Abstract
This paper examines how age intervenes in the APOE ε4 allele’s effects upon the volume and shape morphometrics of the hippocampus and the amygdala in mild cognitive impairment and Alzheimer’s disease. We evaluate the structural morphological differences between the ε4 carriers and non-carriers in two age-dependent subgroups; younger than 75 years (Young-Old) and older than 80 years (Very-Old). While we show that the four structures of interest atrophy significantly in the ε4 carriers, relative to the non-carriers, of the Young-Old group, this effect is not observed in their Very-Old counterparts. The structures in the right hemisphere are found to be more affected by the APOE genotype than those in the left hemisphere and we identify the relevant regions in which significant atrophy occurs to be parts of the basolateral, centromedial, and lateral nucleus subregions of the amygdala and the CA1 and subiculum subregions of the hippocampus. We also observe that the APOE genotype only affects MCI patients that deteriorated to dementia within 3 years while leaving their “non-converting” counterparts unaffected.
Keywords: Alzheimer’s disease, mild cognitive impairment, apolipoprotein E, shape morphometrics, hippocampus, amygdala, age intervention, conversion
1 Introduction
The apolipoprotein E (APOE) gene has been reported to be the major genetic source of common forms of late-onset Alzheimer’s disease (AD) [1]. It has been suggested that the APOE ε4 allele increases the genetic risk and lowers the mean age of onset in AD [1,2]. Furthermore, studies have shown that the APOE ε4 carriers in the AD population have a greater impairment than the non-carriers in terms of global cognitive function, episodic memory, and executive function [3–5]. With that being said, the underlying biological mechanism through which the APOE ε4 allele exerts its effects on AD patients has not yet been fully understood. Atrophy of medial temporal lobe limbic structures, such as the entorhinal cortex, the hippocampus, and the amygdala, has been observed in patients with mild cognitive impairment (MCI) and AD when compared to the normal aging population. Morphological abnormalities of those limbic structures serve as anatomical hallmarks of the Alzheimer dementia on a macroscopic level [6–12]. Based on these observations, one hypothesis arises naturally; a statistical association exists between the APOE genotype and the morphometric phenotype of the medial temporal lobe limbic structures, a topic which has been explored extensively (a detailed review can be found elsewhere [13]). When such an association was considered previously, inconsistencies can be seen between the results reported by individual neuroimaging studies. For instance, it has been suggested that the hippocampal volumes did not differ significantly within either the elderly control group or the AD population on the basis of APOE genotype [14]. Meanwhile, evidence showed that increased rates of hippocampal volume loss in AD were indeed associated with a presence of the APOE ε4 allele [15]. Moreover, one study showed that the APOE ε4 carriers with AD atrophied significantly relative to the non-carriers in terms of the right hippocampal volume and the right amygdalar volume while not differing in terms of the left hippocampal, nor the left amygdalar, volume [16]. Adding further diversity to this question, it has been reported that the two AD subgroups (APOE ε4 carriers and non-carriers) differed in terms of the amygdalar volume (the amygdalar volume was significantly smaller in ε4 carriers than the non-carriers) but not the hippocampal volume [17].
These varying observations on the association between the APOE genotype and the hippocampal and amygdalar morphometrics may be related to an age effect. Indeed, studies have shown that the effect of the APOE ε4 allele on the cognitive function of subjects with MCI or AD diminishes after a specific age [18–21]. Recently, the interaction between age and the APOE genotype was analyzed longitudinally [22,23], in terms of their combined influence on the decline of various cognitive functions and brain structure volumes in healthy control (HC) and AD populations. Evidence to support the role of age in the APOE ε4 allele’s influence on hippocampal and cerebral atrophy was demonstrated in [23]; here the APOE ε4 allele was found to significantly affect only the Young-Old AD population (mean age younger than 75 years old), showing no significant impact on the Very-Old AD population (mean age older than 80 years old).
Existing studies have mainly focused on analyzing the APOE genotype’s effects on the volumes of various brain structures in age-dependent dementia sub-groups (e.g. Young-Old AD and Very-Old AD). A primary concern when examining the APOE effects on the volume of a specific brain structure, such as the hippocampus, is that it does not allow for identification of subregions within the structure that are directly affected. Indeed, inhomogeneity of these effects within a single structure is not accounted for at all. To shed light on this angle, we need approaches that are capable of revealing associations between the APOE genotype and the localized anatomical phenotypes of the structures of interest. With the advent of recent innovations in brain mapping techniques, diffeomorphometry-based statistical shape analysis pipelines have been proposed and applied to various Alzheimer related studies [24,25]. In diffeomorphometry-based statistical analyses, one studies the morphometrics of a set of anatomical shapes by quantitatively analyzing the diffeomorphisms that connect each of those shapes to a common template shape. Herein, we will rely on Large Deformation Diffeomorphic Metric Mapping (LDDMM) as our tool of choice for creating the diffeomorphisms for analysis. The robustness and sensitivity of LDDMM, in detecting and quantifying the shape variations for a range of medial temporal lobe structures in MCI and AD, have been successfully demonstrated in several previous works [24–27].
In this study, our primary goal is to investigate the relationship of both volume and shape of the bilateral amygdalas and hippocampi to the APOE genotype in age-dependent subgroups of MCI and AD. To be specific, we analyze and compare the APOE ε4 allele’s effects in two age-dependent subgroups (younger than 75 years, Young-Old, and older than 80 years, Very-Old) for each of the two disease groups (MCI and AD) in terms of the morphometrics of the amygdala and the hippocampus in both hemispheres. Both the volume and the shape measurements of the two age-dependent disease groups have been normalized to their respective age-matched HC subgroups (Young-Old HC and Very-Old HC) before group comparison so as to subtract the impact of normal aging on the structural atrophy.
It is well known that patients with MCI are at higher risk of developing AD compared to normally aging people [28,29]. Nonetheless, a great heterogeneity exists in MCI patients in terms of their cognitive profiles and clinical progressions. It is not certain that an MCI patient will definitely deteriorate to dementia; some MCI patients convert to AD over time while others remain stable or even revert to being cognitively normal [30,31]. Certain studies have shown that a presence of the APOE ε4 allele is a strong clinical indicator for progression from MCI to AD [32], especially when combined with other types of dementia biomarkers, such as PET measures [33]. In previous work, we have demonstrated a contrast in the shape profiles of the amygdala and the hippocampus between MCI converters (MCI patients who convert to AD within a fixed time, e.g. 3.5 years from baseline) and MCI non-converters (MCI patients who remain stable or revert to normal status within the same fixed period) [25]. The shape morphometrics of deep gray matter structures, especially those of the amygdala and the hippocampus, have been successfully applied to the automated prediction of MCI-to-AD conversion [34]. In light of these previous findings, we aim to also analyze the association between the APOE genotype and the conversion of MCI patients from an amygdalar/hippocampal morphometric perspective. To be specific, we examine how the APOE genotype affects the shape and volume of the amygdala and the hippocampus, individually, in the MCI converter (MCI-C) population and the MCI non-converter (MCI-NC) population. In addition, we investigate whether or not age plays a significant role in the APOE genotype’s effects on each of the two MCI subgroups. For both MCI-C and MCI-NC, we compare the volume and shape of the four structures of interest between the APOE ε4 allele carriers and the non-carriers in the Young-Old group as well as the Very-Old group.
Data for the investigations in this work come from the baseline scans of a subset of subjects in the Alzheimer’s Disease Neuroimaging Initiative (ADNI) study, including a total of 135 HC subjects (93 Young-Old and 42 Very-Old), 276 subjects with MCI (174 Young-Old and 102 Very-Old), out of which 123 have converted to AD within 3 years from baseline while the other 153 did not, and 129 subjects with AD (80 Young-Old and 49 Very-Old).
2 Materials and Methods
2.1 Alzheimer’s Disease Neuroimaging Initiative
Data used in the preparation of this article were obtained from the ADNI database (http://adni.loni.usc.edu/). ADNI was launched in 2003 by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, the Food and Drug Administration, private pharmaceutical companies and non-profit organizations, as a $60 million, 5-year public-private partnership. The primary goal of ADNI has been to test whether serial magnetic resonance imaging, positron emission tomography, other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of MCI and early AD. Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians to develop new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials.
ADNI is a result of the efforts of many co-investigators from a broad range of academic institutions and private corporations. Subjects in ADNI have been recruited from over 50 sites across the U.S. and Canada. ADNI was approved by each local institutional review board. Written informed consent was obtained from each participant.
2.2 Participants
In this study, we included data from 135 HC subjects, 276 subjects with MCI and 129 subjects with AD. In the MCI group, 123 subjects converted to AD (MCI-C) within a follow-up period of 36 months while the other 153 remained MCI or reverted to cognitively normal status (MCI-NC). At baseline, each subject’s age ranged between 55 and 92 years old and none of the participants were depressed (depression was defined as Geriatric Depression scores no smaller than 10 or treatment with medication). The control subjects had Mini-Mental State Examination (MMSE) scores of 25–30 and a clinical dementia rating (CDR) of 0. The MCI subjects had MMSE scores of 23–30, a CDR of 0.5, preserved ability to perform daily living activities, and an absence of dementia. The AD subjects had MMSE scores of 20–28, a CDR of 0.5 or 1.0 and met the criteria for probable AD.
For the purpose of our first analysis, the differentiation of the APOE ε4 allele’s effects by age in individual disease cohorts, we separate each of the three cognitive groupings (HC, MCI, and AD) into subject age dependent subgroups; Young-Old (baseline age, between 55 and 75 years old) and Very-Old (baseline age, between 80 and 92 years old). The result is 93 Young-Old HC (HC-YO) subjects, 42 Very-Old HC (HC-VO) subjects, 174 Young-Old MCI (MCI-YO) subjects, 102 Very-Old MCI (MCI-VO) subjects, 80 Young-Old AD (AD-YO) subjects, and 49 Very-Old AD (AD-VO) subjects. Our reasoning in setting this age criterion is three-fold; firstly, as suggested in [35], individuals over the age of 80 represent the fastest growing segment of our population, the development of AD in the so-called “Very-Old” (i.e. age 80 and above) is a public health problem of increasing magnitude. Secondly, our dataset is a subset of the ADNI database, as is that of [23,36], and for consistency we have followed the criterion for age subgrouping published in those two works. Lastly, to distinguish the two age-dependent groups, some gap must be created and between this intuition and compatibility with other studies using the same criteria we arrive at our thresholds. There are a total of 196 participants that fell between the ages of 76 years and 80 years; 23 HC ε4 carriers, 47 HC non-carriers, 43 MCI ε4 carriers, 41 MCI non-carriers, 31 AD ε4 carriers, and 11 AD non-carriers.
