection between vernalization and freezing tolerance remain unclear. The diverse requirement of vernalization for flowering makes it an ideal choice for studying the relationship between vernalization and freezing tolerance. Here, we isolated and analyzed the spatial and temporal expression patterns of two vernalization related homologous genes, BdVRN1 and BdVRN3 in Bd21, a non-vernalization-requiring accession, and Bd29-1, an accession shown to be vernalization-requiring. We showed that expression of BdVRN1 and BdVRN3 is independent of vernalization in Bd21, but is vernalization dependent in Bd29-1. Moreover, vernalization-induced expression of BdVRN1 appears to precede that of BdVRN3 in Bd29-1. Bd21 RNAi knockdown mutants for BdVRN1 conferred vernalization requirement for flowering, and reduced the expression of BdVRN3. Both Bd29-1 and the BdVRN1 RNAi mutants of Bd21 exhibited reduced freezing tolerance upon vernalization treatment. Cold-responsive genes BdCBF2, BdCBF3, BdCBF5, BdCBF6, and BdDREB2A were all constitutively expressed at a high level in the BdVRN1 RNAi mutants of Bd21. Taken together, our results suggest that expression of BdVRN1 promotes flowering by upregulating BdVRN3, and gaining the competency for flowering reduces freezing tolerance in Brachypodium.
Keywords: vernalization, freezing tolerance, cold acclimation, flowering time, Brachypodium
Introduction
Flowering occurs as a result of a critical growth transition from vegetative to reproductive growth in plants. In plants with a winter growth habit, an exposure to a period of low temperature is essential to flowering induction, a process known as vernalization (Boss et al., 2004; Baurle and Dean, 2006). The long period of time required to fulfill the vernalization requirement assures that reproductive organs, which are more sensitive to chilling/freezing injuries than vegetative tissues, do not form in the middle of the winter (Kim et al., 2004; Balasubramanian et al., 2006; Baurle and Dean, 2006; Trevaskis et al., 2007).
The molecular mechanism of vernalization-induced flowering has been extensively studied in Arabidopsis, wheat, and barley (Amasino, 2005; Izawa, 2007; Schmitz and Amasino, 2007; Distelfeld et al., 2009; Srikanth and Schmid, 2011). FLOWERING LOCUS T (FT) in Arabidopsis is identified to be the “florigen” (Kardailsky et al., 1999; Kobayashi et al., 1999; Notaguchi et al., 2008), which encodes a small globular protein that is able to translocate from the leaves, through the phloem, to the shoot apex where it interacts with a bZIP transcription factor FLOWERING LOCUS D (FD) to activate the expression of the floral meristem identity gene APETALA1 (AP1) for flowering (Abe et al., 2005; Wigge et al., 2005). In wheat and barley, VERNALIZATION 3 (VRN3) and VERNALIZATION 1 (VRN1) genes have been identified to be the homologs of the Arabidopsis FT and AP1, respectively (Danyluk et al., 2003; Murai et al., 2003; Trevaskis et al., 2003; Yan et al., 2003, 2006; Faure et al., 2007).
Previous studies, however, have shown that the vernalization pathways are not conserved between Arabidopsis and temperate cereals (Amasino, 2005; Trevaskis et al., 2007; Dennis and Peacock, 2009; Distelfeld et al., 2009; Greenup et al., 2009; Kim et al., 2009; Andres and Coupland, 2012). In Arabidopsis, FLOWERING LOCUS C (FLC), a MADS-box transcription factor gene, has been identified to be a negative regulator of flowering, which blocks the long-day induction of FT but itself is repressed by vernalization (Michaels and Amasino, 1999; Searle et al., 2006). In temperate cereals, no homologous gene of FLC has been found, but VERNALIZATION 2 (VRN2) gene was identified as a flowering repressor in cereals, which encodes a zinc-finger and CCT (CONSTANS, CONSTANS-LIKE, TOC1) domain protein (ZCCT1) (Yan et al., 2004). VRN2 is induced by long-day and can function like FLC to suppress long-day induction of VRN3 (Yan et al., 2004, 2006). VRN2 has also been reported to repress VRN1 gene, but this repression is relieved by vernalization (Danyluk et al., 2003; Trevaskis et al., 2003; Yan et al., 2003). Vernalization induces the expression of VRN1 which in turn repress VRN2, thereby releasing the repression of VRN3 by VRN2 (Yan et al., 2003, 2004, 2006). VRN3 further elevates the expression of VRN1 forming a signaling feedback loop of VRN1-VRN2-VRN3-VRN1 that regulates flowering (Yan et al., 2004; Dubcovsky et al., 2006; Trevaskis et al., 2006). However, Shimada et al. (2009) proposed an alternative model of vernalization response in wheat. In a mutant that has null alleles of VRN1, the authors could not detect the expression of the VRN3 gene and the mutant remained at the vegetative phase. In this alternative model, VRN1 is directly upstream of VRN3 instead of VRN2 and up-regulates the VRN3 expression with a vernalization treatment. VRN3 in turn represses VRN2, releasing the inhibition of VRN1 by VRN2 (Shimada et al., 2009). The interrelationships among the VRN genes in temperate grasses remain to be investigated.
In general, plants with a vernalization-requiring winter growth habit have better freezing tolerance compared with plants having a spring growth habit that are non-vernalization-requiring, suggesting a link between vernalization requirement and freezing tolerance (Antikainen and Griffith, 1997; Limin and Fowler, 2006; Sandve et al., 2011; Chawade et al., 2012). However, whether and how vernalization is related to freezing tolerance remain to be investigated. Previous findings showed that vernalization pathway and cold acclimation pathway are interconnected (Liu et al., 2002; Seo et al., 2009; Dhillon et al., 2010; Lee et al., 2012). SUPPERSSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1), which encodes a MADS-box transcription factor, has been reported to regulate multiple floral induction pathways including vernalization, photoperiod, and autonomous (Onouchi et al., 2000; Samach et al., 2000). SOC1 may also play an important role on regulation of cold acclimation, a period of exposure to low temperatures that results in a significant increase in freezing tolerance (acquired freezing tolerance). Cold acclimation induces the cold-responsive genes including the C-repeat binding factor (CBFs)/Dehydrate Responsive Element Binding (DREB) and Cold Regulated (COR) (Thomashow, 1999; Chinnusamy et al., 2007; Zhang et al., 2009, 2017; Chinnusamy et al., 2010). The SOC1 knockout mutants increased the expression of cold-responsive genes such as CBFs and COR whereas SOC1 overexpression mutants decreased the expression of these genes in Arabidopsis (Seo et al., 2009). In addition, heterologous expression of the wheat VRN2 (TaVRN2) gene in Arabidopsis delayed flowering and enhanced freezing tolerance due to the accumulation of CBF2, CBF3, and COR genes (Diallo et al., 2010). In barley, genotype with the vrn-1 allele had higher expression of CBF genes than genotypes with the Vrn-1 allele, and CBF transcript abundance decreased after vernalization in the vrn-1 genotype (Dhillon et al., 2010). In wheat, freezing tolerance and transcript abundance of several CBF and COR genes were much lower in a deletion mutant, maintained vegetative phase (mvp) in which VRN1, along with several other genes are deleted (Dhillon et al., 2010). Taken together, these results suggest that VRN1 has a negative effect on the expression of CBF genes that influence freezing tolerance.
To better understand the mechanism of vernalization pathway and its relationship with freezing tolerance in temperate grasses, we used the monocot model plant, Brachypodium distachyon for our study. With its sequenced genome, a small physical stature, self-fertility, a short life cycle, an efficient transformation system and abundant natural variation in flowering habit, Brachypodium is well suited for studying the molecular mechanism of vernalization and its relationship with freezing tolerance (Draper et al., 2001; Vogel et al., 2010).
