Abstract
Human RNPS1 was originally characterized as a pre-mRNA splicing activator in vitro and was shown to regulate alternative splicing in vivo. RNPS1 was also identified as a protein component of the splicing-dependent mRNP complex, or exon-exon junction complex (EJC), and a role for RNPS1 in postsplicing processes has been proposed. Here we demonstrate that RNPS1 incorporates into active spliceosomes, enhances the formation of the ATP-dependent A complex, and promotes the generation of both intermediate and final spliced products. RNPS1 is phosphorylated in vivo and interacts with the CK2 (casein kinase II) protein kinase. Serine 53 (Ser-53) of RNPS1 was identified as the major phosphorylation site for CK2 in vitro, and the same site is also phosphorylated in vivo. The phosphorylation status of Ser-53 significantly affects splicing activation in vitro, but it does not perturb the nuclear localization of RNPS1. In vivo experiments indicated that the phosphorylation of RNPS1 at Ser-53 influences the efficiencies of both splicing and translation. We propose that RNPS1 is a splicing regulator whose activator function is controlled in part by CK2 phosphorylation.
Results from the human genome project have underscored the critical importance of alternative pre-mRNA splicing for the expression of a full complement of proteins from an unexpectedly small set of genes (14, 15, 46). Only 20,000 to 25,000 protein-coding genes are estimated to be present in the human genome, but the human proteome is considerably more diverse and complex due to the extensive use of alternative splicing (reviewed in references 11 and 26). Alternative splicing is often precisely regulated in response to tissue-specific, physiologic, or developmental signals (reviewed in references 7, 21, and 49). Gene-specific regulatory elements and their trans-acting factors have been individually studied. However, the general mechanisms of constitutive and alternative splice site selection are still poorly understood. Through research designed to elucidate the basic mechanisms of alternative splice site selection in mammalian cells, members of the SR and hnRNP A/B protein families have been characterized. Proteins from these two families often have antagonistic effects upon splice site selection, i.e., they cause the stimulation of proximal and distal alternative splice sites, respectively (reviewed in reference 3). Members of the SR and hnRNP A/B protein families also promote opposite effects on splicing activation through binding to an exonic splicing enhancer (ESE) and an exonic splicing silencer (ESS), respectively (reviewed in reference 6).
Pre-mRNA splicing factors such as the SR proteins localize to 20 to 50 irregularly shaped nuclear speckles (also called splicing factor compartments [SFCs]) and to the nucleoplasm (reviewed in reference 30). Transcription and splicing are generally thought to occur on the periphery of and outside of SFCs, with SR proteins shuttling between SFCs and active sites of transcription and splicing in a phosphorylation-dependent manner. The modification of SR proteins by phosphorylation influences their recruitment to sites of transcription, their interaction with other proteins and RNAs, and their nucleocytoplasmic shuttling (reviewed in reference 30). The RS domains of these proteins are specifically phosphorylated by several SR protein kinases, including Clk/Sty1-4 of the LAMMER family of protein kinases and the SR protein kinases SRPK1 and -2 (reviewed in reference 10). The overexpression of these kinases leads to the hyperphosphorylation of SR proteins and the disassembly of SFCs (reviewed in reference 30).
Human RNPS1 was originally purified based on its activation of an alternative distal 3′ splice site in vitro. However, recombinant RNPS1 (r-RNPS1) alone did not discriminate between distal and proximal sites, but rather stimulated splicing in a substrate-independent manner. Thus, RNPS1 was characterized as a general pre-mRNA splicing activator (27). Further investigation of splicing in vivo revealed that RNPS1 interacts with other proteins and promotes alternative splicing in a substrate-specific manner (40). RNPS1 was also discovered as a partner for the p110 isoform of human cyclin-dependent protein kinase 11 (CDK11p110) in vitro and in vivo (22). Furthermore, a recent report suggested that RNPS1 is linked to apoptosis and the concurrent loss of splicing activity as a member of the apoptosis- and splicing-associated protein complex (43).
RNPS1 is also a component of the splicing-dependent mRNP complex, termed the exon-exon junction complex (EJC) (20), which includes several other proteins involved in splicing, mRNA export, and nonsense-mediated mRNA decay (NMD) (reviewed in reference 47). Experimental evidence suggests that the EJC plays a role in linking pre-mRNA splicing and postsplicing processes, such as NMD, mRNA localization, and translation (33, 50; reviewed in reference 23). To date, however, the direct role of RNPS1 per se in postsplicing processes has not been proven.
We further defined the functions of RNPS1 in pre-mRNA splicing and examined the regulation of these functions by phosphorylation. RNPS1 is incorporated into the active splicing complex and enhances the formation of the first ATP-dependent splicing complex, the A complex, which is consistent with the RNPS1 function in splicing regulation. Furthermore, we show here that serine residue 53 (Ser-53) of RNPS1 is phosphorylated by casein kinase II (CK2). CK2 phosphorylates several key cell cycle and transcriptional proteins (36, 42), interacts with multiple signaling pathways, and is proposed to be an important mediator of cell survival. In contrast to the effect of SR protein kinases on SR proteins, we show that the nuclear localization of RNPS1 is not affected by CK2 phosphorylation. Intriguingly, we demonstrate that the site-specific phosphorylation at Ser-53 directly influences the splicing stimulation activity of RNPS1 and also affects mRNA translational activity. We propose a novel mechanism controlling the pre-mRNA splicing activity of RNPS1, which may function as a switch that is capable of regulating alternative splicing of a variety of human genes.
MATERIALS AND METHODS
Preparation of immobilized pre-mRNA containing a biotin-labeled tag.
A biotin-labeled DNA (5′-pCGAACGGACT*GAAT*GGAT*GAAT*GGAT*GAA-3′; biotin-T nucleotides are indicated by T*) was chemically synthesized by Operon Technologies. This biotin-labeled fragment was ligated directly to the 3′ end of the δ-crystallin pre-mRNA by the use of T4 DNA ligase as described previously (31) (Fig. 1A). A GpppG-capped 32P-labeled δ-crystallin pre-mRNA was transcribed in vitro from the pSP14-15 plasmid (41) as described previously (29). The annealing mixture (15 μl), containing 20 pmol of pre-mRNA, 50 pmol of biotin-labeled DNA, and 50 pmol of bridge DNA (5′-TCCGTTCGGGGTACCCG-3′), was incubated at 95°C for 5 min, at 65°C for 10 min, and at room temperature for 10 min. The ligation reaction was performed at 30°C for 3 h by adding 1 μl of 10 mM ATP, 40 U of RNase inhibitor (Promega), and 10 U of T4 DNA ligase (Amersham). The reaction was separated by denaturing 5.5% polyacrylamide gel electrophoresis (PAGE), and the ligated product was eluted from the gel, extracted with Tris-saturated phenol (pH 6.8), and precipitated with ethanol.
FIG. 1.
Analyses of spliceosome complexes and RNA products by in vitro splicing assays with δ-crystallin pre-mRNA. (A) Schematic representation of the preparation of immobilized δ-crystallin pre-mRNA on streptavidin-agarose. B and AV indicate biotin and streptavidin moieties, respectively. (B) Protein analysis of isolated spliceosome complexes by immunoblotting. Incubation times (minutes) of the in vitro splicing reactions are indicated at the top. The positions of protein size markers are indicated on the left. A HeLa cell nuclear extract (5 μl) was analyzed as a control (NE). (C) Analysis of spliceosome complexes by electrophoresis in a 1.5% low-melting-point agarose gel over the time course of splicing (same times as in panel B). A nonspecific hnRNP complex, H, the first ATP-dependent complex, A, and the late complexes B and C (not separated in this gel) are indicated. (D) Analysis of splicing products by denaturing 5.5% PAGE over the time course of splicing (same times as in panel B). The positions of the splicing products are indicated by schematic representations of their structures.
For the immobilization of biotin-tagged pre-mRNA with streptavidin-agarose beads, ∼800 fmol of the biotinylated pre-mRNA was incubated with 80 μl of streptavidin-agarose (Sigma) on ice for 15 min and then rocked gently at 4°C for 1 h in 400 μl of buffer W (12 mM HEPES-NaOH [pH 7.9], 60 mM KCl, 1.5 mM MgCl2, 0.12 mM EDTA, 12% glycerol) containing 0.1% Nonidet P-40, 0.1 mg of glycogen/ml, 1 mg of bovine serum albumin/ml, and 0.1 mg of yeast tRNA/ml. Prior to the splicing assay, the immobilized substrate beads were washed with buffer W four times.
In vitro splicing assays with immobilized pre-mRNA.