All subjects underwent APOE genotyping using DNA extracted from peripheral blood cells. Based on an individual’s APOE allele status, all of the aforementioned six age-diagnosis patient groups were further divided into two genotype subgroups – ε4 carriers (at least one APOE ε4 allele) and ε4 non-carriers (no APOE ε4 allele). It has been suggested that the APOE ε4 allele is associated with an increased risk for AD and an earlier age of AD onset [1] whereas the ε2 allele of APOE is associated with a lower risk [37]. Given this conflict of influence between the ε2 and ε4 alleles (a protective role for the APOE ε2 allele and a detrimental role for the APOE ε4), subjects with the APOE genotype ε4/ε2 were excluded from this study (3 HC, 9 MCI, and 4 AD instances). This further division resulted in: 71 HC-YO ε4 non-carriers, 22 HC-YO ε4 carriers, 33 HC-VO ε4 non-carriers, 9 HC-VO ε4 carriers, 63 MCI-YO ε4 non-carriers, 111 MCI-YO ε4 carriers, 64 MCI-VO ε4 non-carriers, 38 MCI-VO ε4 carriers, 22 AD-YO ε4 non-carriers, 58 AD-YO ε4 carriers, 23 AD-VO ε4 non-carriers, and 26 AD-VO ε4 carriers. The demographic characteristics of each of these groups are listed in Table 1.
Table 1.
The demographic characteristics of each age-categorized subgroup in this study. Within each carrier group, the number of APOE ε4 carriers with only one ε4 allele is reported as “e3/e4”.
| NO. male | Age (years) | MMSE score | CDR-SB score | ||
|---|---|---|---|---|---|
| HC-YO | non-carrier (n=71) | 38 | 72.22 ± 2.21 | 29.03 ± 1.08 | 0.007 ± 0.059 |
| carrier (n=22; e3/e4=20) | 11 | 71.07 ± 3.17 | 29.32 ± 0.78 | 0.045 ± 0.147 | |
|
| |||||
| HC-VO | non-carrier (n=33) | 13 | 84.06 ± 2.59 | 29.06 ± 1.12 | 0.091 ± 0.196 |
| carrier (n=9; e3/e4=9) | 7 | 82.69 ± 2.59 | 28.89 ± 0.93 | 0.000 ± 0.000 | |
|
| |||||
| MCI-YO | non-carrier (n=63) | 43 | 68.11 ± 5.76 | 27.31 ± 1.85 | 1.476 ± 0.820 |
| carrier (n=111; e3/e4=76) | 61 | 69.47 ± 4.25 | 27.14 ± 1.70 | 1.667 ± 0.869 | |
|
| |||||
| MCI-VO | non-carrier (n=64) | 40 | 83.54 ± 2.55 | 26.89 ± 1.58 | 1.484 ± 0.891 |
| carrier (n=38; e3/e4=35) | 29 | 83.21 ± 2.33 | 26.39 ± 1.82 | 1.711 ± 0.942 | |
|
| |||||
| AD-YO | non-carrier (n=22) | 10 | 67.46 ± 5.58 | 23.82 ± 1.76 | 3.886 ± 1.353 |
| carrier (n=58; e3/e4=34) | 32 | 69.06 ± 4.52 | 23.38 ± 1.83 | 4.103 ± 1.385 | |
|
| |||||
| AD-VO | non-carrier (n=23) | 11 | 84.35 ± 2.99 | 23.22 ± 2.26 | 4.369 ± 2.165 |
| carrier (n=26; e3/e4=21) | 18 | 83.34 ± 1.97 | 23.35 ± 2.37 | 4.462 ± 1.702 | |
For the purpose of our second analysis, investigating the APOE ε4 allele’s effects in age-dependent subgroups of the MCI converters and non-converters, we divided both the MCI-C and the MCI-NC group using the same age criterion as above. This results in four subgroups; MCI-C-YO (MCI converters that are younger than 75 years old, a total of 80), MCI-C-VO (MCI converters that are older than 80 years old, a total of 43), MCI-NC-YO (MCI non-converters that are younger than 75 years old, a total of 94), and MCI-NC-VO (MCI non-converters that are older than 80 years old, a total of 59). Based on a further division by the APOE genotype information, we have 60 ε4 carriers and 20 non-carriers in the MCI-C-YO group, 18 ε4 carriers and 25 non-carriers in the MCI-C-VO group, 51 ε4 carriers and 43 non-carriers in the MCI-NC-YO group, 20 ε4 carriers and 39 non-carriers in the MCI-NC-VO group. The demographic information for each of the aforementioned MCI subgroups is tabulated in Table 2.
Table 2.
The demographic information of the two age-categorized subgroups in the MCI converter group and the MCI non-converter group.
| NO. male | Age (years) | MMSE score | CDR-SB score | ||
|---|---|---|---|---|---|
| MCI-C-YO | non-carrier (n=20) | 13 | 67.73 ± 7.02 | 26.95 ± 2.14 | 1.775 ± 0.786 |
| carrier (n=60) | 32 | 69.93 ± 3.71 | 27.17 ± 1.64 | 1.842 ± 0.904 | |
|
| |||||
| MCI-C-VO | non-carrier (n=25) | 15 | 83.32 ± 2.53 | 26.44 ± 1.51 | 1.741 ± 1.072 |
| carrier (n=18) | 13 | 83.27 ± 2.55 | 26.33 ± 1.94 | 1.833 ± 1.163 | |
|
| |||||
| MCI-NC-YO | non-carrier (n=43) | 30 | 68.27 ± 5.16 | 27.47 ± 1.71 | 1.337 ± 0.807 |
| carrier (n=51) | 29 | 68.93 ± 4.81 | 27.11 ± 1.78 | 1.461 ± 0.786 | |
|
| |||||
| MCI-NC-VO | non-carrier (n=39) | 25 | 83.68 ± 2.59 | 27.18 ± 1.59 | 1.321 ± 0.721 |
| carrier (n=20) | 16 | 83.15 ± 2.18 | 26.45 ± 1.76 | 1.601 ± 0.701 | |
2.3 Image processing and volumetric segmentation
The raw data included in this analysis were collected from multiple 1.5T scanners. Only baseline information from the ADNI database was used in this study. The raw Digital Imaging and Communications in Medicine (DICOM) magnetic resonance (MR) images were downloaded from the public ADNI website (http://adni.loni.usc.edu/data-samples/mri/). Locally, the raw MR data were corrected for spatial distortion due to gradient nonlinearity [38] and B1 field inhomogeneity [39]. For each subject, the two T1-weighted images of the baseline scan were rigid-body aligned to one another, averaged to improve the signal-to-noise ratio, and then resampled to be isotropic of 1mm voxel resolution. Volumetric segmentations of the four structures of interest (the bilateral amygdala and hippocampus) were obtained using the segmentation module [40] of the FreeSurfer software (version 4.3.0).
2.4 Shape processing
To extract the shape of each structure of interest, from each individual scan, we took the approach described in our recent study [25]. Briefly, for each subvolume of the brain corresponding to a structure of interest, its bounding surface was obtained by applying an optimized diffeomorphism to the Computational Functional Anatomy (CFA) subcortical template surface [41] of that specific structure. The optimized diffeomorphism was obtained from a multi-channel LDDMM-image registration between the CFA template subvolumes and the scan-specific subvolumes obtained from FreeSurfer [25]. The CFA template surfaces and subvolumes of the four structures of interest were created from manual delineations, ensuring smooth boundary and correct anatomical topology. This surface-generation methodology will generate the “target shapes” whose properties we will examine herein. More details and validation of this methodology can be found in [25].
In shape-based diffeomorphometry, the focus of this study, the morphometrics of target shapes are quantified in terms of the diffeomorphisms that connect a common template shape to those target shapes. A template built from a single scan may not be truly representative of the whole sample. Extreme variations between the template shape and a target shape are likely to cause the subsequent mapping to fail, in which case the diffeomorphism cannot accurately describe the morphological changes between the template and the target shapes. To avoid this potential issue, we created a “population-averaged” template surface, for each structure of interest, from a subset of the baseline surfaces as demonstrated in [25]. All surfaces in that subset were first rigidly aligned to a common spatial position before being used to compute an averaged template surface using the estimation algorithm described in [42]. Constructing an averaged template surface that lies in the center of the population surfaces allows for overall more accurate mappings between the template space and each target space, when compared to using an arbitrary template [41].
After creating the common template surface for each structure, we employed the LDDMM-surface mapping algorithm [43] to map the template surface to each individual subject surface. From each template-to-subject LDDMM-surface mapping, a scalar field is calculated as the log-determinant of the Jacobian of the diffeomorphism. This scalar field is indexed at each vertex of the common template surface, quantifying the factor by which the diffeomorphism expands or shrinks the vertex-based localized surface area in the subject relative to the template in a logarithmic scale; i.e. a positive value corresponds to a localized surface area expansion of the subject relative to the template while a negative value suggests a localized surface area contraction. We shall call this scalar field the deformation marker.
It is worthy of note that to remove the impact of normal aging on the morphometrics of the amygdala and the hippocampus in the two disease groups (MCI and AD), the volumetric and the vertex-based diffeomorphometric measurements of each age-dependent disease subgroup have been normalized with respect to the corresponding age-matched HC subgroup. For example, in any statistical analysis in which the left amygdala volume of the MCI-YO population is used, instead of using the raw values, we have used the z-scores normalized by the left amygdala volume measurement of the HC-YO group. As such, negative values indicate smaller structure volumes or smaller localized shape areas. The z-score is computed as , where x is the raw value, μ is the appropriate mean value from the age-matched HC subgroup, and δ is the corresponding standard deviation. Please note the normalization was not genotype-dependent; for example, we did not use the measurements of the HC-YO ε4 carriers to normalize those of the MCI-YO ε4 carriers and similarly for the AD-YO ε4 carriers et cetera.