In this study, we isolated and analyzed the expression of three putative VRN genes in Brachypodium of either a non-vernalization-requiring (Bd21) or a vernalization-requiring accession (Bd29-1) and studied their freezing tolerance. We showed that BdVRN1 and BdVRN3 are induced by vernalization and are positive regulators of flowering in Bd29-1. The BdVRN2-like gene is likely not involved in the vernalization pathway in Brachypodium. Knockdown of BdVRN1 in Bd21 also reduced the expression of BdVRN3, but had no effect on the expression of the BdVRN2-like gene and resulted in a dramatic non-flowering phenotype, which can be rescued by a vernalization treatment. Meanwhile, enhanced tolerance to freezing stress was observed in the RNAi mutants, accompanied by constitutive expression of several cold-responsive genes, BdCBF2, BdCBF3, BdCBF5, BdCBF6, and DREB2A at high levels. These results suggest that BdVRN1 plays a critical role on flowering in vernalization pathway and its expression negatively affects the regulation of the cold-responsive genes and reduces freezing tolerance in Brachypodium.
Materials and Methods
Plant Materials and Growth Condition
Seeds of B. distachyon Bd21 (PI 254867) and Bd29-1(PI 639818) were sown in 6-inch pots containing Sun Gro Hort soil mix (Bellevue, WA, United States) in the greenhouse at 25°C, 16/8h (day/night) with an irradiance of 450 ± 50 μmol m-2 s-1.
RNAi Vector Construction and Generation of RNAi Mutants
A fragment of BdVRN1 (414 bp) gene was amplified by PCR (primers listed in Supplementary Table S2) with the addition of four bases of CACC at its 3′ end. PCR was performed as follows: 94°C for 5 min; 35 cycles of 94°C for 20 s, 59°C for 30 s, and 72°C for 50 s; final extension at 72°C for 1 min. The PCR product was separated on 1% agarose gel and extracted from the gel. Cleaned gene fragment was cloned into a Gateway® entry vector pENTRTM/D-TOPO® (Life Technologies, Grand Island, NY, United States) containing attL and attR recombination sites according to the manufacturer’s protocol. The Gateway® compatible pANDA vector (Miki and Shimamoto, 2004) containing two cassettes in inverse orientation linked by a small fragment of the GUS gene was used as the destination vector for BdVRN1. The attL × attR reaction is mediated by Gateway® LR ClonaseTM II enzyme mix (Life Technologies, Grand Island, NY, United States). Kanamycin (nptII) and hygromycin (hpt) resistance genes were used for the selection in bacteria and plants, respectively. The final binary vector pANDA::BdVRN1 was verified by Sanger sequencing (Supplementary Figure S1).
The pANDA::BdVRN1 vector was introduced into Agrobacterium tumefaciens C58C1 strain for transformation of Brachypodium following the protocol developed by Vogel and Hill (2008). The selection medium contained 150 mg L-1 Timentin (bioWORLD, Dublin, OH, United States) to suppress Agrobacterium growth and 40 mg L-1 hygromycin B (bioWORLD, Dublin, OH, United States) to kill untransformed calli. Two cycles of selection, each lasting 2 weeks were performed under dark at 23°C. Resistant calli were transferred into the regeneration medium containing Kinetin (KT) at 1 mg L-1 and hygromycin at 10 mg L-1 in a tissue culture chamber at 23°C, 16/8 h (light) with an irradiance of 120 μmol m-2 s-1. Shoots started to appear 7–10 days after the transfer. Rooting took place on a MS medium supplemented with 0.1 mg L-1 NAA and 10 mg L-1 hygromycin. Well-rooted plantlets were carefully moved into 6-inch pots containing Sun Gro Hort soil mix (Bellevue, WA, United States). Plants were grown in a growth chamber at 23°C, 16/8 h (day/night) with an irradiance of 400 ± 30 μmol m-2 s-1.
Transgenic plants were screened by PCR using primers (HPT-F 5′ GAATTCAGCGAGAGCCTG 3′, HPT-R 5′ ACATTGTTGGAGCCGAAA 3′) designed from the sequence of the hygromycin resistant gene present in the pANDA binary vector. Eight independent T0 lines were confirmed by PCR analyses. In two of the independent lines, RNAi-4 and RNAi-12, PCR-positive and negative plants segregated in a 3:1 ratio in the progenies, indicating the integration of the transgene occurred at a single locus in each of the two lines. Homozygous plants of T2 RNAi-4 and RNAi-12 lines were chosen for further analyses.
Cold Acclimation, Vernalization Treatment, and Assessment of Freezing Tolerance
Cold acclimation was done by growing plants in growth chamber at a 4°C and 8/16 h (day/night) with an irradiance of 400 ± 30 μmol m-2 s-1 for 10 days. To determine the vernalization requirement for flowering, 4-week-old plants of Bd 21 or Bd29-1 accession were placed into a growth chamber at 4°C and 8/16 h (day/night) with an irradiance of 400 ± 30 μmol m-2 s-1 for 3, 6, 9, or 12 weeks before they were moved back to a greenhouse (25°C, 16/8 h day/night) with an irradiance of 450 ± 50 μmol m-2 s-1.
Freezing tolerance was evaluated for Bd21, Bd29-1and BdVRN1 RNAi mutants. Five-week-old plants were treated with -5°C and 8/16 h (day/night) for 12 h. Following the freezing treatment, plants were moved into a growth chamber at 4°C for recovery for 1 day and were then transferred into a greenhouse (25°C, 16/8 h day/night) and evaluated for recovery 1 week later. Percentage of plants that survived the freezing test and regrew were recorded.
Gene Expression Analysis by Semiquantitative and Real-Time RT-PCR Analysis
To examine the expression levels of BdVRN genes (Supplementary Table S1) in response to vernalization treatment, 4-week-old plants of each accession were placed into a growth chamber at 4°C and 8/16 h (day/night) with an irradiance of 400 ± 30 μmol m-2 s-1. Leaves and meristem were harvested separately 2, 4, 6, 8, 10, or 12 weeks following the conclusion of the vernalization treatment.
For semiquantitative RT-PCR analysis, total RNA was extracted from leaves or meristems with TRIzol® Reagent (Invitrogen, Carlsbad, CA, United States). Reverse transcription was performed with the SuperScript® III First-Strand Synthesis System for RT-PCR kit (Invitrogen, Carlsbad, CA, United States). PCR was performed with gene specific primers (See Supplementary Table S2 for primer information) with the following program: pre-denaturation at 94°C for 5 min; 25 cycles of denaturation at 94°C for 30 s, primer-annealing at 58°C for 30 s, elongation at 72°C for 50 s, and post elongation at 72°C for 5 min for all genes. The expression level of each gene was normalized to that of the BdGAPDH reference gene.
For Real-Time RT-PCR analysis, total RNA extraction and reverse transcription were done the same way as described for the semiquantitative RT-PCR analysis. Quantitative analyses were carried out on the Eco® Real-Time PCR System (Illumina, Inc., San Diego, CA, United States) using the SYBR® GreenERTM qPCR SuperMix kit (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s instructions. The quantity of PCR products was determined at the end of each cycle by the Eco® Software v4.0 (Illumina, Inc., San Diego, CA, United States). The expression level of each gene was normalized to that of the BdGAPDH gene, and the expression level for each gene in the wild type without treatment was set as 1.0. Primers used for PCR amplification are listed in Supplementary Table S3.