In vitro splicing reaction mixtures (75 μl) consisted of 15 μl of HeLa cell nuclear extract (28) and 10 μl of a slurry containing immobilized δ-crystallin pre-mRNA; reactions were performed under standard conditions as described previously (29), except for the use of 10% (7.5 μl) polyvinyl alcohol. Splicing reactions were gently rocked (splicing does not take place without rocking) at 30°C for 3, 10, 20, 40, or 90 min, and the immobilized substrate beads were washed with buffer W at least four times. Only the excised, biotin-tag-free, lariat intron was released from the beads, whereas lariat intermediates and the final spliced product remained bound to the beads (data not shown).
Immunoblot assays.
After in vitro splicing with immobilized δ-crystallin pre-mRNA, the protein components of the spliceosomes were extracted from the washed beads with 30 μl of sodium dodecyl sulfate-PAGE (SDS-PAGE) sample buffer at 90°C for 5 min. Extracted proteins were separated by 10% SDS-PAGE and analyzed by immunoblot assays using a rabbit anti-RNPS1 antiserum (27), a mouse anti-SF2 antiserum (12), or a rabbit anti U5-116K antibody (a gift from R. Lührmann). The secondary antibodies used were anti-rabbit and anti-mouse immunoglobulin G conjugated to alkaline phosphatase (Promega). BCIP (5-bromo-4-chloro-3-indolylphosphate) and nitroblue tetrazolium reagents were used for detection as described by the manufacturer (Promega).
Analysis of spliceosome complexes and RNA products.
The time course of in vitro splicing was examined as described above, except for the use of nonimmobilized δ-crystallin pre-mRNA. After the splicing reactions (25 μl each), 4 μl of each reaction was used for analyses of spliceosome complexes as described previously (8), with minor modifications. Briefly, 0.8 μl of 4-mg/ml heparin was added to 4 μl of the reaction and kept on ice for 10 min. The mixture (3 μl of each sample) was loaded into a 1.5% low-melting-point agarose gel and electrophoresed with 50 mM Tris-50 mM glycine buffer. The gel was fixed, dried, and analyzed by autoradiography. RNAs were extracted with phenol from the remainder of the splicing reactions (21 μl each) and then were analyzed by denaturing 5.5% PAGE and autoradiography as described previously (29).
A time course study of in vitro splicing to examine the effect of RNPS1 was performed with β-globin pre-mRNAs (29). Splicing reaction mixtures (25 μl) containing 8 μl of HeLa cell cytosolic S100 extract (28) and 1.5 pmol of r-SF2/ASF, with or without baculovirus-expressed r-RNPS1 (10 pmol), were incubated at 30°C for the indicated times under standard conditions as described previously (29). Aliquots (4 μl each) of the splicing reactions were analyzed for the formation of spliceosome complexes as described above. The remainder of each splicing reaction (21 μl) was examined for splicing products by denaturing 9% PAGE (a higher percentage of polyacrylamide facilitates the detection of lariat intermediate products above the pre-mRNA).
Coimmunoprecipitation assays.
Human embryonic kidney (HEK) 293 cells were maintained in Dulbecco's modified Eagle's medium (D-MEM) supplemented with 10% fetal calf serum and 2% l-glutamine and were transfected by the use of JetPEI (Qbiogene). Transfected HEK 293 cells were lysed with a buffer containing 50 mM HEPES-NaOH (pH 7.9), 150 mM NaCl, 0.2 mM EDTA, 0.5% Tween 20, 10 mM β-mercaptoethanol, 10% glycerol, and 1× complete protease inhibitors (Roche). Immunoprecipitations with the anti-FLAG M2 monoclonal antibody (Sigma) and immunoblot analyses with anti-FLAG (Santa Cruz Biotechnology), anti-CK2α (Santa Cruz Biotechnology), anti-RNPS1 (44), and anti-CDK11 P1C (44) polyclonal antibodies were performed as described previously.
In vitro phosphorylation assays.
Preparations of Escherichia coli- and baculovirus-expressed r-RNPS1 were described previously (27). These full-length r-RNPS1 proteins (20 pmol) were incubated in a reaction mixture (25 μl) containing 20 mM Tris-HCl (pH 7.5), 50 mM KCl, 10 mM MgCl2, 0.2 mM ATP, 1 μCi of [γ-32P]ATP (Amersham), and 500 U of recombinant CK2 (New England Biolabs) at 30°C for 30 min. The phosphorylation reactions were stopped by heating at 90°C for 5 min in SDS-PAGE sample buffer. The sample mixture was divided in half and loaded into two 12% SDS-PAGE gels. One gel was electroblotted onto a nitrocellulose membrane, and immunoblot assays were performed with the rabbit anti-RNPS1 antiserum as described above. The other gel was fixed and dried, and the 32P-labeled r-RNPS1 proteins were detected by autoradiography.
Glutathione S-transferase (GST), GST-CTD32-52, and GST-RNPS1-Δ1 (containing amino acids 1 to 136) wild-type (WT) and Ser-53-to-Ala-53 (S53A) mutant proteins were expressed in E. coli and purified as previously described (22, 45). The amounts of substrate proteins were estimated by the Bradford assay and checked with of a Coomassie blue-stained SDS-PAGE gel. CK2 purified from rat livers (Sigma) was used in an in vitro kinase reaction containing 23 pmol of the GST fusion protein, and then the proteins were separated by SDS-PAGE as previously described (45).
In vivo phosphorylation and mapping of RNPS1 phosphopeptides.
HeLa Tet-Off cells (maintained as described above for HEK 293 cells) (9) were transfected by the use of calcium phosphate (Clontech Laboratories) with either a FLAG-RNPS1 wild-type or S53A mutant plasmid (10 μg/dish). After culturing for 20 h, the cells were washed twice with phosphate-free D-MEM (ICN) and then labeled with phosphate-free D-MEM containing 166 μCi of [32P]orthophosphate (ICN), 2% l-glutamine, and 10% fetal calf serum dialyzed with 20 mM Tris-HCl (pH 7.6)-137 mM NaCl (TBS). After incubation at 37°C for 4 h, the cells were washed five times with TBS, lysed in lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% Triton X-100, 5 mM EDTA, 5 mM EGTA, 10 mM NaF, 10 mM β-glycerophosphate, 1 mM sodium orthovanadate, 1 mM benzamidine, and 1× complete protease inhibitors [Roche]), and subjected to immunoprecipitation overnight with anti-FLAG M2 agarose (Sigma). Washed protein-bead complexes were then separated by 10% SDS-PAGE and transferred to an Immobilon-P membrane (Millipore). After autoradiography, the RNPS1 bands were excised, digested at 37°C for 4 h with 50 μg of TPCK-treated trypsin (item code TRTPCK; Worthington) in 50 mM NH4HCO3 (pH 8.0), and processed with a buffer at pH 1.9 as described previously (1). Electrophoresis was performed for 50 min at 1,000 V (horizontal). Thin-layer chromatography was performed overnight in phospho-chromatography buffer (vertical). The dried sheets were analyzed by autoradiography.
Mass spectrometric analysis.
Mass spectrometric analyses to determine the molecular masses of both E. coli- and baculovirus-expressed r-RNPS1 proteins were performed in the Macromolecular Structure Facility at Michigan State University. Mass spectrometric analysis to verify the in vivo phosphorylation at Ser-53 was performed at St. Jude Children's Research Hospital. The purification of native RNPS1 from HeLa cells was performed as described previously (27). Purified RNPS1 was precipitated with trichloroacetic acid, washed with acetone, and digested overnight with endoproteinase Arg-C (Roche). The digest was desalted by use of a C18 ZipTip column (Millipore) and was subjected to matrix-assisted laser desorption ionization-time-of-flight analysis with a Voyager DE-RP mass spectrometer (Applied Biosystems), with α-cyano-4-hydroxycinnamic acid as a matrix. Measurements were performed at an accuracy of ± 200 ppm. An aliquot of the Arg-C peptide mixture was further digested with calf intestinal alkaline phosphatase (New England Biolabs) to remove phosphate residues.
Construction of mutant RNPS1 expression plasmids.
The S53A and S53E mutations were introduced into the RNPS1 plasmids (pFastBac1 and pFlex) by use of a QuikChange site-directed mutagenesis kit (Stratagene), with previously described modifications (48). All plasmid constructs were verified by sequencing that covered the entire open reading frame.
Indirect immunofluorescence assays.
