Hydrogen sulfide (H2S) is a toxic molecule and a recently described gasotransmitter in vertebrates whose function in bacteria is not well understood. In this work, we describe the transcriptomic response of the major human pathogen Staphylococcus aureus to quantified changes in levels of cellular organic reactive sulfur species, which are effector molecules involved in H2S signaling. We show that nitroxyl (HNO), a recently described signaling intermediate proposed to originate from the interplay of H2S and nitric oxide, also induces changes in cellular sulfur speciation and transition metal homeostasis, thus linking sulfide homeostasis to an adaptive response to antimicrobial reactive nitrogen species.
KEYWORDS: hydrogen sulfide, nitric oxide, nitroxyl, persulfide, reactive nitrogen species, reactive sulfur species, transcriptomics
ABSTRACT
Staphylococcus aureus is a commensal human pathogen and a major cause of nosocomial infections. As gaseous signaling molecules, endogenous hydrogen sulfide (H2S) and nitric oxide (NO·) protect S. aureus from antibiotic stress synergistically, which we propose involves the intermediacy of nitroxyl (HNO). Here, we examine the effect of exogenous sulfide and HNO on the transcriptome and the formation of low-molecular-weight (LMW) thiol persulfides of bacillithiol, cysteine, and coenzyme A as representative of reactive sulfur species (RSS) in wild-type and ΔcstR strains of S. aureus. CstR is a per- and polysulfide sensor that controls the expression of a sulfide oxidation and detoxification system. As anticipated, exogenous sulfide induces the cst operon but also indirectly represses much of the CymR regulon which controls cysteine metabolism. A zinc limitation response is also observed, linking sulfide homeostasis to zinc bioavailability. Cellular RSS levels impact the expression of a number of virulence factors, including the exotoxins, particularly apparent in the ΔcstR strain. HNO, like sulfide, induces the cst operon as well as other genes regulated by exogenous sulfide, a finding that is traced to a direct reaction of CstR with HNO and to an endogenous perturbation in cellular RSS, possibly originating from disassembly of Fe-S clusters. More broadly, HNO induces a transcriptomic response to Fe overload, Cu toxicity, and reactive oxygen species and reactive nitrogen species and shares similarity with the sigB regulon. This work reveals an H2S/NO· interplay in S. aureus that impacts transition metal homeostasis and virulence gene expression.
IMPORTANCE Hydrogen sulfide (H2S) is a toxic molecule and a recently described gasotransmitter in vertebrates whose function in bacteria is not well understood. In this work, we describe the transcriptomic response of the major human pathogen Staphylococcus aureus to quantified changes in levels of cellular organic reactive sulfur species, which are effector molecules involved in H2S signaling. We show that nitroxyl (HNO), a recently described signaling intermediate proposed to originate from the interplay of H2S and nitric oxide, also induces changes in cellular sulfur speciation and transition metal homeostasis, thus linking sulfide homeostasis to an adaptive response to antimicrobial reactive nitrogen species.
INTRODUCTION
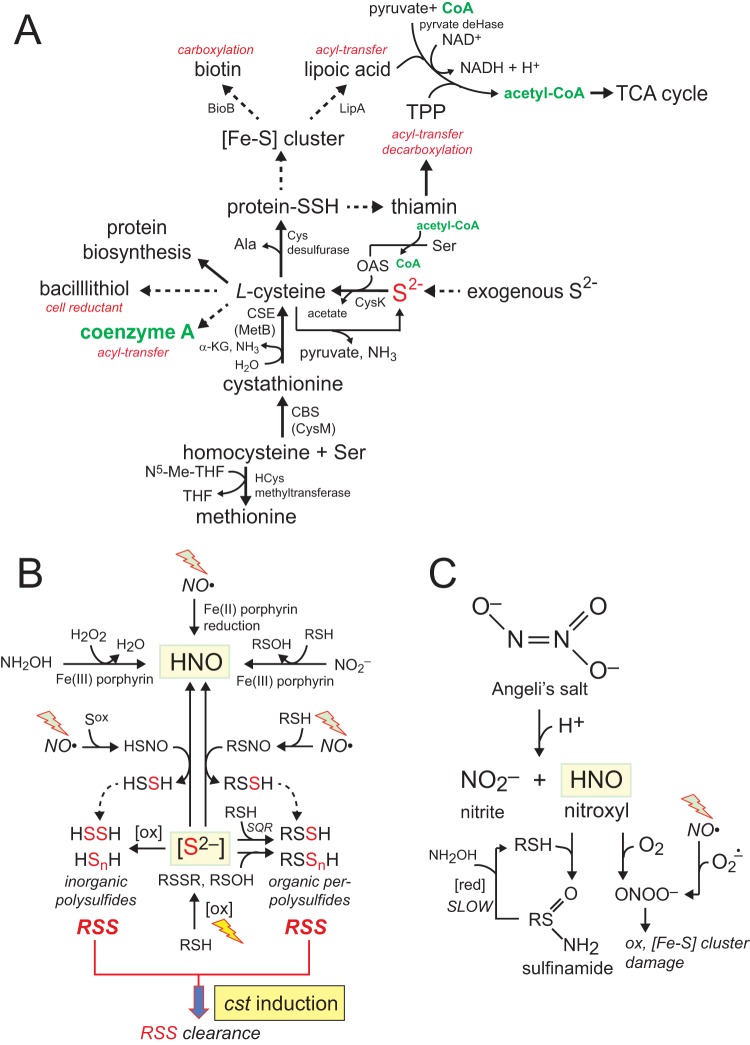
Hydrogen sulfide (H2S) is both a toxic gas and a substrate for the biosynthesis of cysteine, an essential amino acid required for the synthesis of low-molecular-weight (LMW) thiols and the biogenesis of iron-sulfur (Fe-S) proteins (1, 2). Cysteine and H2S are also metabolic precursors for methionine and other sulfur-containing enzyme cofactors and thus significantly impact a wide range of metabolic activities in the cell (Fig. 1A). In bacteria, including the human pathogen Staphylococcus aureus, the cysteine pool is controlled by the global regulator CymR, which impacts sulfur metabolism, virulence gene expression, and survival of the organism both inside and outside the host (3–5). Sulfide (the term which we use here to refer collectively to H2S, HS−, and S2−) can be assimilated from exogenous sulfur sources or produced endogenously primarily by enzymes of the transsulfuration pathway, including cystathionine-β-synthase (CBS) and cystathionine-γ-lyase (CSE) (Fig. 1A). The toxicity of H2S coupled with its beneficial functions suggests a need for sulfide homeostasis, in which the cellular concentrations of sulfide and sulfide-derived reactive sulfur species are tightly regulated.
FIG 1 .
