Abstract
Background
Diagnostic uncertainty in ALS has serious management implications and delays recruitment into clinical trials. Emerging evidence of presymptomatic disease-burden provides the rationale to develop diagnostic applications based on the evaluation of in-vivo pathological patterns early in the disease.
Objectives
To outline and test a diagnostic classification approach based on an array of complementary imaging metrics in key disease-associated anatomical structures.
Methods
Data from 75 ALS patients and 75 healthy controls were randomly allocated in a ‘training’ and ‘validation’ cohort. Spatial masks were created for anatomical foci which best discriminate patients from controls in the ‘training sample’. In a virtual ‘brain biopsy’, data was then retrieved from these key disease-associated brain regions. White matter diffusivity indices, grey matter T1-signal intensity values and basal ganglia volumes were evaluated as predictor variables in a canonical discriminant function.
Results
Following predictor variable selection, a classification specificity of 85.5% and sensitivity of 89.1% was achieved in the training sample and 90% specificity and 90% sensitivity in the validation sample.
Discussion
This study evaluates disease-associated imaging measures in a dummy diagnostic application. Although larger samples will be required for robust validation, the study confirms the potential of multimodal quantitative imaging in future clinical applications.
Keywords: Magnetic resonance imaging, Neuroimaging, Diagnosis, Neurodegeneration, Amyotrophic lateral sclerosis, Motor neuron disease
Graphical abstract
Highlights
-
•
Reliable diagnostic, monitoring and prognostic biomarkers are urgently in ALS.
-
•
Accurate diagnostic classification may be achieved based on MRI metrics.
-
•
Basal ganglia, grey and white matter indices were integrated in a diagnostic model.
-
•
85.5% specificity and 89.1% sensitivity were achieved in the training sample.
-
•
90% specificity and 90% sensitivity were achieved in the validation sample.
1. Introduction
Patients with ALS often describe tripping, intermittent slurred speech, muscle cramping, leg stiffness years before their formal diagnosis. Delays in diagnosis compromises timely enrolment in clinical trials, and limit the neuroprotective potential of new therapeutic agents. It is now also widely recognized that a long presymptomatic phase precedes symptom manifestation (Eisen et al., 2014). The few existing presymptomatic studies ALS confirm that the pathological changes can be captured well before symptom onset (Carew et al., 2011). The diagnosis of ALS remains primarily clinical, despite the unprecedented advances in quantitative neuroimaging and the uniquely specific imaging signature of ALS. After years of descriptive MRI studies in ALS, a number of studies have now emerged outlining diagnostic and prognostic applications based on pattern recognition (Bede and Hardiman, 2014). While methods of machine learning and data mining have been extensively used in marketing, information technology, banking, traffic control and metrology, they have only been recently applied to neurodegeneration (Orru et al., 2012). Many of the pioneering radiology studies come from cancer screening applications developed for the automated detection of malignancies. The automated analysis of imaging data of neurodegenerative conditions poses unique methodological challenges. Age-related variability, overlap of several neurodegenerative conditions, co-existing vascular white matter alterations are just some of the confounders. The most commonly used statistical approaches in neurodegeneration include logistic regression and support vector machines. Discriminant function analysis allows the determination of probability of group membership based on predictor variables. While the assumptions of discriminant analysis may be less flexible than those of logistic regression, it offers relatively high classification accuracy.
We hypothesized that based on the distinct neuroimaging signature of ALS a robust diagnostic framework can be outlined using discriminant function analysis. Our objective was to describe a diagnostic classification approach which incorporates multiple, ALS-defining, cortical, basal ganglia and white matter measures. The overarching hypothesis of this study is that ‘data biopsies’ from blinded data sets can be used to accurately categorize individual participants as patients or controls based on normative and disease-specific training data.
2. Methods
All participants of this prospective biomarker study provided informed consent in accordance with the Medical Ethics Approval of the project (Beaumont Hospital, Dublin, Ireland). ALS patients and controls were randomly allocated in a ‘training group’ and a ‘testing group’ for validation. In total, 55 ALS patients and 55 controls were allocated to the ‘training group’ and 20 patients and 20 controls to the ‘testing group’. Healthy controls have been specifically recruited for this ALS biomarker study. The demographic and clinical profile of study participants are presented in Table 1. The participants of the training and validation cohorts were matched for age, gender and education.
Table 1.