HeLa cells were grown, transfected on coverslips, and then processed with 4% paraformaldehyde 15 h after transfection. Incubations with antibodies were performed for 1 h at 37°C in phosphate-buffered saline containing 10% fetal calf serum. FLAG-tagged RNPS1 or CK2α protein was detected with the anti-FLAG M2 antibody. Endogenous RNPS1 and CK2α were detected with an anti-RNPS1 rabbit antibody (44) and an anti-CK2α goat antibody (Santa Cruz Biotechnology), respectively. Anti-rabbit and anti-mouse secondary antibodies labeled with either fluorescein isothiocyanate (FITC) or Texas Red (Amersham) and anti-goat secondary antibodies labeled with Texas Red (Santa Cruz Biotechnology) were used to detect the corresponding primary antibodies.
In vitro splicing assays with mutant r-RNPS1 proteins.
In vitro splicing assays to examine the effects of the S53A and S53E mutants of RNPS1 were performed with β-globin pre-mRNA as described above (29). Splicing reactions (25 μl each) containing baculovirus-expressed wild-type, S53A, or S53E r-RNPS1 (2 or 10 pmol) were incubated at 30°C for 5 h under standard conditions (29). In vitro splicing assays using HEK 293 whole-cell lysates after transient transfection were performed as described previously (13). Briefly, HEK 293 cells were maintained in D-MEM supplemented with 10% fetal calf serum and 2% l-glutamine. Transfections were performed by the use of JetPEI (Qbiogene). Cells were harvested 24 h after transfection for lysate preparation. Whole-cell lysates containing 250 μg of total protein were used in each splicing reaction along with β-globin pre-mRNA. The extracted RNAs from the splicing reactions were analyzed by denaturing 5.5% PAGE.
In vivo splicing assays with mutant RNPS1 constructs.
HEK 293 cells were maintained and transfected as described above. Cells were harvested 24 h after transfection for use in enzymatic assays. β-Galactosidase and luciferase activities were measured by use of the Dual-Light system (Tropix). All measurements were within the linear range that was established by the use of luciferase and β-galactosidase standards. Immunoblot analyses of cell lysates were performed with the anti-FLAG M2 monoclonal antibody (Sigma), an antiactin polyclonal antibody (Santa Cruz Biotechnology), and an anti-RNPS1 polyclonal antibody as described previously (41). Reverse transcription-PCR (RT-PCR) analysis was performed essentially as described previously (32). Total RNAs (1 or 5 μg) were used for RT reactions (20 μl) with a random hexamer or oligo(dT) primer, followed by purification with RNeasy spin columns (QIAGEN). The reverse transcription products (0.1 or 0.2 μl) were used for PCRs (22 or 23 cycles) with specific primers directed to the β-galactosidase and luciferase sequences (32). The PCR products were analyzed by 6% PAGE. Ethidium bromide-stained gels ware analyzed by quantitation with an AlphaImager 2000 instrument (Alpha Innotech).
RESULTS
RNPS1 is incorporated into active spliceosomes.
RNPS1 exists with spliced mRNAs as a component of the EJC (20). However, the function of RNPS1 as a splicing activator indicates that RNPS1 is also a component of functional spliceosomes (27). To further elucidate the dynamic association of RNPS1 with active spliceosomes, we employed a solid-phase in vitro splicing assay using an immobilized δ-crystallin pre-mRNA. The 3′ end of the pre-mRNA was tethered to solid beads (streptavidin-agarose), and thus active spliceosomes at different stages were isolated by extensive washing of the beads without any contamination of the crude nuclear extract (Fig. 1A). The spliceosomal protein components were analyzed by immunoblotting over the time course of splicing (Fig. 1B). For control splicing factors, we chose an snRNP protein (116-kDa protein of U5 snRNP) and an SR protein (SF2/ASF). The generation of spliceosomes and RNA products (with nonimmobilized δ-crystallin pre-mRNA) was simultaneously monitored under the same assay conditions (Fig. 1C and D). We observed almost equivalent kinetics of splicing for immobilized and nonimmobilized δ-crystallin pre-mRNAs by analysis of the RNA products (data not shown; Fig. 1D).
Using the anti-U5-116K antibody, we observed that U5 snRNP initiated incorporation into spliceosomes after 10 min of incubation and that incorporation gradually increased up to 40 min (Fig. 1B). U5 snRNP was barely observed after 3 min of incubation (data not shown), whereas at 90 min, the bulk of the U5 snRNP was already dissociated from the spliceosomes. No splicing products were observed at 10 min, but pre-splicing H complexes and the first ATP-dependent A complexes were detected (Fig. 1C). Since this splicing assay was performed in the presence of ATP, we could not detect the early ATP-independent E complex (8). The A complex gradually converted to B and C complexes between 10 and 40 min, followed by their dissociation at 90 min, consistent with the accumulation of spliced mRNA (Fig. 1D). Since the relative contents of specific spliceosomes were markedly distinctive at different time points (Fig. 1C), it appears that the results of the immunoblot analysis of tethered proteins represent the protein components of specific spliceosomes, even though each splicing complex was not further purified (Fig. 1B).
We found that RNPS1 is fully incorporated into spliceosomes after 10 min of incubation and is stably retained in the complexes until 90 min (Fig. 1B). As a control, we used an immobilized intronless δ-crystallin substrate which was not spliced and thus did not form active spliceosomes. We found that this substrate is very stable in the splicing reaction; nevertheless, we barely detected RNPS1 with this tethered substrate over the course of the experiment (10 to 90 min) (data not shown). Therefore, our detection of RNPS1 with the tethered δ-crystallin substrate was due to a splicing-dependent association. The pattern of RNPS1 association with spliceosomes was very similar to that of SF2/ASF but was different from that of U5 snRNP (Fig. 1B). Since we previously showed that RNPS1 and SF2/ASF synergistically activate splicing in vitro (27), this observation is consistent with a functional interaction between RNPS1 and SF2/ASF. Consistent with RNPS1 being a component of the EJC, our detection of RNPS1 at 90 min may reflect the fact that a portion of RNPS1 is associated with the EJC containing spliced mRNA, which can also be tethered to streptavidin-agarose beads (Fig. 1A). SF2/ASF was also detected at 90 min, suggesting a possible association of SF2/ASF with spliced mRNA as a nucleocytoplasmic shuttling protein (Fig. 1B) (4).
RNPS1 enhances formation of the A complex and activates splicing in vitro.
We previously demonstrated that RNPS1 cooperates with SR proteins, e.g., SF2/ASF, and stimulates the in vitro splicing of several different pre-mRNA substrates (27). To induce maximal splicing stimulation through the addition of (r-RNPS1), we performed in vitro splicing of β-globin pre-mRNA with a limited amount of r-SF2/ASF in a HeLa cell cytosolic S100 extract, which barely contains SR and RNPS1 proteins (27). We examined the effect of the RNPS1 addition on the formation of spliceosome complexes and on the generation of splicing products in the same in vitro reaction over the time course of the experiment (Fig. 2A and B).
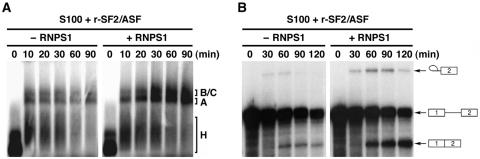
FIG. 2.
Effect of RNPS1 over the time course of in vitro splicing with β-globin pre-mRNA. Splicing reactions including constant amounts of HeLa cell S100 extract plus r-SF2/ASF, with (+) or without (−) baculovirus-expressed r-RNPS1 (10 pmol), were incubated at 30°C for the indicated times (minutes). (A) Analysis of spliceosome complexes by electrophoresis in a 1.5% low-melting-point agarose gel. The complexes H, A, and B/C (not separated in this gel) are indicated (see the legend to Fig. 1C). The ratios of complex A amounts to complex H amounts were quantitated from the scanned digital image, and the results are as follows (ratio of −RNPS1 versus ratio of +RNPS1 at each time point): 0.20 versus 0.16 (10 min), 0.14 versus 0.25 (20 min), 0.17 versus 0.32 (30 min), 0.19 versus 0.45 (60 min), and 0.39 versus 0.51 (90 min). (B) Analysis of splicing products by denaturing 9% PAGE. The positions of the splicing products are indicated by schematic representations of their structures.
When RNPS1 was added, we observed a significantly increased accumulation of the A complex after 20 min, followed by an enhanced generation of B and C complexes after 30 to 90 min of incubation (Fig. 2A; see the legend for the quantitation of complexes H and A). Accordingly, we detected more production of the lariat intermediate, which is the product of the first catalytic step of splicing, at 30 min (Fig. 2B). The enhanced production of the lariat intermediate lasted up to 120 min and was followed by a marked accumulation of the final spliced mRNA beginning at 60 min. The significant accumulation of the lariat intermediate and of mature mRNA indicated that RNPS1 enhances both the first and the second step of splicing. With this assay system, however, we could not strictly determine whether the observed splicing stimulation was merely a consequence of the enhanced generation of the A complex or if RNPS1 also promoted either of the catalytic splicing steps.