Results
Brachypodium Accessions Vary in Vernalization Requirement for Flowering
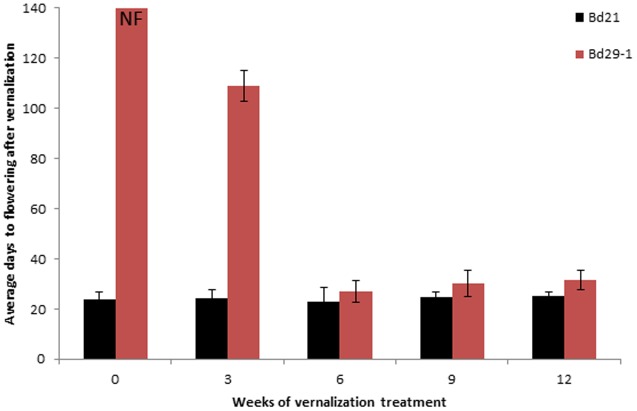
Time to flower was determined for accessions Bd29-1and Bd21 with or without vernalization treatment. Bd21 started flowering within 24 ± 3 days, regardless of the vernalization treatment (Figure 1), indicating vernalization treatment had no significant effect on accelerating flowering in Bd21. In contrast, without vernalization treatment Bd29-1 did not flower even after 140 days when record-taking was stopped. When subjected to a 3-week vernalization treatment, Bd29-1 started flowering after 109 ± 6.2 day. A 6-week vernalization treatment of Bd29-1 reduced the time to flower to just 27 ± 4.2 days. Longer than 6 weeks of vernalization treatment did not reduce the number of days to flowering further. Clearly, accession Bd29-1 requires vernalization treatment to flower.
FIGURE 1.
Average number of days required for flowering for the non-vernalization-requiring accession Bd21 and the vernalization-requiring accession Bd29-1 after vernalization treatment. Four-week-old seedlings (30 plants for each accession) were either not vernalized or vernalized for 3, 6, 9, or 12 weeks at 4°C and 8/16 h (day/night) in a growth chamber. After vernalization treatment, plants were transferred to a greenhouse (25°C, 16/8 h day/night). NF denotes no flowering observed. Average number of days to flowering indicate the number of days required for flowering following a vernalization treatment. Error bars represent the standard deviation from independent plants.
Vernalization Requirement of Brachypodium Is Strongly Associated with the Expression of BdVRN Genes
Three genes of VRN1, VRN2, and VRN3 that regulate the vernalization requirement in wheat or barley have been characterized. Previous comparative genomic analysis showed that Brachypodium has a VRN1 homolog, Bradi1g08340 (BdVRN1), located in a position co-linear to the rice (OsMADS14) and wheat (TaVRN-1) homologous genes, and a VRN3 homolog, Bradi1g48830 (BdVRN3). Although there was no apparent homologous VRN2-like gene in Brachypodium, Bradi3g10010 is a member of the group IV CCT gene, an intermediate between the barley HvCO9 and cereal VRN2 genes in the phylogenetic tree (Higgins et al., 2010). For these reasons, we designated Bradi3g10010 as BdVRN2.
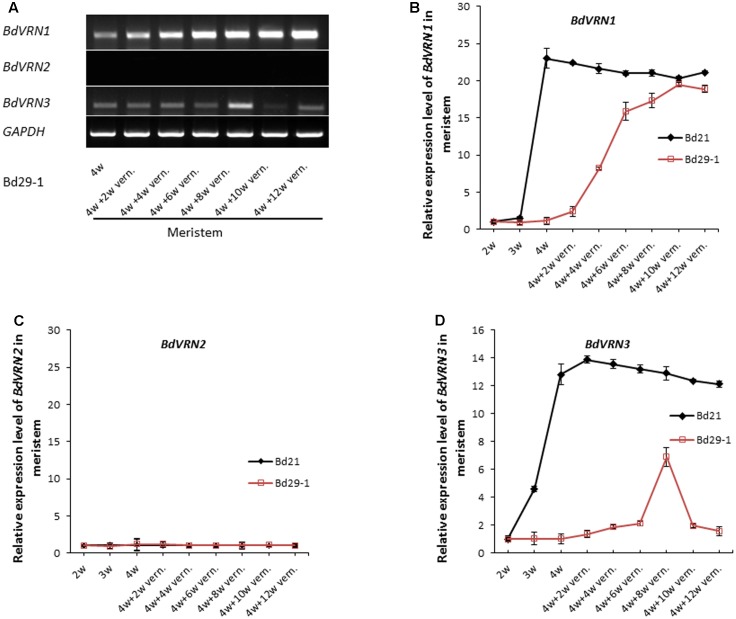
We next assayed the expression of the BdVRN1, 2 and 3 genes in both Bd21 and Bd29-1 with or without vernalization treatment. Without vernalization treatment, expression of BdVRN1 expression remained at a low level in leaves and the meristem during the first 3 weeks following seed germination for both Bd21 and Bd29-1 (Figure 2A and Supplementary Figure S2). Four weeks after germination, BdVRN1 expression was dramatically increased by 20-fold in the meristem in Bd21, whereas it remained unchanged in Bd29-1 (Figures 2A,B). Following various length of vernalization treatments, expression of BdVRN1 in the meristem of Bd21 remained at a high level, whereas in Bd29-1 it was gradually increased to a level similar to that of Bd21 after 6 weeks of vernalization treatment (Figures 2A,B).
FIGURE 2.
Expression of BdVRN genes in meristems of the non-vernalization-requiring Bd 21 and vernalization-requiring Bd29-1 accessions. (A) BdVRN gene expression in meristems of 4-week-old seedlings of Bd29-1 that were vernalized for 2, 4, 6, 8, 10, or 12 weeks. BdGAPDH was used as loading control. (B–D) Relative expression levels of BdVRN1, BdVRN2, and BdVRN3 genes in meristems of Bd21 and Bd29-1. Prior to vernalization, plants were grown at a greenhouse (25°C, 16/8 h day/night). Four-week-old seedlings were transferred to a growth chamber at 4°C and 8/16 h (day/night) for vernalization treatment. Meristems were harvested after 2, 4, 6, 8, 10, or 12 weeks of vernalization treatment. Error bars represent the standard deviation from independent plants.
BdVRN2 was expressed at a high level in leaves (Supplementary Figure S2A) but was expressed at a very low level in the meristem of both Bd21 and Bd29-1 (Figure 2C). Semiquantitative RT-PCR showed that BdVRN2 was highly expressed in leaves of Bd21 with or without vernalization and its expression in Bd29 appeared to increase slightly after a 2-week vernalization; but longer vernalization did not increase further (Supplementary Figure S2B). Interestingly, the expression of BdVRN2 in the meristem is not influenced by vernalization in both Bd21 and Bd29-1 (Figure 2C and Supplementary Figure S2B). In Bd21, expression of BdVRN3 in the meristem increased continuously before vernalization treatment took place, and did not change significantly by vernalization treatment (Figure 2D). In contrast, in Bd29-1, the expression of BdVRN3 in the meristem remained at a low level without vernalization, but increased gradually with vernalization treatment and increased by sixfolds when it reached the peak following a 8-week vernalization (Figure 2D).
