Abstract
Formation of the mature 3′ ends of the vast majority of cellular mRNAs occurs through cleavage and polyadenylation and requires a cleavage and polyadenylation specificity factor (CPSF) containing, among other proteins, CPSF-73 and CPSF-100. These two proteins belong to a superfamily of zinc-dependent β-lactamase fold proteins with catalytic specificity for a wide range of substrates including nucleic acids. CPSF-73 contains a zinc-binding histidine motif involved in catalysis in other members of the β-lactamase superfamily, whereas CPSF-100 has substitutions within the histidine motif and thus is unlikely to be catalytically active. Here we describe two previously unknown human proteins, designated RC-68 and RC-74, which are related to CPSF-73 and CPSF-100 and which form a complex in HeLa and mouse cells. RC-68 contains the intact histidine motif, and hence it might be a functional counterpart of CPSF-73, whereas RC-74 lacks this motif, thus resembling CPSF-100. In HeLa cells RC-68 is present in both the cytoplasm and the nucleus whereas RC-74 is exclusively nuclear. RC-74 does not interact with CPSF-73, and neither RC-68 nor RC-74 is found in a complex with CPSF-160, indicating that these two proteins form a separate entity independent of the CPSF complex and are likely involved in a pre-mRNA processing event other than cleavage and polyadenylation of the vast majority of cellular pre-mRNAs. RNA interference-mediated depletion of RC-68 arrests HeLa cells early in G1 phase, but surprisingly the arrested cells continue growing and reach the size typical of G2 cells. RC-68 is highly conserved from plants to humans and may function in conjunction with RC-74 in the 3′ end processing of a distinct subset of cellular pre-mRNAs encoding proteins required for G1 progression and entry into S phase.
In metazoans, there are two distinct mechanisms of 3′ end processing of pre-mRNAs that lead to formation of mature mRNAs. The vast majority of pre-mRNAs are processed at the 3′ end by a coupled cleavage and polyadenylation reaction (9, 57, 71, 78). However, the replication-dependent histone pre-mRNAs are processed by a one-step mechanism that involves only a cleavage reaction (13, 40, 47). Both mechanisms require two cis-acting elements in the pre-mRNA and involve different sets of trans-acting factors.
In mammalian pre-mRNAs, the cleavage and polyadenylation reaction occurs about 20 nucleotides downstream of a highly conserved polyadenylation signal, AAUAAA. The AAUAAA sequence is recognized by cleavage and polyadenylation specificity factor 160 (CPSF-160) (32, 49), which exists in a stable complex with three other proteins, CPSF-100, CPSF-73, and CPSF-30 (2, 4, 31, 33, 48). The CPSF complex also contains a fifth component, Fip1, which may be only loosely associated with the other subunits or which may exist in nonequimolar amounts in the complex (34). The CPSF complex is required for both cleavage and polyadenylation. About 30 nucleotides downstream of the cleavage site there is a weakly conserved GU-rich element, which is recognized by a three-subunit cleavage stimulation factor (CstF) required for cleavage but not poly(A) addition (38). Two poorly characterized multisubunit complexes, referred to as cleavage factor I (CF I) and CF II, are also required for cleavage but not polyadenylation (59, 78). Efficient cleavage and polyadenylation additionally require poly(A) polymerase (49).
Mammalian replication-dependent histone pre-mRNAs are cleaved 5 nucleotides downstream of a highly conserved stem-loop structure consisting of a 6-bp stem and a 4-nucleotide loop. This processing results in the generation of mature histone mRNAs that end with the stem-loop followed by an ACCCA single-stranded sequence. This unusual 3′ end plays a key role at the posttranscriptional level in coordinating histone synthesis with DNA replication (40). A smaller group of histone mRNAs transcribed from the replacement variant histone genes are synthesized throughout the cell cycle (52). These mRNAs are generated by a standard cleavage and polyadenylation mechanism. The stem-loop in replication-dependent histone pre-mRNA is recognized by the stem-loop binding protein (SLBP) (74), also known as the hairpin binding protein (39). About 15 nucleotides downstream of the cleavage site there is a short purine-rich sequence called the histone downstream element (HDE). This downstream sequence is recognized by the U7 snRNP, which contains an ∼60-nucleotide U7 snRNA (46). Interaction of the U7 snRNP with the histone pre-mRNA is mediated by base pairing between the HDE and the 5′ end of U7 snRNA (66). The U7 snRNP, in place of the D1 and D2 Sm proteins present in the spliceosomal snRNPs, contains two Sm-like proteins, Lsm10 (55) and Lsm11 (54). Lsm11 interacts with ZFP 100, a 100-kDa zinc finger protein that bridges the U7 snRNP with the stem-loop/SLBP complex (12, 54).
An important goal of recent studies on 3′ end processing is determining the identity of a protein that cleaves the phosphodiester bond in each type of pre-mRNA. Based on structural and sequence analyses, it has been recently proposed that the cleavage preceding polyadenylation may be catalyzed by CPSF-73 (1, 7). CPSF-73 belongs to a superfamily of metal-dependent proteins with the β-lactamase fold that includes proteins with a broad range of biological activities and substrate specificities (10). Among members of the β-lactamase superfamily are three recently characterized zinc-dependent proteins with either proven or predicted endonuclease activity: ELAC2, also referred to as tRNase Z, required for 3′ end maturation of eukaryotic tRNAs (21, 62, 67) and linked to a hereditary form of prostate cancer (68); SNM 1, involved in DNA cross-link repair (20); and Artemis, involved in V(D)J recombination and DNA double-strand break repair (36, 45). SNM 1 and Artemis, the two DNA-specific proteins, share a domain with CPSF-73 and CPSF-100, called the β-CASP domain (7). The majority of proteins of the β-lactamase superfamily, including CPSF-73, contain the conserved zinc binding motif HXHXDH (where X indicates any amino acid), which plays a central role in catalysis (42, 73). CPSF-73 exists in the five-subunit CPSF complex with another member of the β-lactamase superfamily, CPSF-100. CPSF-100, in contrast to CPSF-73, has substitutions within the histidine motif and thus is unlikely to play a direct role in catalysis. Involvement of CPSF-73 in cleaving pre-mRNAs prior to addition of the poly(A) tail is supported by recent studies demonstrating that a protein with a molecular size consistent with that of CPSF-73 can be UV-cross-linked to pre-mRNA in the vicinity of the cleavage site (60).
Here we describe the identification and preliminary characterization of two unknown members of the β-lactamase protein superfamily, RC-68 and RC-74, which have sequence similarity with CPSF-73 and CPSF-100 and which belong to the β-CASP family. RC-68 and RC-74 interact with each other in HeLa and mouse cells but do not associate with subunits of the CPSF complex. We demonstrate that depletion of RC-68 by RNA interference (RNAi) results in the arrest of HeLa cells in G1 phase, suggesting a link between this putative RNA processing factor and cell cycle regulation.
MATERIALS AND METHODS
Cloning. Full-length cDNAs encoding CPSF-73, CPSF-100, RC-68, and RC-74 were obtained from ATCC (Manassas, Va.) and subcloned into appropriate vectors with PCR-generated fragments containing the desired restriction sites.
Subcellular localization.
The cDNAs for CPSF-73, RC-68, and RC-74 were inserted in frame into pEGFP-N1 vector (BD Biosciences) upstream of the region encoding green fluorescent protein (GFP) and transfected into HeLa cells with Lipofectamine 2000 (Invitrogen). The intracellular distribution of the fusion proteins was visualized by fluorescence microscopy 24 h after transfection. For immunostaining, HeLa cells stably expressing the hemagglutinin (HA)-tagged RC-68 or RC-74 were fixed with 3.7% formaldehyde and permeabilized with 0.5% Triton X-100. The tagged proteins were detected with an anti-HA antibody and a Cy3-conjugated goat anti-mouse antibody.
Histone pre-mRNA processing.
Preparation of mammalian and Drosophila melanogaster nuclear extracts and histone pre-mRNA processing were carried out as previously described (15, 17, 19, 41).
RNAi experiments.
Expression of RC-68 was downregulated in HeLa cells by a double-hit protocol, as described previously (70). Cells were collected 48 h after the second small interfering RNA (siRNA) transfection. Chemically synthesized siRNAs were obtained from Dharmacon (Lafayette Colo.) and had the following sequences of the top strand: 5′AGCACAUCAAGGCCUUCGAdTdT3′ (RC-68 specific 1), 5′ACGAAAAGAACAUGGUCAUdTdT3′ (RC-68 specific 2), and 5′GGUCCGGCUCCCCCAAAUGdTdT3′ (control). A portion of cells were lysed in a buffer containing 150 mM NaCl, 50 mM Tris-HCl (pH 8), 10 mM sodium azide, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 1× protease inhibitor mixture (Sigma), and 0.5% NP-40 and analyzed by Western blotting. The remaining cells were fixed with 70% ethanol and stained with propidium iodide, and 104 cells were analyzed for their DNA content by flow cytometry with a FACscan and the Summit software (Cytomation, Inc.).
The yeast two-hybrid system.