CK2 interacts with RNPS1 in mammalian cell lysates.
Several lines of evidence suggest that significant posttranslational modification of the RNPS1 protein takes place (27). The molecular mass of RNPS1 as computed from its cDNA is 34,208 Da. However, HeLa cell-purified RNPS1, r-RNPS1 translated by the use of wheat germ extract, and r-RNPS1 expressed in baculovirus-infected insect cells had an apparent molecular mass of ∼50 kDa by SDS-PAGE analysis. In contrast, E. coli-expressed r-RNPS1, which lacks splicing stimulation activity, had a mobility of ∼42 kDa. Moreover, 32P incorporation into r-RNPS1 expressed in insect cells indicated that this protein is phosphorylated in vivo (data not shown).
Previously, we reported that the CDK11p110 protein kinase interacts with both RNPS1 and CK2 (22, 45). Therefore, we examined whether RNPS1 interacts with CK2 by immunoprecipitation. CK2 is a heterodimer composed of two catalytic subunits (α and/or α′) responsible for protein kinase activity and two regulatory β subunits (reviewed in reference 37). Neither our anti-RNPS1 antibodies nor the commercially available anti-CK2α antibodies worked well for immunoprecipitation. Therefore, we used epitope-tagged forms of RNPS1 and CK2α for immunoprecipitation analyses. We found that endogenous RNPS1 coimmunoprecipitates with wild-type FLAG-tagged CK2 and its kinase-dead form expressed transiently in HEK 293 cells (Fig. 3A). Endogenous CK2 and CDK11p110 were also found in immunoprecipitated FLAG-RNPS1 wild-type, S53A, and S53E complexes (Fig. 3B; see below for more information about the RNPS1 mutants). CK2 and RNPS1 were coimmunoprecipitated from HeLa and Jurkat cell lysates as well (data not shown). These results suggest that CK2 robustly interacts with RNPS1 in mammalian cells.
FIG. 3.
Coimmunoprecipitation of RNPS1 and CK2. (A) HEK 293 cells expressing either wild-type FLAG-CK2 (WT) or a kinase-dead mutant (KD) (or empty FLAG vector-transfected control cells) were lysed and immunoprecipitated with the anti-FLAG M2 antibody and detected by immunoblotting with the indicated antibodies. The untransfected lysate was also directly assayed. (B) HEK 293 cells expressing either the FLAG-RNPS1 wild-type (WT), S53A, or S53E protein (or empty FLAG vector-transfected control cells) were lysed and immunoprecipitated with the anti-FLAG M2 antibody and detected by immunoblotting with the indicated antibodies. The untransfected lysate was also directly assayed. The positions of the protein size markers (in kilodaltons) are indicated.
CK2 phosphorylates Ser-53 of RNPS1 in vitro.
We previously found that immunoprecipitated CDK11p110 complexes phosphorylate RNPS1 but that this activity is not affected when a kinase-inactive CDK11p110 expression construct is used as a control (data not shown). This result indicates that a CDK11p110-associated kinase phosphorylates RNPS1, similar to what was previously observed for phosphorylation of the C-terminal domain (CTD) of the largest subunit of RNA polymerase (RNAP) II (45). We therefore determined whether RNPS1 serves as a substrate for CK2, which is a CDK11p110-associated kinase. As expected, CK2 efficiently phosphorylated E. coli-expressed r-RNPS1 (∼42 kDa) in vitro and shifted its gel mobility to ∼47 to 48 kDa (Fig. 4A). In contrast, insect cell r-RNPS1, which is already phosphorylated in vivo (see above), was labeled with far less 32P and retained its relative mobility in the gel (∼50 kDa).
FIG. 4.

In vitro phosphorylation of RNPS1 with CK2. (A) E. coli- or baculovirus-expressed r-RNPS1 was incubated with (+) or without (−) CK2 in the presence of [γ-32P]ATP. The proteins were separated by 12% SDS-PAGE and detected by immunoblotting (top) and autoradiography (bottom). The positions of protein size markers are indicated on the left. E. coli-expressed r-RNPS1 (in the absence of CK2) contained two polypeptides (∼42 and ∼43 kDa; not apparent in this figure). Mass spectrometry analysis revealed that the smaller, ∼42-kDa protein (measured as 33,053 ± 330 Da) had a possible C-terminal truncation around His 295 (±1 or 2 amino acids). Accordingly, phosphorylated E. coli r-RNPS1 (in the presence of CK2) separated as a doublet (∼47 and ∼48 kDa; visible in this figure). (B) E. coli-expressed recombinant proteins GST (control), wild-type (WT) and mutant (S53A) GST-RNPS1-Δ1, and GST-CTD32-52 were incubated with CK2 in the presence of [γ-32P]ATP. The proteins were separated by 12% SDS-PAGE and detected by autoradiography. The GST-CTD32-52 substrate contained three consensus CK2 sites, whereas the GST-RNPS1-Δ1-WT substrate contained one consensus CK2 phosphorylation site, which was reflected in the intensities of radioactive signals in which equimolar amounts of substrate proteins were loaded.
Multiple potential serine/threonine phosphorylation sites in RNPS1 were identified by a computer search (data not shown). However, a perfect CK2 consensus phosphorylation site exists uniquely at Ser-53 of RNPS1. Therefore, we prepared an S53A mutated form of r-RNPS1 (r-RNPS1-S53A), in which phosphorylation can no longer take place at amino acid 53. Mutant proteins, expressed in either E. coli or insect cells, were poorly phosphorylated by CK2 (Fig. 4B; data not shown). In contrast to CK2, several other kinases that target RNPS1, such as Clk-1, strongly phosphorylated the mutant r-RNPS1-S53A protein (data not shown). These results indicate that Ser-53 serves as the major site of CK2 phosphorylation of RNPS1, although a minor CK2 phosphorylation site(s) may also exist within the RNPS1 polypeptide. The experiments performed thus far indicated that RNPS1 is targeted by several other protein kinases (data not shown; see below). For this study, we focused on the CK2 phosphorylation of RNPS1 since we found that this specific phosphorylation is functionally important for splicing.
Ser-53 of RNPS1 is phosphorylated in vivo.
We used two different techniques to evaluate the phosphorylation of RNPS1 Ser-53 in vivo. One evaluation involved mass spectrometric analysis, for which native endogenous RNPS1 purified from HeLa cells was subjected to digestion with the endoproteinase Arg-C. The Arg-C peptides were then analyzed by mass spectrometry both before and after treatment with alkaline phosphatase. Presumably because of the poor ionization efficiency of this segment, no signal was observed at the average m/z value, 1,746.8, corresponding to the expected mass of the singly phosphorylated peptide 43-57 (SKDKGATKESSEKDR). However, a strong signal appeared at the average m/z value, 1,666.7, after alkaline phosphatase treatment. This value is in close agreement with the m/z value of 1,666.8 expected for the dephosphorylated form of the RNPS1 peptide 43-57. The very low levels of this signal prior to alkaline phosphatase treatment and its absence in mock phosphatase digests (without a substrate) indicate that a large proportion of the RNPS1 molecules contain phosphate within this segment in vivo. These results indicate that one or more of the four Ser/Thr residues (Ser-43, Thr-49, Ser-52, or Ser-53) in this Arg-C peptide are phosphorylated in endogenous RNPS1.
To further pinpoint the phosphorylation site of RNPS1 in vivo, we transiently transfected HeLa cells with expression constructs encoding either the wild-type or S53A mutant FLAG-tagged RNPS1 protein and then labeled these cells with 32P during growth. The FLAG-tagged proteins were immunoprecipitated from the in vivo-labeled cell lysates, digested with trypsin, and analyzed by two-dimensional thin-layer chromatography (Fig. 5). At least 10 distinct 32P-labeled tryptic spots were generated from wild-type RNPS1. However, one tryptic spot (number 2) was repeatedly absent from the analysis when the S53A mutant was used. According to a proteolytic peptide mobility plotting program, the position of this spot correlates well with the theoretical position of the tryptic phosphopeptide that includes Ser-53 relative to the other possible RNPS1 tryptic phosphopeptides. It is theoretically possible that Ser-52 is the phosphorylation site on the tryptic peptide 51-55 (ESSEK) and that the phosphorylation of Ser-52 requires the presence of Ser-53 in vivo. However, it is more likely that Ser-53 is the residue targeted by CK2 because Ser-53 (but not Ser-52) shows a much closer match to the CK2 consensus phosphorylation site. The amounts of 32P incorporated into several other tryptic phosphopeptides within the mutant RNPS1 protein appeared to vary relative to those incorporated into their wild-type counterparts, suggesting that the in vivo phosphorylation of RNPS1 by other protein kinases may be slightly enhanced when Ser-53 is phosphorylated. Taken together with the in vitro CK2 phosphorylation data, these data show that Ser-53 is phosphorylated in vivo by CK2.