Knockdown of BdVRN1 in Bd21 Conferred Vernalization Requirement
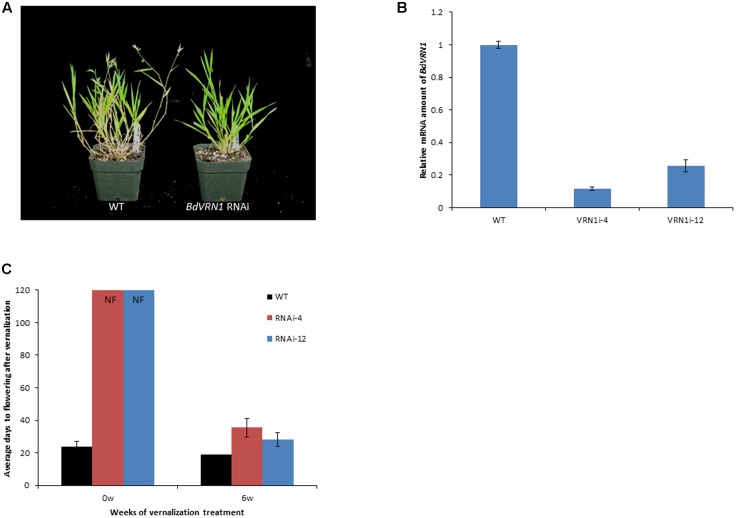
To determine whether BdVRN1 indeed regulates flowering time, RNAi mutants of BdVRN1 were created for Bd21. Eight independent T0 transgenic lines were confirmed by PCR analysis and two of them, RNAi-4 and RNAi-12 did not flower without vernalization (Figures 3A,C). Real-Time RT-PCR showed that the BdVRN1 transcripts were reduced to 0.12 ± 0.02 in RNAi-4 and 0.26 ± 0.04 in RNAi-12 (Figure 3B). After a 6-week vernalization treatment, RNAi-4 and RNAi-12 mutants started flowering after 35.5 ± 5.6 days and 28.2 ± 8.3 days, respectively, compared to 19.2 ± 5.4 days for the wild type. The phenotypic change of Bd21 from non-vernalization-requiring to vernalization-requiring in the BdVRN1 knockdown mutants strongly indicate that BdVRN1 is of paramount importance to flowering induction in Brachypodium.
FIGURE 3.
(A) Wild type (WT) (left) and BdVRN1 RNAi mutant plant (right) 7 weeks after seed germination. (B) Real-Time RT-PCR analysis of the BdVRN1 expression level in the WT, knockdown mutants of BdVRN1 RNAi-4 and RNAi-12. (C) Average number of days required for flowering for the WT Bd21 and RNAi mutants with or without a 6-week vernalization treatment. Four-week-old seedlings (30 plants for each accession) were either not vernalized or vernalized for 6 weeks at 4°C and 8/16 h (day/night) in a growth chamber. After the vernalization treatment, plants were transferred to a greenhouse (25°C, 16/8 h day/night). NF denotes non-flowering phenotype. The average number of days to flowering indicates the number of days passed before flowering occurs following a vernalization treatment. Error bars represent the standard deviation from independent plants.
Knockdown of BdVRN1 in Bd21 Reduced the Expression of BdVRN1 and BdVRN3, But Not BdVRN2
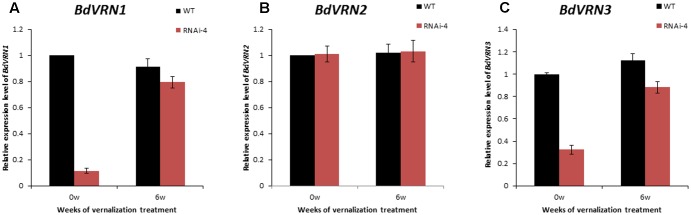
To better understand how BdVRN1 affects flowering at the molecular level, we analyzed the expression patterns of BdVRN1, BdVRN2, and BdVRN3 in the RNAi-4 knockdown mutant. The expression of BdVRN1 was significantly suppressed in 4-week-old seedlings of the RNAi mutant, but it increased significantly following a 6-week vernalization treatment, although it is still only half of the value observed for the wild type without cold acclimation (Figure 4A). Moreover, the expression level of BdVRN3 also decreased in the BdVRN1 mutants, but its expression was also elevated by vernalization treatment (Figure 4C). However, the expression of BdVRN2 was not affected in the mutant (Figure 4B). These results indicated that upregulation of BdVRN1 is positively correlated with BdVRN3, but not with BdVRN2.
FIGURE 4.
Expression analysis of BdVRN1 (A), BdVRN2 (B), and BdVRN3 (C) genes in the WT (Bd21) and BdVRN1 RNAi mutant. The relative expression levels of each gene were determined by Real-Time RT-PCR in leaf tissues. Prior to vernalization, plants were grown at a greenhouse (25°C, 16/8 h day/night). Four-week-old seedlings were then transferred to a growth chamber at 4°C (8/16 h day/night) for 0 or 6 weeks. BdGAPDH is a loading control. Error bars represent the standard deviation from independent plants.
Decreased BdVRN1 Expression Is Associated with Increased Freezing Tolerance Accompanied by Increased Expression of Cold-Responsive Genes
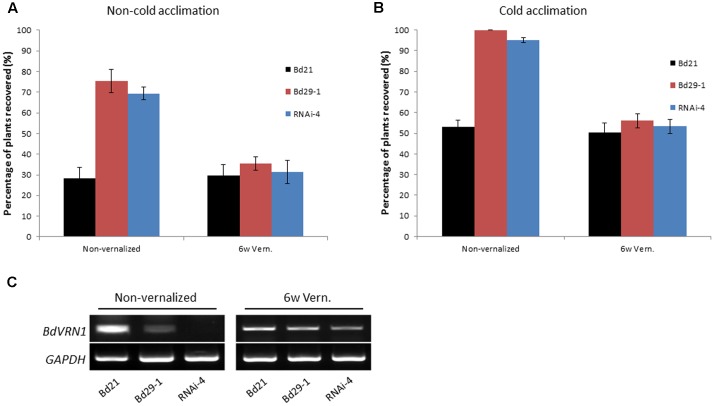
To determine the relationship between the vernalization requirement and freezing tolerance, we compared the freezing tolerance of Bd29-1 and Bd21. Without a vernalization treatment, Bd29-1 showed a much higher freezing tolerance than did Bd21 based on the percentage of plants that survived the freeze-thaw-regrowth test. When plants of both accessions were vernalized for 6 weeks, the recovery rate of Bd21 was nearly unchanged whereas that of Bd29-1 greatly decreased (Figure 5A). With a cold acclimation, both Bd21 and Bd29-1 had an increase in recovery rate compared with their non-cold acclimated counterparts (Figure 5B), which clearly indicated that both accessions have a well-defined cold acclimation mechanism to increase freezing tolerance upon an exposure to low temperatures. The result that vernalization treatment reduced the freezing tolerance of Bd29-1, but not Bd21suggested that vernalization requirement is negatively correlated with freezing tolerance. Furthermore the BdVRN1 RNAi-4 mutant that gained vernalization requirement is also much more freezing tolerant than that of the wild type Bd21 with or without cold acclimation (Figures 5A,B). Semiquantitative RT-PCR experiments confirmed that without vernalization treatment Bd29-1 and the Bd21 RNAi-4 mutant had lower BdVRN1 expression levels than the wild type Bd21, but a 6-week vernalization treatment increased the expression levels of BdVRN1 in both Bd29-1 and the BdVRN1 RNAi-4 mutant approaching to that in Bd21 (Figure 5C). This increase in BdVRN1 expression coincides with the reduced freezing tolerance.
FIGURE 5.
Freezing tolerance of vernalized or non-vernalized Bd21 and Bd29-1 with or without cold acclimation. (A) Freezing tolerance of non-vernalized or vernalized plants of Bd21, Bd29-1, and BdVRN1 mutant RNAi-4 without cold acclimation. Freezing tolerance was assayed by subjecting plants to –5°C and 8/16 h (day/night) for 12 h in a growth chamber. Plants were then moved back into a greenhouse (25°C, 16/8 h day/night) and were examined for recovery a week later. The non-vernalized plants were 5-week-old. (B) Freezing tolerance of non-vernalized or vernalized plants of Bd21, Bd29-1, and RNAi-4 mutant following a 10 day cold acclimation. (C) Semiquantitative RT-PCR of BdVRN1 gene in leaves from vernalized or non-vernalized plants of Bd21, Bd29-1, and RNAi-4 mutant. Leave tissues were collected before assessing freezing tolerance for each accession. Error bars represent the standard deviation from independent plants.