A HeLa cDNA library was screened against RC-74 fused to the GAL4 DNA binding domains as described previously (14). The transformants were plated on selective plates containing 2.5 mM 3-aminotriazole (3-AT), and fast-growing colonies were subsequently tested on plates containing up to 100 mM 3-AT.
Cell synchronization.
HeLa cells were synchronized by a double thymidine block and collected at different time points after release from the block, as described previously (75).
Antibodies.
Rabbit antibodies against the C-terminal peptides of mouse RC-68 (SFLTTLLKNGLPQAPS) and mouse RC-74 (LRVRLRDLVLRFLQKF) were generated. Anti-RC-68 and anti-RC-74 were affinity purified on a Sulfolink column (Pierce) as recommended by the manufacturer. Each antibody recognizes both the mouse and human proteins.
Coimmunoprecipitation of RC-74 and RC-68.
The full-length cDNA for human RC-74 was cloned into a pcDNA 3-HA vector and stably expressed in HeLa cells as a fusion protein with two HA tags on the N terminus (HA/RC-74). The cells from 10 15-cm-diameter plates (about 1 ml of packed cell volume) were lysed in 10 ml of the NP-40 lysis buffer (see above), and cell debris was removed by centrifugation at 12,000 g for 10 min. The lysate was incubated for 3 h with 20 μl of a monoclonal anti-HA antibody (Covance), followed by a 2-h incubation with 40 μl of protein G-Sepharose beads (Amersham). The beads were washed for 2 h with the NP-40 lysis buffer and divided into two equal portions, which were used for Western blotting with either the anti-HA antibody or the affinity-purified anti-RC-68. To immunoprecipitate the HA/RC-74 from a nuclear extract, HeLa cells expressing HA/RC-74 were collected from 20 15-cm-diameter plates and used to prepare nuclear extract as previously described (15, 41). The nuclear extract was not dialyzed against buffer D (20 mM HEPES-KOH [pH 7.9], 100 mM KCl, 0.5 mM dithiothreitol, 0.2 mM EDTA, 20% glycerol); instead it was adjusted to 100 mM KCl by adding a buffer with the same composition as buffer D but lacking KCl. The nuclear extract was divided in two portions, and each was used for immunoprecipitation with either anti-HA or anti-FLAG (Sigma) as a negative control. The subsequent steps were carried out as described above with the difference that protein G-Sepharose beads were washed with buffer D instead of NP-40 lysis buffer. Mouse RC-68 was precipitated from 250 μl of a dialyzed nuclear extract from mouse myeloma cells with 25 μl of the anti-RC-68 antibody. The immunoprecipitates collected on protein A-agarose beads were tested for the presence of RC-74 and CPSF-160 by Western blotting.
RESULTS
Identification of RC-68 and RC-74.
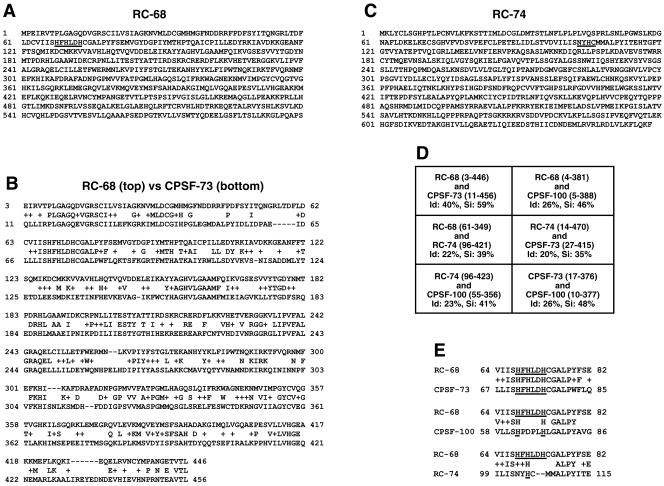
The hydrolytic activity of several zinc-dependent proteins with the β-lactamase fold on nucleic acids brought our attention to previously uncharacterized members of this family. By searching the available databases, we identified two unknown human proteins of 68 and 74 kDa with sequence similarity to CPSF-73 and CPSF-100, designated here RC-68 and RC-74 (for proteins of 68 and 74 kDa related to CPSF subunits). RC-68 (NP_060341, AAH07978, CAB66747) contains 600 amino acids (Fig. 1A) and is 40% identical to CPSF-73 throughout the first 450 amino acids (Fig. 1B) and 25% identical to CPSF-100 throughout the first 400 amino acids. RC-68, like CPSF-73, contains within the N-terminal region the sequence HFHLDH, conforming to the histidine motif consensus, HXHXDH, which in other proteins of the β-lactamase superfamily is involved in zinc binding and catalysis (73). The sequences of the first 450 amino acids in RC-68 and CPSF-73 have been highly conserved during evolution, consistent with the N-terminal domain in both proteins being involved in catalysis. For example, the N-terminal domain in human RC-68 is 80% identical to the Drosophila orthologue (NP_651721, AAF56931, AAL28645) and 55% identical to the recently described Arabidopsis thaliana orthologue (NP_178282, AAN87883) designated AtCPSF73-II (76).
FIG. 1.
RC-68 and RC-74 are members of the β-lactamase protein superfamily related to CPSF-73 and CPSF-100. (A) Amino acid sequence of RC-68 with the conserved zinc binding motif underlined. (B) Pairwise alignment of RC-68 (top) and CPSF-73 (bottom) with BLASTP. (C) Amino acid sequence of RC-74 with the degenerate histidine motif underlined. (D) Identities (Id) and similarities (Si) between the indicated regions of the four members of the β-lactamase superfamily. (E) Alignment of RC-68 (top) with the three other members of the β-lactamase superfamily (bottom) within the region containing the histidine motif or its degenerate form.
RC-74 (NP_060720, BAA91867, AAH25267) is a 658-amino-acid polypeptide (Fig. 1C) and within the N-terminal region has about 20% identity to RC-68, CPSF-73, and CPSF-100 (Fig. 1D). RC-74, like CPSF-100, contains mutations within the histidine motif, suggesting that it is inactive in catalysis (Fig. 1E). RC-74 has been conserved during evolution to a significantly lesser extent than CPSF-73 and RC-68. Human RC-74 has 51% identity to the Drosophila orthologue (NP_ 648838, AAL13803, AAF49538) and 26% identify to the Arabidopsis orthologue (NP 187409) throughout most of the protein. Interestingly, the lack of the histidine motif is an evolutionarily conserved feature of all RC-74 orthologues.
Intracellular localization of RC-68 and RC-74.
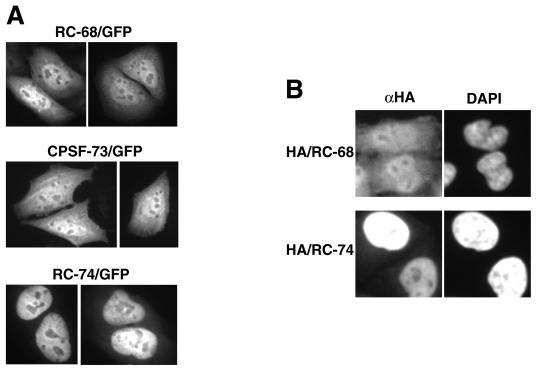
To determine the in vivo localization of RC-68 and RC-74, each protein was fused at the C terminus to GFP and transiently expressed in HeLa cells. The intracellular distribution of the fusion proteins was analyzed by fluorescence microscopy. For comparison, we also determined the intracellular localization of CPSF-73 fused to GFP. The RC-68 fusion protein was present in both the nucleus and the cytoplasm, with a higher concentration detected in the nucleus (Fig. 2A). Interestingly, CPSF-73 fused to GFP was also detected in both cellular compartments. In contrast, the RC-74 fusion protein was exclusively nuclear (Fig. 2A). To confirm these results, we transfected HeLa cells with a cDNA construct encoding either RC-68 or RC-74 fused at the N terminus with two HA tags. The intracellular distribution of the stably expressed tagged proteins was determined by immunostaining with the anti-HA antibody and the Cy3-conjugated goat anti-mouse antibody. The HeLa cells were simultaneously stained with DAPI (4′,6′-diamidino-2-phenylindole) to visualize nuclei. The intracellular localization of the HA-tagged proteins was virtually identical with the localization of the GFP fusion proteins: HA-tagged RC-68 was detected in both cellular compartments, and HA-tagged RC-74 was detected only in the nuclei (Fig. 2B).
FIG. 2.
Intracellular localization of RC-68, CPSF-73, and RC-74. (A) RC-68, CPSF-73, and RC-74 were expressed in HeLa cells as fusion proteins with the C-terminal GFP (top), and the intracellular distribution of the fusion proteins was analyzed by fluorescence microscopy. (B) Intracellular distribution of the HA-tagged RC-68 (top) and RC-74 (bottom) detected by immunostaining. The nuclei were visualized by staining with DAPI. αHA, anti-HA.
Interaction between RC-74 and RC-68.