FIG. 5.
In vivo phosphorylation of RNPS1 at Ser-53. HeLa Tet-Off cells transiently expressing wild-type (WT) and mutant (S53A) FLAG-RNPS1 were labeled with [32P]orthophosphate, and the tryptic phosphopeptides of these proteins were analyzed by electrophoresis and thin-layer chromatography. The arrows indicate the directions of electrophoresis (horizontal) and chromatography (vertical). The phosphopeptides (circled with numbers) were detected by autoradiography.
Nuclear localization of RNPS1 is not altered by phosphorylation at Ser-53.
The phosphorylation of SR proteins by Clk/Sty and SRPK kinases typically alters their predominant subcellular localization from an intense nuclear speckled pattern to a diffuse nucleoplasmic distribution (reviewed in reference 30). To determine whether CK2 phosphorylation of RNPS1 at Ser-53 affects its nuclear localization, we performed indirect immunofluorescence microscopy analysis. First, we examined whether FLAG-RNPS1 colocalizes with endogenous CK2. FLAG-tagged RNPS1 (green due to FITC) and endogenous CK2α (red due to Texas Red) colocalized exclusively in the nucleus in HeLa cells, and both exhibited similar nuclear speckle and diffuse nucleoplasmic staining patterns (Fig. 6A to C). Colocalization of RNPS1 with the cytoplasmic portion of CK2α was not detected.
FIG. 6.
Subcellular localization of RNPS1 and CK2 proteins, which was assayed by indirect immunofluorescence microscopy. For all panels, HeLa Tet-Off cells were transfected and processed for indirect immunofluorescence. Green images correspond to RNPS1, and red images correspond to CK2α. (A to C) Nuclear colocalization of FLAG-RNPS1 and endogenous CK2α in nuclear speckles and the nucleoplasm. FLAG-RNPS1 was detected with the anti-FLAG M2 monoclonal antibody followed by a FITC-conjugated anti-mouse antibody (A). CK2α was detected with an anti-CK2α goat antibody followed by a Texas Red-conjugated anti-goat antibody (B). These two images were superimposed, and the yellow areas indicate colocalization of RNPS1 and CK2α (C). (D to F) Nuclear localization of wild-type FLAG-RNPS1 (D) and the FLAG-RNPS1 mutants S53A (E) and S53E (F). These FLAG-RNPS1 proteins were detected with the anti-FLAG M2 antibody as described above. (G to I) Subcellular localization of endogenous RNPS1 in untransfected cells (bottom) and in FLAG-CK2 wild-type holoenzyme-transfected cells (top). Endogenous RNPS1 was detected with an anti-RNPS1 rabbit antibody followed by a FITC-conjugated anti-rabbit antibody (G). FLAG-CK2α was detected with the anti-FLAG M2 antibody followed by a Texas Red-conjugated anti-mouse antibody (H). The cells were also stained with DAPI (4′,6-diamidino-2-phenylindole), which detects nuclear DNA (I). (J to L) Subcellular localization of endogenous RNPS1 in untransfected cells (bottom) and in FLAG-CK2 kinase-inactive holoenzyme-transfected cells (top). The antibodies used for panels J, K, and L were the same as those used for panels G, H, and I, respectively.
Next, we compared the nuclear localization pattern of FLAG-RNPS1 (wild type) with those of RNPS1 mutants to examine the effect of Ser-53 phosphorylation in RNPS1 on its subcellular localization. Both the S53A (which cannot be phosphorylated) and S53E (which structurally mimics phosphoserine) mutant proteins exhibited patterns of nuclear immunofluorescence that were quite similar to that of wild-type RNPS1 (Fig. 6D to F). Furthermore, ectopic expression of either the active CK2 holoenzyme or a kinase-inactive CK2 holoenzyme (containing a D156A mutation in the α subunit) did not affect the nuclear localization pattern of endogenous RNPS1 (Fig. 6G to L). These data indicate that the nuclear localization of RNPS1 is not influenced by either its phosphorylation at Ser 53 or its associated CK2 kinase activity.
Splicing stimulation activity of RNPS1 is repressed by mutations of Ser-53 in vitro.
Since RNPS1 was previously characterized as a splicing activator in vitro, we examined whether Ser-53 phosphorylation is important for splicing stimulation. The r-RNPS1-S53A and -S53E mutant proteins were prepared by use of a baculovirus expression system since E. coli-expressed r-RNPS1 has no splicing stimulation activity (27). In vitro splicing assays with the β-globin pre-mRNA revealed that both the RNPS1-S53A and -S53E mutant proteins significantly reduced, but did not fully abolish, splicing stimulation (Fig. 7A). Given the evidence that the S53A and S53E mutant proteins exhibit protein interaction and nuclear localization characteristics identical to those of wild-type RNPS1 (Fig. 3 and 6), it is unlikely that the loss of splicing stimulation activity in these mutants was due to a disruption of their proper folding. These results indicate that bona fide Ser-53 phosphorylation is important for the splicing stimulation activity of RNPS1 in vitro.
FIG. 7.
Effect of mutation at Ser-53 of RNPS1 on its splicing stimulation activity in vitro. (A) Splicing reactions with β-globin pre-mRNA included constant amounts of HeLa cell S100 extract plus r-SF2/ASF (0.072 μM) and various amounts of baculovirus-expressed wild-type (WT) or mutant (S53A or S53E) r-RNPS1 (0, 0.08, and 0.4 μM). Pre-mRNAs and spliced mRNAs are indicated by schematic representations of their structures. (B) Splicing reactions with β-globin pre-mRNA included HEK 293 whole-cell lysates (250 μg of total protein) transiently expressing WT, S53A, or S53E FLAG-RNPS1 or an empty FLAG vector. A splicing reaction using a HeLa nuclear extract (N.E.) was included as a positive control. Pre-mRNAs and spliced mRNAs are indicated as described above.
Since the transient expression of FLAG-RNPS1 and its mutants in HEK 293 cells was used to analyze protein-protein interactions and the subcellular localization of RNPS1 and CK2 (Fig. 3 and 6), we also used HEK 293 cell lysates to examine in vitro splicing activity after the expression of FLAG-RNPS1 wild-type, S53A, and S53E proteins. Whole-cell lysates were prepared 24 h after transfection, and in vitro splicing assays were performed with the β-globin pre-mRNA. The expression of wild-type RNPS1 greatly enhanced the accumulation of spliced β-globin mRNA, whereas the expression of RNPS1-S53A and -S53E marginally enhanced the splicing activity compared to expression of the control FLAG vector (Fig. 7B). These results were quite consistent with those of in vitro splicing assays using purified r-RNPS1 proteins in HeLa cell S100 extracts (Fig. 7A). We concluded that RNPS1 functions best to stimulate pre-mRNA splicing in vitro when the amino acid at position 53 is a serine which can be reversibly modified by phosphorylation. Artificially imposing either a nonphosphorylated (S53A) or a constitutively phosphorylated (S53E) state at Ser-53 greatly diminishes the splicing stimulation activity of RNPS1 in vitro.
Phosphorylation of RNPS1 at Ser-53 influences efficiency of both splicing and translation in vivo.
The significance of the RNPS1 Ser-53 phosphorylation site for splicing stimulation activity was further confirmed in vivo by use of a double reporter system that allows the quantitation of changes in the ratio of spliced to unspliced mRNA in mammalian cells (32). Splicing activities were quantitated by use of a reporter construct (pTN24) from which any unspliced transcripts yield a protein with only β-galactosidase activity, whereas spliced transcripts yield a fusion protein with both β-galactosidase and luciferase activities. Therefore, the ratio of luciferase to β-galactosidase activities in the cell lysate provides a measurement of the proportion of spliced mRNA at the protein level.