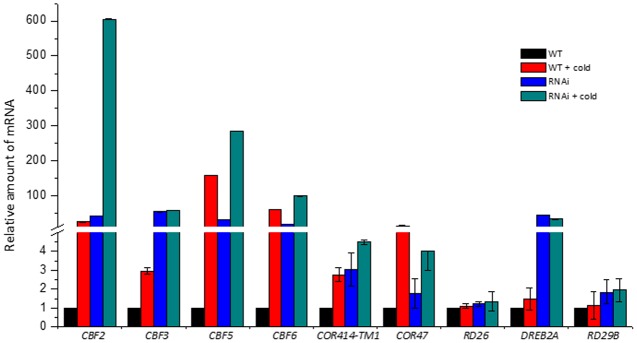
The regulation of gene expression in response to cold is quite complex with more than a 1000 genes are cold induced, but a critical group of them are organized into a cascade controlled through a regulatory hub involving cold-inducible CBF genes (Chinnusamy et al., 2007; Thomashow, 2010). The CBF genes encode members of the AP2/ERF family of transcription factors, which bind to the CRT/DRE regulatory elements in target genes of the CBF regulon and increase freezing tolerance (Jaglo-Ottosen et al., 1998; Liu et al., 1998; Catala et al., 2003; Dubouzet et al., 2003; Ito et al., 2006; Xiong and Fei, 2006; Lata and Prasad, 2011). For example, CBF1 in Arabidopsis is a transcriptional activator for the COR genes, such as COR47 and COR414-TM1, which are involved in cold and dehydration responses (Xiong et al., 2002; Sakuma et al., 2006; Griffith et al., 2007). DREB2A is a transcription factor induced by dehydration in Arabidopsis, rice, soybean, and maize (Simpson et al., 2003; Sakuma et al., 2006; Qin et al., 2007; Cui et al., 2011; Mizoi et al., 2013). The RD (RESPONSIVE TO DESICCATION) genes, for example RD26 and RD29B, are induced by cold or drought stress through the regulation of the transcription factor DREBs (Yamaguchi-Shinozaki and Shinozaki, 1994; Nakashima et al., 2006; Msanne et al., 2011). To further characterize the relationship between BdVRN1 gene and freezing tolerance, we analyzed the expression of some important cold-responsive genes by real-time RT-PCR in both the wild type Bd21 and the BdVRN1 RNAi-4 mutants without vernalization treatment.
Previously, we isolated eight CBF genes (CBF1 Bradi4g35630, CBF2 Bradi4g35620, CBF3 Bradi4g35650, CBF4 Bradi4g35570, CBF5 Bradi4g35580, CBF6 Bradi4g35590, CBF7 Bradi4g35600, and CBF8 Bradi4g35650) from Brachypodium that are tandemly arranged in chromosome 4 and are all cold induced (not published). Here we compared their expression with four other important cold-responsive genes between the wild type Bd21 and the BdVRN1 RNAi-4 mutants. We showed that CBF2, CBF3, CBF5, CBF6, COR414-TM1, and COR47 were highly induced by 3- to 600-fold in both the wild type and the BdVRN1 RNAi-4 mutants after a mere 24 h of cold treatment (Figure 6). More interestingly, CBF2, CBF3, CBF5, CBF6, and DREB2A were highly expressed in the BdVRN1 RNAi-4 mutant without cold induction, and the constitutive expression of CBF3 and DREB2A in the RNAi mutant was even higher than that of the cold-treated wild type. On the other hand, RD26 and RD29B genes showed no noticeable changes in the RNAi mutant, compared to the wild type.
FIGURE 6.
Real-Time RT-PCR analysis of the cold-responsive genes in the WT and RNAi mutant with or without cold treatment. Leaves were harvested 24 h after a 4°C cold treatment. The expression level of each gene was normalized to that of BdGAPDH gene, and the expression level for each gene in the WT without cold treatment was set to 1.0. Error bars represent the standard deviation from independent plants.
Discussion
In this study, we examined the requirement for vernalization in Brachypodium accessions Bd29-1 and Bd21 and discovered that while Bd21 does not require vernalization for flowering, Bd29-1 requires vernalization. To understand the molecular basis underlining the differences in vernalization requirement between the two accessions, we isolated homologs of the three known cereal vernalization genes, BdVRN1, BdVRN2, and BdVRN3 from both accessions, and analyzed their temporal expression patterns in leaves and the meristem. It was shown that the vernalization requirement is closely correlated with the expression patterns of the BdVRN1 and BdVRN3 genes, but not with the BdVRN2 gene. Bd21 started flowering in approximately 7 weeks following seed germination, accompanied by accumulations of both BdVRN1 and BdVRN3 transcripts in the meristem regardless of vernalization treatment. In contrast, a minimum of 3-week vernalization is required for Bd29-1 to gain competence for flowering while 6 weeks or more significantly reduced the number of days for vernalized plants to flower. Accumulation of BdVRN1 appears to precede the accumulation of BdVRN3. However, the expression of BdVRN2 was not correlated to vernalization treatment in both Bd21 and Bd29-1. Therefore, we speculated that BdVRN1 and BdVRN3 positively regulate vernalization response of the vernalization-requiring accession Bd29-1. It is consistent with the results of a previous study that overexpression of FT1 (VRN3) caused extremely early flowering during shoot regeneration and downregulation of FT1 by RNA interference (RNAi) resulted in non-flowering Brachypodium (Lv et al., 2014). The loss-of-function mutants exhibited vernalization-requiring phenotype, similar to Bd29-1. Gene expression analysis showed that the expression of not only BdVRN1 but also BdVRN3 were suppressed in the mutants, however, expression of both were elevated by vernalization treatment. The expression of BdVRN2 was not affected in the RNAi mutants with or without vernalization treatment, suggesting BdVRN2 may not be a true homolog, or is not involved in flowering in Brachypodium. This is different from Ream et al. (2014) who reported that BdVRN2 is induced during cold.
Our result is consistent with previous studies in wheat and barley that both VRN1 and VRN3 are induced by vernalization with a quantitative effect on the timing of flowering initiation (Danyluk et al., 2003; Murai et al., 2003; Yan et al., 2003, 2006). Different hypotheses have been proposed on the relationship between VRN1 and VRN3 in temperate cereals. Li and Dubcovsky (2008) posited VRN3 regulate the VRN1 expression in the vernalization pathway, while others suggested that VRN1 is upstream of VRN3 (Sasani et al., 2009; Shimada et al., 2009). Our results largely agree with the idea that VRN1 is induced by vernalization and up-regulates the expression of VRN3.
Comparative genomic analysis suggested that VRN2 gene has been lost in the Bd21 genome, similar to rice, which has no requirement for vernalization (Higgins et al., 2010). In our work, we chose a CCT gene grouped between HvCO9 and VRN2 in a phylogenetic tree as BdVRN2. We showed that BdVRN2 was stably expressed in Bd21, which is consistent with the idea by comparative genomic analysis that this is a loss-of-function BdVRN2 in Bd21. However, the BdVRN2 was also stably expressed independent of the vernalization both in Bd29-1 and the BdVRN1 RNAi mutants of Bd21, suggesting that BdVRN2 is not involved in the vernalization pathway. This is different from wheat or barley, in which VRN2 was characterized as a repressor of flowering and is regulated by both photoperiod and vernalization. Our result suggested that there are evolutionary difference between Brachypodium and temperate cereals. Interestingly a recent study identified a FLC homolog in Brachypodium that is a vernalization-regulated repressor (Sharma et al., 2016).