Sulfide (S2−) homeostasis and small-molecule chemistry that couples reactive sulfur species (RSS) and reactive nitrogen species (RNS). (A) Sulfide (S2−, HS−, and H2S) and O-acetyl-l-serine (OAS) are substrates for cysteine synthase (CysK) to form l-cysteine. l-Cysteine is the biosynthetic precursor to the major cellular reducing thiol in S. aureus, bacillithiol (BSH), and to coenzyme A (CoASH), which plays important roles in acyl-transfer reactions. l-Cysteine is also the precursor to other major sulfur-containing cofactors, including [Fe-S] clusters, biotin, lipoic acid, and thiamine pyrophosphate (TPP). Lipoic acid and TPP function in the pyruvate dehydrogenase (pyruvate deHase) complex (upper right), used to synthesize acetyl-CoA, which is fed into the tricarboxylic acid (TCA) cycle and other cellular processes. S2– can be accumulate in cells either from exogenous sulfide sources or endogenously via the activity of two enzymes in a transsulfuration pathway, cystathionine β-synthase (CBS) and cystathionine γ-lyase (CSE), which converts homocysteine to l-cysteine. (B) Nitroxyl (HNO) defines an intersection of sulfide, LMW thiols (RSH), reactive sulfur species (RSS), reactive oxygen species (ox), and nitric oxide (NO·). Sulfide can lead to the accumulation of inorganic polysulfides (HSnH) and of organic persulfides (n = 1) and polysulfides (RSSnH), collectively termed reactive sulfur species (RSS), for which there is evidence in mammalian cells (16) and bacterial cells (26). These RSS are sensed by CstR (29), leading to the upregulation of a sulfide oxidation system that includes a canonical sulfide:quinone oxidoreductase (SQR) (26, 81). The RSS can also be derived from exogenous or endogenous NO·, which reacts with oxidized sulfur species (Sox, RSox) to create the nitrosothiol thionitrous acid (HSNO) and organic nitrosothiols (RSNO), which in turn react with S2– to make HNO (82-86). HNO can also be made via 1-electron reduction of NO· or other transformations (top of figure; note that not all possible reactions are shown [85]). (C) Angeli’s salt (AS) is an HNO donor (34) which, at pH values between 4 and 8, decomposes to HNO (pKa, 11.4) (87) and nitrite (NO2−). HNO further reacts with O2 to create the potent oxidant peroxynitrite (ONOO−), which in turn can react with S2– of [Fe-S] clusters to give perthionitrite (SSNO−) (83), thus leading to the decomposition of protein-bound [Fe-S] clusters. SSNO− is unstable in aqueous solution and may regenerate HNO (83). HNO is also a highly reactive electrophile at neutral pH that reacts with protein thiols to form sulfinamides in aqueous solution (56); this reaction is in competition with HNO dimerization to form N2O and H2O (66) (not shown).
H2S freely passes through cell membranes, unlike its deprotonated conjugate bases (HS− and S2−), and this property underscores recent suggestions that H2S, like nitric oxide (NO·) and carbon monoxide (CO), is a vertebrate gasotransmitter that functions as an endothelium-derived vasorelaxer (6, 7). Recent findings in mammalian cells suggest that H2S and NO· functionally interact to form nitroxyl (HNO), the one-electron reduced and protonated form of NO·, which controls the activity of the sensory chemoreceptor channel TRPA1 (8–10) (Fig. 1B). Likewise, in bacterial systems, H2S (11) and NO· (12) are reported to act synergistically to protect a number of bacterial strains against antibiotic stress, a finding that originates with the bacterially encoded nitric oxide synthases which are generally protective against immune oxidative attack (13, 14). In vitro experiments firmly establish that this cross talk between H2S and NO· leads to the formation of HNO as a primary species, as well as of thiol persulfides (RSS−) and organic (RSSn−) and inorganic (HSn−) polysulfides (n > 1) as bioactive products (Fig. 1B) (15). These per- and polysulfide species, collectively termed reactive sulfur species (RSS), are reportedly maintained in relatively high concentrations (0.01 to 0.1 mM) in mammalian cells and are proposed to function as true small-molecule signaling species in an H2S signaling pathway (16).
Although our understanding of the biological impact of HNO is not yet complete, HNO appears to be characterized by a chemical reactivity and biogenesis profile that is distinct from that of NO·. It is thought that the reduction of NO· by specific Fe(II)-heme and high-spin Fe(II) or Mn(II) complexes makes them potential sources of endogenous HNO in living cells (17). HNO is a highly reactive and short-lived molecule that self-quenches via dimer formation to yield nitrous oxide (N2O) (18). However, prior to self-quenching, this potent electrophile is capable of reacting with nucleophilic thiolate groups to form sulfinamides (Fig. 1C) (19, 20). In addition, while HNO reacts only slowly with molecular oxygen at neutral pH to generate peroxynitrite (ONOO−) (21), ONOO– is formed from NO· and superoxide anion at diffusion-controlled rates (Fig. 1C) (22, 23). Thus, the presence of multiple reactive nitrogen species (RNS) and reactive oxygen species (ROS) at the host-pathogen interface may lead to the formation of ONOO−, which is a strong oxidant that causes lipid peroxidation, nitration of aromatic residues, thiol oxidation, and disassembly of Fe-S clusters (24) (Fig. 1C).
There is emerging evidence that endogenously produced sulfide and a pool of RSS and perhaps downstream products (Fig. 1B) protect cells against the effects of ROS (16, 25, 26). Indeed, proteome S-sulfhydration might constitute a reservoir of persulfidated cysteines while driving up- and downregulation of metabolic pathways to provide protection against oxidative stress (16, 25, 27). In this model, cells must be capable of both biosynthesis and clearance of RSS to maintain sulfide homeostasis. In previous work, we discovered and characterized a novel per- and polysulfide-sensing dithiol-containing repressor from the major nosocomial pathogen S. aureus, named CstR (for "CsoR-like sulfurtransferase repressor") (28, 29). CstR represses transcription of cstA, cstB, and sqr, which together encode an H2S oxidation system (S2– to thiosulfate, S2O32−) (26, 30, 31). Sulfide-responsive and RSS-sensing repressors that are functionally analogous to CstR and yet are structurally unrelated are also found and have recently been characterized in Gram-negative bacteria, suggesting that sulfide homeostasis may be more widespread than previously anticipated (32, 33).
In this work, we employed transcriptomic and organic persulfide metabolite profiling approaches to investigate the cellular response of S. aureus to sodium sulfide and a commonly used HNO donor, Angeli’s salt (AS) (34) (Fig. 1C), added exogenously to cells. We found that sulfide treatment induces the cst operon as expected (29) and strongly downregulates a subset of genes regulated by CymR, the master regulator of cysteine biosynthesis (3, 5). We further show that AS treatment bears some similarity to sulfide stress, a finding directly attributable to HNO and not nitrite (NO2−) (29). Both sulfide stress and HNO also result in a significant zinc starvation response, while the ΔcstR strain, characterized by lower levels of RSS, shows significant repression of the expression of immunomodulators and superantigen toxins, some of which are controlled by the global virulence regulator MgrA (35). The implications of these findings for the biological pathway(s) influenced by sulfide/RSS signaling and H2S/HNO interplay in virulence gene expression and transition metal bioavailability are discussed.
RESULTS AND DISCUSSION
Transcriptomic analysis of the effects of exogenous sulfide stress and a ΔcstR strain.