HEK 293 cells were cotransfected with pTN24 and expression constructs encoding FLAG-RNPS1 (wild type), FLAG-RNPS1-S53A, and FLAG-RNPS1-S53E. FLAG-9G8 (an SR protein) and an empty FLAG vector (the expression vector used for RNPS1 constructs) were used as controls. The cells were harvested 24 h after transfection and then assayed for luciferase and β-galactosidase activities, and the splicing activities were quantitated relative to that of cells transfected with the empty vector (Fig. 8A). The FLAG-RNPS1 constructs expressed equivalent amounts of protein, which was confirmed by immunoblot analysis with an anti-FLAG antibody (Fig. 8B). The relative increase in RNPS1 protein abundance between empty vector (endogenous RNPS1 only)- and FLAG-RNPS1-expressing cells was monitored by immunoblot analysis with an anti-RNPS1 antibody (Fig. 8C). Transient expression of the SR protein 9G8 and of wild-type RNPS1 resulted in ∼1.6- and ∼1.9-fold increases in the splicing activity, respectively, whereas expression of the RNPS1-S53A mutant increased the splicing activity ∼1.7-fold (Fig. 8A). In contrast, expression of the RNPS1-S53E mutant increased the splicing activity ∼2.9-fold.
FIG. 8.
Effect of mutation at Ser-53 of RNPS1 on its splicing stimulation activity in vivo. (A) Wild-type (WT) and mutant FLAG-RNPS1 (S53A and S53E), FLAG-9G8 (positive control), and an empty FLAG expression vector (negative control) were cotransfected with the splic-ing reporter plasmid pTN24 into HEK 293 cells. The ratios of luciferase to β-galactosidase activities (reflecting the extent of splicing in transfected cells in vivo) were calculated and expressed as relative splicing activities (the activity of the FLAG vector control was set at 1.0). The results shown for WT, S53A, and S53E RNPS1 represent five independent transfection experiments in which three plates of cells were transfected and each lysate was measured in quadruplicate. The results shown for 9G8 represent two independent transfection experiments which were also measured in quadruplicate. The average deviations are indicated by bars. (B) Immunoblot analysis of equal volumes of cell lysates used for in vivo splicing assays to verify equivalent expression from the three plates of transfected cells. An anti-FLAG antibody was used for the detection of FLAG-RNPS1. An anti-actin antibody was used to detect endogenous actin as a control. (C) Immunoblot analysis of lysates from empty FLAG vector- and FLAG-RNPS1 (WT, S53A, and S53E)-transfected cells. An anti-RNPS1 antibody was used to detect both endogenous and overexpressed FLAG-RNPS1 proteins. (D) RT-PCR analysis of lysates from empty FLAG vector- and FLAG-RNPS1 (WT, S53A, and S53E)-transfected cells. The RT-PCR products were separated by 6% PAGE and detected by ethidium bromide staining. The β-galactosidase/luciferase pre-mRNA (unspliced) and mRNA (spliced) are indicated. (E) Quantitation of the RT-PCR products analyzed in panel D. The values represent the ratios of spliced (Spl) to unspliced (Unspl) products and are expressed relative to the FLAG vector control (set at 1.0). The results shown for WT, S53A, and S53E RNPS1 represent three of the five independent transfection experiments described for panel A, with each lysate being assayed four times.
Since RNPS1, as a component of the EJC, is reported to enhance the translation of spliced mRNA (33, 50), the increased luciferase activity in FLAG-RNPS1-expressing cells could be due to enhanced translation of the mRNA for the β-galactosidase/luciferase fusion protein. Therefore, we also purified RNAs from the cells used for double reporter assays and examined the levels of spliced mRNA by RT-PCR (Fig. 8D). Quantitation of unspliced and spliced products revealed an ∼1.4-fold increased splicing activity (compared to the empty vector control) after the expression of wild-type and S53E mutant RNPS1. A smaller increase in the splicing activity (∼1.2-fold) was observed after the expression of RNPS1-S53A (Fig. 8E). Interestingly, all of these values were markedly less than corresponding values from reporter assays that reflected both splicing and translation of the β-galactosidase/luciferase fusion mRNA (Fig. 8A). Taken together, these data suggest that phosphorylation at Ser-53 not only stimulates splicing activity but also enhances translational activity.
DISCUSSION
The splicing factor RNPS1 exists in active spliceosomes.
Evidence from several laboratories suggests that human RNPS1 participates in several aspects of RNA processing and regulation, including a potential role in the postsplicing processes of mRNA export and NMD, in cooperation with other EJC components (20, 24). However, the role of human RNPS1 in mRNA export is controversial, and instead, it has been suggested that RNPS1 facilitates the 3′ end processing of mRNA and enhances its translational activity (33, 50). The direct function of RNPS1 in these postsplicing processes remains to be elucidated. As for the characterization of RNPS1 as a splicing factor, we have recently demonstrated that RNPS1 and its associated factors cooperatively regulate the alternative splicing of various pre-mRNAs in vivo (40). The experiments described here provide further mechanistic insights into the role of RNPS1 in splicing regulation.
Several analyses of protein components that are specifically associated with spliceosome complexes have been reported (reviewed in reference 16). A recent mass spectrometric analysis of functional spliceosomes successfully detected RNPS1 together with many known splicing factors (52). However, since a bulk spliceosome mixture was examined, the association of RNPS1 exclusively with the EJC or with earlier stages of the spliceosomes was not resolved. Here we demonstrated that RNPS1 is present not only in the EJC, as previously demonstrated, but also in active splicing complexes. It is plausible that RNPS1 is stably retained on spliced mRNAs after dissociation of the spliceosomes to generate the EJC together with other EJC components. While this article was in preparation, an analysis of the stepwise EJC assembly revealed that RNPS1 is associated with pre-mRNAs and that it also exists in the active spliceosome C complex (17, 39). Moreover, RNPS1 was also shown to be associated with the largest subunit of RNAP II (44). These results support our conclusion and are also consistent with RNPS1's functions as a splicing activator and regulator (27, 40).
Possible mechanism of splicing stimulation by RNPS1.
Since we did not perform a spliceosome analysis in the absence of ATP, we could not determine the effect of RNPS1 on ATP-independent E complex formation, which precedes the generation of the ATP-dependent A complexes. Under active splicing conditions in the presence of ATP, however, we did indeed observe a significant increase in the generation of A complexes caused by RNPS1 which was subsequently followed by the accumulation of intermediate and mature spliced mRNA products. These observations provide potentially interesting insights into the mechanism of RNPS1-mediated splicing stimulation.
During the first ATP-dependent A complex formation, the binding of U2 snRNP to the branch site is stabilized, along with the binding of U2AF65 to the downstream pyrimidine tract (reviewed in references 18 and 38). Recently, we identified pinin (also known as DRS and memA) and the SR protein p54 as RNPS1 interacting factors, and we verified their protein-protein interactions in vitro and in vivo (40). We demonstrated that pinin and p54 specifically bind to distinct domains of RNPS1, i.e., the RNA recognition motif (RRM) and the serine-rich domain, respectively. Therefore, in theory, RNPS1 may be capable of interacting with these two factors simultaneously. Intriguingly, it was previously suggested that pinin associates with the U2 snRNP proteins (SF3a and SF3b) and that p54 can directly interact with U2AF65 (2, 51). SF3a/b proteins are required for the assembly of the A complex as well as for a stable association of U2 snRNP with the branch site region (reviewed in references 18 and 38). In view of these observations, we hypothesize that RNPS1 stimulates the recruitment of U2 snRNP and U2AF65 via interactions with pinin and p54, respectively, and that, consequently, the formation of the A complex is enhanced.
Regulation of RNPS1 splicing activity by phosphorylation.
RNPS1 has intrinsic activity as a splicing activator in vitro (27), and it regulates alternative splicing both negatively and positively through interactions with associated factors in vivo (40). If RNPS1 is a versatile splicing regulator for a wide variety of alternatively spliced genes, then it is physiologically important to control its activity in response to tissue-specific and developmentally regulated signals. Here we have presented evidence that supports a molecular signaling mechanism governing the activation of RNPS1, which enables a modulation of splicing activity. We report that RNPS1 interacts with CK2, and we identified Ser-53 as a major site in RNPS1 for CK2 phosphorylation in vitro. Importantly, we also demonstrated that Ser-53 is phosphorylated in vivo. We observed a significant loss of splicing activation with either the RNPS1-S53A or -S53E mutant compared to that of wild-type RNPS1 in vitro, which was not a consequence of RNPS1 mislocalization within the nucleus or of a loss of protein-protein interactions. However, the effects of these mutations differed somewhat in vivo, for which we used a β-galactosidase/luciferase assay system to measure splicing activities after expression of the different RNPS1 forms. We observed equivalent splicing enhancements at the mRNA level with wild-type RNPS1 and the S53E mutant, which potentially mimics constitutive phosphorylation at Ser-53. It is likely that in vivo splicing involves more dynamic interactions than those occurring in vitro, and therefore the combined contributions of expressed mutant RNPS1 and endogenous RNPS1 probably allow for enhanced splicing activities in vivo.