2.5 Statistical analysis
For inter-group comparisons, we used a linear model as described in [25]
| (1) |
where yk (s) is the z-transformed deformation marker for subject s at vertex k on the template surface, g(s) is a binary group variable, and Xcov(s) denotes the covariate information of subject s included in the analysis. In this study, we covaried for sex, and the estimated total intracranial volume (eTCV). The eTCV values were automatically computed in FreeSurfer. The noise structure εk(s) is modeled as a Gaussian process . The parameters were obtained from maximum-likelihood estimation, for details of which see [24].
To check for differences between any two groupings, in terms of the z-transformed localized surface area at vertex k, we tested the null hypothesis against the general hypothesis . The complete null hypothesis is simultaneously for all k. The test statistic Fk, computed for each vertex k, is defined as , where RSS0 is the residual sum of squares under the null hypothesis, and RSS is the residual sum of squares under the general hypothesis. Since we have performed the hypothesis testing at multiple vertices simultaneously, we need to correct for multiple comparisons and thus the familywise error rate (FWER) was controlled at a level of 0.05 with the maximum statistic defined as F* = maxk Fk. The statistical significance of a group difference is quantified by a p-value obtained from Fisher’s method of randomization. To be specific, p-values were computed based on non-parametric permutation tests by randomizing the model residuals. Details on the implementation of our permutation testing can be found in [26]. Briefly, Monte Carlo simulations were used to generate 40,000 uniformly distributed random permutations thus giving rise to a collection of F* statistics, one from each permutation. The p-value is then given by the fraction of permutations in the collection that have F* values larger than the value obtained from the true groupings. The set of vertices for which the null hypothesis is not valid is estimated to be D = {k: Fk ≥ q*}, where q* is the 95th percentile of the F* statistic [44]. The group difference in terms of the z-score of the localized shape area at vertex k is quantified as −βk,1, so that greater atrophy in the latter group of a comparison induces a higher positive value. To make group-pairwise comparison of the z-scores of the volume measurements of each structure, we used the same statistical model as in Eq. (1), wherein yk(s) is replaced by the z-transformed structure volume measurement of subject s. Given that there are a total of 4 structures of interest, the Bonferroni correction was adopted at the structure level so that statistical significance is achieved only when the structure-level p-value satisfies p < 0.0125.
In this study, we adopt a coarse-to-fine evaluation strategy. For the first analysis, we compare the z-scores of the volume and shape measurements of each of the four structures between the ε4 carriers and non-carriers within MCI and AD separately, followed by comparisons between the ε4 carriers and non-carriers, in terms of the z-scores of the morphometric measures, for each of the four groups (MCI-YO, MCI-VO, AD-YO, and AD-VO). For the second analysis, we compare the z-transformed volume and diffeomorphometric-based shape measurements of those four structures, between the ε4 carriers and non-carriers within MCI-C and MCI-NC, followed by z-score comparisons between the two APOE genotype groups within each of the four groups (MCI-C-YO, MCI-C-VO, MCI-NC-YO, and MCI-NC-VO).
The z-transformed volume measurements for all groups involved in this study, relative to their age-matched healthy control counterparts, are listed in the Appendix (see Supplementary Table 1).
2.6 Template surface partition
To examine how the APOE genotype’s effect manifests itself at a subregional level, we divide our template surface for each of the four structures into subregions using the approach we detailed in our previous work [25]. This partition was achieved using pre-delineated surfaces of high-field segmentations (obtained from a 7 Tesla scanner with an image voxel resolution of 0.8mm) and a transfer of the boundary definitions of those subregions to our population template surfaces. Both the left and right amygdala were partitioned into four subregions; the basolateral, the basomedial, the centromedial, and the lateral nucleus. Each of the bilateral hippocampi was also partitioned into four subregions; CA1, CA2, CA3 combined with the dentate gyrus, and the subiculum.
3 Results
3.1 Alzheimer’s disease
3.1.1 Volume analysis
According to our analysis, there exists statistically significant volumetric reduction, in the ε4 carriers relative to the non-carriers in the AD group, of the right hippocampus but not for the other three structures of interest. The p-values of those comparisons are tabulated in Table 3. As for the two age-dependent AD subgroups (AD-YO and AD-VO), significant group differences between the ε4 carriers and their non-carrier counterparts (carriers < non-carriers) are observed for all of the four structures within the Young-Old AD group (all p-values smaller than 0.0125, see Table 3), whereas none of the four Very-Old AD structures have exhibited differences between the two APOE groups in terms of their volumetric measurements (all p-values larger than 0.0125, see Table 3 for detailed values).
Table 3.
The p-values and the mean z-score between-group differences (listed inside the parenthesis) obtained from both the volume and the shape analysis of contrasts between the ε4 carriers and non-carriers within the entire AD group, the Young-Old AD (AD-YO) group, and the Very-Old AD (AD-VO) group respectively. Bold italic typesetting indicates that the group difference in that comparison is statistically significant after multiple comparison correction at a level of p = 0.0125. For the mean z-score difference, the more positive, the more atrophy there is in the latter group, and the more negative, the more atrophy there is in the former group.
| AD-e3 vs. AD-e4 | AD-YO-e3 vs. AD-YO-e4 | AD-VO-e3 vs. AD-VO-e4 | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| volume | shape | volume | shape | volume | shape | |
| lhi | 0.0134 (0.526) | 0.2530 (0.279) | 0.0098 (0.897) | 0.0895 (0.460) | 0.2085 (0.074) | 0.2185 (0.234) |
| rhi | 0.0050 (0.556) | 0.0235 (0.319) | 0.0000 (1.111) | 0.0012 (0.644) | 0.9795 (−0.235) | 0.9995 (−0.057) |
| lam | 0.1580 (0.250) | 0.0017 (0.198) | 0.0101 (0.616) | 0.0026 (0.478) | 0.7680 (−0.166) | 0.6695 (0.053) |
| ram | 0.0583 (0.310) | 0.0391 (0.295) | 0.0002 (0.887) | 0.0007 (0.754) | 0.4165 (−0.542) | 0.4240 (−0.226) |
Key: lhi – left hippocampus, rhi – right hippocampus, lam – left amygdala, ram – right amygdala.
3.1.2 Shape analysis
From our vertex-based shape analysis upon testing the complete null hypothesis, we find significant group difference between the two APOE genotype subgroups of the entire AD group only for the left amygdala (p = 0.0101) but not the right amygdala nor the hippocampus in either hemisphere (see Table 3 for the p-value of each group comparison). The surface map demonstrating the left amygdalar differences that are statistically significant, between AD-e3 (the ε4 non-carriers) and AD-e4 (the ε4 carriers), in terms of the localized shape area is presented in panel A) of Figure 1. The amount of surface area with significant atrophy in AD-e4 relative to AD-e3 is 26.71mm2 for the left amygdala, accounting for 3.94% of the entire structure surface.
Figure 1.
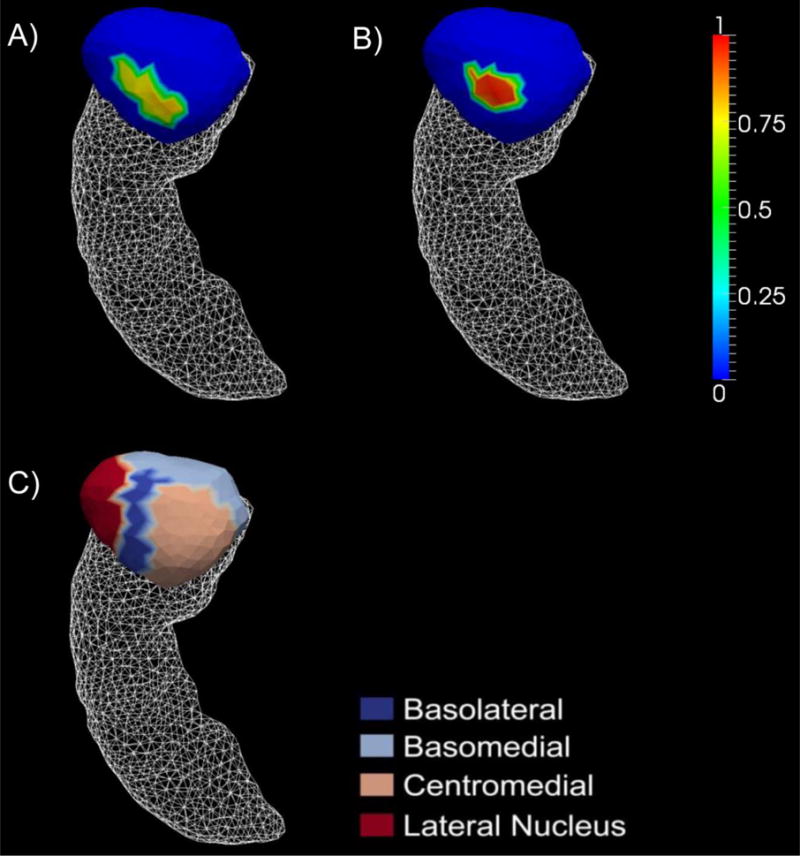
Panel A) represents the vertex-wise significant shape differences of the left amygdala between the ε4 carriers and the non-carriers of the entire AD group whereas panel B) represents the differences when restricted to the AD-YO group. The color bar values are the differences in z-score of the shape-based diffeomorphometry. Panel C) represents a division of the left amygdala template surface into four compatible subregions: the basolateral, basomedial, centromedial, and lateral nucleus.
Our shape-based diffeomorphometric comparison results of the ε4 carriers and the non-carriers for AD-YO and AD-VO are very consistent with the volumetric results. For the two APOE genotype groups of AD-VO, we did not observe any significant shape difference in any of the four structures while for the comparisons within AD-YO, we found significant shape atrophy in the ε4 carriers for the right hippocampus and the left and right amygdala. The p-values obtained from all the AD shape comparisons are listed in Table 3 for the purpose of a direct comparison with the volume analysis results. The surface map illustrating the significant e3-versus-e4 right hippocampal shape differences, within the AD-YO group, is shown in panel A) of Figure 2. The AD-YO genotype subgroup differences of the left amygdalar shape are illustrated in panel B) of Figure 1. The right amygdalar shape differences, between the carriers and the non-carriers within AD-YO, are presented in panel A) of Figure 3. The amount of regions with statistically significant surface area reductions in AD-YO-e4 relative to AD-YO-e3, for the right hippocampus, the left amygdala, and the right amygdala, are respectively 449.87mm2 (25.14% of the entire surface), 18.71mm2 (2.76% of the entire surface), and 406.28mm2 (51.85% of the entire surface).