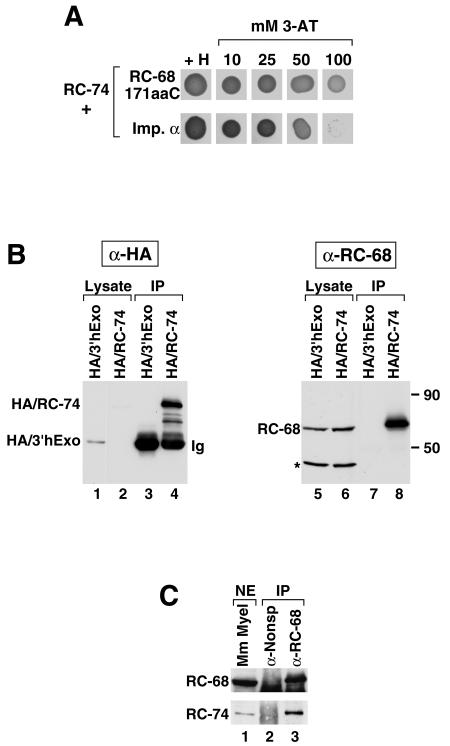
To gain some insight into possible functions of RC-74, we searched for its interacting protein partners. We constructed a yeast strain expressing a fusion protein consisting of the GAL4 DNA binding domain and the full-length RC-74 and carried out a yeast two-hybrid screen with a HeLa cDNA library. Out of 4 million yeast transformants we isolated more than 10 rapidly growing colonies, all of which expressed C-terminal fragments of RC-68. The shortest and the longest RC-68 inserts encoded the last 124 and 214 amino acids, respectively, while the intermediate-size insert encoded a 171-amino-acid portion of RC-68. In addition to the various C-terminal portions of RC-68, we also isolated several yeast clones expressing full-length importin α, which together with importin β is involved in importing proteins from the cytoplasm to the nucleus (26, 27). All the RC-68 clones grew rapidly in the presence of 100 mM 3-AT, indicative of a very strong interaction, while the clones containing importin α grew at a lower rate on 50 mM 3-AT and did not grow in the presence of 100 mM 3-AT (Fig. 3A). We conclude that the last 124 amino acids of RC-68 are sufficient to mediate interaction with RC-74.
FIG. 3.
RC-68 and RC-74 interact in the yeast two-hybrid system and in mammalian cells. (A) Two yeast clones isolated in the yeast two-hybrid system as interacting with the full-length RC-74 as bait, one expressing the last 171 amino acids of RC-68 (top row) and the other expressing full-length importin α (bottom row), were tested for growth in the presence of histidine or increasing concentrations of 3-AT. (B) Cell lysates from HeLa cells stably expressing HA-tagged 3′hExo (lanes 1 and 5) or HA-tagged RC-74 (lanes 2 and 6) were incubated with the anti-HA (α-HA) antibody and protein G-Sepharose. The immunoprecipitates (IP) were tested by Western blotting for the presence of the HA-tagged proteins with anti-HA (lanes 3 and 4) or for the presence of RC-68 with an antibody against the C-terminal peptide (lanes 7 and 8). The amounts of the lysates analyzed in lanes 1 and 2 and 5 and 6 represent 0.25% of the input used for immunoprecipitation. Migration of molecular size markers of 90 and 50 kDa is indicated at the right. Asterisk, 48-kDa protein that cross-reacts with the antibody against RC-68. (C) A dialyzed nuclear extract (NE) from mouse myeloma cells (lane 1) was incubated with either a nonspecific antibody (lane 2) or with anti-RC-68 (lane 3), and the precipitates were tested by Western blotting for the presence of RC-68 (top) or RC-74 (bottom) with anti-RC-68 or anti-RC-74, respectively. The amount of the nuclear extract analyzed in lane 1 represents 5% of the input used for immunoprecipitation. A band visible below RC-68 in the precipitated samples (lanes 2 and 3) represents the heavy chain of immunoglobulin, which interacts with the secondary antibody.
To test whether RC-68 and RC-74 interact in mammalian cells, we used a coimmunoprecipitation assay. Since the rabbit antibody against the C-terminal peptide of RC-74 was inefficient in precipitating the protein from cell extracts, we used HeLa cells stably expressing the HA-tagged RC-74 and the anti-HA antibody. The cells were lysed in 0.5% NP-40 lysis buffer, and the lysate was incubated with the anti-HA antibody. The HA immunoprecipitates were divided into two equal portions and tested by Western blotting for the presence of either the HA-tagged RC-74, with the anti-HA antibody, or endogenous RC-68, with a rabbit antibody against the C-terminal peptide of RC-68. As a negative control we used HeLa cells stably expressing HA-tagged 3′hExo (16). The anti-RC-68 antibody detects RC-68 migrating at 65 kDa and a cross-reacting protein migrating at 48 kDa (Fig. 3B, lanes 5 and 6). Western blotting with the anti-HA antibody indicates that HA/3′hExo was expressed at about a threefold-higher level than HA/RC-74 (Fig. 3B, lanes 1 and 2, respectively; a band corresponding to the HA/RC-74 is more visible on longer exposures). Judging from the amount of HA/RC-74 and HA/3′hExo remaining in the supernatants after incubation of the lysates with anti-HA and protein G-Sepharose, the efficiency of immunoprecipitation of each protein was about 50% (not shown). The anti-HA antibody did not precipitate detectable amounts of endogenous RC-68 from the cell extract containing HA-tagged 3′hExo (Fig. 3B, lane 7). The same antibody incubated with cell extract containing HA-tagged RC-74 coprecipitated significant amounts of RC-68 but not the cross-reacting protein migrating at 48 kDa (Fig. 3B, lane 8). We used a reciprocal approach and tested the ability of the anti-RC-68 antibody to coprecipitate RC-74 from a nuclear extract prepared from mouse myeloma cells. As shown in Fig. 3C the antibody specifically precipitated RC-68 from the mouse nuclear extract (top, lane 3) and coprecipitated significant amounts of RC-74 (bottom, lane 3). A nonspecific antibody used as a negative control was unable to precipitate any detectable amounts of each protein (Fig. 3C, lane 2). Similar results were obtained with nuclear extracts from HeLa cells (Fig. 4C, lane 2). These results, together with the results of the yeast two-hybrid screen, demonstrate that RC-68 and RC-74 interact with each other and form a stable complex in the nuclei of mammalian cells.
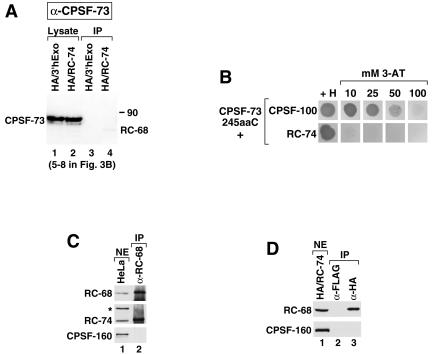
FIG. 4.
RC-68 and RC-74 do not associate with subunits of the CPSF complex. (A) The blot shown in Fig. 3B (lanes 5 to 8) was stripped and incubated with anti-CPSF-73. A weak band corresponding to RC-68 and visible in lane 4 is due to incomplete stripping of the anti-RC-68 from the blot. IP, immunoprecipitates. (B) The ability of the last 245 amino acids of CPSF-73 fused to the GAL4 DNA binding domain to interact with the full-length CPSF-100 or RC-74 was assayed with a directed two-hybrid system in the presence of histidine or in the absence of histidine plus increasing concentrations of 3-AT. (C) A nondialyzed nuclear extract (NE) from HeLa cells (lane 1) was incubated with anti-RC-68 (lane 2), and the precipitates were tested for the presence of RC-68 (top), RC-74 (middle), and CPSF-160 (bottom) with anti-RC-68 (α-RC-68), anti-RC-74, and anti-CPSF-160, respectively. Asterisk, HeLa protein cross-reacting with anti-RC-74. The amount of the nuclear extract analyzed in lane 1 represents 5% of the input used for immunoprecipitation. (D) A nondialyzed nuclear extract from HeLa cells stably expressing the HA/RC-74 fusion protein (lane 1) was incubated with either anti-FLAG (lane 2) or anti-HA (lane 3), and the precipitates were tested for the presence of RC-68 (top) and CPSF-160 (bottom) with anti-RC-68 and anti-CPSF-160, respectively. The amount of the nuclear extract analyzed in lane 1 represents 2.5% of the input used for immunoprecipitation.
RC-68 and RC-74 do not interact with CPSF-160.
To determine the specificity of the interaction between RC-74 and RC-68, we incubated the blot shown in Fig. 3B (lanes 5 to 8) with an antibody against CPSF-73 (kindly provided by D. Bentley, University of Colorado). The anti-CPSF-73 specifically recognizes CPSF-73 migrating at about 75 kDa in HeLa lysates expressing either HA/RC-74 or HA/3′hExo (Fig. 4A, lanes 1 and 2). However, CPSF-73 was not detected in the HA precipitates containing HA tagged RC-74 (Fig. 4A, lane 4), indicating that these two proteins do not interact in HeLa cells.