The residual in vitro splicing stimulation activities of the Ser-53 mutant proteins suggested that other minor phosphorylation sites, including potential minor CK2 phosphorylation sites as well as phosphorylation sites of other protein kinases, are also involved in maintaining the minimal splicing activity of RNPS1. In support of this notion, there was still a small discrepancy in mobility by SDS-PAGE between CK2-phosphorylated E. coli r-RNPS1 and baculovirus-expressed r-RNPS1. A mass spectrometric analysis of baculovirus-expressed r-RNPS1 (∼38,700 Da; compare to 34,208 Da for the computed molecular mass) indicated that additional modifications other than phosphorylation occur at multiple sites. We have ruled out the possibility of glycosylation (27) and are still investigating other potential posttranslational modifications. It is likely that other RNPS1 protein modifications also influence its activity.
Potential translational regulation mediated by phosphorylation of RNPS1.
As a component of the EJC, RNPS1 was demonstrated to enhance the translation of spliced mRNAs (29, 47). Interestingly, our results from in vivo double reporter enzymatic assays indicated a potential involvement of Ser-53 phosphorylation in translational activity. We observed a marked increase in luciferase activity after the expression of wild-type RNPS1 and RNPS1-S53E and a lesser increase in the amount of spliced mRNA encoding the luciferase protein. The difference between the enhancement of luciferase activity and that of the mRNA level was especially pronounced after the expression of RNPS1-S53E. The in vivo splicing efficiency observed by a reporter protein assay is the consequence of more global phenomena, including postsplicing processes such as mRNA transport and translation, which are closely linked with splicing in the gene expression network (reviewed in references 25 and 34). We concluded that RNPS1 phosphorylation at Ser-53 is important for modulating RNPS1 function, not only as a splicing regulator, but also as a component of the EJC.
Our results are reminiscent of those of other studies indicating that changes in the phosphorylation status of SR proteins regulate their functional activities. Recent studies demonstrated that hyperphosphorylated SF2/ASF and 9G8 were recruited to pre-mRNAs and promoted spliceosome assembly, whereas hypophosphorylated forms of these proteins promoted the downstream splicing step of transesterification and interactions with the mRNA export receptor TAP/NXF1 (5, 13, 19). Our in vitro and in vivo pre-mRNA splicing results also indicated that the differential phosphorylation of RNPS1 modulates its pre-mRNA splicing activity in the nucleus and its mRNA translational activity in the cytoplasm. Future experiments will determine whether the phosphorylation state of RNPS1 in the EJC affects RNPS1 activities downstream of pre-mRNA splicing.
Involvement of CK2 in global regulation of transcription and RNA processing.
Our results provide the first evidence that CK2 protein kinase activity directly influences pre-mRNA splicing in vitro and in vivo through site-specific phosphorylation of a splicing factor. Numerous signaling pathways in the cytoplasm and the nucleus are influenced by CK2 kinase activity. Several of the nuclear CK2 substrates are proteins that are known to regulate transcriptional events, including the CTD of the largest RNAP II subunit, cyclin H, and FCP1 (TFIIF-dependent CTD phosphatase 1) (36, 45). CK2 phosphorylation of cyclin H is important for full cyclin H/CDK7/Mat1 kinase activity with a CTD peptide as a substrate (42). In Xenopus laevis cell extracts, CK2 phosphorylation also enhances the phosphatase activity of FCP1 and binding to the RAP74 subunit of TFIIF, thereby promoting sequential transcriptional cycles (35). Thus, CK2 phosphorylation of its various protein substrates may promote or modulate transcriptional and RNA processing activities through diverse mechanisms involving the catalytic activation of enzymes. Interestingly, we found that RNPS1 also interacts with the largest RNAP II subunit and/or RNAP II-containing protein complexes (44). Recently, the importance of dynamic posttranslational modification of the RNAP II CTD in functional coupling between transcription and pre-mRNA processing has been well documented (reviewed in references 25 and 34). The data herein suggest that through phosphorylation of RNPS1, CK2 influences the splicing events associated with the RNAP II CTD.
Acknowledgments
We thank J. Grenet, S. Bothner, T. Venkataraman, and A. Mishra for their excellent technical assistance; C. Naeve for DNA primers and sequencing; J. F. Leykam for the analysis of mass spectrometry; R. Lührmann for the anti-U5-116K antibody; A. R. Krainer for the anti-SF2/ASF (mAb96) antibody; M. L. Hastings for HeLa cell S100 extracts; and I. C. Eperon for the pTN24 plasmid. We are grateful to M. J. Moore for helpful advice on the RNA joining technique; M. P. Deutscher, K. Ohe, D. M. Helfman, and J. M. Lahti for their helpful discussions and critical readings of the manuscript; and J. F. Cáceres for communicating results prior to publication.
A.M. was supported by the L. P. Markey Trust funds, an institutional research grant (IRG-98-277-04) from the American Cancer Society, and a Florida Biomedical Research Program grant (BM031) from the Florida Department of Health. V.J.K. was supported by a National Institutes of Health (NIH) grant (5R01 GM44088-13). V.J.K. and C.A.S. were also supported by an NIH Cancer Center grant awarded to SJCRH (P30 CA21765-25) as well as by support from the American Lebanese Syrian Associated Charities (ALSAC). E.S. was supported by a postdoctoral fellowship from the Uehara Memorial Biological Science Foundation, a Jichi Medical School Young Investigator Award, and a grant-in-aid for young scientists (B) from the Ministry of Education, Culture, Science and Technology of Japan. A.M. is a research member of the Sylvester Comprehensive Cancer Center.
Footnotes
We dedicate this article to the memory of our colleague and mentor, Vincent J. Kidd, who suddenly passed away in May 2004. Through his research on CDK11 signaling and cell death pathways, he made significant contributions to the fields of cell cycle, transcription-splicing, apoptosis, and neuroblastoma.
REFERENCES
- 1.Boyle, W. J., P. van der Geer, and T. Hunter. 1991. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 201:110-149. [DOI] [PubMed] [Google Scholar]
- 2.Brandner, J. M., S. Reidenbach, C. Kuhn, and W. W. Franke. 1998. Identification and characterization of a novel kind of nuclear protein occurring free in the nucleoplasm and in ribonucleoprotein structures of the “speckle” type. Eur. J. Cell Biol. 75:295-308. [DOI] [PubMed] [Google Scholar]
- 3.Cáceres, J. F., and A. R. Krainer. 1997. Mammalian pre-mRNA splicing factors, p. 174-212. In A. R. Krainer (ed.), Eukaryotic mRNA processing. IRL Press, Oxford, United Kingdom.