Previous work showed that addition of sodium sulfide to S. aureus cells aerobically grown to the early log phase resulted in the transient induction of the cst operon and massive accumulation of organic LMW persulfides in cells (26, 29). In contrast, unregulated expression of genes in a cst operon in a ΔcstR stain reduced the levels of these cellular RSS below those seen with wild-type cells (see below). In order to understand the impact of cellular sulfide and RSS concentrations on global gene expression, we carried out a transcriptomic analysis of mid-log S. aureus strain Newman treated with 0.2 mM NaHS for 10 min versus an untreated ΔcstR strain and of both relative to untreated wild-type cells (see Table S1A and Fig. S1 in the supplemental material). Totals of 38 genes and 37 genes, respectively, exhibited an increase or decrease in expression of more than 3.0-fold (1 standard deviation from the mean induction level; see Table S1A) in sulfide-treated cells and in untreated wild-type cells (Tables 1 and 2, respectively). These genes included cstA, cstB, and sqr, all direct targets of CstR regulation (29), as well as a number of enzymes and regulators involved in sugar (glpF, marR, gapB, scrR, gntK, and gntR) and amino acid (putA) metabolism.
TABLE 1 .
List of genes upregulated in response to NaHS stress (≥3-fold) or by Angeli’s salt (≥10-fold)
| Locus tag (Newman) |
Locus tag (N315) |
Gene name |
Fold induction (S2−) |
Fold induction (Angeli’s salt)a |
Function | Plus CP treatmentb (fold increase)? |
ΔcstR regulation? |
|---|---|---|---|---|---|---|---|
| NWMN_0026 | SA0080 | tauE | 5.5 | Putative sulfonate/thiosulfate (TS) efflux | Yes | ||
| NWMN_0027 | SA0082 | cstA | 6.1 | 11.5 | Multidomain sulfurtransferasea | Yes | |
| NWMN_0028 | SA0083 | cstB | 5.8 | 7.5 | Persulfide dioxygenase-sulfur transferaseb | Yes | |
| NWMN_0029 | SA0084 | sqr | 4.9 | 6.4 | Sulfide-quinone reductaseb | Yes | |
| NWMN_0047 | SA0098 | 15.0 | Hypothetical | ||||
| NWMN_0048 | SA0099 | 14.1 | Hypothetical | ||||
| NWMN_0071 | SA0122 | 2.0c | 12.2 | Acetoin reductase | |||
| NWMN_0113 | SA0162 | aldA | 4.8 | 56.6 | Aldehyde dehydrogenase-like | Yes | |
| NWMN_0134 | SA0184 | 4.4 | 7.2 | Hypothetical (NWMN_0134–0137; carbohydrate metabolism) | |||
| NWMN_0151 | SA0206 | 8.6 | 7.5 | Hypothetical (NWMN_0151–0154 operon) | Yes | ||
| NWMN_0162 | SA0218 | pflB | 8.1 | Formate acetyltransferase | |||
| NWMN_0163 | SA0219 | 19.1 | Formate-lyase activating enzyme | ||||
| NWMN_0173 | SA0229 | 2.2 | 7.5 | Hypothetical | |||
| NWMN_0174 | SA0230 | 2.1 | 16.0 | Hypothetical | Yes (3.4) | ||
| NWMN_0175 | SA0231 | 2.2 | 20.7 | Flavohemoprotein | Yes (2.9) | ||
| NWMN_0330 | SA0326 | 8.0 | Glyoxalase-like protein | ||||
| NWMN_0331 | SA0327 | 9.2 | Luciferase-like monooxygenase | ||||
| NWMN_0332 | SA0328 | 9.2 | NADH-dependent flavin mononucleotide (FMN) reductase (like NWMN_2421) | ||||
| NWMN_0371 | SA0365 | 5.7* | Alkyl hydroperoxide reductase subunit F | ||||
| NWMN_0372 | SA0366 | 5.8* | Alkyl hydroperoxide reductase subunit C | ||||
| NWMN_0373 | SA0367 | 13.7 | Nitroreductase family protein | ||||
| NWMN_0410 | SA0396 | lpl7nm | 4.0 | Tandem lipoprotein (NWMN_0410–0411 operon) | |||
| NWMN_0417 | SA0410 | cobW | 7.2 | Putative COG0523 GTPase | |||
| NWMN_0418 | SA0411 | ndhF | 3.6 | 6.6 | Putative NADH dehydrogenase subunit 5 | ||
| NWMN_0484 | SA0480 | ctsR | 16.3 | Transcriptional regulator CtsR (stress induced) | |||
| NWMN_0485 | SA0481 | 20.6 | UvrB/UvrC motif-containing protein | ||||
| NWMN_0486 | SA0482 | 16.1 | ATP:guanido phosphotransferase | ||||
| NWMN_0487 | SA0483 | clpC | 12.7 | ClpC protease, ATP-binding subunit | |||
| NWMN_0587 | SA0572 | 15.5 | Hypothetical | ||||
| NWMN_0669 | SA0655 | fruA | 14.2 | Fructose permease (NWMN_0667–0669, frc operon) | |||
| NWMN_0815 | SA0806 | 17.5 | 8.4 | NADH-dependent pyridine nucleotide disulfide oxidoreductase (short-chain dehydrogenase/reductase [SDR]) | Yes (229) | ||
| NWMN_0826 | SA0817 | 13.1 | NADH-dependent flavin oxidoreductase | ||||
| NWMN_0845 | SA0835 | 28.2 | ClpB protease, ATP binding subunit | ||||
| NWMN_0900 | 3.2 | Hypothetical (57 residues) | |||||
| NWMN_1084 | 3.5 | −4.1 | Anti-protein (44 residues) | ||||
| NWMN_1207 | SA1140 | glpF | 4.3 | 2.1 | Glycerol uptake facilitator protein | Yes | |
| NWMN_1224 | 3.4 | Hypothetical (82 residues) | |||||
| NWMN_1246 | SA1170 | katA | 2.5 | 26.8 | Catalase | ||
| NWMN_1247 | rpmG2 | 6.6 | 3.6 | Non-Zn-containing paralog rpmG | Yes (2.0) | ||