Figure 2.
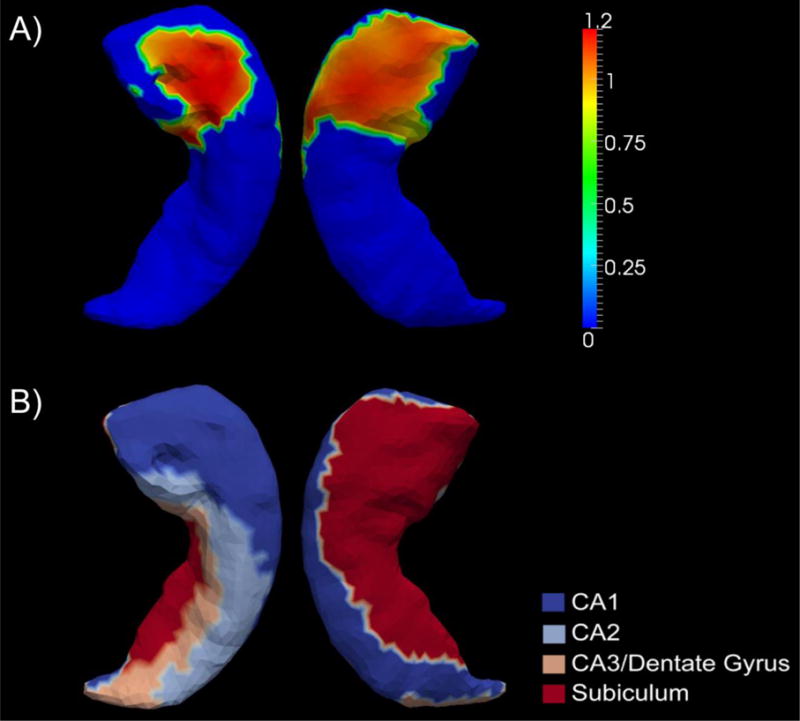
Panel A) demonstrates the significant group differences in terms of z-score of the shape diffeomorphometric measurements, of the right hippocampus, between the two APOE genotypes in AD-YO. Panel B) represents an anatomical partition of the right hippocampus template surface into four subregions – CA1, CA2, CA3 combined with the dentate gyrus, and the subiculum.
Figure 3.
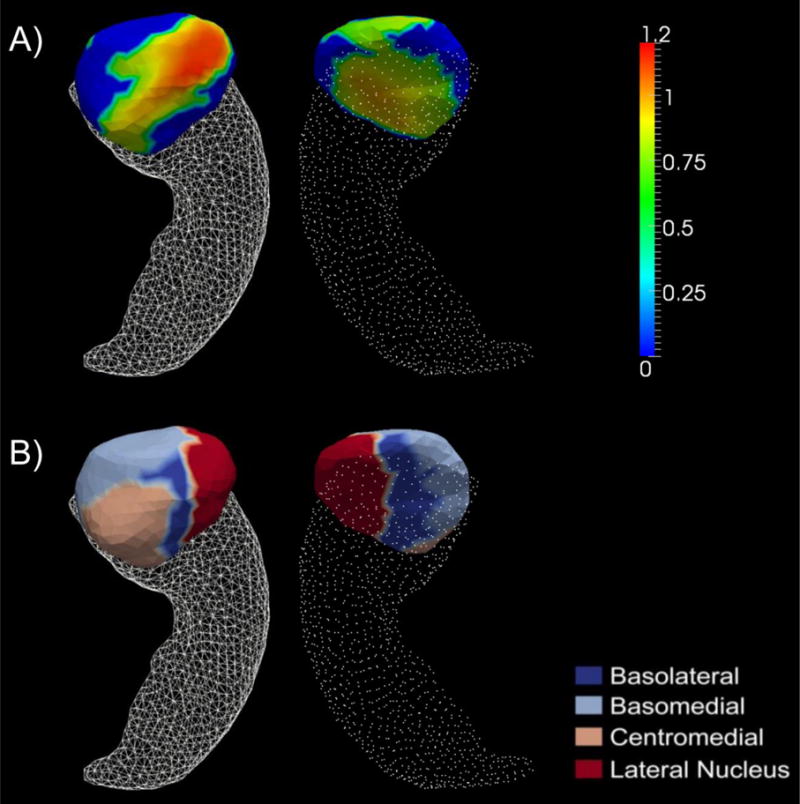
Panel A) shows the significant group differences in terms of z-score of the shape diffeomorphometry of the right amygdala between the two APOE genotypes in AD-YO. Panel B) represents an anatomical partition of the right amygdala template surface into four subregions: basolateral, basomedial, centromedial, and lateral nucleus.
3.2 Mild cognitive impairment
For the MCI population, to account for the inhomogeneity induced by the conversion status, we investigated two separate analyses. Initially we treated MCI as a single entity and examined whether age plays a role in the APOE genotype’s effects on structural morphometry. To be specific, we first compared the volume and shape measurements of the four structures between MCI-e3 (non-carriers in the entire MCI group) and MCI-e4 (carriers in the entire MCI group), and then between MCI-YO-e3 (non-carriers in the Young-Old MCI group) and MCI-YO-e4 (carriers in the Young-Old MCI group), and finally between MCI-VO-e3 (non-carriers in the Very-Old MCI group) and MCI-VO-e4 (carriers in the Very-Old MCI group). For convenience, we will refer to this as our “Type I MCI analysis”. For the next investigation, we divided MCI into two subgroups based on the conversion status – MCI-C and MCI-NC. From this, we investigated whether the APOE genotype status affects those two groups and then how age factors into such effects. Specifically, we first compared the volume and shape of the four structures for MCI-C-e3 (non-carriers in the MCI converters) versus MCI-C-e4 (carriers in the MCI converters) and MCI-NC-e3 (non-carriers in the MCI non-converters) versus MCI-NC-e4 (carriers in the MCI non-converters). We then refined by age and compared MCI-C-YO-e3 (non-carriers in the Young-Old MCI converters) versus MCI-C-YO-e4 (carriers in the Young-Old MCI converters), MCI-C-VO-e3 (non-carriers in the Very-Old MCI converters) versus MCI-C-VO-e4 (carriers in the Very-Old MCI converters), MCI-NC-YO-e3 (non-carriers in the Young-Old MCI non-converters) versus MCI-NC-YO-e4 (carriers in the Young-Old MCI non-converters), and MCI-NC-VO-e3 (non-carriers in the Very-Old MCI non-converters) versus MCI-NC-YO-e4 (non-carriers in the Very-Old MCI non-converters). For convenience, we will refer to this as our “Type II MCI analysis”.
3.2.1 Type I MCI analysis
3.2.1.1 Volume analysis
In comparing the volume of the four structures between MCI-e3 and MCI-e4, we detected statistically significant group differences (MCI-e4 < MCI-e3) for the bilateral hippocampi and the right amygdala but not for the left amygdala. This finding was re-iterated in our subsequent comparison of the two APOE genotype groups of the MCI-YO population. However, for the MCI-VO population, no volumetric difference was observed between the ε4 carriers and non-carriers in any of the four structures. Details on the p-values obtained from these volumetric Type I MCI group comparisons are listed in Table 4.
Table 4.
The p-values and the mean z-score between-group differences (listed inside the parenthesis) obtained from both the volume and the shape analysis of contrasts between the ε4 carriers and non-carriers within the entire MCI group, the Young-Old MCI (MCI-YO) group, and the Very-Old MCI (MCI-VO) group respectively. Bold italic typesetting indicates the group difference in that comparison is statistically significant after multiple comparison correction at a level of p = 0.0125. For the mean z-score difference, the more positive, the more atrophy there is in the latter group, and the more negative, the more atrophy there is in the former group.
| MCI-e3 vs. MCI-e4 | MCI-YO-e3 vs. MCI-YO-e4 | MCI-VO-e3 vs. MCI-VO-e4 | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| volume | shape | volume | shape | volume | shape | |
| lhi | 0.0031 (0.466) | 0.0382 (0.184) | 0.0022 (0.736) | 0.0111 (0.279) | 0.6015 (0.102) | 0.8980 (0.078) |
| rhi | 0.0006 (0.546) | 0.0016 (0.318) | 0.0007 (0.750) | 0.0104 (0.362) | 0.4850 (0.166) | 0.5145 (0.203) |
| lam | 0.1025 (0.288) | 0.3265 (0.169) | 0.1160 (0.395) | 0.1810 (0.227) | 0.5380 (0.124) | 0.6255 (0.121) |
| ram | 0.0008 (0.509) | 0.0000 (0.404) | 0.0033 (0.583) | 0.0016 (0.382) | 0.2445 (0.215) | 0.1370 (0.327) |
Key: lhi – left hippocampus, rhi – right hippocampus, lam – left amygdala, ram – right amygdala.
3.2.1.2 Shape analysis
For comparison between MCI-e3 and MCI-e4, in terms of the shape-based diffeomorphometrics of each of the four structures, significant group differences (MCI-e4 < MCI-e3) were observed in the right hippocampus (p = 0.0016) and the right amygdala (p = 0.0000). Group differences in the shape of the left hippocampus are almost statistically significant (p = 0.0382). No significant difference was detected for the left amygdalar shape (p = 0.3265).
In comparing the four structures between the carriers and the non-carriers within MCI-YO and MCI-VO, our hypothesis testing of the shape measurements yielded the same conclusions as that of the volume ones. Specifically, the shape diffeomorphometrics of the left hippocampus, the right hippocampus, and the right amygdala were all found to differ significantly between the carriers and the non-carriers of the MCI-YO population but not the shape of the left amygdala, which is also the case for the volume measurements. In the MCI-VO population, no significant group difference, in terms of the shape of any of the four structures, was detected between the two APOE genotype groups, an observation which is mirrored in our volumetric results. All p-values for our diffeomorphometric Type I MCI analysis are tabulated in Table 4.