While it has been known for over 10 years that CPSF-73 and CPSF-100 exist together in the CPSF complex (2, 4, 31, 33, 48), it has not been previously established whether the two proteins directly interact with each other or exist in the same complex through interaction with other CPSF subunits. CPSF-73 and RC-68 are 40% identical throughout the first 450 amino acids (Fig. 1B) but do not have any sequence similarity within the C-terminal region. The yeast two-hybrid system revealed that the C-terminal region of RC-68 strongly interacts with the full-length RC-74, raising the possibility that the same region of CPSF-73 is involved in interaction with CPSF-100. We tested this hypothesis by using the directed yeast two-hybrid system. A portion of CPSF-73 encompassing amino acids 440 to 684 (the last few amino acids of the N-terminal domain and the entire C-terminal region) was expressed in yeast cells as a fusion protein with the N-terminal GAL4 activation domain and tested for interaction with the full-length CPSF-100 or RC-74, each fused to the GAL4 DNA binding domain. CPSF-73 has a longer C terminus than RC-68; thus the tested fragment of CPSF-73 consisted of 245 amino acids rather than 171 amino acids as does the C-terminal region of RC-68. Since the full-length cDNA for human CPSF-100 was not available, we instead cloned mouse CPSF-100, which is nearly identical to the human protein. The strength of the interaction was determined by analyzing the ability of yeast cells expressing both fusion proteins to grow on different concentrations of 3-AT. As shown in Fig. 4B, the interaction between the C-terminal region of CPSF-73 and the full-length CPSF-100 resulted in rapid growth of yeast cells in the presence of 25 mM 3-AT and slower growth in the presence of 50 mM AT. The highest concentration of 3-AT (100 mM) almost completely inhibited growth, indicating that the interaction between the two subunits of the CPSF complex is not as strong as the interaction between RC-74 and the C-terminal domain of RC-68, which allows yeast cells to rapidly grow under the restrictive conditions of 100 mM 3-AT. Shortening the length of the C-terminal region of CPSF-73 from 245 to 206 amino acids reduced the strength of the interaction but did not abolish it (not shown). Importantly, the yeast cells expressing the last 245 amino acids of the CPSF-73 and the full-length RC-74 were not able to grow in the presence of 10 mM 3-AT, confirming that these two proteins do not interact with each other (Fig. 4B).
We considered the possibility that the RC-68/RC-74 dimer is exchangeable with the CPSF-73/CPSF-100 dimer and by interacting with CPSF-160 forms an alternative CPSF complex involved in the 3′ end processing of a subset of pre-mRNAs. To test this hypothesis, we prepared nuclear extract from HeLa cells and used anti-RC-68 to precipitate RC-68 and associated proteins. We used a nuclear extract rather than a whole-cell lysate made with NP-40 to assure that processing factors present in HeLa cells remain intact. Since it has been reported that prolonged dialysis against buffer D containing 0.2 mM EDTA leads to gradual loss of activity, which could indicate disruption of processing factors (60), the nuclear extract was not dialyzed and instead adjusted to 100 mM KCl by adding buffer D lacking any salt. The anti-RC-68 antibody precipitated significant amounts of RC-68 from the nuclear extract (Fig. 4C, top, lane 2). As detected by Western blotting RC-74 was coprecipitated with RC-68 (Fig. 4C, middle, lane 2). We tested the same immunoprecipitates for the presence of CPSF-160 using anti-CPSF-160. The antibody (kindly provided by W. Keller, University of Basel) detected CPSF-160 in an aliquot of the nuclear extract from HeLa cells (Fig. 4C, bottom, lane 1) but not in the RC-68 precipitates collected from a 20-fold-larger amount of the extract (Fig. 4C, bottom, lane 2). We repeated this experiment with a dialyzed mouse nuclear extract and also did not observe any CPSF-160 in the RC-68 precipitates (not shown). Since it was possible that the anti-RC-68 very inefficiently precipitates the RC-68/RC-74 dimer when associated with CPSF-160, we also prepared a nondialyzed nuclear extract from HeLa cells stably expressing the HA tagged RC-74 and used the anti-HA to precipitate HA/RC-74 and associated proteins. As a negative control in the immunoprecipitation we used an anti-FLAG antibody. The anti-HA and the anti-FLAG precipitates were tested for the presence of RC-68 and CPSF-160 by Western blotting with antibodies specific to each protein. Only background amounts of each protein were detected in the anti-FLAG immunoprecipitates (Fig. 4D, lane 2). However, the anti-HA specifically coprecipitated RC-68 but not CPSF-160, further supporting the notion that the RC-68/RC-74 dimer does not form a complex with this CPSF subunit (Fig. 4D, lane 3). We conclude that collectively these experiments argue against the possibility that RC-68 and RC-74 form a complex with CPSF-160.
Depletion of RC-68 in HeLa cells by siRNA.
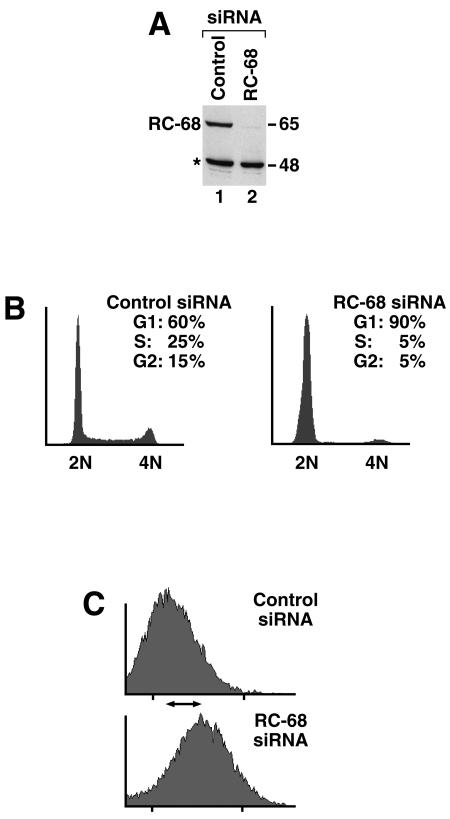
We used RNAi to reduce the in vivo level of RC-68 in HeLa cells to determine whether it plays an essential role in the cell. A specific siRNA directed to the RC-68 mRNA reproducibly reduced expression of RC-68 in HeLa cells to about 5% of its normal level (Fig. 5A and 6A, lane 2) but did not affect the level of the cross-reacting protein migrating at 48 kDa. During the entire treatment time the cells showed very limited cell division and by the end of the treatment had ceased to divide. A second siRNA targeted to a different region of the RC-68 mRNA produced a comparable reduction of RC-68 expression, leading to the same phenotypic changes (not shown), while a control siRNA had no effect (Fig. 5A and 6A, lane 1). As determined by flow cytometry, at least 90% of cells treated with the specific siRNA were arrested in G1 while HeLa cells treated with a control siRNA displayed the typical cell cycle profile, with about 60% of cells in G1 (Fig. 5B). Visually, cells treated with RC-68 siRNAs appeared significantly larger than control siRNA-treated cells. Flow cytometry analysis demonstrated that the G1-arrested cells were the size of G2 cells (Fig. 5C), indicating that depletion of RC-68 does not prevent cell growth although it does block entry into S phase.
FIG. 5.
RNAi-mediated downregulation of RC-68 results in arrest of HeLa cells in G1 phase. (A) RC-68 expression in HeLa cells treated with either a control siRNA (lane 1) or a specific siRNA targeted to RC-68 (lane 2). Asterisk, 48-kDa protein that cross-reacts with the antibody against RC-68. (B) Analysis of HeLa cells treated with either a control siRNA (left) or a specific siRNA targeted to RC-68 (right) by flow cytometry. x and y axes, DNA content and cell number, respectively. The percentages of cells in each phase of the cell cycle are indicated. (C) Shift in the average sizes of cells in the control population and in the population of cells depleted of RC-68. x and y axes, forward scatter (indicative of the cell size) and cell number, respectively.
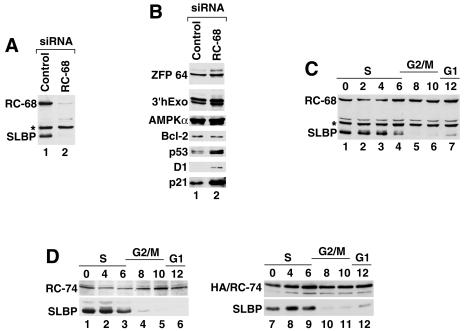
FIG. 6.
(A and B) Expression of selected proteins in RC-68-depleted HeLa cells. Cell lysates prepared from HeLa cells treated with either the control siRNA (lane 1) or the siRNA specific to RC-68 (lane 2) were analyzed by Western blotting with various antibodies. Asterisk, 48-kDa protein interacting with anti-RC-68. (C and D) Whole-cell extracts from HeLa cells synchronized by a double thymidine block were analyzed by Western blotting for RC-68 (C, lanes 1 to 7), RC-74 (D, lanes 1 to 6), or the HA-tagged RC-74 (D, lanes 7 to 12) with anti-RC-68, anti-RC-74, or anti-HA, respectively. Cell cycle regulation of SLBP was analyzed with anti-SLBP.