| NWMN_1248 | SA1171 | rpsN2 | 9.1 | 4.2 | Non-Zn-containing paralog rpsN (S14) | Yes (27.3) | |
| NWMN_1415 | SA1339 | marR | 6.3 | 4.5 | Putative PurR/LacI family repressor | Yes | |
| NWMN_1483 | SA1409 | dnaJ | 9.1 | Molecular chaperone DnaK (hsp70) | |||
| NWMN_1484 | SA1410 | grpE | 8.9 | Heat shock protein GrpE | |||
| NWMN_1485 | SA1411 | hrcA | 11.1 | Heat-inducible transcriptional repressor | |||
| NWMN_1580 | SA1510 | gapB | 3.7 | 3.9 | glyceraldehyde-3-phosphate dehydrogenase 2 | ||
| NWMN_1658 | SA1585 | putA | 5.8 | 17.1 | Proline dehydrogenase | Yes | |
| NWMN_1709 | bsaG | 15.2 | ABC transporter protein: lantibiotic | ||||
| NWMN_1710 | bsaE | 9.5 | ABC transporter protein: lantibiotic | ||||
| NWMN_1711 | bsaF | 8.6 | ABC transporter protein: lantibiotic | ||||
| NWMN_1929 | SA1814 | dapE | 9.5 | Succinyl-diaminopimelate desuccinylase (dinuclear) | |||
| NWMN_1949 | SA1847 | scrR | 3.1 | 4.0 | Sucrose operon repressor | ||
| NWMN_2026 | SA1924 | 11.3 | Aldehyde dehydrogenase family | ||||
| NWMN_2043 | SA1941 | 25.8 | Dps; nonheme Fe-containing ferritin | Yes (3.0) | |||
| NWMN_2044 | SA1942 | 8.0 | Hypothetical; predicted disulfide oxidoreductase | Yes (2.9) | |||
| NWMN_2048 | SA1946 | 19.1 | Hypothetical | ||||
| NWMN_2059 | SA1962 | mtlA | 14.7 | Mannitol-specific IIA component (2057–2060 operon) | |||
| NWMN_2060 | SA1963 | mtlD | 15.4 | Mannitol-1-phosphate 5-dehydrogenase | |||
| NWMN_2074 | SAS074 | 6.3 | 5.9 | Hypothetical | Yes | ||
| NWMN_2086 | SA1984 | 15.1 | Alkaline shock protein 23 | ||||
| NWMN_2087 | SA1985 | 18.3 | Hypothetical (79 residues; COG5547) | ||||
| NWMN_2088 | SA1986 | 11.8 | Hypothetical | ||||
| NWMN_2091 | SA1989 | 10.9 | Hypothetical (quinone oxidoreductase) | ||||
| NWMN_2109 | SA2006 | 12.2 | Truncated MHC class II analog protein | ||||
| NWMN_2180 | SA2075 | 13.3 | Formate dehydrogenase accessory protein | ||||
| NWMN_2209 | SA2101 | 10.7 | Hypothetical | ||||
| NWMN_2210 | SA2102 | 5.15 | Formate dehydrogenase-like | ||||
| NWMN_2229 | SA2119 | 15.2 | Hypothetical | ||||
| NWMN_2273 | SA2161 | 13.6 | Acetyltransferase, GNAT family protein | ||||
| NWMN_2274 | SA2162 | 13.6 | Pyridine nucleotide-disulfide oxidoreductase (TrxB-like) | Yes (3.4) | |||
| NWMN_2282 | SA2170 | 12.3 | Hypothetical | ||||
| NWMN_2306 | SA2184 | 32.0 | Zinc-binding lipoprotein, AdcA-like | Yes (12.1) | |||
| NWMN_2359 | SA2250 | cntE | 5.6 | Major facilitator superfamily (MFS) | Yes (66.1) | ||
| NWMN_2360 | SA2251 | cntF | 7.2 | ABC transporter; cobalt-nickel | Yes (108) | ||
| NWMN_2361 | SA2252 | cntD | 7.9 | ABC transporter; cobalt-nickel | Yes (107) | ||
| NWMN_2362 | SA2253 | cntC | 9.0 | ABC transporter; cobalt-nickel | Yes (104) | ||
| NWMN_2363 | SA2254 | cntB | 6.5 | 3.7 | ABC transporter; cobalt-nickel | Yes (101) | |
| NWMN_2364 | SA2255 | cntA | 6.6 | 5.0 | ABC transporter; cobalt-nickel | Yes (199) | |
| NWMN_2365 | SA2256 | cntM | 13.8 | 10.0 | Hypothetical (NWMN_2367–2365 operon) | Yes (504) | |
| NWMN_2366 | SA2257 | cntL | 12.1 | 15.7 | Hypothetical, epimerase-like | Yes (445) | |
| NWMN_2367 | SA2258 | cntK | 11.2 | 14.2 | Hypothetical | Yes (346) | |
| NWMN_2368 | SA2259 | 8.9 | Hypothetical | ||||
| NWMN_2369 | SA2260 | 15.1 | Short-chain dehydrogenase | ||||
| NWMN_2402 | SA2294 | gntK | 3.1 | 6.0 | Gluconate kinase | Yes | |
| NWMN_2403 | SA2295 | gntR | 6.9 | 5.5 | Gluconate operon repressor | Yes | |
| NWMN_2414 | SA2304 | fbp | 2.1 | 12.2 | Fructose-1,6-bisphosphatase | ||
| NWMN_2434 | SA2323 | 5.0 | Hypothetical | ||||
| NWMN_2435 | SA2324 | 11.0 | Hypothetical | ||||
| NWMN_2436 | SA2325 | 10.5 | Hypothetical | ||||
| NWMN_2456 | SA2343 | 3.0 | Hypothetical (63 residues) | ||||
| NWMN_2457 | SA2344 | copA | 68.3 | Cu(I)-specific P-type ATPase efflux transporter6 | |||
| NWMN_2458 | SA2345 | copZ | 26.1 | Cu(I) chaperone6 | |||
| NWMN_2461 | SA2348 | ctrM | 5.6 | Squalene synthase | |||
| NWMN_2462 | SA2349 | ctrN | 9.0 | Squalene synthase | |||
| NWMN_2463 | SA2350 | 13.6 | Glycosyl transferase, group 2 family protein7 | ||||
| NWMN_2464 | SA2351 | crtI | 4.6 | Phytoene dehydrogenase | |||
| NWMN_2479 | SA2366 | 6.8 | Amidohydrolase family protein | ||||
| NWMN_2480 | SA2367 | 11.5 | α/β hydrolase family protein | ||||
| NWMN_2481 | SA2368 | 2.4d | 1.7 | Putative COG0523 GTPase | Yes (43.2) | ||
| NWMN_2482 | SA2369 | 4.0 | 2.6 | Hypothetical | Yes (83.1) | ||
| NWMN_2483 | SA2370 | 5.5 | 5.0 | FAD-dependent pyridine nucleotide disulfide oxidoreductase | Yes (152) |
TABLE 2 .