The demographic and clinical profile of participants.
| Diagnosis | Training cohort |
Validation cohort |
p - value | ||
|---|---|---|---|---|---|
| ALS | Healthy controls | ALS | Healthy controls | ||
| n | 55 | 55 | 20 | 20 | |
| Age (Mean/St.Dev.) |
59.745/9.4579 | 60.309/9.1528 | 63.850/8.1323 | 57.700/10.2346 | 0.202 |
| Gender (Male/Female) |
34/21 | 28/27 | 14/6 | 9/11 | 0.278 |
| ALSFRS-r (Mean/St.Dev.) |
38.29/5.398 | 37.80/5.854 | 0.734 | ||
| Disease duration (Mean/St.Dev.) |
23.65/7.399 | 24.65/6.409 | 0.596 | ||
2.1. Methods overview
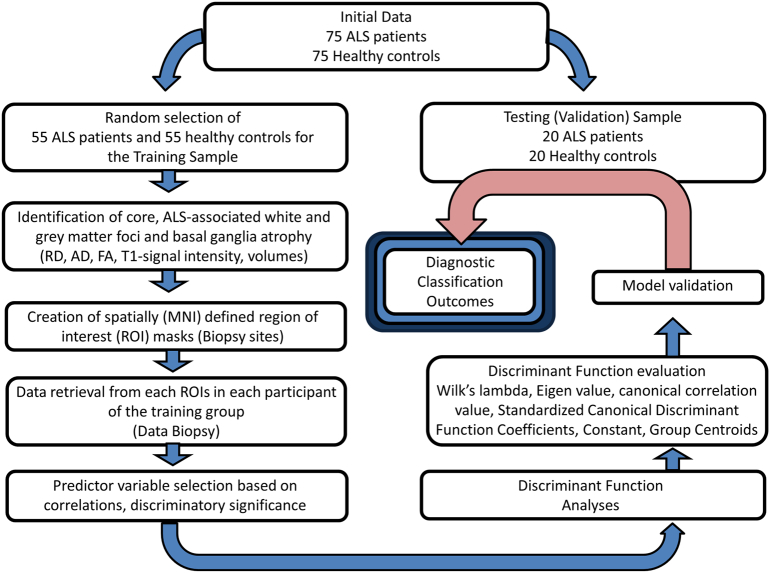
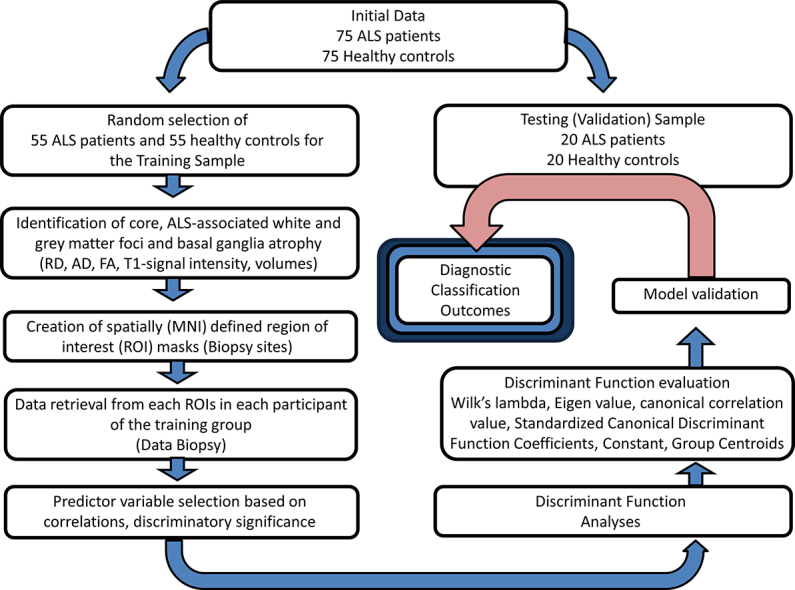
The schematic overview of the development of the diagnostic protocol is presented in Fig. 1. First, comparative analyses were carried out between the patients and controls of the training group to establish core ALS-associated patterns of degeneration. Whole-brain tract-based spatial statistics (TBSS), voxel based morphometry and basal ganglia volumetrics were carried out correcting for age and gender, the methods of which have been described previously described (Bede et al., 2013a, Bede et al., 2015, Bede et al., 2013b, Bede et al., 2013c). Additionally, the volumes of brain stem, left and right thalamus, caudate, pallidum, hippocampus, amygdala, putamen and accumbens nuclei were compared between the controls and ALS patients of the training group correcting for age. The main ‘disease-defining’ anatomical structures identified by these comparisons included the corticospinal tracts (CST), corpus callosum (CC), primary motor cortex (PMC), thalamus, caudate nuclei, accumbens nuclei, amygdala and the hippocampus. Accordingly, region-of-interest (ROIs) anatomical maps were created for these structures, including voxels which showed statistically significant differences between patients and controls in the corticospinal tracts, corpus callosum, and precentral gyrus.