It is well observed that plants with vernalization requirement for flowering typically have better freezing tolerance than their non-vernalization-requiring close relatives (spring wheat vs. winter wheat or spring barley vs. winter barley), but the molecular link between them remains unclear. Previous works suggested that there might be a negative correlation between freezing tolerance and VRN1 transcript in wheat (Liu et al., 2002; Danyluk et al., 2003; Limin and Fowler, 2006). The expression of TaVRT-1, a homolog of AP1/VRN1 in wheat, was positively associated with vernalization treatment, and was negatively associated with the accumulation of COR genes and freezing tolerance (Danyluk et al., 2003). Expression studies showed that high levels of constitutive expression of some CBF genes in winter wheat cultivars confers a higher freezing tolerance compared to spring cultivars (Kobayashi et al., 2005; Badawi et al., 2007). In addition, the VRN1 deletion mutant, mvp, increased freezing tolerance along with an increased expression level of several CBF and COR genes (Dhillon et al., 2010). A subsequent study found that the deletion of the mvp mutant include other genes besides VRN1 such as AGLG1, CYS, and PHYC (Distelfeld and Dubcovsky, 2010). In the present study we showed that the vernalization-requiring Bd29-1 accession is more freezing tolerant than the non-vernalization-requiring Bd21, which corresponds to the different transcript levels of BdVRN1 between the two accessions. Significantly, when we treated the Bd29-1 and Bd21with vernalization for 6 weeks, the freezing tolerance of Bd29-1 was greatly reduced, but that of Bd21 remained the same. In order to exclude other factors in regulating freezing tolerance, we compared the freezing tolerance between the non-vernalization-requiring Bd21 and its VRN1 RNAi mutants which requires vernalization for flowering. Our results showed that freezing tolerance was enhanced in BdVRN1 knockdown mutants relative to the wild type. Vernalization treatment of the RNAi mutants reduced their freezing tolerance. Moreover, several key cold-responsive genes, BdCBF2, BdCBF3, BdCBF5, BdCBF6, and DREB2A, are constitutively expressed at high levels in the knockdown mutants. Whether BdVRN1 directly regulates the freezing tolerance pathway or function through interaction with other genes remains to be determined.
Author Contributions
SF designed the experiment; YF carried out the experiment; YF, SF, and YY analyzed the data; and YF, SF, and YY wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank John Vogel at USDA-ARS for kindly providing seeds of Brachypodium distachyon. We thank the Plant Science Institute at Iowa State University, the Consortium for Plant Biotechnology Research Inc. (GO12026-322 to SF and YY), the National Institute of Food and Agriculture of the US Department of Agriculture (2013-33522-21091 to SF), and the United States Golf Association for their support.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01107/full#supplementary-material
References
- Abe M., Kobayashi Y., Yamamoto S., Daimon Y., Yamaguchi A., Ikeda Y., et al. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309 1052–1056. 10.1126/science.1115983 [DOI] [PubMed] [Google Scholar]
- Amasino R. M. (2005). Vernalization and flowering time. Curr. Opin. Biotechnol. 16 154–158. 10.1016/j.copbio.2005.02.004 [DOI] [PubMed] [Google Scholar]
- Andres F., Coupland G. (2012). The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13 627–639. 10.1038/nrg3291 [DOI] [PubMed] [Google Scholar]
- Antikainen M., Griffith M. (1997). Antifreeze protein accumulation in freezing-tolerant cereals. Physiol. Plant. 99 423–432. 10.1034/j.1399-3054.1997.990310.x [DOI] [Google Scholar]
- Badawi M., Danyluk J., Boucho B., Houde M., Sarhan F. (2007). The CBF gene family in hexaploid wheat and its relationship to the phylogenetic complexity of cereal CBFs. Mol. Genet. Genomics 277 533–554. 10.1007/s00438-006-0206-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S., Sureshkumar S., Lempe J., Weigel D. (2006). Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genet 2:e106 10.1371/journal.pgen.0020106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baurle I., Dean C. (2006). The timing of developmental transitions in plants. Cell 125 655–664. 10.1016/j.cell.2006.05.005 [DOI] [PubMed] [Google Scholar]
- Boss P. K., Bastow R. M., Mylne J. S., Dean C. (2004). Multiple pathways in the decision to flower: enabling, promoting, and resetting. Plant Cell 16(Suppl.), S18–S31. 10.1105/tpc.015958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catala R., Santos E., Alonso J. M., Ecker J. R., Martinez-Zapater J. M., Salinas J. (2003). Mutations in the Ca2+/H+ transporter CAX1 increase CBF/DREB1 expression and the cold-acclimation response in Arabidopsis. Plant Cell 15 2940–2951. 10.1105/Tpc.015248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawade A., Linden P., Brautigam M., Jonsson R., Jonsson A., Moritz T., et al. (2012). Development of a model system to identify differences in spring and winter oat. PLoS ONE 7:e29792 10.1371/journal.pone.0029792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V., Zhu J., Zhu J. K. (2007). Cold stress regulation of gene expression in plants. Trends Plant Sci. 12 444–451. 10.1016/j.tplants.2007.07.002 [DOI] [PubMed] [Google Scholar]
- Chinnusamy V., Zhu J. K., Sunkar R. (2010). Gene regulation during cold stress acclimation in plants. Methods Mol. Biol. 639 39–55. 10.1007/978-1-60761-702-0_3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui M., Zhang W., Zhang Q., Xu Z., Zhu Z., Duan F., et al. (2011). Induced over-expression of the transcription factor OsDREB2A improves drought tolerance in rice. Plant Physiol. Biochem. 49 1384–1391. 10.1016/j.plaphy.2011.09.012 [DOI] [PubMed] [Google Scholar]
- Danyluk J., Kane N. A., Breton G., Limin A. E., Fowler D. B., Sarhan F. (2003). TaVRT-1, a putative transcription factor associated with vegetative to reproductive transition in cereals. Plant Physiol. 132 1849–1860. 10.1104/pp.103.023523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis E. S., Peacock W. J. (2009). Vernalization in cereals. J. Biol. 8:57 10.1186/jbiol156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon T., Pearce S. P., Stockinger E. J., Distelfeld A., Li C., Knox A. K., et al. (2010). Regulation of freezing tolerance and flowering in temperate cereals: the VRN-1 connection. Plant Physiol. 153 1846–1858. 10.1104/pp.110.159079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diallo A., Kane N., Agharbaoui Z., Badawi M., Sarhan F. (2010). Heterologous expression of wheat VERNALIZATION 2 (TaVRN2) gene in Arabidopsis delays flowering and enhances freezing tolerance. PLoS ONE 5:e8690 10.1371/journal.pone.0008690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distelfeld A., Dubcovsky J. (2010). Characterization of the maintained vegetative phase deletions from diploid wheat and their effect on VRN2 and FT transcript levels. Mol. Genet. Genomics 283 223–232. 10.1007/s00438-009-0510-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distelfeld A., Li C., Dubcovsky J. (2009). Regulation of flowering in temperate cereals. Curr. Opin. Plant Biol. 12 178–184. 10.1016/j.pbi.2008.12.010 [DOI] [PubMed] [Google Scholar]