To determine whether the cells were arrested early or late in G1 phase, we analyzed an extract from cells depleted of RC-68 for the presence of SLBP, a cell cycle-regulated protein involved in controlling histone mRNA metabolism. SLBP accumulates during late G1 phase and is degraded at the end of S phase (75). Since the concentration of SLBP in the cells depleted of RC-68 was also reduced over 95% (Fig. 6A, lane 2), we conclude that the cells were arrested early in G1 phase. The levels of two proteins constitutively expressed during the cell cycle, a novel zinc finger protein migrating at 90 kDa (our unpublished results) and 3′hExo, migrating at 50 kDa (16), were comparable to those in the control cells (Fig. 6B). Among five proteins tested with commercially available antibodies, the levels of Bcl-2 and AMPKα were also unaffected by depletion of RC-68 while p53, p21, and cyclin D1, normally expressed at the highest level in G1 cells (64), were clearly more abundant in the treated cells (Fig. 6B).
To determine whether cell cycle arrest of HeLa cells depleted of RC-68 is a consequence of cell cycle regulation of RC-68 itself, we analyzed the level of this protein in whole-cell and nuclear lysates prepared from synchronized HeLa cells. HeLa cells were collected at different time points after release from the double thymidine block and tested by Western blotting for the presence of RC-68 and SLBP. We determined that RC-68 is expressed at similar levels throughout the cell cycle (Fig. 6C and data not shown). As expected, the level of SLBP remained constant as cells progressed through S phase and rapidly declined at the S/G2 boundary. We also tested expression of RC-74 during the cell cycle by measuring the level of both the endogenous protein using anti-RC-74 (Fig. 6D, lanes 1 to 6) and stably expressed HA-tagged RC-74 using the anti-HA antibody (Fig. 6D, lanes 7 to 12). Expression of this protein also did not change significantly during the cell cycle, while SLBP showed its normal profile of expression, with the highest accumulation in S phase.
DISCUSSION
RC-68 and RC-74 form a complex separate from the CPSF complex.
We identified two unknown human proteins, RC-68 and RC-74, with high similarity to CPSF-73 and CPSF-100 and demonstrated that they form a complex in HeLa cells and mouse myeloma cells. In HeLa cells RC-74 has virtually exclusive nuclear localization whereas RC-68, in addition to being present in the nucleus, is also present in the cytoplasm. All four proteins belong to the β-lactamase superfamily of metal-dependent hydrolases. RC-68 contains the histidine motif typical of all active proteins of the superfamily and is most similar to CPSF-73, while RC-74 lacks the motif and hence seems to be a counterpart of CPSF-100. The existence of two complexes in human cells, CPSF-73/CPSF-100 and RC-68/RC-74, each consisting of a protein containing the typical histidine motif and another protein lacking this motif, is striking and suggests that this structural organization plays an important role in the function of each complex. It is possible that, while CPSF-73 and RC-68 are predicted to contain the active site within each complex, they may require association with another protein of the β-lactamase fold, in which the histidine motif has been disrupted, to adopt a unique structural arrangement important for catalysis. Interestingly, ELAC2, a β-lactamase fold protein involved in 3′ end processing of eukaryotic pre-tRNAs, contains two related sequences in a single polypeptide, one containing the catalytically active histidine motif and the other lacking the histidine motif but retaining similarity in the flanking sequences (68). Further studies will be required to determine the functional significance of such an organization and the role played in each complex by the inactive β-lactamase domain. The orthologues of RC-68 and RC-74 are easily identifiable in all known genomes of plants and animals, but not in fungi. It is likely that the interaction of RC-68 and RC-74 is not limited to human cells and also occurs in all organisms where the two proteins exist.
Using the yeast two-hybrid system we demonstrated that the region of RC-68 interacting with RC-74 is located within the last 124 amino acids. The corresponding region of CPSF-73 is involved in its interaction with CPSF-100 although formation of a stronger complex between the two proteins may require additional portions of CPSF-73 and/or contribution from the three remaining CPSF subunits. The C-terminal regions in RC-68 and CPSF-73 do not have any sequence similarity, and we did not observe any interaction between RC-74 and the C-terminal region of CPSF-73 in the directed yeast two-hybrid system. In addition, we did not detect CPSF-73 in the immunoprecipitates of the HA-tagged RC-74. These results demonstrate that RC-74 and CPSF-73 do not form a mixed pair and also suggest that formation of such a pair between CPSF-100 and RC-68 is unlikely. Immunoprecipitation experiments using both anti-RC-68 and anti-HA demonstrated that the RC-68/RC-74 dimer does not exist in a complex with the largest subunit of the CPSF complex, CPSF-160. Altogether, these results indicate that RC-68 and RC-74 form a specific complex that is independent of the CPSF subunits and that likely plays a role in a process other than the cleavage and polyadenylation of typical cellular pre-mRNAs.
RC-68 is required for cell cycle progression.
Depletion of the intracellular pool of RC-68 by RNAi arrested HeLa cells in G1 phase. HeLa cells are known for their inherent inability to arrest the cell cycle due to frequent activation of alternative bypass pathways. Therefore, the rapid arrest of HeLa cells due to depletion of RC-68 was unexpected and demonstrated that RC-68 plays a critical role in cell cycle regulation. The most likely hypothesis is that reduction of the cellular concentration of RC-68 activates a checkpoint response that allows the RC-68-depleted cells to complete the cell cycle but that prevents them from progressing beyond G1 phase. Unlike cells arrested by aphidicolin or the double thymidine block (75), the cells depleted of RC-68 did not contain SLBP, suggesting that they had arrested relatively early in G1. Even more unexpectedly, depletion of RC-68 did not inhibit growth of HeLa cells, which reached the size typical of G2 cells. Normally, cell growth is required for subsequent progression through the cell cycle, and in cultured cells growth and cell cycle progression are coupled (29). A similar phenotype in which cell growth is uncoupled from cell division was previously observed in untransformed liver cells, with conditional deletion of 40S ribosomal protein S6 (69). Depletion of RC-68 resulted in the accumulation of the G1 regulatory proteins, cyclin D1 and the cyclin-dependent kinase (cdk) inhibitor, p21 (64). The increased level of cyclin D1 is consistent with the finding that in Drosophila this protein is primarily involved in cell growth and not cell cycle regulation (11, 23, 43). There are also reports that cyclin D is required for growth of hepatocytes (50) and for cardiac hypertrophy (6). The increased level of the cdk inhibitor p21 in the RC-68-depleted cells is consistent with the failure of these cells to progress through G1 and enter S phase. Interestingly, RNAi-mediated silencing of CPSF-73 did not affect the cell cycle of the treated cells but instead significantly reduced their growth rate, further indicating that CPSF-73 and RC-68 play different roles in the cell (8).
Possible roles for RC-68 and RC-74.
RC-68 and CPSF-73, in the first 450 amino acids, have 40% identity and 60% similarity, and both contain the histidine motif and so-called β-CASP domain typical of a small group of proteins of the β-lactamase superfamily acting on nucleic acids (7). This group includes Artemis, a protein involved in V(D)J recombination and DNA double-strand break repair (45). Artemis in vitro displays 5′-to-3′ exonuclease activity and in complex with the DNA-dependent protein kinase catalytic subunit acquires endonuclease activity (36). Mutations of the critical residues of the histidine motif and the β-CASP domain predicted to play a role in metal binding and catalysis abolish the in vivo function of Artemis and endonuclease activity in vitro (53, 56). Some of these residues were found to also be essential for the in vivo activity of CPSF-73 (60) and another member of the β-CASP family, SNM1, involved in DNA interstrand cross-link repair (35). Although the definitive evidence is missing, the presence of the histidine motif and the β-CASP domain (7) and recent cross-linking studies (60) strongly implicate CPSF-73 as the 3′ endonuclease cleaving pre-mRNAs prior to addition of the poly(A) tail.
The cellular roles for RC-68 and RC-74 remain unknown, and it is possible that the RC-68/RC-74 complex functions in a process unrelated to RNA metabolism and that the involvement in cell cycle regulation is the only role played by RC-68. However, the strong similarity of RC-68 and RC-74 to CPSF-73 and CPSF-100 suggests that the two new proteins play a role equivalent to that of the CPSF complex and form a distinct endonuclease involved in the 3′ end processing of a subset of pre-mRNAs encoding proteins required for cell cycle progression. There are at least three groups of potential pre-mRNA substrates for RC-68 and RC-74. First, the two proteins may be a part of an alternative CPSF complex containing CPSF-160 and involved in 3′ end processing of a special subset of the AAUAAA-containing pre-mRNAs. However, since our experiments did not detect any CPSF-160 in immunoprecipitates containing both RC-68 and RC-74, we consider this hypothesis rather unlikely. The second group of potential substrates for RC-68 and RC-74 could consist of pre-mRNAs lacking the AAUAAA polyadenylation signal or its sequence variants recognizable by CPSF-160. These pre-mRNAs are found in mammalian cells with a frequency much higher than previously anticipated and may represent as many as 10% of all cellular pre-mRNAs (3, 28, 37, 77). RC-68 and RC-74 could form a complex with a functional equivalent of CPSF-160 capable of recognizing some structural features present in the pre-mRNAs lacking AAUAAA. The third possibility is that the RC-68/RC-74 complex is involved in 3′ end processing of replication-dependent histone pre-mRNAs and therefore plays an essential role in synthesis of histone proteins.