List of genes downregulated in response to NaHS stress (≥3-fold) or by Angeli’s salt (≥5-fold)
| Locus tag (Newman) |
Locus tag (N315) |
Gene name |
Fold repression (S2−) |
Fold repression (Angeli’s salt)a |
Function | CymR regulon?b |
ΔcstR regulation? |
|---|---|---|---|---|---|---|---|
| NWMN_0115 | SA0165 | 16.9 | 4.2 | Hypothetical | + | ||
| NWMN_0116 | SA0166 | 34.0 | 5.7 | ABC transporter: putative TauB (sulfonate) | + | ||
| NWMN_0117 | SA0167 | 11.5 | 5.1 | ABC transporter: putative TauA | + | ||
| NWMN_0118 | SA0168 | 13.0 | 6.6 | ABC transporter: putative TauC | + | ||
| NWMN_0119 | SA0169 | 13.7 | 3.6 | Putative CiaA: NAD-dependent formate dehydrogenase | + | ||
| NWMN_0143 | SA0198 | 11.9 | 3.1 | ABC transporter: peptide (ATPase)c | + | −/+ downd | |
| NWMN_0144 | SA0199 | 9.4 | 11.1 | ABC transporter: peptide (permease)c | − | ||
| NWMN_0145 | SA0200 | 11.8 | 9.4 | ABC transporter: peptide (permease)c | − | −/+ downd | |
| NWMN_0146 | SA0201 | rlp | 6.0 | 4.3 | Putative RGD-containing lipoprotein | − | |
| NWMN_0374 | SA0368 | tcyP | 7.5 | 4.4 | Putative cystine transporter homolog | + | |
| NWMN_0391 | SA0385 | ssl4nm | 5.1 | Superantigen-like protein 4 | − | Down | |
| NWMN_0400 | ssl11nm | 3.3 | 5.4 | Superantigen-like protein 11 | − | −/+ downd | |
| NWMN_0401 | 3.4 | 7.1 | Hypothetical | − | −/+ downd | ||
| NWMN_0423 | SA0417 | 2.3e | 5.4 | Sodium-dependent symporter | − | −/+ upd | |
| NWMN_0424 | SA0418 | cysM | 5.7 | Cystathionine-β-synthase (CBS) | + | ||
| NWMN_0425 | SA0419 | metB | 8.5 | Cystathionine-γ-lyase (CSE) | + | ||
| NWMN_0426 | SA0420 | 13.0 | 11.6 | ABC transporter: peptide (ATPase) | − | −/+ downd | |
| NWMN_0427 | SA0421 | 13.9 | 11.9 | ABC transporter: peptide (permease) | − | −/+ downd | |
| NWMN_0428 | SA0422 | 9.9 | 4.1 | ABC transporter: peptide (substrate binding) | − | −/+ downd | |
| NWMN_0475 | SA0471 | cysK | 10.4 | Cysteine synthase (OAS + H2S → acetate + Cys) | + | ||
| NWMN_0557 | SA0551 | 7.5 | Up 4.6 | Putative pyridine nucleotide disulfide oxidoreductase | − | ||
| NWMN_0558 | SA0552 | 16.6 | Rrf2 family repressor (related to B. subtilis SaiR) | − | |||
| NWMN_0655 | SA0641 | mgrA | 5.1 | MarR family repressor MgrA | − | ||
| NWMN_0757 | 2.3e | 28.8 | Secreted von Willebrand factor-binding protein | − | −/+ downd | ||
| NWMN_0759 | 3.0 | Hypothetical | − | −/+ downd | |||
| NWMN_0766 | SA0751 | 4.2 | Hypothetical | − | |||
| NWMN_0813 | SA0804 | 5.8 | Sodium/proton antiporter family protein | − | |||
| NWMN_0831 | SA0821 | argH | 3.0 | Argininosuccinate lyase (Arg biosynthesis) | − | −/+ downd | |
| NWMN_0832 | SA0822 | argG | 6.2 | Argininosuccinate synthase (Arg biosynthesis) | − | −/+ downd | |
| NWMN_0907 | SA0890 | 6.5 | Hypothetical | − | |||
| NWMN_1047 | SA0983 | isdG | 3.1f | Heme-degrading monooxygenase | − | ||
| NWMN_1066 | SA1000 | 5.1 | Hypothetical (fibrinogen binding) | − | |||
| NWMN_1068 | SA1002 | 5.6 | Hypothetical | − | |||
| NWMN_1069 | SA1003 | 3.4 | Hypothetical (fibrinogen binding) | − | |||
| NWMN_1070 | SA1004 | 6.9 | Hypothetical (fibrinogen binding) | − | |||
| NWMN_1352 | SA1275 | 5.0 | Hypothetical | − | |||
| NWMN_1749 | SA1674 | tcyC | 1.8e | 7.1 | ABC transporter: cystine (ATPase)e | + | Down |
| NWMN_1750 | SA1675 | tycB | 1.6e | 6.7 | ABC transporter: cystine (permease)e | + | Down |
| NWMN_1751 | SA1676 | tycA | 1.2e | 4.8 | ABC transporter: cystine (cystine binding)e | + | |
| NWMN_1877 | SA1755 | chp | 32.7 | Chemotaxis-inhibiting protein | − | ||
| NWMN_1951 | SA1849 | 9.8 | 4.4 | TusA-like (SirA/YedF/YeeD) protein | + | ||
| NWMN_1952 | SA1850 | 14.2 | 5.0 | Putative thiosulfate (TS) importer | + | ||
| NWMN_2049 | SA1949 | czrA | 15.9 | Zinc-specific repressor (ArsR family) | − | ||
| NWMN_2050 | SA1950 | czrB | 14.0 | Zinc cation diffusion facilitator (CDF) transporter | − | ||
| NWMN_2075 | SA1976 | 11.8 | Hypothetical | − | |||
| NWMN_2186 | SA2080 | ydbM | 9.3 | Putative CiaA; acyl (butyryl)-CoA dehydrogenase | + | ||
| NWMN_2199 | SA2093 | 2.1e | 4.2 | Secretory antigen precursor SsaA | − | ||
| NWMN_2200 | SA2094 | nhaC | 8.4 | Sodium/proton antiporter family, NhaC | − | ||
| NWMN_2201 | SA2095 | 2.1e | 9.5 | Dehydrogenase family protein | − | ||
| NWMN_2203 | 6.5 | Secretory antigen precursor SsaA | − | ||||
| NWMN_2265 | SA2153 | 5.5 | Hypothetical | − | |||
| NWMN_2311 | SA2200 | 6.1 | ABC transporter: amino acid (ATPase) | − | |||
| NWMN_2312 | SA2201 | 5.3 | ABC transporter: amino acid (permease) | − | |||
| NWMN_2313 | SA2202 | 10.1 | ABC transporter: amino acid (substrate binding) | − | |||
| NWMN_2317 | SA2206 | sbi | 5.0 | Immunoglobulin G-binding protein | − | ||
| NWMN_2470 | SA2357 | 5.2 | Hypothetical (regulatory protein) | − | |||
| NWMN_2472 | SA2359 | 7.0 | Hypothetical (just upstream of dtr) | − | |||
| NWMN_2473 | SA2360 | 5.4 | Hypothetical (just upstream of dtr) | − | −/+ downd | ||
| NWMN_2577 | SA2471 | hisG | 7.9 | ATP phosphoribosyltransferase C subunit (His biosynthesis) | − | ||
| NWMN_2578 | SA2472 | hisZ | 6.0 | ATP phosphoribosyltransferase R subunit (His biosynthesis) | − |
Fold induction shown if S2− induction is ≥3.0-fold.
CymR regulon as determined previously on TSB plus 2.0 mM cysteine (3).
Proposed to be involved in glutathione (GSH) assimilation and degradation (3).
Approximately 2-fold repressed (downregulated [down]) or activated (upregulated [up]) in the ΔcstR strain (see Tables 3 and 4 for a list of genes differentially expressed in the ΔcstR strain), with adjusted P value of ≤0.05.
At least 2-fold induction with NaHS (see Table S1A; adjusted P value of ≤0.05) in HNO induction is significant.
The entire isd operon is modestly (≥2-fold) repressed (see Table S1A for a complete listing of these genes).