Fig. 1.
A schematic outline of model development, testing and validation.
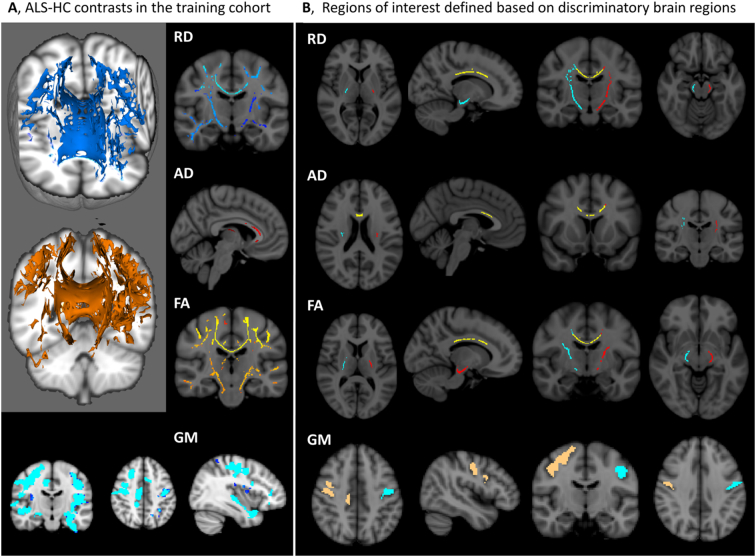
Imaging data of both the training and testing groups have been spatially co-registered to the MNI reference system and averaged MR metrics were retrieved from each participant in the above ROIs. In summary, our standardized ‘virtual brain biopsy’ consisted of retrieving quantitative MRI metrics from key, ALS-associated brain regions in each participant which were then evaluated in a discriminant function model. The outcomes of the initial comparisons and the atlas-based ROI masks created based on these contrasts are presented in Fig. 2.
Fig. 2.
A, Outcomes of initial contrasts between ALS patients and controls in the ‘training group’ highlighting key, disease-associated brain regions at p < 0.05 corrected for age and gender. B, Region-of-interest (ROI) masks created based on the initial comparisons incorporating statistically significant voxels in the left corticospinal tract (red), right corticospinal tract (turquoise), corpus callosum (yellow), left primary motor cortex (turquoise) and right primary motor cortex (gold). – Contrasts and ROI masks are shown for radial diffusivity values (RD), axial diffusivity values (AD), fractional anisotropy values (FA) and grey matter (GM) signal intensity. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.2. MRI pulse sequence descriptions
MR data were acquired on a 3 Tesla Philips Achieva system with a gradient strength of 80 mT/m and slew rate of 200 T/m/s using an 8-channel receive-only head coil. T1-weighted images were obtained using a three-dimensional inversion recovery prepared spoiled gradient recalled echo (IR-SPGR) sequence with FOV = 256 × 256 × 160 mm, spatial resolution = 1 mm3, TR/TE = 8.5/3.9 ms, TI = 1060 ms, flip angle = 8°, SENSE factor = 1.5. DTI images were acquired using a spin-echo planar imaging (SE-EPI) sequence with a 32-direction Stejskal-Tanner diffusion encoding scheme: FOV = 245 × 245 × 150 mm, 60 slices with no interslice gap, TR/TE = 7639/59 ms, SENSE factor = 2.5, b-values = 0, 1100 s/mm2, with SPIR fat suppression and dynamic stabilization in an acquisition time of 5 min 41 s.
2.3. MRI processing pipelines
Raw diffusion tensor imaging data sets underwent eddy current corrections, motion correction and brain-tissue extraction using FMRIB's software library (FSL) (Smith et al., 2004). A diffusion tensor model was then fitted, generating maps of axial diffusivity (AD), radial diffusivity (RD) and fractional anisotropy (FA). Tract-based spatial statistics (TBSS) and permutation-based nonparametric inference was used for group comparisons in a study-specific white matter template applying the threshold-free cluster enhancement (TFCE) method (Smith et al., 2006, Smith and Nichols, 2009). The comparisons of the ALS patients and healthy controls of the ‘training group’ were corrected for age and gender and statistical significance was set at p < 0.05 FWE.