Do RC-68 and RC-74 function as the endonuclease in 3′ end processing of histone pre-mRNAs?
Since synthesis of histones is required for entry into S phase (22, 51), the hypothesis that RC-68 and RC-74 are responsible for cleaving replication-dependent histone pre-mRNAs provides a logical explanation for G1 arrest upon depletion of RC-68. The 3′ end cleavage coupled to polyadenylation and 3′ end cleavage of histone pre-mRNAs, despite many differences in composition of processing signals and trans-acting factors, have several striking similarities, suggesting involvement of related cleavage factors. These similarities include preferred cleavage of both pre-mRNA types after a CA between two closely spaced sequence elements and subsequent generation of a family of downstream products differing in the location of the 5′ end rather than a single product with the 5′ end corresponding to the cleavage site (24, 61, 63, 71). In 3′ end processing of histone pre-mRNAs generation of these downstream products results from activity of a 5′-to-3′ exonuclease associated with the processing machinery (72), and the same mechanism may be responsible for trimming the downstream cleavage product in 3′ end processing of nonhistone pre-mRNAs. As mentioned above, Artemis can act as both an endonuclease and a 5′-to-3′ exonuclease, and it is possible that this rather unusual activity is also shared by the structurally related CPSF-73 and RC-68 and accounts for the postprocessing trimming of the downstream cleavage product.
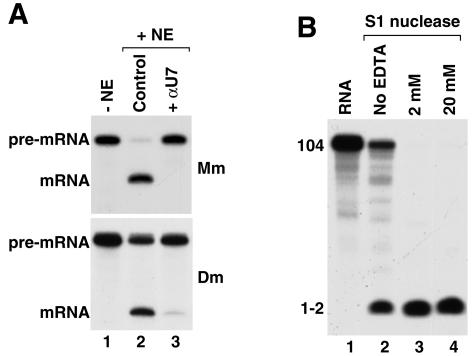
The most important similarity between cleavage that precedes polyadenylation and cleavage of histone pre-mRNAs is generation of a hydroxyl group at the 3′ end of the upstream cleavage product typical of metal-mediated catalysis and the resistance of both reactions to EDTA (17, 24, 25, 30, 44, 61, 63, 71). These features indicate that catalysis in both cases is independent of magnesium and might instead depend on zinc ions. Metal-independent ribonucleases resistant to EDTA, such as RNase A, generate a phosphate at the 3′ end (5). Interestingly, while cleavage preceding polyadenylation is inhibited by concentrations of EDTA higher than 5 mM (30, 60), 3′ end processing of histone pre-mRNAs in mammalian (25) and Drosophila nuclear extracts (17, 18) is extremely refractory to EDTA and proceeds in the presence of 20 mM EDTA (Fig. 7A, lane 2) and can tolerate up to 40 mM EDTA without significant loss of activity (not shown). While the different resistances to EDTA of 3′ end processing of histone pre-mRNAs and 3′ cleavage coupled to polyadenylation could reflect general differences in composition of the respective processing complexes, the failure of 20 mM EDTA to affect 3′ end processing of histone pre-mRNA was difficult to reconcile with the potential role for zinc in catalysis. To address this concern, we tested whether S1 nuclease, a known zinc-dependent enzyme that cleaves phosphodiester bonds in RNA and DNA substrates and generates 3′ OH in the cleavage products (65), is also resistant to high concentrations of EDTA. As shown in Fig. 7B, the ability of S1 nuclease to degrade RNA was in fact stimulated by the presence of 2 and 20 mM EDTA (lanes 3 and 4). The activity of calf intestinal phosphatase, another zinc-dependent enzyme which hydrolyzes phosphoester bonds and generates 3′ OH, was also unaffected by 20 mM EDTA (not shown). These results demonstrated that zinc ions are very tightly bound by both enzymes and are consistent with catalysis of 3′ end processing of histone pre-mRNAs by a zinc-dependent enzyme. We used the anti-RC-68 antibody to directly test whether RC-68 is required for 3′ end processing of histone pre-mRNAs in vitro. However, in spite of using various conditions of immunodepletion, we were unable to sufficiently remove RC-68 from the nuclear extract. The partial depletion did not affect histone pre-mRNA processing.
FIG. 7.
The endonuclease involved in 3′ end processing of histone pre-mRNAs and S1 nuclease are resistant to EDTA. (A) 3′ end processing of histone pre-mRNA in a mouse (Mm; top, lanes 2 and 3) or a Drosophila (Dm) nuclear extract (NE; bottom, lanes 2 and 3) in the presence of 20 mM EDTA. Lane 1, mouse H2a-614 (top) or Drosophila H3 histone pre-mRNA (bottom) labeled at the 5′ end with 32P; lane 2, control 3′ end processing; lane 3, processing in the presence of the specific anti-U7 (αU7) 2′-O-methyl oligonucleotide that blocks the 5′ end of U7 snRNA, thus inhibiting cleavage. (B) Digestion of a 5′ end-labeled 104-nucleotide RNA by S1 nuclease in the presence of the indicated concentrations of EDTA. The band at the bottom of the gel is a mono- or dinucleotide.
Given the essential role of RC-68 in cell cycle progression in HeLa cells it was not surprising that depletion of the Caenorhabditis elegans RC-68 orthologue by RNAi leads to an early lethal phenotype (our unpublished results with E. McCarthy, B. Meier, and Y. Clejan). It has recently been shown that the Arabidopsis orthologue of human RC-68, AtCPSF73-II, is also essential for development (76). Intuitively, the presence of the RC-68 in plants seems to contradict its involvement in the processing of histone pre-mRNAs. In plants, histone mRNAs are polyadenylated and are believed to be generated through the regular cleavage and polyadenylation mechanism (58, 78). However, the mechanism of cleavage and polyadenylation in plants is poorly understood, and processing signals in pre-mRNAs are only weakly conserved, suggesting that in plants histone pre-mRNAs might comprise a separate class of cellular pre-mRNAs that in spite of undergoing cleavage and polyadenylation requires a unique set of trans-acting factors. Future studies should provide important clues to understanding the function of the RC-68/RC-74 complex in both plants and animals and links of RC-68 to cell cycle progression in human cells
Acknowledgments
Z.D. and W.F.M. were supported by NIH grant GM29832. E.J.W. was supported by NCI training grant T32-CA09156 and the Cottrell Foundation.
We thank H. Kelkar (UNC Bioinformatics Center) for help in the database analysis; B. Stefanovic (Florida State University, Tallahassee) for the HeLa cDNA library; and J. Spychala (UNC, Chapel Hill), D. Bentley (University of Colorado), and W. Keller (University of Basel) for antibodies. We are very grateful to E. McCarthy, B. Meier, and Y. Clejan (UNC, Chapel Hill) for RNAi experiments with C. elegans. We also thank L. Levinger (York College/CUNY, Jamaica, N.Y.) for many interesting discussions and critical reading of the manuscript.