We will now turn to a diagrammatic point of view by using surface maps to demonstrate group differences in terms of the vertex-based z-scores of the structural localized shape areas. Because the amount of surface regions showing significant area atrophy in MCI-YO-e4 relative to MCI-YO-e3, in the left hippocampus, is of area 11.57mm2, accounting for only 0.63% of the entire left hippocampus surface area, we will not display the corresponding shape difference map. Panel (a) of Figure 4 maps vertex-wise statistically significant genotype differences in z-scored diffeomorphometrics of the right hippocampus in the MCI population (MCI-e3 versus MCI-e4). Such significant vertices account for an area of 326.83mm2, being 18.26% of the entire right hippocampus surface area. The surface map of the right hippocampus, for the comparisons of MCI-YO-e3 and MCI-YO-e4, is demonstrated in panel (b) of Figure 4, wherein the amount of affected surface area is 52.2mm2 accounting for 2.92% of the entire structural surface area. Figure 5 proceeds in a similar fashion, this time illustrating vertex-wise significant shape differences between the right amygdala of MCI-e3 and MCI-e4, panel (a), and of MCI-YO-e3 and MCI-YO-e4, panel (b). The amount of right amygdala surface area revealing significant group difference between MCI-e3 and MCI-e4 is 437.73mm2, accounting for 55.87% of the entire structural surface area. The corresponding numbers for the MCI-YO genotype comparison are 90.39mm2 and 11.54% respectively.
Figure 4.
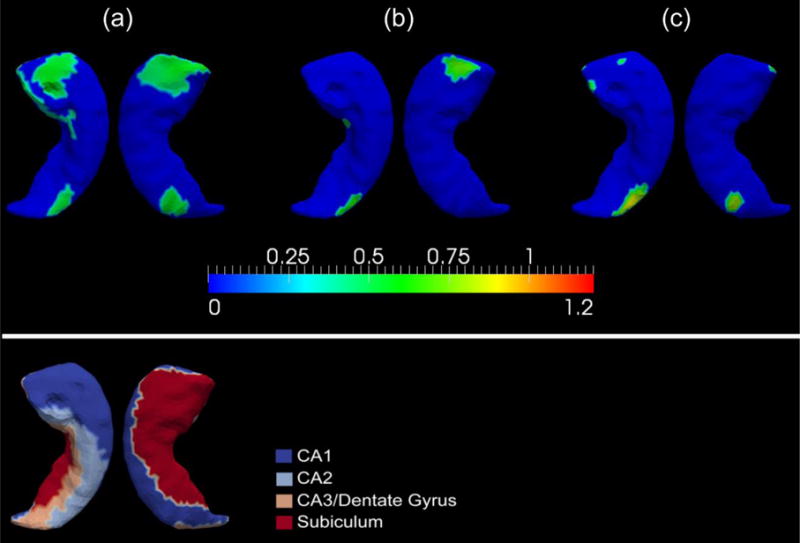
Panels (a), (b), and (c) in the top part of this figure represent the significant atrophy in the APOE ε4 carriers relative to their non-carrier counterpart, at each vertex of the right hippocampus, for the entire MCI group, the MCI-YO group, and the MCI-C group respectively. The color bar scale shows the differences in terms of z-score. The bottom part of this figure displays the four-subregion partition of the right hippocampal template surface – CA1, CA2, CA3 combined with the dentate gyrus, and the subiculum.
Figure 5.
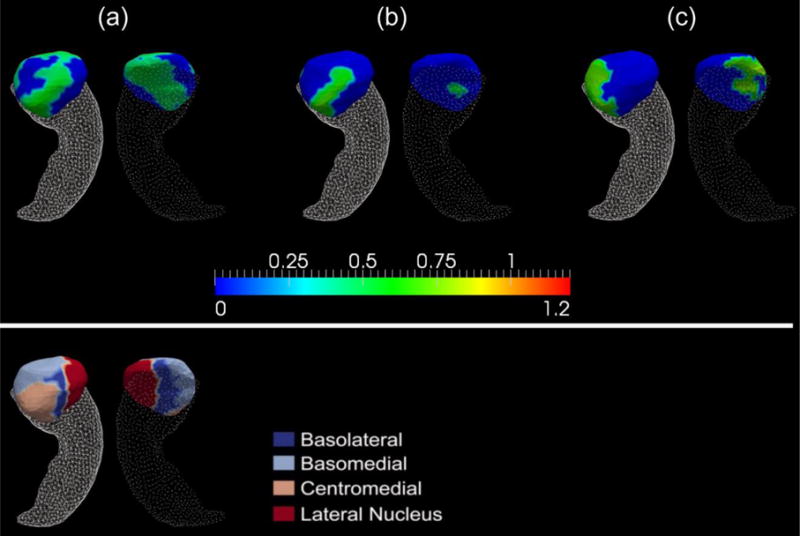
Top: Panels (a), (b), and (c) show the significant shape differences of the right amygdala between the two APOE genotypes for the entire MCI group, the MCI-YO group, and the MCI-C group respectively. The color bar scale shows the differences in z-score of the ε4 carriers relative to the non-carriers (the more positive, the more atrophy there is in the carriers). Bottom: the four-subregion partition of the right amygdala template surface – basolateral, basomedial, centromedial, and lateral nucleus.
3.2.2 Type II MCI analysis
3.2.2.1 Volume analysis
The p-values obtained in comparing the four structure volumes between the ε4 carriers and the non-carriers within MCI-C, MCI-C-YO, and MCI-C-VO are all listed in Table 5. According to that table, there is clear indication that the APOE ε4 allele makes a difference to the volumes of both the right amygdala and the right hippocampus of the entire MCI converter population while not affecting the volumes of the other two structures. After introducing the age variable, we did not observe any group difference, between the two APOE genotype groups of the Very-Old MCI converter population, for any of the four structures of interest. However, significant left hippocampal volume reduction was detected in the carriers compared with the non-carriers of the Young-Old MCI converters. For the volumetric analysis of the MCI non-converters, we did not detect any genotype effects in any of the three groups (MCI-NC, MCI-NC-YO, and MCI-NC-VO). The exact p-values are listed in Supplementary Table 2 of the Appendix.
Table 5.
The p-values and the mean z-score between-group differences (listed inside the parenthesis) obtained from both the volume and the shape analysis of contrasts between the ε4 carriers and non-carriers within respectively the entire MCI converter (MCI-C) group, the Young-Old MCI-C (MCI-C-YO) group, and the Very-Old MCI-C (MCI-C-VO) group. Bold italic typesetting indicates the group difference in that comparison is statistically significant after multiple comparison correction at a level of p = 0.0125. For the mean z-score difference, the more positive, the more atrophy there is in the latter group, and the more negative, the more atrophy there is in the former group.
| MCI-C-e3 vs. MCI-C-e4 | MCI-C-YO-e3 vs. MCI-C-YO-e4 | MCI-C-VO-e3 vs. MCI-C-VO-e4 | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| volume | shape | volume | shape | volume | shape | |
| lhi | 0.0148 (0.511) | 0.4820 (0.192) | 0.0084 (0.863) | 0.0750 (0.334) | 0.5135 (0.282) | 0.9490 (0.052) |
| rhi | 0.0043 (0.630) | 0.0038 (0.405) | 0.0374 (0.695) | 0.0295 (0.347) | 0.1265 (0.669) | 0.3395 (0.454) |
| lam | 0.0243 (0.455) | 0.1425 (0.318) | 0.0965 (0.481) | 0.0895 (0.322) | 0.3040 (0.672) | 0.7055 (0.212) |
| ram | 0.0003 (0.696) | 0.0059 (0.555) | 0.0757 (0.548) | 0.0840 (0.396) | 0.0704 (0.811) | 0.1235 (0.628) |
Key: lhi – left hippocampus, rhi – right hippocampus, lam – left amygdala, ram – right amygdala.
3.2.2.2 Shape analysis
In the analysis of the shape diffeomorphometric measurements of the four structures, we observed results consistent with those from the volumetric comparisons of MCI-C-e3 and MCI-C-e4; significant shape differences (MCI-C-e4 < MCI-C-e3) were detected for the right hippocampus and the right amygdala but not for the two structures on the left hemisphere (see Table 5 for detailed p-values). However, in analyzing whether age matters in terms of the APOE ε4 allele’s effects on the shape morphometrics of the four structures in MCI-C, we found no such age influence as was observed in the entire MCI population. To be specific, there is no statistically significant group difference between the carriers and the non-carriers of both the MCI-C-YO and the MCI-C-VO with respect to the shape diffeomorphometrics of each of the four structures. The corresponding p-values are listed in Table 5. For the MCI non-converter population, no APOE genotype effects were observed in any of the structure shapes of MCI-NC, MCI-NC-YO, or MCI-NC-VO, which is entirely consistent with the corresponding volume results. The p-values of the MCI-NC shape analysis can be found in Supplementary Table 2.
The surface maps of the shape diffeomorphometric differences of the right hippocampus and the right amygdala, between MCI-C-e3 and MCI-C-e4, are shown in panel (c) of Figure 4 and Figure 5 respectively. The total surface areas spanned by vertices with significant shape differences is respectively 58.46mm2 (accounting for 3.27% of the entire structural surface area) and 208.07mm2 (accounting for 26.56% of the entire structural surface area).
4 Discussion
In this study, we have analyzed how age impacts the APOE ε4 allele’s effects on the volumes and shapes of the amygdala and the hippocampus in both hemispheres in MCI and AD. Considering the heterogeneity in the MCI population brought about by the conversion status (a subject may deteriorate to dementia or remain stable), we also investigated whether there is any APOE genotype effect in the MCI converters and non-converters and whether age exerts any influence in the APOE genotype’s effects on the four structures of interest in those two MCI subgroups. Instead of directly analyzing the volumes and shape areas, we worked with the z-scores of the measurements normalized by those of the age-matched healthy control counterparts so as to reduce the impact of normal aging.
According to the results of our analysis on those four structures that play an important role in cognitive memory function, the age range was found to be a key element in the APOE ε4 allele’s influence on their volume and shape morphometrics in both the MCI and the AD population. To be specific, for the MCI and AD patients that are older than 80 years of age there were no significant group differences between the APOE ε4 carriers and the non-carriers in the z-transformed volumes and shape areas of the four aforementioned structures. However, significant group differences were detected between the carriers and non-carriers in those two patient groups that are younger than 75 years old (see Table 3 and Table 4).