Grey matter analyses were carried out using voxel based morphometry (VBM) tool box if FMRIB's software library (FSL) (Douaud et al., 2007). Following brain extraction, and tissue-type segmentation, grey matter partial volume images were aligned to the Montreal Neurological Institute 152 standard space. The grey matter partial volume estimates were non-linearly co-registered to a study-specific template, modulated by a Jacobian field warp and smoothed with an isotropic Gaussian kernel with a sigma of 3 mm. The TFCE method and permutation-based nonparametric inference was used for the comparisons of the ALS patients and healthy controls of the ‘training group’, which were corrected for age and gender and statistical significance was set at p < 0.05 FWE.
2.4. Basal ganglia volumes
Volumes of subcortical structures were estimated using the subcortical segmentation and registration tool FIRST, part of the FMRIB's Software Library (FSL). T1-weighted structural data first underwent brain extraction and the resulting images were individually verified. FSL-FIRST uses a two-stage affine registration algorithm of the input T1 data sets to MNI152 (Montreal Neurological Institute 152) standard space; first a standard 12 degrees of freedom registration, then a 12 degrees of freedom registration to a MNI152 sub-cortical mask. A model-based approach is used by FSL-FIRST for the segmentation of subcortical structures. The models incorporated in FIRST are constructed from manually segmented images from 336 subjects provided by the Center for Morphometric Analysis (CMA), MGH, Boston. Registration and segmentation were individually reviewed and visually verified for all subjects. Following registration and segmentation, subcortical mesh and volumetric outputs were generated by FSL-FIRST using automatic boundary corrections.
2.5. Region of interest maps
The grey matter region of interest (ROI) for the left and right primary motor cortex was created based on the Harvard-Oxford probabilistic atlas(Desikan et al., 2006) and defined based on voxels of the primary motor cortex which showed statistically significant differences at p < 0.05 between controls and patients in the ‘training’ cohort. White matter ROIs were defined based on the ICBM-DTI-81 white-matter labels atlas (Mori et al., 2008, Oishi et al., 2008). White matter ROIs were defined based on statistically significant differences in AD, RD and FA between the controls and ALS patients of the ‘training cohort’ in left corticospinal tract, right corticospinal tract and corpus callosum. While fractional anisotropy (FA) was evaluated, radial diffusivity (RD) and axial diffusivity (AD) were regarded as superior predictor variable candidates. This is because, AD (λ1) and RD ((λ2 + λ3) / 2) are mathematically independent white matter metrics and thought to reflect on axonal and myelin integrity respectively (Budde et al., 2009, Song et al., 2005).
The initial list of candidate predictor variables included RD, AD and FA in the left and right corticospinal tract and corpus callosum ROIs, T1-signal intensity in the left and right motor cortex ROIs, the bilaterally averaged volumes the thalamus and caudate, left and right hippocampus, nucleus accumbens, amygdala, brain stem and total intracranial volume (ICV). Candidate predictor variables were excluded from the model based on tests of equality of group means if Wilk's lambda was not statistically significant. Predictor variables were also excluded if they were highly correlated with each other in the pooled within-groups matrices. Based on the evaluation of candidate predictors, the final variables in the model included the AD and RD of the left and right CST and CC, left and right PMC ROIs, and volumes of the left and right hippocampus and nucleus accumbens, the left amygdala, and thalamus and caudate volumes (Table 2). The constant, canonical discriminant function coefficients and group centroids were calculated based on these variables. No leave-one-out cross-validation was performed in the training group, as separate-groups covariance matrixes were used. Classification accuracy was tested in the ‘training’ group and in the independent validation sample separately to appraise the discriminatory power and generalizability of the model.
Table 2.
Final predictor variables in the discriminant model following candidate predictor variable evaluation. RD – radial diffusivity, AD – axial diffusivity.