REFERENCES
- 1.Aravind, L. 1999. An evolutionary classification of the metallo-beta-lactamase fold proteins. In Silico Biol. 1:69-91. [PubMed] [Google Scholar]
- 2.Barabino, S. M. L., W. Hübner, A. Jenny, L. Minvielle-Sebastia, and W. Keller. 1997. The 30-kD subunit of mammalian cleavage and polyadenylation specificity factor and its yeast homolog are RNA-binding zinc finger proteins. Genes Dev. 11:1703-1716. [DOI] [PubMed] [Google Scholar]
- 3.Beaudoing, E., S. Freier, J. R. Wyatt, J. M. Claverie, and D. Gautheret. 2000. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 10:1001-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bienroth, S., E. Wahle, C. Suter-Crazzolara, and W. Keller. 1991. Purification of the cleavage and polyadenylation factor involved in the 3′-processing of messenger RNA precursors. J. Biol. Chem. 266:19768-19776. [PubMed] [Google Scholar]
- 5.Breslow, R., and W. H. Chapman, Jr. 1996. On the mechanism of action of ribonuclease A: relevance of enzymatic studies with a p-nitrophenylphosphate ester and a thiophosphate ester. Proc. Natl. Acad. Sci. USA 93:10018-10021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Busk, P. K., J. Bartkova, C. C. Strom, L. Wulf-Andersen, R. Hinrichsen, T. E. Christoffersen, L. Latella, J. Bartek, S. Haunso, and S. P. Sheikh. 2002. Involvement of cyclin D activity in left ventricle hypertrophy in vivo and in vitro. Cardiovasc. Res. 56:64-75. [DOI] [PubMed] [Google Scholar]
- 7.Callebaut, I., D. Moshous, J. P. Mornon, and J. P. De Villartay. 2002. Metallo-beta-lactamase fold within nucleic acids processing enzymes: the beta-CASP family. Nucleic Acids Res. 30:3592-3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calzado, M. A., R. Sancho, and E. Munoz. 2004. Human immunodeficiency virus type 1 Tat increases the expression of cleavage and polyadenylation specificity factor 73-kilodalton subunit modulating cellular and viral expression. J. Virol. 78:6846-6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colgan, D. F., and J. L. Manley. 1997. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 11:2755-2766. [DOI] [PubMed] [Google Scholar]
- 10.Daiyasu, H., K. Osaka, Y. Ishino, and H. Toh. 2001. Expansion of the zinc metallo-hydrolase family of the beta-lactamase fold. FEBS Lett. 503:1-6. [DOI] [PubMed] [Google Scholar]
- 11.Datar, S. A., H. W. Jacobs, A. F. De la Cruz, C. F. Lehner, and B. A. Edgar. 2000. The Drosophila cyclin D-Cdk4 complex promotes cellular growth. EMBO J. 19:4543-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dominski, Z., J. A. Erkmann, X. Yang, R. Sanchez, and W. F. Marzluff. 2002. A novel zinc finger protein is associated with U7 snRNP and interacts with the stem-loop binding protein in the histone pre-mRNP to stimulate 3′-end processing. Genes Dev. 16:58-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dominski, Z., and W. F. Marzluff. 1999. Formation of the 3′ end of histone mRNA. Gene 239:1-14. [DOI] [PubMed] [Google Scholar]
- 14.Dominski, Z., and W. F. Marzluff. 2001. Three-hybrid screens for RNA-binding proteins: proteins binding the 3′ end of histone mRNA. Methods Mol. Biol. 177:291-318. [DOI] [PubMed] [Google Scholar]
- 15.Dominski, Z., J. Sumerel, R. J. Hanson, and W. F. Marzluff. 1995. The polyribosomal protein bound to the 3′ end of histone mRNA can function in histone pre-mRNA processing. RNA 1:915-923. [PMC free article] [PubMed] [Google Scholar]
- 16.Dominski, Z., X. Yang, H. Kaygun, and W. F. Marzluff. 2003. A 3′ exonuclease that specifically interacts with the 3′ end of histone mRNA. Mol. Cell. 12:295-305. [DOI] [PubMed] [Google Scholar]
- 17.Dominski, Z., X. Yang, C. S. Raska, C. S. Santiago, C. H. Borchers, R. J. Duronio, and W. F. Marzluff. 2002. 3′ end processing of Drosophila histone pre-mRNAs: requirement for phosphorylated dSLBP and coevolution of the histone pre-mRNA processing system. Mol. Cell. Biol. 22:6648-6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dominski, Z., X. C. Yang, M. Purdy, and W. F. Marzluff. 2003. Cloning and characterization of the Drosophila U7 small nuclear RNA. Proc. Natl. Acad. Sci. USA 100:9422-9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dominski, Z., L.-X. Zheng, R. Sanchez, and W. F. Marzluff. 1999. The stem-loop binding protein facilitates 3′ end formation by stabilizing U7 snRNP binding to the histone pre-mRNA. Mol. Cell. Biol. 19:3561-3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dronkert, M. L., J. de Wit, M. Boeve, M. L. Vasconcelos, H. Van Steeg, T. L. Tan, J. H. Hoeijmakers, and R. Kanaar. 2000. Disruption of mouse SNM1 causes increased sensitivity to the DNA interstrand cross-linking agent mitomycin C. Mol. Cell. Biol. 20:4553-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dubrovsky, E. B., V. A. Dubrovskaya, L. Levinger, S. Schiffer, and A. Marchfelder. 2004. Drosophila RNase Z processes mitochondrial and nuclear pre-tRNA 3′ ends in vivo. Nucleic Acids Res. 32:255-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ewen, M. E. 2000. Where the cell cycle and histones meet. Genes Dev. 14:2265-2270. [DOI] [PubMed] [Google Scholar]
- 23.Frei, C., and B. A. Edgar. 2004. Drosophila cyclin D/Cdk4 requires Hif-1 prolyl hydroxylase to drive cell growth. Dev. Cell 6:241-251. [DOI] [PubMed] [Google Scholar]
- 24.Furger, A., A. Schaller, and D. Schümperli. 1998. Functional importance of conserved nucleotides at the histone RNA 3′ processing site. RNA 4:246-256. [PMC free article] [PubMed] [Google Scholar]
- 25.Gick, O., A. Krämer, W. Keller, and M. L. Birnstiel. 1986. Generation of histone mRNA 3′ ends by endonucleolytic cleavage of the pre-mRNA in a snRNP-dependent in vitro reaction. EMBO J. 5:1319-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorlich, D. 1997. Nuclear protein import. Curr. Opin. Cell Biol. 9:412-419. [DOI] [PubMed] [Google Scholar]
- 27.Gorlich, D., and U. Kutay. 1999. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15:607-660. [DOI] [PubMed] [Google Scholar]
- 28.Graber, J. H., C. R. Cantor, S. C. Mohr, and T. F. Smith. 1999. In silico detection of control signals: mRNA 3′-end-processing sequences in diverse species. Proc. Natl. Acad. Sci. USA 96:14055-14060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grewal, S. S., and B. A. Edgar. 24 April 2003, posting date. Controlling cell division in yeast and animals: does size matter? J. Biol. 2:5. [Online.] http://jbiol.com/content/2/2/5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirose, Y., and J. L. Manley. 1997. Creatine phosphate, not ATP, is required for 3′ end cleavage of mammalian pre-mRNA in vitro. J. Biol. Chem. 272:29636-29642. [DOI] [PubMed] [Google Scholar]
- 31.Jenny, A., H.-P. Hauri, and W. Keller. 1994. Characterization of cleavage and polyadenylation specificity factor and cloning of its 100-kilodalton subunit. Mol. Cell. Biol. 14:8183-8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jenny, A., and W. Keller. 1995. Cloning of cDNAs encoding the 160 kDa subunit of the bovine cleavage and polyadenylation specificity factor. Nucleic Acids Res. 23:2629-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jenny, A., L. Minvielle-Sebastia, P. J. Preker, and W. Keller. 1996. Sequence similarity between the 73-kilodalton protein of mammalian CPSF and a subunit of yeast polyadenylation factor I. Science 274:1514-1517. [DOI] [PubMed] [Google Scholar]
- 34.Kaufmann, I., G. Martin, A. Friedlein, H. Langen, and W. Keller. 2004. Human Fip1 is a subunit of CPSF that binds to U-rich RNA elements and stimulates poly(A) polymerase. EMBO J. 23:616-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li, X., and R. E. Moses. 2003. The beta-lactamase motif in Snm1 is required for repair of DNA double-strand breaks caused by interstrand crosslinks in S. cerevisiae. DNA Repair 2:121-129. [DOI] [PubMed] [Google Scholar]
- 36.Ma, Y., U. Pannicke, K. Schwarz, and M. R. Lieber. 2002. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell 108:781-794. [DOI] [PubMed] [Google Scholar]
- 37.MacDonald, C. C., and J. L. Redondo. 2002. Reexamining the polyadenylation signal: were we wrong about AAUAAA? Mol. Cell. Endocrinol. 190:1-8. [DOI] [PubMed] [Google Scholar]
- 38.MacDonald, C. C., J. Wilusz, and T. Shenk. 1994. The 64-kilodalton subunit of the CstF polyadenylation factor binds to pre-mRNAs downstream of the cleavage site and influences cleavage site location. Mol. Cell. Biol. 14:6647-6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin, F., A. Schaller, S. Eglite, D. Schümperli, and B. Müller. 1997. The gene for histone RNA hairpin binding protein is located on human chromosome 4 and encodes a novel type of RNA binding protein. EMBO J. 16:769-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marzluff, W. F., and R. J. Duronio. 2002. Histone mRNA expression: multiple levels of cell cycle regulation and important developmental consequences. Curr. Opin. Cell Biol. 14:692-699. [DOI] [PubMed] [Google Scholar]
- 41.Marzluff, W. F., M. L. Whitfield, Z. Dominski, and Z.-F. Wang. 1997. Identification of the protein that interacts with the 3′ end of histone mRNA, p. 163-193. In J. D. Richter (ed.), mRNA formation and function. Academic Press, New York, N.Y.