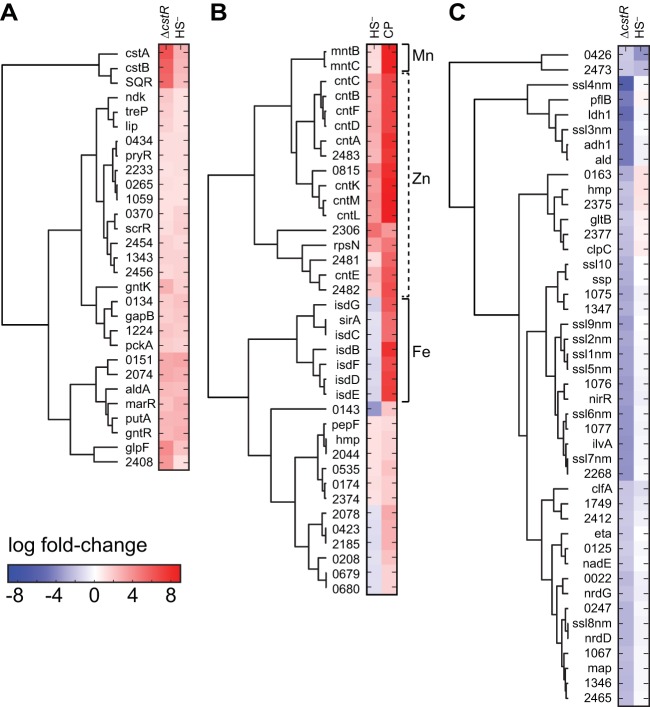
qRT-PCR validation of the RNAseq results. (A) Subset of the genes (expressed as S. aureus strain Newman locus tags, NWMN_wxyz) induced (compare to Table 1) or repressed (Table 2) upon treatment with 0.2 mM Na2S at 10 min posttreatment relative to unstressed cells. (B) Subset of genes induced (compare to Table 3) and repressed (Table 4) in the unstressed, early log-phase ΔcstR strain relative to the isogenic wild-type strain cultured under the same conditions. (C) Subset of the genes induced (compare to Table 1) and repressed (Table 2) upon treatment with 0.2 mM AS at 10 min posttreatment relative to unstressed cells. Download FIG S1, EPS file, 1.1 MB (1.1MB, eps) .
Copyright © 2017 Peng et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Complete transcriptomic data and sequences of qRT-PCR primers used in this study. (A) Excel file listing all genes whose expression is altered in sulfide-treated cells (sheets 1 to 2, corresponding to Fig. 4 and Tables 1 and 2, main text), in the ΔcstR strain (sheets 3 to 4, corresponding to Fig. 4 and Tables 3 and 4, main text), in AS-treated cells (sheets 5 to 6, corresponding to Fig. 6 and Tables 1 and 2, main text), and in CP-treated cells (sheets 7 to 8, corresponding to Fig. 3, main text) that meet the minimal acceptance criteria (2-fold change in expression; adjusted P value of ≤0.05). (B) A list of all the primers used in the qRT-PCR experiments. Download TABLE S1, XLSX file, 0.1 MB (103KB, xlsx) .
Copyright © 2017 Peng et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A subset of genes that were upregulated in both the sulfide-stressed wild-type and ΔcstR cultures (Table 3) relative to untreated wild-type S. aureus cultures included aldA (NWMN_0113), encoding an uncharacterized aldehyde dehydrogenase, and hypothetical operons beginning with NWMN_0134 (NWMN_0134 to NWMN_0137) and NWMN_0151 (NWMN_0151 to NWMN_0154), the latter of which is associated with changes in carbohydrate metabolism and uptake in other Staphylococcus strains. Although some of these sulfide-inducible genes are also induced in the ΔcstR strain, none would appear to be direct targets of CstR regulation since they lack clearly identifiable cstR operators upstream (28) in the immediately adjacent intergenic regions, a finding supported by our clustering analysis (Fig. 2A, bottom). These data, taken collectively, suggest that CstR likely directly regulates the expression of a single operon, cst, with other genes induced by sulfide treatment likely indirectly influenced by metabolic changes in the cell, which includes RSS.
TABLE 3 .
List of genes significantly (≥3.5-fold) upregulated in the ΔcstR strain relative to the wild-type strain
| Locus tag (Newman) |
Locus tag (N315) |
Gene name |
Fold induction |
Function | Sulfide stress?a |
|---|---|---|---|---|---|
| NWMN_0025 | SA0079 | 15.7 | Hypothetical | − | |
| NWMN_0026 | SA0080 | tauE | 43.0 | Putative sulfonate/TS effluxer | −/+b |
| NWMN_0027 | SA0082 | cstA | 70.2 | Multidomain sulfurtransferasea | + |
| NWMN_0028 | SA0083 | cstB | 46.5 | Persulfide dioxygenase-sulfur transferaseb | + |
| NWMN_0029 | SA0084 | sqr | 40.2 | Sulfide-quinone reductaseb | + |
| NWMN_0031 | SA0087 | 5.8 | Hypothetical | − | |
| NWMN_0113 | SA0162 | aldA | 5.3 | Aldehyde dehydrogenase-like | + |
| NWMN_0151 | SA0206 | 7.7 | Putative ABC sugar transporter (NWMN_0151–0154) | − | |
| NWMN_0329 | SA0325 | 5.2 | Glycerol-3-phosphate transporter | − | |
| NWMN_1207 | SA1140 | glpF | 16.7 | Glycerol uptake facilitator protein | + |
| NWMN_1224 | 4.1 | Hypothetical (82 residues) | − | ||
| NWMN_1378 | SA1301 | ndk | 3.9 | Nucleotide diphosphate kinase | − |
| NWMN_1415 | SA1339 | marR | 4.2 | Maltose operon repressor | + |
| NWMN_1658 | SA1585 | putA | 5.6 | Proline dehydrogenase | + |
| NWMN_1674 | SA1601 | 4.1 | Camphor resistance protein CrcB | − | |
| NWMN_1681 | SA1609 | pckA | 3.5 | Phosphoenolpyruvate carboxykinase | − |
| NWMN_2074 | SAS074 | 7.4 | Hypothetical (conserved; 86 residues) | − | |
| NWMN_2318 | SA2207 | hlgA | 5.2 | Gamma-hemolysin component A | − |
| NWMN_2402 | SA2294 | gntK | 6.1 | Gluconate kinase | + |
| NWMN_2403 | SA2295 | gntR | 5.4 | Gluconate operon repressor | + |
| NWMN_2408 | SA2300 | 12.0 | Hypothetical; putative transporter protein (glucaronic acid) | − | |
| NWMN_2510 | SA2406 | 6.2 | Glycine betaine aldehyde dehydrogenase GbsA | − | |
| NWMN_2513 | SA2408 | 3.7 | Putative choline transporter | − |
See Table 1.
At least 2-fold induction with NaHS (see Table S1A; adjusted P value of ≤0.05).
FIG 2 .
Clustering analysis and RNAseq transcriptomic analysis of Staphylococcus aureus strain Newman. Genes that change expression significantly in pairwise comparisons of results of sulfide (HS−) treatment versus the ΔcstR strain (upregulated genes only) (A) or of sulfide treatment versus CP treatment (all genes) (B) are indicated. (C) A list of genes in the ΔcstR strain (Table 4) that are significantly downregulated compared to their expression under sulfide stress conditions. Genes are clustered according to similarities in changes in expression in each pair of experiments.