| Predictor variables after candidate predictor assessment and selection |
|---|
| Average RD in the “right RD corticospinal tract ROI” |
| Average RD in the “left RD corticospinal tract ROI” |
| Average RD in the “RD corpus callosum ROI” |
| Average AD in the “right AD corticospinal tract ROI” |
| Average AD in the “left AD corticospinal tract ROI” |
| Average AD in the “AD corpus callosum ROI” |
| T1-signal intensity in the “right motor cortex ROI” |
| T1-signal intensity in the “left motor cortex ROI” |
| Averaged volume of the left and right thalamus |
| Averaged volume of the left and right caudate |
| Volume of the left hippocampus |
| Volume of the right hippocampus |
| Volume of the left nucleus accumbens |
| Volume of the right nucleus accumbens |
| Volume of the left amygdala |
3. Results
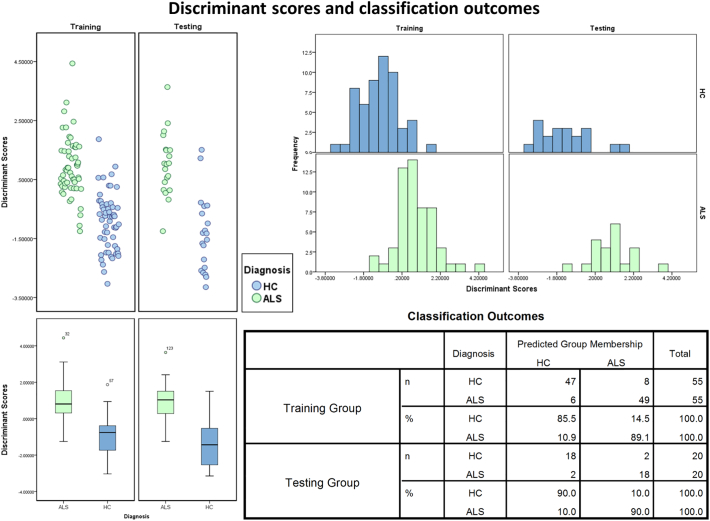
The canonical discriminant function reached an Eigen value of 0.871, a canonical correlation value of 0.682, Wilks lambda of 0.534, Chi-square 62.983 and a significance of < 0.001. The model showed an overall classification sensitivity of 89.1% and specificity of 85.5% in the ‘training’ group. In the ‘testing sample’ both sensitivity and specificity reached 90%. Classification function outcomes, histograms and scatter plots of discriminant score distributions are shown in Fig. 3.
Fig. 3.
Classification outcomes: scatter plots, box plots and histograms of discriminant score distributions in the training and testing cohort. Healthy controls are shown in blue, patients with ALS in green. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
Neuroimaging in ALS has been very successful in characterizing group-level, disease-specific changes (Agosta et al., 2010, Turner et al., 2012, Prell and Grosskreutz, 2013, Bede et al., 2016), and also in describing phenotype (Bede et al., 2013a, Machts et al., 2015, Floeter et al., 2014) and genotype-specific (Cistaro et al., 2014, Floeter et al., 2016) neuroimaging signatures. More recently, ALS MRI studies captured presymptomatic imaging alterations (Carew et al., 2011, Benatar and Wuu, 2012, Menke et al., 2016). Despite the significant advances in group-level, cross-sectional (Muller et al., 2016) and longitudinal analyses (Menke et al., 2014, Agosta et al., 2009a) in neurodegenerative conditions, the interpretation of single data sets remains methodologically challenging and a striking paucity of such studies persists in ALS (Welsh et al., 2013, Foerster et al., 2013, Ben Bashat et al., 2011, Schuster et al., 2016). From a biomarker perspective, the meaningful interpretation of data from single individuals is paramount for the development of viable clinical applications (Agosta et al., 2010, Turner et al., 2013). In ALS, the gap between the number of ‘group-level’ descriptive studies and ‘individual-level’ classification studies continues to widen. Admittedly, ‘individual-level’ analyses require large training and validation data sets, robust multicenter validation, but even small pilot studies are surprisingly scarce in ALS.
The main limitation of the study lies in its sample size; however it indicates that discriminant function analyses provide relatively accurate diagnostic classification. The classification protocol outlined in this study should to be replicated in larger data sets and tested using data from multiple MRI platforms. Large imaging repositories such as that of the neuroimaging society of ALS (NISALS) are invaluable resources to test and validate such frameworks on multi-site data (Turner et al., 2011). While our patients had relatively high ALSFRS-r, indicating early stage disease, their disease duration was relatively long. The accurate classification of patients with significant disability or long disease duration says relatively little about the sensitivity of a proposed diagnostic algorithm. The validation of classification algorithms should ideally be carried out using data from patients scanned immediately after their diagnoses or presymptomatic mutation carriers. In this study, we have outlined a binary classification approach integrating multiple imaging indices from multiple anatomical sites. However, in order to simulate real-life diagnostic dilemmas, multinomial (multiclass) follow-up studies are required to evaluate algorithms distinguishing between ALS patients, healthy individual and mimic conditions. Such studies should ideally include conditions which are pathologically, clinically or radiologically similar to ALS, for example hereditary spastic paraplegia (HSP), primary lateral sclerosis (PLS), post-polio or Kennedy's patients.(Querin et al., 2017) Another limitation of the study is that no quantitative spinal cord MR indices were included. Spinal measures are likely to improve diagnostic classification further as they have been previously shown to be sensitive longitudinal and prognostic markers in ALS (Agosta et al., 2009b, El Mendili et al., 2015, El Mendili et al., 2014, Bede et al., 2012).