In the entire AD population (without age-based stratification), there is no statistically significant group difference between the two APOE genotype groups in terms of both the volume and the shape of the left hippocampus although the volumetric difference is nearly significant (p = 0.0134). In contrast, the volume of the right hippocampus was found to differ significantly between the ε4 carriers and the non-carriers (p = 0.005) while the shape difference was nearly significant (p = 0.0235). This clearly suggests that the APOE genotype affects the hippocampus of patients with AD in an asymmetric fashion. This finding is in good agreement with those reported in several other studies [16,45]. Certain studies have also demonstrated an important association between the APOE genotype and the asymmetry of the hippocampal pattern in AD [46]. In this study, the volume of the amygdala in either hemisphere, of the entire AD group, was not found to differ significantly depending on the APOE genotype whereas the shape of the left amygdala differed significantly (p = 0.0101) and the shape of the right amygdala differed nearly significantly (p = 0.0391). This observation indicates that our vertex-based shape diffeomorphometrics of the bilateral amygdalas are more sensitive than their volume counterparts, which has also been observed in our other studies [25,47]. As shown in panel (A) of Figure 1, the locations on the left amygdala revealing significant shape differences between AD-e3 and AD-e4 belong to the basolateral, centromedial, and lateral nucleus subregions, with the affected area comprising 3.94% of the entire left amygdala surface. Such an investigation of the APOE genotype’s effects at each vertex of the amygdala surfaces is, as far as we know, the first of its kind ever reported.
After dividing the entire AD population into two categories based on each subject’s age (Young-Old AD and Very-Old AD), we found that the APOE genotype affects only the amygdala and the hippocampus of the Young-Old AD group but not the Very-Old AD one. According to Table 3, there is significant volumetric difference, between the carriers and the non-carriers of the AD-YO population, for each of the four structures. The shapes of the right hippocampus, the left amygdala, and the right amygdala were all found to differ significantly between AD-YO-e3 and AD-YO-e4 whereas the shape of the left hippocampus differed nearly significantly (p = 0.0895).
According to the surface map shown in panel (B) of Figure 1, the locations on the left amygdala revealing significant shape differences between AD-YO-e3 and AD-YO-e4 are part of the basolateral and centromedial subregions. The comparison demonstrated in Figure 1 clearly suggests that the magnitude of the group difference between the two APOE genotypes within the Young-Old AD group, at the same vertex location, is much stronger than that within the entire AD group.
According to Figure 2, the locations on the right hippocampus showing significant shape group differences between AD-YO-e3 and AD-YO-e4 occur in the CA1 and subiculum subregions. The observation that the CA1 and subiculum of the hippocampus are the most affected by the APOE ε4 genotype agrees well with those from other studies [48–50]. The CA1 field and the subiculum of the hippocampus are generally recognized as the subregions that are affected the earliest and also the most severely by the pathology of AD [25,51–54]. This may imply that the APOE ε4 genotype is more likely to exert influences on subregions that are more vulnerable to the AD pathology. Previous studies [16,55,56] have observed significant effects of the APOE ε4 genotype on the right hippocampal volume. That said, as a novel contribution, we found that this selective influence of the APOE genotype on the right hippocampus is specific to Young-Old AD patients and specific to CA1 and subiculum subregions.
In our shape based comparison of the right amygdala between AD-YO-e3 and AD-YO-e4, as presented in Figure 3, we found that a majority of the right amygdala (51.85% of the entire right amygdala surface) atrophied in the carriers relative to the non-carriers with almost the entire basolateral, centromedial, and the lateral nucleus being affected. In keeping with our left amygdala findings, the basomedial subregion was mostly unaffected. This further supports our previous statement that the APOE ε4 genotype affects a given structure in the right hemisphere more than it does in the left. Furthermore, to the best of our knowledge, this is the first study observing an age-specificity in the effect of the APOE ε4 genotype on subregional atrophy of the bilateral amygdalas in patients with AD.
In summary, our aforementioned observations in AD-YO, along with our finding of no statistical difference in the AD-VO group, indicates that the APOE ε4 allele mainly affects the hippocampal and amygdalar morphology in Young-Old AD patients. A similar observation has been reported in other studies as well though not always on a subregional level, in terms of not only brain structural morphology but also cognitive functions [23,36].
For the entire MCI population, as illustrated in Table 4, significant volumetric atrophy of the left hippocampus, the right hippocampus, and the right amygdala has been detected in the ε4 carriers relative to the non-carriers. However, for the shape measurements, only those differences in the right hippocampus and the right amygdala were statistically significant while those in the left hippocampus were nearly significant (p = 0.0382). This agrees with our finding in the AD population which identifies an asymmetric pattern (R > L in terms of the APOE genotype’s effect). As shown in panel (a) of Figure 4, the locations on the right hippocampus exhibiting significant group difference between MCI-e3 and MCI-e4 mainly belong to the CA1 and subiculum subregions along with a very small part of CA2, the total amount of which accounts for 18.26% of the entire right hippocampal surface area. The observation that the right hippocampal subregion affected by the APOE genotype in MCI is primarily CA1 and the subiculum confirms our previous conclusion that the APOE ε4 genotype mainly affects those subregions that are sensitive to the pathology of Alzheimer dementia. Regarding the shape atrophy of the right amygdala in MCI-e4 relative to MCI-e3, as we can see in Figure 5, the majority of the right amygdala atrophied significantly, covering almost the entire basolateral, centromedial, and lateral nucleus, accounting for 55.87% of the total structural surface area. In addition to the hippocampal and amygdalar morphology, studies have shown that the APOE genotype impacts the cognitive functions in the MCI population as well [57].
After using the age thresholding to divide the MCI population into two groups – MCI-YO and MCI-VO, we again observed that the effect of the APOE ε4 genotype manifests in MCI-YO but not at all in MCI-VO; significant volume and shape atrophy of the bilateral hippocampi and the right amygdala were observed in the ε4 carriers compared to the non-carriers only within the MCI-YO group, not MCI-VO (see Table 4). According to the surface maps of the right hippocampus (panel (b) in Figure 4) and the right amygdala (panel (b) in Figure 5), the portion of vertices showing significant group difference between the two APOE genotype groups within MCI-YO is less than those within the entire MCI group (panel (a) in the same figure) for both structures, which contrasts with our findings in the AD case wherein significant group difference for the shapes of those two structures occur only in the Young-Old subgroup but not the entire AD group. This may have been induced by an inhomogeneity in the MCI population, which brings us to our Type II MCI analysis.
According to our results from the Type II MCI analysis, the APOE ε4’s effects on the morphology of the right hippocampus and the right amygdala in MCI only exist for MCI patients who converted to AD within 3 years from their baseline, not for those who remained MCI or reverted to cognitively normal over the same period (see Table 5 and Supplementary Table 2). The atrophy of the left hippocampus and the left amygdala in the ε4 carriers relative to the non-carriers in MCI-converters is nearly significant (p = 0.0148 for the left hippocampus; p = 0.0243 for the left amygdala). This finding, to some degree, suggests that the brain atrophy pattern of MCI-C resembles that of AD whereas the atrophy pattern of MCI-NC resembles that of normal aging. Such a conclusion has also been reported in other imaging based studies [25,58].
According to the surface maps illustrating the vertex-by-vertex shape differences, between MCI-C-e3 and MCI-C-e4, of the right hippocampus (panel (c) of Figure 4), the main region affected by the APOE ε4 allele in MCI-C is CA1 and CA2. Those are part of the regions that are also affected in the entire MCI population (panel (a) of the same figure). However, for the right amygdala (panel (c) of Figure 5), the main regions affected by the APOE genotype in MCI-C belong to the basolateral, basomedial, and centromedial amygdala, which is not entirely a subset of those affected in the entire MCI population (particularly the basomedial subregion). This observation again confirms the heterogeneity in the MCI population and an urgent need for more refined categorization within the MCI population. A combination of various assessment techniques, e.g. neurological testing, APOE genotyping, and neuroimaging, may indeed aid early prognosis.
Following our procedure by dividing both MCI-C and MCI-NC into two age-dependent subgroups (Young-Old and Very-Old), we did not observe an age effect in terms of the APOE genotype’s influence on the morphology of those four structures (see Table 5 and Supplementary Table 2). This implies that, even though age intervenes in the APOE genotype’s effects on the MCI population, such an effect is not perceptible within a fixed conversion status grouping.
We now give mention to one noticeable aspect of this study. The subgrouping of the MCI subjects follows the criterion published in our previous works [27,34]; that is, an MCI subject is categorized as MCI-C if that subject converted to AD within 36 months from their baseline and MCI-NC otherwise. Our primary aim of this study is to explore whether the APOE genotype makes a difference, and whether age interacts, in the MCI-to-AD conversion. This MCI subgrouping criteria are different from both the standard subtyping methodology of categorizing MCI into amnestic MCI and non-amnestic MCI and the recently proposed more accurate MCI subtyping approach [59,60]. One natural extension of this study is to examine the influence of the APOE genotype and age on the hippocampal and amygdalar morphometry (both volume and shape) in more accurately categorized MCI subgroups such as the four different MCI subgroups (dysnomic, amnestic, dysexecutive, and “normal”) as suggested in [60].
Our current study has focused on the hippocampus and the amygdala. Another medial temporal lobe structure of great interest is the entorhinal cortex, which has been shown to be severely affected by the pathology of Alzheimer disease [61]. Actually, it has been suggested that the Alzheimer disease starts at the entorhinal cortex [26,62]. It is thus natural to envisage future investigations into the effects of age and APOE genotype on the volume and shape morphology of the entorhinal cortex in patients with MCI and AD. Another important extension to the current study will be to evaluate whether and how age intervenes in the rates of change in both volume and shape of the four targeted structures by using longitudinal samples and time series based sequential shape diffeomorphometrics.
In closing, we have analyzed how age modulates the APOE genotype’s influence on the volume and shape morphometrics of the hippocampus and the amygdala in both hemispheres based on a large sample of 540 subjects. Contributions and novel findings from this study can be summarized as: 1) the influence that the APOE genotype exerts on the amygdala and the hippocampus was only observed in Young-Old patients with MCI and AD, not the Very-Old ones; 2) the genotype-specific atrophy patterns observed in the MCI subjects resembles those of the AD population; 3) in all such groupings, when an effect is present, the structures in the right hemisphere are affected more by the APOE genotype than those in the left hemisphere; 4) selective effects were observed on the right hippocampus with the CA1 and the subiculum subregions being affected the most; 5) for the right amygdala, the basolateral, centromedial, and lateral nucleus subregions are affected the most while the basomedial is less affected; 6) further refinement showed that the APOE genotype only affects MCI converters, not the MCI non-converters; 7) age dependence was not observed in the APOE genotype’s influence when restricting to subgroups of the MCI population defined by conversion status.