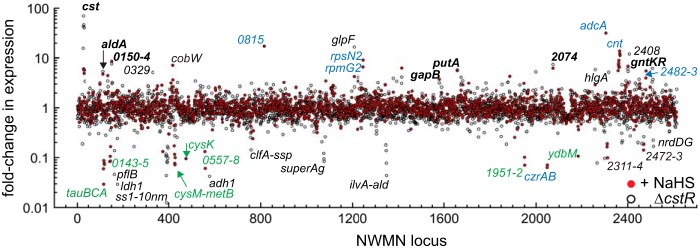
Comparing this list of sulfide-induced genes relative to the untreated control wild-type strain with results of other transcriptomics experiments using the S. aureus transcript regulatory network analysis tool (SATRAT) (36) reveals that a majority of these genes were oppositely (down)regulated by oxidative stress induced by short-time-scale exogenously added hydrogen peroxide (Fig. S2A and C) (37) and hypochlorous acid. In addition, the ratio of reduced to oxidized bacillithiol (BSH), one of the major LMW thiols in S. aureus (see below), increased upon sulfide treatment relative to untreated cells (Fig. S3), which is opposite what is expected under conditions of oxidative stress. These data collectively reveal that the transcriptomic response induced by changes in cellular RSS is opposite that induced by this potent oxidant (Table S1A). Thus, RSS and ROS transcriptional programs operate independently in the cell, consistent with what is known about the inducer specificity of CstR relative to that of PerR, the major ROS-sensing repressor in S. aureus (29, 38).
Analysis of all transcriptomic (RNAseq) changes in the sulfide-stressed (HS−) and AS-treated cells that showed significant changes in other stress-treated cells. These stresses included oxidative stress (ROS) (10 mM H2O2) (A), nitrosative stress (RNS) (150 μM GSNO [S-nitrosoglutathione]) (B), and combined ROS/RNS stress (10 mM H2O2 plus 150 μM GSNO) (C) (37). The cutoff is 2-fold change. Gene names are shown (where known) on the right. Note that only genes that are significantly regulated by either sulfide or AS stress or both sulfide and AS stress and under other stress conditions are shown. (A) The 90 genes shown represent the 45% of the transcriptome that is affected by acute-phase ROS, corresponding to 56% of upregulated and 18% of downregulated genes that change in the same direction. (B) The 43 genes shown represent the 41% of the transcriptome that is affected by GSNO, corresponding to 37% of upregulated and 19% of downregulated genes that change in the same direction. (C) The 123 genes shown represent the 43% of the transcriptome that is affected by the combined ROS/RNS stress, corresponding to 33% of upregulated and 36% of downregulated genes that change in the same direction. Download FIG S2, EPS file, 2.6 MB (2.6MB, eps) .
Copyright © 2017 Peng et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The ratio of reduced to oxidized bacillithiol (BSH/BSSB) in the sulfide-stressed (HS–) and HNO-treated cells compared to untreated cells. Sulfide treatment increased BSH/BSSB levels significantly, but the ratio remained unchanged under AS treatment. The experiments were done in triplicate, with standard deviations shown as error bars. Statistical significance relative to untreated cells was established by using a paired t test (***, P < 0.001; n.s., no significant change). Download FIG S3, EPS file, 0.6 MB (615.9KB, eps) .
Copyright © 2017 Peng et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Exogenous sulfide induces a zinc limitation transcriptomic response.
In addition to the transcriptomic response to exogenous sulfide treatment described above, exogenous sulfide stress also induces a pronounced zinc limitation response in S. aureus that is similar to that mediated by the antimicrobial protein calprotectin (CP) (Fig. 2B and Fig. 3). For example, sulfide treatment induces strong upregulation of the zinc uptake repressor (Zur) regulon (39) (Table 1; Table S1A), while downregulating the expression of czrAB (zntRA [40]), encoding the Zn efflux regulator and cation diffusion facilitator transporter (Table 2) (41, 42). Sulfide treatment also leads to upregulation of mntABC, controlled by the Mn-sensing repressor MntR (43), but no measurable Fe limitation response, in contrast to CP treatment (Fig. 2B). Genes that are upregulated by both exogenous sulfide treatment and CP treatment and linked to transition metal homeostasis include NWMN_2481, encoding a putative COG0523 G3E family GTPase linked to zinc homeostasis in other organisms (44, 45); two genes encoding uncharacterized pyridine nucleotide disulfide oxidoreductases (NWMN_0815 and NWMN_2370); rpmG2 and rpsN2, encoding two non-zinc-containing paralogs of ribosomal proteins (46); the adcA gene encoding a zinc-binding lipoprotein (NWMN_2306); and the entire cnt gene cluster, cntA to cntF (cntA-F) (Table 1) (Fig. 2B and Fig. 3). cntA-F is reported to encode a Co/Ni uptake system that is expressed upon metal limitation (47), while cntKLM encodes the biosynthetic machinery that produces a broad-spectrum nicotianamine-like metallophore that is capable of scavenging metals from the environment (48).
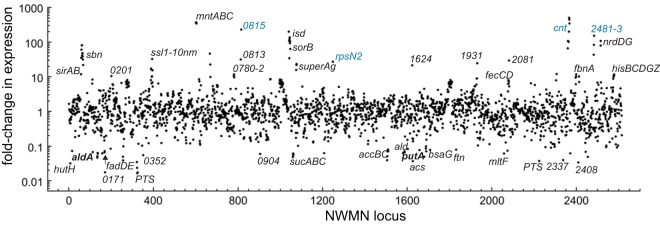
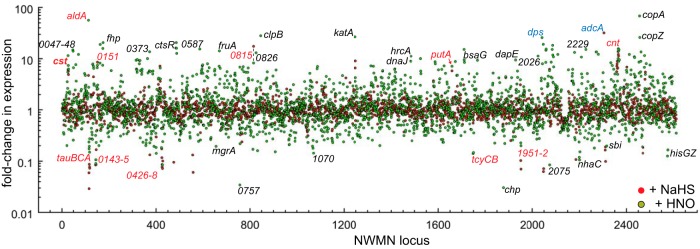
FIG 3 .
RNAseq transcriptomic analysis of wild-type Staphylococcus aureus strain Newman treated with calprotectin (CP). The fold change in expression for each locus tag is indicated (see Table S1A for a complete list of genes induced ≥2.0-fold and with adjusted P values of ≤0.05). Gene names are indicated where known; otherwise, the locus identifier (NWMN_wxyz [where "wxyz" represents the locus number]) is indicated. Names in blue are genes also induced by sulfide (Fig. 4).
Induction of a zinc starvation response by application of exogenously added sulfide can be traced to the ability of bisulfide (HS−) salts to form stable coordination complexes with transition metals, leading to their precipitation from solution and making them nonbioavailable (49). Analysis of the growth medium before and after addition of 0.2 mM Na2S reveals an approximately 10-fold decrease in total Zn levels, with relatively smaller reductions in Cu(II) and Ni and no change in Mn and Fe (Fig. S4A). This suggests that increased levels of endogenous sulfide in the cytoplasm may lead to a reduction in the bioavailability of intracellular transition metals, particularly Zn, by chelation.