A range of statistical methods are available to classify blinded data in neurodegeneration, and each method is associated with unique advantages and limitations. Despite the relatively restrictive assumptions of discriminant function analyses, the advantage of this approach is that it readily integrates multiple measures, and provides diagnostic probabilities in addition to discriminant scores. Until robust, multi-center classification studies are published in ALS, it remains to be seen which approach is best suited to reliably capture early pathological changes in vivo. The concept of ROI-based, spatial reference system guided ‘data biopsies’ is also applicable to longitudinal analyses and potentially for the development of future monitoring markers.
Ethics approval and consent to participate
All participants provided written informed consent in accordance to the Medical Ethics approval of the research project (Ethics (Medical Research) Committee - Beaumont Hospital, Dublin, Ireland).
Funding
The study was supported by the Health Research Board (HRB-Ireland), the Irish Institute of Clinical Neuroscience IICN − Novartis Ireland Research Grant, the Iris O'Brien Foundation, the Perrigo Clinician-Scientist Research Fellowship, the Research Motor Neuron (RMN-Ireland) Foundation and the EU-Joint Programme for Neurodegeneration (JPND) SOPHIA project. The sponsors of the study had no role in study design, writing of the report or decision to submit this work for publication.
Competing interests
Peter Bede, Parameswaran M. Iyer, Eoin Finegan, Taha Omer reports no disclosures.
Prof. Hardiman has received speaking honoraria from Janssen Cilag, Biogen Idec, Sanofi Aventis and Merck-Serono, she has been a member of advisory panels for Biogen Idec, Allergen, Cytokinetics, Ono Pharmaceuticals and Sanofi Aventis.
Authors' contributions
All authors contributed to data collection, study design, analyses and drafting the manuscript.
Acknowledgements
We are grateful for the generosity and kindness of all of our patients and their caregivers who have kindly participated in this study.
Contributor Information
Peter Bede, Email: bedep@tcd.ie.
Eoin Finegan, Email: finegane@tcd.ie.
Orla Hardiman, Email: orla@hardiman.net.
References
- Agosta F., Gorno-Tempini M.L., Pagani E. Longitudinal assessment of grey matter contraction in amyotrophic lateral sclerosis: a tensor based morphometry study. Amyotroph. Lateral Scler. 2009;10(3):168–174. doi: 10.1080/17482960802603841. [DOI] [PubMed] [Google Scholar]
- Agosta F., Rocca M.A., Valsasina P. A longitudinal diffusion tensor MRI study of the cervical cord and brain in amyotrophic lateral sclerosis patients. J. Neurol. Neurosurg. Psychiatry. 2009;80(1):53–55. doi: 10.1136/jnnp.2008.154252. [DOI] [PubMed] [Google Scholar]
- Agosta F., Chio A., Cosottini M. The present and the future of neuroimaging in amyotrophic lateral sclerosis. AJNR Am. J. Neuroradiol. 2010;31(10):1769–1777. doi: 10.3174/ajnr.A2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bede P., Hardiman O. Lessons of ALS imaging: pitfalls and future directions - a critical review. Neuroimage Clin. 2014;4:436–443. doi: 10.1016/j.nicl.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bede P., Bokde A.L., Byrne S. Spinal cord markers in ALS: diagnostic and biomarker considerations. Amyotroph. Lateral Scler. 2012;13(5):407–415. doi: 10.3109/17482968.2011.649760. [DOI] [PubMed] [Google Scholar]
- Bede P., Bokde A., Elamin M. Grey matter correlates of clinical variables in amyotrophic lateral sclerosis (ALS): a neuroimaging study of ALS motor phenotype heterogeneity and cortical focality. J. Neurol. Neurosurg. Psychiatry. 2013;84(7):766–773. doi: 10.1136/jnnp-2012-302674. [DOI] [PubMed] [Google Scholar]
- Bede P., Elamin M., Byrne S. Basal ganglia involvement in amyotrophic lateral sclerosis. Neurology. 2013;81(24):2107–2115. doi: 10.1212/01.wnl.0000437313.80913.2c. [DOI] [PubMed] [Google Scholar]