Supplementary Material
Acknowledgments
This work is partially supported by NIH R01 EB000975, NIH P41 RR15241, and R01 EB008171.
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott; Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Amorfix Life Sciences Ltd.; AstraZeneca; Bayer HealthCare; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. This research was also supported by NIH grants P30 AG010129 and K01 AG030514.
Anders M. Dale is a founder and holds equity in CorTechs Labs, Inc, and also serves on its Scientific Advisory Board. Michael I. Miller owns an equal share in Anatomyworks LLC.
Footnotes
The terms of this arrangement have been reviewed and approved by the Johns Hopkins University, as well as the University of California, San Diego in accordance with their conflict of interest policies.
References
- 1.Corder E, Saunders A, Strittmatter W, Schmechel D, Gaskell P, Small G, Roses A, Haines J, Pericak-Vance M. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 2.Sando S, Melquist S, Cannon A, Hutton M, Sletvold O, Saltvedt I, White L, Lydersen S, Aasly J. APOE epsilon4 lowers age at onset and is a high risk factor for Alzheimer’s disease; A case control study from central Norway. BMC Neurology. 2008;8:9. doi: 10.1186/1471-2377-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lehtovirta M, Soininen H, Helisalmi S, Mannermaa A, Helkala EL, Hartikainen P, Hanninen T, Ryynanen M, Riekkinen PJ. Clinical and neuropsychological characteristics in familial and sporadic Alzheimer’s disease: relation to apolipoprotein E polymorphism. Neurology. 1996;46:413–419. doi: 10.1212/wnl.46.2.413. [DOI] [PubMed] [Google Scholar]
- 4.van der Vlies AE, Pijnenburg YA, Koene T, Klein M, Kok A, Scheltens P, van der Flier WM. Cognitive impairment in Alzheimer’s disease is modified by APOE genotype. Dement Geriatr Cogn Disord. 2007;24:98–103. doi: 10.1159/000104467. [DOI] [PubMed] [Google Scholar]
- 5.Wolk DA, Dickerson BC, Alzheimer’s Disease Neuroimaging Initiative Apolipoprotein E (APOE) genotype has dissociable effects on memory and attentional-executive network function in Alzheimer’s disease. Proc Natl Acad Sci U S A. 2010;107:10256–10261. doi: 10.1073/pnas.1001412107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cuénod C, Denys A, Michot J, Jehenson P, Forette F, Kaplan D, Syrota A, Boller F. Amygdala atrophy in Alzheimer’s disease: an in vivo magnetic resonance imaging study. Arch Neurol. 1993;50:941–945. doi: 10.1001/archneur.1993.00540090046009. [DOI] [PubMed] [Google Scholar]
- 7.Du AT, Schuff N, Amend D, Laakso MP, Hsu YY, Jagust WJ, Yaffe K, Kramer JH, Reed B, Norman D, Chui HC, Weiner MW. Magnetic resonance imaging of the entorhinal cortex and hippocampus in mild cognitive impairment and Alzheimer’s disease. Journal of Neurology, Neurosurgery & Psychiatry. 2001;71:441–447. doi: 10.1136/jnnp.71.4.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hyman B, Van Hoesen G, Damasio A, Barnes C. Alzheimer’s disease: cell-specific pathology isolates the hippocampal formation. Science. 1984;225:1168–1170. doi: 10.1126/science.6474172. [DOI] [PubMed] [Google Scholar]
- 9.Juottonen K, Laakso MP, Partanen K, Soininen H. Comparative MR Analysis of the Entorhinal Cortex and Hippocampus in Diagnosing Alzheimer Disease. American Journal of Neuroradiology. 1999;20:139–144. [PubMed] [Google Scholar]
- 10.Price JL, Ko AI, Wade MJ, Tsou SK, McKeel DW, Morris JC. Neuron number in the entorhinal cortex and CA1 in preclinical Alzheimer disease. Arch Neurol. 2001;58:1395–1402. doi: 10.1001/archneur.58.9.1395. [DOI] [PubMed] [Google Scholar]
- 11.Hopper MW, Vogel FS. The limbic system in Alzheimer’s disease. A neuropathologic investigation. Am J Pathol. 1976;85:1–20. [PMC free article] [PubMed] [Google Scholar]
- 12.Nestor PJ, Fryer TD, Smielewski P, Hodges JR. Limbic hypometabolism in Alzheimer’s disease and mild cognitive impairment. Ann Neurol. 2003;54:343–351. doi: 10.1002/ana.10669. [DOI] [PubMed] [Google Scholar]
- 13.Cherbuin N, Leach LS, Christensen H, Anstey KJ. Neuroimaging and APOE genotype: a systematic qualitative review. Dement Geriatr Cogn Disord. 2007;24:348–362. doi: 10.1159/000109150. [DOI] [PubMed] [Google Scholar]
- 14.Jack CR, Petersen RC, Xu YC, O’Brien PC, Waring SC, Tangalos EG, Smith GE, Ivnik RJ, Thibodeau SN, Kokmen E. Hippocampal atrophy and apolipoprotein E genotype are independently associated with Alzheimer’s disease. Ann Neurol. 1998;43:303–310. doi: 10.1002/ana.410430307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schuff N, Woerner N, Boreta L, Kornfield T, Shaw LM, Trojanowski JQ, Thompson PM, Jack CR, Weiner MW, the Alzheimer’s, Disease Neuroimaging Initiative MRI of hippocampal volume loss in early Alzheimer’s disease in relation to ApoE genotype and biomarkers. Brain. 2009;132:1067–1077. doi: 10.1093/brain/awp007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lehtovirta M, Laakso MP, Soininen H, Helisalmi S, Mannermaa A, Helkala EL, Partanen K, Ryynanen M, Vainio P, Hartikainen P. Volumes of hippocampus, amygdala and frontal lobe in Alzheimer patients with different apolipoprotein E genotypes. Neuroscience. 1995;67:65–72. doi: 10.1016/0306-4522(95)00014-a. [DOI] [PubMed] [Google Scholar]
- 17.Basso M, Gelernter J, Yang J, MacAvoy MG, Varma P, Bronen RA, van Dyck CH. Apolipoprotein E epsilon4 is associated with atrophy of the amygdala in Alzheimer’s disease. Neurobiol Aging. 2006;27:1416–1424. doi: 10.1016/j.neurobiolaging.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 18.Bondi MW, Houston WS, Salmon DP, Corey-Bloom J, Katzman R, Thal LJ, Delis DC. Neuropsychological deficits associated with Alzheimer’s disease in the very-old: discrepancies in raw vs. standardized scores. J Int Neuropsychol Soc. 2003;9:783–795. doi: 10.1017/S1355617703950119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, Myers RH, Pericak-Vance MA, Risch N, van Duijn CM. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease: a meta-analysis. JAMA. 1997;278:1349–1356. [PubMed] [Google Scholar]
- 20.Small BJ, Rosnick CB, Fratiglioni L, Bäckman L. Apolipoprotein E and Cognitive Performance: A Meta-Analysis. Psychol Aging. 2004;19:592–600. doi: 10.1037/0882-7974.19.4.592. [DOI] [PubMed] [Google Scholar]
- 21.Juva K, Verkkoniemi A, Viramo P, Polvikoski T, Kainulainen K, Kontula K, Sulkava R. APOE e4 does not predict mortality, cognitive decline, or dementia in the oldest old. Neurology. 2000;54:412–412. doi: 10.1212/wnl.54.2.412. [DOI] [PubMed] [Google Scholar]
- 22.Holland D, Desikan RS, Dale AM, McEvoy LK. Higher Rates of Decline for Women and Apolipoprotein E e4 Carriers. American Journal of Neuroradiology. 2013;34:2287–2293. doi: 10.3174/ajnr.A3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang YL, Fennema-Notestine C, Holland D, McEvoy LK, Stricker NH, Salmon DP, Dale AM, Bondi MW, Alzheimer’s Disease Neuroimaging Initiative APOE interacts with age to modify rate of decline in cognitive and brain changes in Alzheimer’s disease. Alzheimers Dement. 2014;10:336–348. doi: 10.1016/j.jalz.2013.05.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller MI, Younes L, Ratnanather JT, Brown T, Trinh H, Postell E, Lee DS, Wang MC, Mori S, O’Brien R, Albert M, BIOCARD Research Team The diffeomorphometry of temporal lobe structures in preclinical Alzheimer’s disease. Neuroimage Clin. 2013;3:352–360. doi: 10.1016/j.nicl.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang X, Holland D, Dale AM, Younes L, Miller MI, for the Alzheimer’s Disease Neuroimaging Initiative Shape abnormalities of subcortical and ventricular structures in mild cognitive impairment and Alzheimer’s disease: Detecting, quantifying, and predicting. Hum Brain Mapp. 2014;35:3701–3725. doi: 10.1002/hbm.22431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Younes L, Albert M, Miller MI, BIOCARD Research Team Inferring changepoint times of medial temporal lobe morphometric change in preclinical Alzheimer’s disease. Neuroimage Clin. 2014;5:178–187. doi: 10.1016/j.nicl.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang X, Holland D, Dale AM, Younes L, Miller MI, for the Alzheimer’s Disease Neuroimaging Initiative The diffeomorphometry of regional shape change rates and its relevance to cognitive deterioration in mild cognitive impairment and Alzheimer’s disease. Hum Brain Mapp. 2015 doi: 10.1002/hbm.22758. n/a-n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeCarli C. Mild cognitive impairment: prevalence, prognosis, aetiology, and treatment. The Lancet Neurology. 2003;2:15–21. doi: 10.1016/s1474-4422(03)00262-x. [DOI] [PubMed] [Google Scholar]
- 29.Petersen RC, Doody R, Kurz A, Mohs RC, Morris JC, Rabins PV, Ritchie K, Rossor M, Thal L, Winblad B. Current concepts in mild cognitive impairment. Arch Neurol. 2001;58:1985–1992. doi: 10.1001/archneur.58.12.1985. [DOI] [PubMed] [Google Scholar]
- 30.Larrieu S, Letenneur L, Orgogozo JM, Fabrigoule C, Amieva H, Le Carret N, Barberger–Gateau P, Dartigues JF. Incidence and outcome of mild cognitive impairment in a population-based prospective cohort. Neurology. 2002;59:1594–1599. doi: 10.1212/01.wnl.0000034176.07159.f8. [DOI] [PubMed] [Google Scholar]