- Draper J., Mur L. A., Jenkins G., Ghosh-Biswas G. C., Bablak P., Hasterok R., et al. (2001). Brachypodium distachyon. A new model system for functional genomics in grasses. Plant Physiol. 127 1539–1555. 10.1104/pp.010196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubcovsky J., Loukoianov A., Fu D., Valarik M., Sanchez A., Yan L. (2006). Effect of photoperiod on the regulation of wheat vernalization genes VRN1 and VRN2. Plant Mol. Biol. 60 469–480. 10.1007/s11103-005-4814-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubouzet J. G., Sakuma Y., Ito Y., Kasuga M., Dubouzet E. G., Miura S., et al. (2003). OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 33 751–763. 10.1046/j.1365-313X.2003.01661.x [DOI] [PubMed] [Google Scholar]
- Faure S., Higgins J., Turner A., Laurie D. A. (2007). The FLOWERING LOCUS T-like gene family in barley (Hordeum vulgare). Genetics 176 599–609. 10.1534/genetics.106.069500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenup A., Peacock W. J., Dennis E. S., Trevaskis B. (2009). The molecular biology of seasonal flowering-responses in Arabidopsis and the cereals. Ann. Bot. 103 1165–1172. 10.1093/aob/mcp063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith M., Timonin M., Wong A. C., Gray G. R., Akhter S. R., Saldanha M., et al. (2007). Thellungiella: an Arabidopsis-related model plant adapted to cold temperatures. Plant Cell Environ. 30 529–538. 10.1111/j.1365-3040.2007.01653.x [DOI] [PubMed] [Google Scholar]
- Higgins J. A., Bailey P. C., Laurie D. A. (2010). Comparative genomics of flowering time pathways using Brachypodium distachyon as a model for the temperate grasses. PLoS ONE 5:e10065 10.1371/journal.pone.0010065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y., Katsura K., Maruyama K., Taji T., Kobayashi M., Seki M., et al. (2006). Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol. 47 141–153. 10.1093/Pcp/Pci230 [DOI] [PubMed] [Google Scholar]
- Izawa T. (2007). Adaptation of flowering-time by natural and artificial selection in Arabidopsis and rice. J. Exp. Bot. 58 3091–3097. 10.1093/jxb/erm159 [DOI] [PubMed] [Google Scholar]
- Jaglo-Ottosen K. R., Gilmour S. J., Zarka D. G., Schabenberger O., Thomashow M. F. (1998). Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280 104–106. 10.1126/science.280.5360.104 [DOI] [PubMed] [Google Scholar]
- Kardailsky I., Shukla V. K., Ahn J. H., Dagenais N., Christensen S. K., Nguyen J. T., et al. (1999). Activation tagging of the floral inducer FT. Science 286 1962–1965. 10.1126/science.286.5446.1962 [DOI] [PubMed] [Google Scholar]
- Kim D. H., Doyle M. R., Sung S., Amasino R. M. (2009). Vernalization: winter and the timing of flowering in plants. Annu. Rev. Cell Dev. Biol. 25 277–299. 10.1146/annurev.cellbio.042308.113411 [DOI] [PubMed] [Google Scholar]
- Kim H. J., Hyun Y., Park J. Y., Park M. J., Park M. K., Kim M. D., et al. (2004). A genetic link between cold responses and flowering time through FVE in Arabidopsis thaliana. Nat. Genet. 36 167–171. 10.1038/ng1298 [DOI] [PubMed] [Google Scholar]
- Kobayashi F., Takumi S., Kume S., Ishibashi M., Ohno R., Murai K., et al. (2005). Regulation by Vrn-1/Fr-1 chromosomal intervals of CBF-mediated Cor/Lea gene expression and freezing tolerance in common wheat. J. Exp. Bot. 56 887–895. 10.1093/jxb/eri081 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y., Kaya H., Goto K., Iwabuchi M., Araki T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286 1960–1962. 10.1126/science.286.5446.1960 [DOI] [PubMed] [Google Scholar]
- Lata C., Prasad M. (2011). Role of DREBs in regulation of abiotic stress responses in plants. J. Exp. Bot. 62 4731–4748. 10.1093/Jxb/Err210 [DOI] [PubMed] [Google Scholar]
- Lee J. H., Kim J. J., Kim S. H., Cho H. J., Kim J., Ahn J. H. (2012). The E3 ubiquitin ligase HOS1 regulates low ambient temperature-responsive flowering in Arabidopsis thaliana. Plant Cell Physiol. 53 1802–1814. 10.1093/pcp/pcs123 [DOI] [PubMed] [Google Scholar]
- Li C., Dubcovsky J. (2008). Wheat FT protein regulates VRN1 transcription through interactions with FDL2. Plant J. 55 543–554. 10.1111/j.1365-313X.2008.03526.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limin A. E., Fowler D. B. (2006). Low-temperature tolerance and genetic potential in wheat (Triticum aestivum L.): response to photoperiod, vernalization, and plant development. Planta 224 360–366. 10.1007/s00425-006-0219-y [DOI] [PubMed] [Google Scholar]
- Liu J., Gilmour S. J., Thomashow M. F., Van Nocker S. (2002). Cold signalling associated with vernalization in Arabidopsis thaliana does not involve CBF1 or abscisic acid. Physiol. Plant. 114 125–134. 10.1034/j.1399-3054.2002.1140117.x [DOI] [PubMed] [Google Scholar]
- Liu Q., Kasuga M., Sakuma Y., Abe H., Miura S., Yamaguchi-Shinozaki K., et al. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10 1391–1406. 10.1105/tpc.10.8.1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv B., Nitcher R., Han X., Wang S., Ni F., Li K., et al. (2014). Characterization of FLOWERING LOCUS T1 (FT1) gene in brachypodium and wheat. PLoS ONE 9:e94171 10.1371/journal.pone.0094171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S. D., Amasino R. M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11 949–956. 10.1105/tpc.11.5.949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki D., Shimamoto K. (2004). Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol. 45 490–495. 10.1093/pcp/pch048 [DOI] [PubMed] [Google Scholar]
- Mizoi J., Ohori T., Moriwaki T., Kidokoro S., Todaka D., Maruyama K., et al. (2013). GmDREB2A;2 a canonical DEHYDRATION-RESPONSIVE ELEMENT-BINDING PROTEIN2-type transcription factor in soybean, is posttranslationally regulated and mediates dehydration-responsive element-dependent gene expression. Plant Physiol. 161 346–361. 10.1104/pp.112.204875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Msanne J., Lin J., Stone J. M., Awada T. (2011). Characterization of abiotic stress-responsive Arabidopsis thaliana RD29A and RD29B genes and evaluation of transgenes. Planta 234 97–107. 10.1007/s00425-011-1387-y [DOI] [PubMed] [Google Scholar]
- Murai K., Miyamae M., Kato H., Takumi S., Ogihara Y. (2003). WAP1 a wheat APETALA1 homolog, plays a central role in the phase transition from vegetative to reproductive growth. Plant Cell Physiol. 44 1255–1265. 10.1093/pcp/pcg171 [DOI] [PubMed] [Google Scholar]
- Nakashima K., Fujita Y., Katsura K., Maruyama K., Narusaka Y., Seki M., et al. (2006). Transcriptional regulation of ABI3- and ABA-responsive genes including RD29B and RD29A in seeds, germinating embryos, and seedlings of Arabidopsis. Plant Mol. Biol. 60 51–68. 10.1007/s11103-005-2418-5 [DOI] [PubMed] [Google Scholar]
- Notaguchi M., Abe M., Kimura T., Daimon Y., Kobayashi T., Yamaguchi A., et al. (2008). Long-distance, graft-transmissible action of Arabidopsis FLOWERING LOCUS T protein to promote flowering. Plant Cell Physiol. 49 1645–1658. 10.1093/pcp/pcn154 [DOI] [PubMed] [Google Scholar]