- Bede P., Bokde A.L., Byrne S. Multiparametric MRI study of ALS stratified for the C9orf72 genotype. Neurology. 2013;81(4):361–369. doi: 10.1212/WNL.0b013e31829c5eee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bede P., Elamin M., Byrne S. Patterns of cerebral and cerebellar white matter degeneration in ALS. J. Neurol. Neurosurg. Psychiatry. 2015;86(4):468–470. doi: 10.1136/jnnp-2014-308172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bede P., Iyer P.M., Schuster C. The selective anatomical vulnerability of ALS: ‘disease-defining’ and ‘disease-defying’ brain regions. Amyotroph. Lateral Scler. Frontotemporal. Degener. 2016:1–10. doi: 10.3109/21678421.2016.1173702. [DOI] [PubMed] [Google Scholar]
- Ben Bashat D., Artzi M., Tarrasch R. A potential tool for the diagnosis of ALS based on diffusion tensor imaging. Amyotroph. Lateral Scler. 2011;12(6):398–405. doi: 10.3109/17482968.2011.582646. [DOI] [PubMed] [Google Scholar]
- Benatar M., Wuu J. Presymptomatic studies in ALS: rationale, challenges, and approach. Neurology. 2012;79(16):1732–1739. doi: 10.1212/WNL.0b013e31826e9b1d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budde M.D., Xie M., Cross A.H. Axial diffusivity is the primary correlate of axonal injury in the experimental autoimmune encephalomyelitis spinal cord: a quantitative pixelwise analysis. J. Neurosci. 2009;29(9):2805–2813. doi: 10.1523/JNEUROSCI.4605-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carew J.D., Nair G., Andersen P.M. Presymptomatic spinal cord neurometabolic findings in SOD1-positive people at risk for familial ALS. Neurology. 2011;77(14):1370–1375. doi: 10.1212/WNL.0b013e318231526a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cistaro A., Pagani M., Montuschi A. The metabolic signature of C9ORF72-related ALS: FDG PET comparison with nonmutated patients. Eur. J. Nucl. Med. Mol. Imaging. 2014 doi: 10.1007/s00259-013-2667-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan R.S., Segonne F., Fischl B. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. NeuroImage. 2006;31(3):968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Douaud G., Smith S., Jenkinson M. Anatomically related grey and white matter abnormalities in adolescent-onset schizophrenia. Brain. 2007;130(Pt 9):2375–2386. doi: 10.1093/brain/awm184. [DOI] [PubMed] [Google Scholar]
- Eisen A., Kiernan M., Mitsumoto H. Amyotrophic lateral sclerosis: a long preclinical period? J. Neurol. Neurosurg. Psychiatry. 2014;85(11):1232–1238. doi: 10.1136/jnnp-2013-307135. [DOI] [PubMed] [Google Scholar]
- El Mendili M.M., Cohen-Adad J., Pelegrini-Issac M. Multi-parametric spinal cord MRI as potential progression marker in amyotrophic lateral sclerosis. PLoS One. 2014;9(4) doi: 10.1371/journal.pone.0095516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Mendili M.M., Chen R., Tiret B. Fast and accurate semi-automated segmentation method of spinal cord MR images at 3T applied to the construction of a cervical spinal cord template. PLoS One. 2015;10(3) doi: 10.1371/journal.pone.0122224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floeter M.K., Katipally R., Kim M.P. Impaired corticopontocerebellar tracts underlie pseudobulbar affect in motor neuron disorders. Neurology. 2014;83(7):620–627. doi: 10.1212/WNL.0000000000000693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floeter M.K., Bageac D., Danielian L.E. Longitudinal imaging in C9orf72 mutation carriers: relationship to phenotype. Neuroimage Clin. 2016;12:1035–1043. doi: 10.1016/j.nicl.2016.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foerster B.R., Dwamena B.A., Petrou M. Diagnostic accuracy of diffusion tensor imaging in amyotrophic lateral sclerosis: a systematic review and individual patient data meta-analysis. Acad. Radiol. 2013;20(9):1099–1106. doi: 10.1016/j.acra.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machts J., Loewe K., Kaufmann J. Basal ganglia pathology in ALS is associated with neuropsychological deficits. Neurology. 2015;85(15):1301–1309. doi: 10.1212/WNL.0000000000002017. [DOI] [PubMed] [Google Scholar]