- 31.Luis CA, Loewenstein DA, Acevedo A, Barker WW, Duara R. Mild cognitive impairment: Directions for future research. Neurology. 2003;61:438–444. doi: 10.1212/01.wnl.0000080366.90234.7f. [DOI] [PubMed] [Google Scholar]
- 32.Petersen RC, Smith GE, Ivnik RJ, et al. APolipoprotein e status as a predictor of the development of alzheimer’s disease in memory-impaired individuals. JAMA. 1995;273:1274–1278. [PubMed] [Google Scholar]
- 33.Mosconi L, Perani D, Sorbi S, Herholz K, Nacmias B, Holthoff V, Salmon E, Baron J, De Cristofaro MTR, Padovani A, Borroni B, Franceschi M, Bracco L, Pupi A. MCI conversion to dementia and the APOE genotype: A prediction study with FDG-PET. Neurology. 2004;63:2332–2340. doi: 10.1212/01.wnl.0000147469.18313.3b. [DOI] [PubMed] [Google Scholar]
- 34.Tang X, Holland D, Dale AM, Younes L, Miller MI. Baseline Shape Diffeomorphometry Patterns of Subcortical and Ventricular Structures in Predicting Conversion of Mild Cognitive Impairment to Alzheimer’s Disease. J Alzheimers Dis. 2014 doi: 10.3233/JAD-141605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bondi MW, Houston WS, Salmon DP, Corey-Bloom J, Katzman R, Thal LJ, Delis DC. Neuropsychological deficits associated with Alzheimer’s disease in the very-old: discrepancies in raw vs. standardized scores. J Int Neuropsychol Soc. 2003;9:783–795. doi: 10.1017/S1355617703950119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stricker NH, Chang YL, Fennema-Notestine C, Delano-Wood L, Salmon DP, Bondi MW, Dale AM, Alzheimer’s Disease Neuroimaging Initiative Distinct profiles of brain and cognitive changes in the very old with Alzheimer disease. Neurology. 2011;77:713–721. doi: 10.1212/WNL.0b013e31822b0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Corder EH, Saunders AM, Risch NJ, Strittmatter WJ, Schmechel DE, Gaskell PC, Jr, Rimmler JB, Locke PA, Conneally PM, Schmader KE. Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat Genet. 1994;7:180–184. doi: 10.1038/ng0694-180. [DOI] [PubMed] [Google Scholar]
- 38.Jovicich J, Czanner S, Greve D, Haley E, van der Kouwe A, Gollub R, Kennedy D, Schmitt F, Brown G, Macfall J, Fischl B, Dale A. Reliability in multi-site structural MRI studies: effects of gradient non-linearity correction on phantom and human data. Neuroimage. 2006;30:436–443. doi: 10.1016/j.neuroimage.2005.09.046. [DOI] [PubMed] [Google Scholar]
- 39.Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. Medical Imaging, IEEE Transactions on. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- 40.Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale AM. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- 41.Qiu A, Brown T, Fischl B, Ma J, Miller MI. Atlas Generation for Subcortical and Ventricular Structures With Its Applications in Shape Analysis. Image Processing, IEEE Transactions on. 2010;19:1539–1547. doi: 10.1109/TIP.2010.2042099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma J, Miller MI, Younes L. A bayesian generative model for surface template estimation. Int J Biomed Imaging. 2010;2010:974957. doi: 10.1155/2010/974957. Epub 2010 Sep 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vaillant M, Glaunès J. Surface matching via currents. Information Processing in Medical Imaging. 2005:381–392. doi: 10.1007/11505730_32. [DOI] [PubMed] [Google Scholar]
- 44.Nichols T, Hayasaka S. Controlling the familywise error rate in functional neuroimaging: a comparative review. Statistical Methods in Medical Research. 2003;12:419–446. doi: 10.1191/0962280203sm341ra. [DOI] [PubMed] [Google Scholar]
- 45.Lind J, Larsson A, Persson J, Ingvar M, Nilsson L, Bäckman L, Adolfsson R, Cruts M, Sleegers K, Van Broeckhoven C, Nyberg L. Reduced hippocampal volume in non-demented carriers of the apolipoprotein E ε4: Relation to chronological age and recognition memory. Neurosci Lett. 2006;396:23–27. doi: 10.1016/j.neulet.2005.11.070. [DOI] [PubMed] [Google Scholar]
- 46.Geroldi C, Laakso MP, DeCarli C, Beltramello A, Bianchetti A, Soininen H, Trabucchi M, Frisoni GB. Apolipoprotein E genotype and hippocampal asymmetry in Alzheimer’s disease: a volumetric MRI study. J Neurol Neurosurg Psychiatry. 2000;68:93–96. doi: 10.1136/jnnp.68.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller MI, Younes L, Ratnanather JT, Brown T, Trinh H, Lee DS, Tward D, Mahon PB, Mori S, Albert M. Amygdalar atrophy in symptomatic Alzheimer’s disease based on diffeomorphometry: the BIOCARD cohort. Neurobiol Aging. 2015;36(Supplement 1):S3–S10. doi: 10.1016/j.neurobiolaging.2014.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Khan W, Westman E, Jones N, Wahlund LO, Mecocci P, Vellas B, Tsolaki M, Kloszewska I, Soininen H, Spenger C, Lovestone S, Muehlboeck JS, Simmons A, for the AddNeuroMed consortium and for the Alzheimer’s Disease Neuroimaging Initiative Automated Hippocampal Subfield Measures as Predictors of Conversion from Mild Cognitive Impairment to Alzheimer’s Disease in Two Independent Cohorts. Brain Topogr. 2014 doi: 10.1007/s10548-014-0415-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pievani M, Galluzzi S, Thompson PM, Rasser PE, Bonetti M, Frisoni GB. APOE4 is associated with greater atrophy of the hippocampal formation in Alzheimer’s disease. Neuroimage. 2011;55:909–919. doi: 10.1016/j.neuroimage.2010.12.081. [DOI] [PubMed] [Google Scholar]
- 50.Kerchner GA, Berdnik D, Shen JC, Bernstein JD, Fenesy MC, Deutsch GK, Wyss-Coray T, Rutt BK. APOE e4 worsens hippocampal CA1 apical neuropil atrophy and episodic memory. Neurology. 2014;82:691–697. doi: 10.1212/WNL.0000000000000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Braak H, Braak E. Staging of Alzheimer’s disease-related neurofibrillary changes. Neurobiol Aging. 1995;16:271–8. doi: 10.1016/0197-4580(95)00021-6. discussion 278–84. [DOI] [PubMed] [Google Scholar]
- 52.Schonheit B, Zarski R, Ohm TG. Spatial and temporal relationships between plaques and tangles in Alzheimer-pathology. Neurobiol Aging. 2004;25:697–711. doi: 10.1016/j.neurobiolaging.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 53.Wang L, Miller JP, Gado MH, McKeel DW, Rothermich M, Miller MI, Morris JC, Csernansky JG. Abnormalities of hippocampal surface structure in very mild dementia of the Alzheimer type. Neuroimage. 2006;30:52–60. doi: 10.1016/j.neuroimage.2005.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rossler M, Zarski R, Bohl J, Ohm TG. Stage-dependent and sector-specific neuronal loss in hippocampus during Alzheimer’s disease. Acta Neuropathol. 2002;103:363–369. doi: 10.1007/s00401-001-0475-7. [DOI] [PubMed] [Google Scholar]
- 55.Agosta F, Vossel KA, Miller BL, Migliaccio R, Bonasera SJ, Filippi M, Boxer AL, Karydas A, Possin KL, Gorno-Tempini ML. Apolipoprotein E epsilon4 is associated with disease-specific effects on brain atrophy in Alzheimer’s disease and frontotemporal dementia. Proc Natl Acad Sci U S A. 2009;106:2018–2022. doi: 10.1073/pnas.0812697106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Geroldi C, Pihlajamaki M, Laakso MP, DeCarli C, Beltramello A, Bianchetti A, Soininen H, Trabucchi M, Frisoni GB. APOE-epsilon4 is associated with less frontal and more medial temporal lobe atrophy in AD. Neurology. 1999;53:1825–1832. doi: 10.1212/wnl.53.8.1825. [DOI] [PubMed] [Google Scholar]
- 57.Farlow MR, He Y, Tekin S, Xu J, Lane R, Charles HC. Impact of APOE in mild cognitive impairment. Neurology. 2004;63:1898–1901. doi: 10.1212/01.wnl.0000144279.21502.b7. [DOI] [PubMed] [Google Scholar]
- 58.Fan Y, Batmanghelich N, Clark CM, Davatzikos C. Spatial patterns of brain atrophy in MCI patients, identified via high-dimensional pattern classification, predict subsequent cognitive decline. Neuroimage. 2008;39:1731–1743. doi: 10.1016/j.neuroimage.2007.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bondi MW, Edmonds EC, Jak AJ, Clark LR, Delano-Wood L, McDonald CR, Nation DA, Libon DJ, Au R, Galasko D, Salmon DP. Neuropsychological criteria for mild cognitive impairment improves diagnostic precision, biomarker associations, and progression rates. J Alzheimers Dis. 2014;42:275–289. doi: 10.3233/JAD-140276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Edmonds EC, Delano-Wood L, Clark LR, Jak AJ, Nation DA, McDonald CR, Libon DJ, Au R, Galasko D, Salmon DP, Bondi MW, Alzheimer’s Disease Neuroimaging Initiative Susceptibility of the conventional criteria for mild cognitive impairment to false-positive diagnostic errors. Alzheimers Dement. 2015;11:415–424. doi: 10.1016/j.jalz.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Hoesen GW, Hyman BT, Damasio AR. Entorhinal cortex pathology in Alzheimer’s disease. Hippocampus. 1991;1:1–8. doi: 10.1002/hipo.450010102. [DOI] [PubMed] [Google Scholar]
- 62.Khan UA, Liu L, Provenzano FA, Berman DE, Profaci CP, Sloan R, Mayeux R, Duff KE, Small SA. Molecular drivers and cortical spread of lateral entorhinal cortex dysfunction in preclinical Alzheimer’s disease. Nat Neurosci. 2014;17:304–311. doi: 10.1038/nn.3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