- Onouchi H., Igeno M. I., Perilleux C., Graves K., Coupland G. (2000). Mutagenesis of plants overexpressing CONSTANS demonstrates novel interactions among Arabidopsis flowering-time genes. Plant Cell 12 885–900. 10.1105/tpc.12.6.885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin F., Kakimoto M., Sakuma Y., Maruyama K., Osakabe Y., Tran L. S., et al. (2007). Regulation and functional analysis of ZmDREB2A in response to drought and heat stresses in Zea mays L. Plant J. 50 54–69. 10.1111/j.1365-313X.2007.03034.x [DOI] [PubMed] [Google Scholar]
- Ream T. S., Woods D. P., Schwartz C. J., Sanabria C. P., Mahoy J. A., Walters E. M., et al. (2014). Interaction of photoperiod and vernalization determines flowering time of Brachypodium distachyon. Plant Physiol. 164 694–709. 10.1104/pp.113.232678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma Y., Maruyama K., Qin F., Osakabe Y., Shinozaki K., Yamaguchi-Shinozaki K. (2006). Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc. Natl. Acad. Sci. U.S.A. 103 18822–18827. 10.1073/pnas.0605639103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A., Onouchi H., Gold S. E., Ditta G. S., Schwarz-Sommer Z., Yanofsky M. F., et al. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288 1613–1616. 10.1126/science.288.5471.1613 [DOI] [PubMed] [Google Scholar]
- Sandve S. R., Kosmala A., Rudi H., Fjellheim S., Rapacz M., Yamada T., et al. (2011). Molecular mechanisms underlying frost tolerance in perennial grasses adapted to cold climates. Plant Sci. 180 69–77. 10.1016/j.plantsci.2010.07.011 [DOI] [PubMed] [Google Scholar]
- Sasani S., Hemming M. N., Oliver S. N., Greenup A., Tavakkol-Afshari R., Mahfoozi S., et al. (2009). The influence of vernalization and daylength on expression of flowering-time genes in the shoot apex and leaves of barley (Hordeum vulgare). J. Exp. Bot. 60 2169–2178. 10.1093/jxb/erp098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz R. J., Amasino R. M. (2007). Vernalization: a model for investigating epigenetics and eukaryotic gene regulation in plants. Biochim. Biophys. Acta 1769 269–275. 10.1016/j.bbaexp.2007.02.003 [DOI] [PubMed] [Google Scholar]
- Searle I., He Y., Turck F., Vincent C., Fornara F., Krober S., Amasino R. A., et al. (2006). The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 20 898–912. 10.1101/gad.373506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo E., Lee H., Jeon J., Park H., Kim J., Noh Y. S., et al. (2009). Crosstalk between cold response and flowering in Arabidopsis is mediated through the flowering-time gene SOC1 and its upstream negative regulator FLC. Plant Cell 21 3185–3197. 10.1105/tpc.108.063883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N., Ruelens P., Dhauw M., Maggen T., Dochy N., Torfs S., et al. (2016). A flowering locus C homolog is a vernalization-regulated repressor in Brachypodium and is cold-regulated in wheat. Plant Physiol. 173 1301–1315. 10.1104/pp.16.01161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada S., Ogawa T., Kitagawa S., Suzuki T., Ikari C., Shitsukawa N., et al. (2009). A genetic network of flowering-time genes in wheat leaves, in which an APETALA1/FRUITFULL-like gene, VRN1 is upstream of FLOWERING LOCUS T. Plant J. 58 668–681. 10.1111/j.1365-313X.2009.03806.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson S. D., Nakashima K., Narusaka Y., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2003). Two different novel cis-acting elements of erd1 a clpA homologous Arabidopsis gene function in induction by dehydration stress and dark-induced senescence. Plant J. 33 259–270. 10.1046/j.1365-313X.2003.01624.x [DOI] [PubMed] [Google Scholar]
- Srikanth A., Schmid M. (2011). Regulation of flowering time: all roads lead to Rome. Cell Mol. Life Sci 68 2013–2037. 10.1007/s00018-011-0673-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow M. F. (1999). PLANT COLD ACCLIMATION: freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 571–599. 10.1146/annurev.arplant.50.1.571 [DOI] [PubMed] [Google Scholar]
- Thomashow M. F. (2010). Molecular basis of plant cold acclimation: insights gained from studying the CBF cold response pathway. Plant Physiol. 154 571–577. 10.1104/pp.110.161794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevaskis B., Bagnall D. J., Ellis M. H., Peacock W. J., Dennis E. S. (2003). MADS box genes control vernalization-induced flowering in cereals. Proc. Natl. Acad. Sci. U.S.A. 100 13099–13104. 10.1073/pnas.1635053100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevaskis B., Hemming M. N., Dennis E. S., Peacock W. J. (2007). The molecular basis of vernalization-induced flowering in cereals. Trends Plant Sci. 12 352–357. 10.1016/j.tplants.2007.06.010 [DOI] [PubMed] [Google Scholar]
- Trevaskis B., Hemming M. N., Peacock W. J., Dennis E. S. (2006). HvVRN2 responds to daylength, whereas HvVRN1 is regulated by vernalization and developmental status. Plant Physiol. 140 1397–1405. 10.1104/pp.105.073486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J., Hill T. (2008). High-efficiency Agrobacterium-mediated transformation of Brachypodium distachyon inbred line Bd21-3. Plant Cell Rep. 27 471–478. 10.1007/s00299-007-0472-y [DOI] [PubMed] [Google Scholar]
- Vogel J. P., Garvin D. F., Mockler T. C., Schmutz J., Rokhsar D., Bevan M. W., et al. (2010). Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463 763–768. 10.1038/nature08747 [DOI] [PubMed] [Google Scholar]
- Wigge P. A., Kim M. C., Jaeger K. E., Busch W., Schmid M., Lohmann J. U., et al. (2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309 1056–1059. 10.1126/science.1114358 [DOI] [PubMed] [Google Scholar]
- Xiong L., Lee H., Ishitani M., Zhu J. K. (2002). Regulation of osmotic stress-responsive gene expression by the LOS6/ABA1 locus in Arabidopsis. J. Biol. Chem. 277 8588–8596. 10.1074/jbc.M109275200 [DOI] [PubMed] [Google Scholar]
- Xiong Y., Fei S. Z. (2006). Functional and phylogenetic analysis of a DREB/CBF-like gene in perennial ryegrass (Lolium perenne L.). Planta 224 878–888. 10.1007/s00425-006-0273-5 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Shinozaki K. (1994). A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6 251–264. 10.1105/tpc.6.2.251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Fu D., Li C., Blechl A., Tranquilli G., Bonafede M., et al. (2006). The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc. Natl. Acad. Sci. U.S.A. 103 19581–19586. 10.1073/pnas.0607142103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Loukoianov A., Blechl A., Tranquilli G., Ramakrishna W., SanMiguel P., et al. (2004). The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303 1640–1644. 10.1126/science.1094305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Loukoianov A., Tranquilli G., Helguera M., Fahima T., Dubcovsky J. (2003). Positional cloning of the wheat vernalization gene VRN1. Proc. Natl. Acad. Sci. U.S.A. 100 6263–6268. 10.1073/pnas.0937399100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Fei S.-Z., Liu P., Ji T., Peng J., Frei U., et al. (2017). Transcriptome changes in response to cold acclimation in perennial ryegrass as revealed by a cross-species microarray analysis. Crop Sci. 57 1–13. 10.2135/cropsci2016.04.0252 [DOI] [Google Scholar]
- Zhang C., Fei S.-Z., Warnke S., Li L., Hannapel D. (2009). Identification of genes associated with cold acclimation in perennial ryegrass. J. Plant Physiol. 166 1436–1445. 10.1016/j.jplph.2009.03.001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.