- Menke R.A., Korner S., Filippini N. Widespread grey matter pathology dominates the longitudinal cerebral MRI and clinical landscape of amyotrophic lateral sclerosis. Brain. 2014;137(Pt 9):2546–2555. doi: 10.1093/brain/awu162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menke R.A., Proudfoot M., Wuu J. Increased functional connectivity common to symptomatic amyotrophic lateral sclerosis and those at genetic risk. J. Neurol. Neurosurg. Psychiatry. 2016;87(6):580–588. doi: 10.1136/jnnp-2015-311945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S., Oishi K., Jiang H. Stereotaxic white matter atlas based on diffusion tensor imaging in an ICBM template. NeuroImage. 2008;40(2):570–582. doi: 10.1016/j.neuroimage.2007.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H.P., Turner M.R., Grosskreutz J. A large-scale multicentre cerebral diffusion tensor imaging study in amyotrophic lateral sclerosis. J. Neurol. Neurosurg. Psychiatry. 2016;87(6):570–579. doi: 10.1136/jnnp-2015-311952. [DOI] [PubMed] [Google Scholar]
- Oishi K., Zilles K., Amunts K. Human brain white matter atlas: identification and assignment of common anatomical structures in superficial white matter. NeuroImage. 2008;43(3):447–457. doi: 10.1016/j.neuroimage.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orru G., Pettersson-Yeo W., Marquand A.F. Using support vector machine to identify imaging biomarkers of neurological and psychiatric disease: a critical review. Neurosci. Biobehav. Rev. 2012;36(4):1140–1152. doi: 10.1016/j.neubiorev.2012.01.004. [DOI] [PubMed] [Google Scholar]
- Prell T., Grosskreutz J. The involvement of the cerebellum in amyotrophic lateral sclerosis. Amyotroph. Lateral Scler. Frontotemporal. Degener. 2013;14(7–8):507–515. doi: 10.3109/21678421.2013.812661. [DOI] [PubMed] [Google Scholar]
- Querin G., Soraru G., Pradat P.F. Kennedy disease (X-linked recessive bulbospinal neuronopathy): a comprehensive review from pathophysiology to therapy. Rev. Neurol. 2017;173(5):326–337. doi: 10.1016/j.neurol.2017.03.019. [DOI] [PubMed] [Google Scholar]
- Schuster C., Hardiman O., Bede P. Development of an automated MRI-based diagnostic protocol for amyotrophic lateral sclerosis using disease-specific pathognomonic features: a quantitative disease-state classification study. PLoS One. 2016;11(12) doi: 10.1371/journal.pone.0167331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.M., Nichols T.E. Threshold-free cluster enhancement: addressing problems of smoothing, threshold dependence and localisation in cluster inference. NeuroImage. 2009;44(1):83–98. doi: 10.1016/j.neuroimage.2008.03.061. [DOI] [PubMed] [Google Scholar]
- Smith S.M., Jenkinson M., Woolrich M.W. Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage. 2004;23(Suppl. 1):208–219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- Smith S.M., Jenkinson M., Johansen-Berg H. Tract-based spatial statistics: voxelwise analysis of multi-subject diffusion data. NeuroImage. 2006;31(4):1487–1505. doi: 10.1016/j.neuroimage.2006.02.024. [DOI] [PubMed] [Google Scholar]
- Song S.K., Yoshino J., Le T.Q. Demyelination increases radial diffusivity in corpus callosum of mouse brain. NeuroImage. 2005;26(1):132–140. doi: 10.1016/j.neuroimage.2005.01.028. [DOI] [PubMed] [Google Scholar]
- Turner M.R., Grosskreutz J., Kassubek J. Towards a neuroimaging biomarker for amyotrophic lateral sclerosis. Lancet Neurol. 2011;10(5):400–403. doi: 10.1016/S1474-4422(11)70049-7. [DOI] [PubMed] [Google Scholar]
- Turner M.R., Agosta F., Bede P. Neuroimaging in amyotrophic lateral sclerosis. Biomark. Med. 2012;6(3):319–337. doi: 10.2217/bmm.12.26. [DOI] [PubMed] [Google Scholar]
- Turner M.R., Hardiman O., Benatar M. Controversies and priorities in amyotrophic lateral sclerosis. Lancet Neurol. 2013;12(3):310–322. doi: 10.1016/S1474-4422(13)70036-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh R.C., Jelsone-Swain L.M., Foerster B.R. The utility of independent component analysis and machine learning in the identification of the amyotrophic lateral sclerosis diseased brain. Front. Hum. Neurosci. 2013;7:251. doi: 10.3389/fnhum.2013.00251. [DOI] [PMC free article] [PubMed] [Google Scholar]