- 42.Melino, S., C. Capo, B. Dragani, A. Aceto, and R. Petruzzelli. 1998. A zinc-binding motif conserved in glyoxalase II, beta-lactamase and arylsulfatases. Trends Biochem. Sci. 23:381-382. [DOI] [PubMed] [Google Scholar]
- 43.Meyer, C. A., H. W. Jacobs, S. A. Datar, W. Du, B. A. Edgar, and C. F. Lehner. 2000. Drosophila Cdk4 is required for normal growth and is dispensable for cell cycle progression. EMBO J. 19:4533-4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moore, C. L., and P. A. Sharp. 1985. Accurate cleavage and polyadenylation of exogenous RNA substrate. Cell 41:845-855. [DOI] [PubMed] [Google Scholar]
- 45.Moshous, D., I. Callebaut, R. De Chasseval, B. Corneo, M. Cavazzana-Calvo, F. Le Deist, I. Tezcan, O. Sanal, Y. Bertrand, N. Philippe, A. Fischer, and J. P. De Villartay. 2001. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell 105:177-186. [DOI] [PubMed] [Google Scholar]
- 46.Mowry, K. L., and J. A. Steitz. 1987. Identification of the human U7 snRNP as one of several factors involved in the 3′ end maturation of histone premessenger RNA's. Science 238:1682-1687. [DOI] [PubMed] [Google Scholar]
- 47.Muller, B., and D. Schumperli. 1997. The U7 snRNP and the hairpin binding protein: key players in histone mRNA metabolism. Semin. Cell Dev. Biol. 8:567-576. [DOI] [PubMed] [Google Scholar]
- 48.Murthy, K. G., and J. L. Manley. 1992. Characterization of the multisubunit cleavage-polyadenylation specificity factor from calf thymus. J. Biol. Chem. 267:14804-14811. [PubMed] [Google Scholar]
- 49.Murthy, K. G. K., and J. L. Manley. 1995. The 160-kD subunit of human cleavage polyadenylation specificity factor coordinates pre-mRNA 3′-end formation. Genes Dev. 9:2672-2683. [DOI] [PubMed] [Google Scholar]
- 50.Nelsen, C. J., D. G. Rickheim, M. M. Tucker, L. K. Hansen, and J. H. Albrecht. 2003. Evidence that cyclin D1 mediates both growth and proliferation downstream of TOR in hepatocytes. J. Biol. Chem. 278:3656-3663. [DOI] [PubMed] [Google Scholar]
- 51.Nelson, D. M., X. Ye, C. Hall, H. Santos, T. Ma, G. D. Kao, T. J. Yen, J. W. Harper, and P. D. Adams. 2002. Coupling of DNA synthesis and histone synthesis in S-phase independent of cyclin/cdk2 activity. Mol. Cell. Biol. 22:7459-7472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Osley, M. A. 1991. The regulation of histone synthesis in the cell cycle. Annu. Rev. Biochem. 60:827-861. [DOI] [PubMed] [Google Scholar]
- 53.Pannicke, U., Y. Ma, K. P. Hopfner, D. Niewolik, M. R. Lieber, and K. Schwarz. 2004. Functional and biochemical dissection of the structure-specific nuclease ARTEMIS. EMBO J. 23:1987-1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pillai, R. S., M. Grimmler, G. Meister, C. L. Will, R. Luhrmann, U. Fischer, and D. Schumperli. 2003. Unique Sm core structure of U7 snRNPs: assembly by a specialized SMN complex and the role of a new component, Lsm11, in histone RNA processing. Genes Dev. 17:2321-2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pillai, R. S., C. L. Will, R. Lührmann, D. Schümperli, and B. Müller. 2001. Purified U7 snRNPs lack the Sm proteins D1 and D2 but contain Lsm10, a new 14 kDa Sm D1-like protein. EMBO J. 20:5470-5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Poinsignon, C., D. Moshous, I. Callebaut, R. De Chasseval, I. Villey, and J. P. De Villartay. 2004. The metallo-{beta}-lactamase/{beta}-CASP domain of Artemis constitutes the catalytic core for V(D)J recombination. J. Exp. Med. 199:315-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Proudfoot, N. J., A. Furger, and M. J. Dye. 2002. Integrating mRNA processing with transcription. Cell 108:501-512. [DOI] [PubMed] [Google Scholar]
- 58.Rothnie, H. M. 1996. Plant mRNA 3′-end formation. Plant Mol. Biol. 32:43-61. [DOI] [PubMed] [Google Scholar]
- 59.Ruegsegger, U., K. Beyer, and W. Keller. 1996. Purification and characterization of human cleavage factor Im involved in the 3′ end processing of messenger RNA precursors. J. Biol. Chem. 271:6107-6113. [DOI] [PubMed] [Google Scholar]
- 60.Ryan, K., O. Calvo, and J. L. Manley. 2004. Evidence that polyadenylation factor CPSF-73 is the mRNA 3′ processing endonuclease. RNA 10:565-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scharl, E. C., and J. A. Steitz. 1994. The site of 3′ end formation of histone messenger RNA is a fixed distance from the downstream element recognized by the U7 snRNP. EMBO J. 13:2432-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schiffer, S., S. Rösch, and A. Marchfelder. 2002. Assigning a function to a conserved group of proteins: the tRNA 3′-processing enzymes. EMBO J. 21:2769-2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sheets, M. D., P. Stephenson, and M. P. Wickens. 1987. Products of in vitro cleavage and polyadenylation of simian virus 40 late pre-mRNAs. Mol. Cell. Biol. 7:1518-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sherr, C. J., and J. M. Roberts. 1999. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 13:1501-1512. [DOI] [PubMed] [Google Scholar]
- 65.Shishido, K., and N. Habuka. 1986. Purification of S1 nuclease to homogeneity and its chemical, physical and catalytic properties. Biochim. Biophys. Acta 884:215-218. [DOI] [PubMed] [Google Scholar]
- 66.Strub, K., G. Galli, M. Busslinger, and M. L. Birnstiel. 1984. The cDNA sequences of the sea urchin U7 small nuclear RNA suggest specific contacts between histone mRNA precursor and U7 RNA during RNA processing. EMBO J. 3:2801-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Takaku, H., A. Minagawa, M. Takagi, and M. Nashimoto. 2003. A candidate prostate cancer susceptibility gene encodes tRNA 3′ processing endoribonuclease. Nucleic Acids Res. 31:2272-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tavtigian, S. V., J. Simard, D. H. Teng, V. Abtin, M. Baumgard, A. Beck, N. J. Camp, A. R. Carillo, Y. Chen, P. Dayananth, M. Desrochers, M. Dumont, J. M. Farnham, D. Frank, C. Frye, S. Ghaffari, J. S. Gupte, R. Hu, D. Iliev, T. Janecki, E. N. Kort, K. E. Laity, A. Leavitt, G. Leblanc, J. McArthur-Morrison, A. Pederson, B. Penn, K. T. Peterson, J. E. Reid, S. Richards, M. Schroeder, R. Smith, S. C. Snyder, B. Swedlund, J. Swensen, A. Thomas, M. Tranchant, A. M. Woodland, F. Labrie, M. H. Skolnick, S. Neuhausen, J. Rommens, and L. A. Cannon-Albright. 2001. A candidate prostate cancer susceptibility gene at chromosome 17p. Nat. Genet. 27:172-180. [DOI] [PubMed] [Google Scholar]
- 69.Volarevic, S., M. J. Stewart, B. Ledermann, F. Zilberman, L. Terracciano, E. Montini, M. Grompe, S. C. Kozma, and G. Thomas. 2000. Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science 288:2045-2047. [DOI] [PubMed] [Google Scholar]
- 70.Wagner, E. J., and M. A. Garcia-Blanco. 2002. RNAi-mediated PTB depletion leads to enhanced exon definition. Mol. Cell 10:943-949. [DOI] [PubMed] [Google Scholar]
- 71.Wahle, E., and W. Keller. 1996. The biochemistry of polyadenylation. Trends Biochem. Sci. 21:247-250. [PubMed] [Google Scholar]
- 72.Walther, T. N., K. T. Wittop, D. Schümperli, and B. Muller. 1998. A 5′-3′ exonuclease activity involved in forming the 3′ products of histone pre-mRNA processing in vitro. RNA 4:1034-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang, Z., W. Fast, A. M. Valentine, and S. J. Benkovic. 1999. Metallo-beta-lactamase: structure and mechanism. Curr. Opin. Chem. Biol. 3:614-622. [DOI] [PubMed] [Google Scholar]
- 74.Wang, Z.-F., M. L. Whitfield, T. I. Ingledue, Z. Dominski, and W. F. Marzluff. 1996. The protein which binds the 3′ end of histone mRNA: a novel RNA-binding protein required for histone pre-mRNA processing. Genes Dev. 10:3028-3040. [DOI] [PubMed] [Google Scholar]
- 75.Whitfield, M. L., L.-X. Zheng, A. Baldwin, T. Ohta, M. M. Hurt, and W. F. Marzluff. 2000. Stem-loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol. Cell. Biol. 20:4188-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xu, R., X. Ye, and Q. Q. Li. 2004. AtCPSF73-II gene encoding an Arabidopsis homolog of CPSF 73 kDa subunit is critical for early embryo development. Gene 324:35-45. [DOI] [PubMed] [Google Scholar]
- 77.Zarudnaya, M. I., I. M. Kolomiets, A. L. Potyahaylo, and D. M. Hovorun. 2003. Downstream elements of mammalian pre-mRNA polyadenylation signals: primary, secondary and higher-order structures. Nucleic Acids Res. 31:1375-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhao, J., L. Hyman, and C. Moore. 1999. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63:405-445. [DOI] [PMC free article] [PubMed] [Google Scholar]