- 4.Cáceres, J. F., G. R. Screaton, and A. R. Krainer. 1998. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev. 12:55-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao, W., S. F. Jamison, and M. A. Garcia-Blanco. 1997. Both phosphorylation and dephosphorylation of ASF/SF2 are required for pre-mRNA splicing in vitro. RNA 3:1456-1467. [PMC free article] [PubMed] [Google Scholar]
- 6.Cartegni, L., S. L. Chew, and A. R. Krainer. 2002. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 3:285-298. [DOI] [PubMed] [Google Scholar]
- 7.Cooper, T. A., and W. Mattox. 1997. The regulation of splice-site selection, and its role in human disease. Am. J. Hum. Genet. 61:259-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Das, R., and R. Reed. 1999. Resolution of the mammalian E complex and the ATP-dependent spliceosomal complexes on native agarose mini-gels. RNA 5:1504-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gossen, M., and H. Bujard. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA 89:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graveley, B. R. 2000. Sorting out the complexity of SR protein functions. RNA 6:1197-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graveley, B. R. 2001. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 17:100-107. [DOI] [PubMed] [Google Scholar]
- 12.Hanamura, A., J. F. Cáceres, A. Mayeda, B. R. Franza, Jr., and A. R. Krainer. 1998. Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA 4:430-444. [PMC free article] [PubMed] [Google Scholar]
- 13.Huang, Y., T. A. Yario, and J. A. Steitz. 2004. A molecular link between SR protein dephosphorylation and mRNA export. Proc. Natl. Acad. Sci. USA 101:9666-9670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.International Human Genome Sequencing Consortium. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 15.International Human Genome Sequencing Consortium. 2004. Finishing the euchromatic sequence of the human genome. Nature 431:931-945. [DOI] [PubMed] [Google Scholar]
- 16.Jurica, M. S., and M. J. Moore. 2003. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell 12:5-14. [DOI] [PubMed] [Google Scholar]
- 17.Kataoka, N., and G. Dreyfuss. 2004. A simple whole-cell lysate system for in vitro splicing reveals a stepwise-assembly of the exon-exon junction complex. J. Biol. Chem. 279:7009-7013. [DOI] [PubMed] [Google Scholar]
- 18.Krämer, A. 1996. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu. Rev. Biochem. 65:367-409. [DOI] [PubMed] [Google Scholar]
- 19.Lai, M. C., and W. Y. Tarn. 2004. Hypophosphorylated ASF/SF2 binds TAP and is present in messenger ribonucleoproteins. J. Biol. Chem. 279:31745-31749. [DOI] [PubMed] [Google Scholar]
- 20.Le Hir, H., E. Izaurralde, L. E. Maquat, and M. J. Moore. 2000. The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 19:6860-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez, A. J. 1998. Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu. Rev. Genet. 32:279-305. [DOI] [PubMed] [Google Scholar]
- 22.Loyer, P., J. H. Trembley, J. M. Lahti, and V. J. Kidd. 1998. The RNP protein, RNPS1, associates with specific isoforms of the p34cdc2-related PITSLRE protein kinase in vivo. J. Cell Sci. 111:1495-1506. [DOI] [PubMed] [Google Scholar]
- 23.Lykke-Andersen, J. 2001. mRNA quality control: marking the message for life or death. Curr. Biol. 11:R88-R91. [DOI] [PubMed] [Google Scholar]
- 24.Lykke-Andersen, J., M. D. Shu, and J. A. Steitz. 2001. Communication of the position of exon-exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science 293:1836-1839. [DOI] [PubMed] [Google Scholar]
- 25.Maniatis, T., and R. Reed. 2002. An extensive network of coupling among gene expression machines. Nature 416:499-506. [DOI] [PubMed] [Google Scholar]
- 26.Maniatis, T., and B. Tasic. 2002. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature 418:236-243. [DOI] [PubMed] [Google Scholar]
- 27.Mayeda, A., J. Badolato, R. Kobayashi, M. Q. Zhang, E. M. Gardiner, and A. R. Krainer. 1999. Purification and characterization of human RNPS1: a general activator of pre-mRNA splicing. EMBO J. 18:4560-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayeda, A., and A. R. Krainer. 1999. Preparation of HeLa cell nuclear and cytosolic S100 extracts for in vitro splicing. Methods Mol. Biol. 118:309-314. [DOI] [PubMed] [Google Scholar]
- 29.Mayeda, A., and A. R. Krainer. 1999. Mammalian in vitro splicing assays. Methods Mol. Biol. 118:315-321. [DOI] [PubMed] [Google Scholar]
- 30.Misteli, T. 2000. Cell biology of transcription and pre-mRNA splicing: nuclear architecture meets nuclear function. J. Cell Sci. 113:1841-1849. [DOI] [PubMed] [Google Scholar]
- 31.Moore, M. J., and C. C. Query. 1998. Uses of site-specifically modified RNAs constructed by RNA ligation, p. 75-108. In C. W. J. Smith (ed.), RNA-protein interactions: a practical approach. IRL Press, Oxford, United Kingdom.
- 32.Nasim, M. T., H. M. Chowdhury, and I. C. Eperon. 2002. A double reporter assay for detecting changes in the ratio of spliced and unspliced mRNA in mammalian cells. Nucleic Acids Res. 30:e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nott, A., H. Le Hir, and M. J. Moore. 2004. Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Dev. 18:210-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orphanides, G., and D. Reinberg. 2002. A unified theory of gene expression. Cell 108:439-451. [DOI] [PubMed] [Google Scholar]
- 35.Palancade, B., M. F. Dubois, and O. Bensaude. 2002. FCP1 phosphorylation by casein kinase 2 enhances binding to TFIIF and RNA polymerase II carboxyl-terminal domain phosphatase activity. J. Biol. Chem. 277:36061-36067. [DOI] [PubMed] [Google Scholar]
- 36.Payne, J. M., P. J. Laybourn, and M. E. Dahmus. 1989. The transition of RNA polymerase II from initiation to elongation is associated with phosphorylation of the carboxyl-terminal domain of subunit IIa. J. Biol. Chem. 264:19621-19629. [PubMed] [Google Scholar]
- 37.Pinna, L. A. 2002. Protein kinase CK2: a challenge to canons. J. Cell Sci. 115:3873-3878. [DOI] [PubMed] [Google Scholar]
- 38.Reed, R., and L. Palandjian. 1997. Spliceosome assembly, p. 103-129. In A. R. Krainer (ed.), Eukaryotic mRNA processing. IRL Press, Oxford, United Kingdom.
- 39.Reichert, V. L., H. Le Hir, M. S. Jurica, and M. J. Moore. 2002. 5′ exon interactions within the human spliceosome establish a framework for exon junction complex structure and assembly. Genes Dev. 16:2778-2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakashita, E., S. Tatsumi, D. Werner, H. Endo, and A. Mayeda. 2004. Human RNPS1 and its associated factors: a versatile alternative pre-mRNA splicing regulator in vivo. Mol. Cell. Biol. 24:1174-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sawa, H., M. Ohno, H. Sakamoto, and Y. Shimura. 1988. Requirement of ATP in the second step of the pre-mRNA splicing reaction. Nucleic Acids Res. 16:3157-3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schneider, E., S. Kartarius, N. Schuster, and M. Montenarh. 2002. The cyclin H/cdk7/Mat1 kinase activity is regulated by CK2 phosphorylation of cyclin H. Oncogene 21:5031-5037. [DOI] [PubMed] [Google Scholar]
- 43.Schwerk, C., J. Prasad, K. Degenhardt, H. Erdjument-Bromage, E. White, P. Tempst, V. J. Kidd, J. L. Manley, J. M. Lahti, and D. Reinberg. 2003. ASAP, a novel protein complex involved in RNA processing and apoptosis. Mol. Cell. Biol. 23:2981-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trembley, J. H., D. Hu, L. C. Hsu, C. Y. Yeung, C. Slaughter, J. M. Lahti, and V. J. Kidd. 2002. PITSLRE p110 protein kinases associate with transcription complexes and affect their activity. J. Biol. Chem. 277:2589-2596. [DOI] [PubMed] [Google Scholar]
- 45.Trembley, J. H., D. Hu, C. A. Slaughter, J. M. Lahti, and V. J. Kidd. 2003. Casein kinase 2 interacts with cyclin-dependent kinase 11 (CDK11) in vivo and phosphorylates both the RNA polymerase II carboxyl-terminal domain and CDK11 in vitro. J. Biol. Chem. 278:2265-2270. [DOI] [PubMed] [Google Scholar]
- 46.Venter, J. C., M. D. Adams, E. W. Myers, P. W. Li, R. J. Mural, G. G. Sutton, H. O. Smith, M. Yandell, C. A. Evans, R. A. Holt, J. D. Gocayne, P. Amanatides, R. M. Ballew, D. H. Huson, J. R. Wortman, Q. Zhang, C. D. Kodira, X. H. Zheng, L. Chen, M. Skupski, et al. 2001. The sequence of the human genome. Science 291:1304-1351. [DOI] [PubMed] [Google Scholar]
- 47.Wagner, E., and J. Lykke-Andersen. 2002. mRNA surveillance: the perfect persist. J. Cell Sci. 115:3033-3038. [DOI] [PubMed] [Google Scholar]
- 48.Wang, W., and B. A. Malcolm. 1999. Two-stage PCR protocol allowing introduction of multiple mutations, deletions and insertions using QuikChange site-directed mutagenesis. BioTechniques 26:680-682. [DOI] [PubMed] [Google Scholar]
- 49.Wang, Y.-C., M. Selvakumar, and D. M. Helfman. 1997. Alternative pre-mRNA splicing, p. 242-279. In A. R. Krainer (ed.), Eukaryotic mRNA processing. IRL Press, Oxford, United Kingdom.
- 50.Wiegand, H. L., S. Lu, and B. R. Cullen. 2003. Exon junction complexes mediate the enhancing effect of splicing on mRNA expression. Proc. Natl. Acad. Sci. USA 100:11327-11332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang, W. J., and J. Y. Wu. 1996. Functional properties of p54, a novel SR protein active in constitutive and alternative splicing. Mol. Cell. Biol. 16:5400-5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou, Z., L. J. Licklider, S. P. Gygi, and R. Reed. 2002. Comprehensive proteomic analysis of the human spliceosome. Nature 419:182-185. [DOI] [PubMed] [Google Scholar]