Metal analysis of the HHWm growth medium before and after treatment with 0.2 mM NaHS (A) and 0.2 mM AS (B). ICP-MS was used to quantify the concentrations of transition metal ions in the HHWm growth medium (gray bars), HHWm plus chloramphenicol plus 0.2 mM thiosulfate (TS) as the sole sulfur source (red bars), or HHWm plus chloramphenicol plus 0.2 mM thiosulfate plus 0.2 mM NaHS or AS (light blue bars). Sulfide treatment led to a significant decrease in the level of bioavailable (soluble) Zn (15.2 µM to 1.2 µM), with correspondingly small changes in the levels of Cu (approximately 2-fold) and Ni (less than 2-fold). AS treatment led to a significant decrease in the level of bioavailable (soluble) Zn (18.2 µM to 5.9 µM), with a correspondingly small change in the level of Ni (1.8-fold). Download FIG S4, EPS file, 2.7 MB (2.8MB, eps) .
Copyright © 2017 Peng et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Part of the CymR regulon is repressed by exogenous sulfide treatment.
The suite of genes that are strongly (≥3.0-fold) repressed under conditions of exogenous sulfide stress are largely limited to a subset of CymR-regulated genes (5, 50), defined by differential expression in a cymR mutant S. aureus strain, strain N315 (Table 2; Fig. 4) (3). CymR is the master regulator of cysteine metabolism, repressing the expression of genes that lead to cysteine biosynthesis, and thus controls the response of the organism to a sulfur source. Increased exogenous sulfide levels lead to increased levels of intracellular sulfide (29) and LMW thiol persulfides (Fig. 5C). These sulfide-repressed genes include the operon NWMN_0115 to NWMN_0119, which is thought to encode a taurine (sulfonate) importer (NWMN_0116 to NWMN_0118); an operon encoding an uncharacterized ABC transporter, which is proposed to be involved in glutathione assimilation (NWMN_0143 to NWMN_0146) (3); a gene encoding a cysteine synthase (cysK); and genes encoding two enzymes of the transsulfuration pathway, CBS (encoded by cysM) and CSE (metB), that allow sulfur assimilation from the major human thiol, homocysteine. In addition, the expression levels of genes encoding methionine (NWMN_0246 to NWMN_0428) and cystine (NWMN_0374 and NWMN_1749 to NWMN_1751) ABC transporters and an operon (NWMN_1951 to NWMN_1952) encoding a TusA-like sulfurtransferase and a putative thiosulfate importer are also repressed (3). Loss of cymR in a cymR mutant results in overexpression of the CymR regulon, which is a transcriptomic response that is opposite the repression of the CymR regulon that we observed with exogenous sulfide. Two other genes that are strongly repressed and not part of the CymR regulon are NWMN_0557 and NWMN_0558, encoding a putative flavin adenine dinucleotide (FAD)-dependent oxidoreductase (candidate dihydrolipoamide dehydrogenases) and an Rrf2 family repressor (distinct from cymR; NWMN_1528 [5]) that is related to SaiR, which was recently characterized in Bacillus subtilis (51). SaiR regulates the expression of Spx, an activator of the response to toxic oxidants; this is consistent with the orthogonal nature of the cellular response to RSS relative to ROS (see above).
FIG 4 .
RNAseq transcriptomic analysis of Staphylococcus aureus strain Newman. Cells were either treated with 0.2 mM NaHS (red filled circles) or left untreated (ΔcstR strain) (black open symbols); data are expressed relative to the untreated wild-type strain results. The fold change in expression for each locus tag (NWMN_wxyz) is indicated (see Tables 1 to 4 for partial lists of these genes and Table S1A for a complete list). Gene names are indicated where known. Black bold type is used to represent genes that change expression in sulfide-treated cells, in Angeli’s salt (AS; nitroxyl)-treated cells, and in the ΔcstR strain; light blue type is used to represent genes observed to change in the calprotectin-treated samples; green type is used to represent genes of the CymR regulon (3).
FIG 5 .
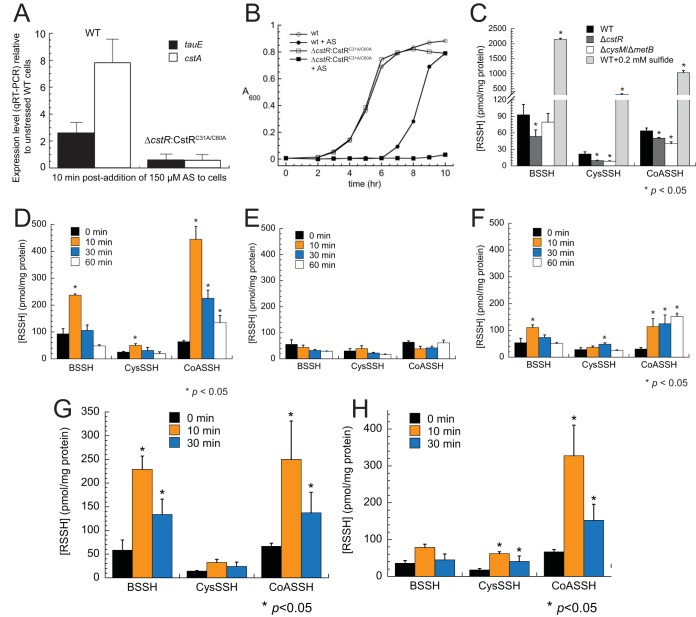
Angeli’s salt (AS) induces expression of the cst operon in a CstR-dependent manner and gives rise to a transient increase in endogenous LMW thiol persulfide levels, attributable to HNO. (A and B) AS induces cst operon expression as measured by qRT-PCR (A) and gives rise to a measurable growth phenotype when cells express an inactive CstR, C31A/C60A CstR (29) (B). WT, wild type. (C) LMW persulfide levels can be manipulated by genetic background and exogenous sulfide exposure. (D and E) AS (D) causes increased levels of LMW persulfides under aerobic conditions but nitrite (E) does not, indicating that HNO induces a transient increase in cellular RSS in S. aureus. (F) Levels of LMW persulfides in a ΔcysM ΔmetB strain were also transiently increased by AS. (G and H) Both AS treatment under microaerophilic conditions (G) and ONOO– treatment under aerobic conditions (H) cause an increase in the cellular accumulation of LMW persulfides. Error bars represent standard deviations of results of triplicate biological experiments, with statistical significance relative to the results seen with untreated wild-type cells (C) or to the results seen with each of the endogenous LMW persulfides at 0 min (D to H) established using paired t tests (*, P ≤ 0.05). Note that the quantitations of [RSSH] for t = 0 min in panels C, D, E, and H differ slightly from one another, reflecting the culture-to-culture variability of the measurements. Over all 12 replicates, [BSSH] = 77.1 ± 10.5 pmol/mg protein, [CysSSH] = 22.9 ± 2.6 pmol/mg protein, and [CoASSH] = 64.3 ± 3.0 pmol/mg protein, values fully consistent with previously published findings (26).
Unregulated cst operon expression results in repression of staphylococcal toxin genes.
Further consideration of the genes that are downregulated in the ΔcstR strain (Table 4) revealed uniform repression of the expression of staphylococcal exotoxins, encoded by genes NWMN_0388 to NWMN_0397 (ss1nm to ss10nm). There is also repression of additional toxin genes, including NWMN_1075 to NWMN_1077 and the gene encoding exfoliative toxin A (eta), a major histocompatibility complex (MHC) class II analog that impacts T-cell function and proliferation, and a gene encoding a formyl peptide receptor-like 1 inhibitory factor which inhibits the activation of neutrophils and monocytes. In addition, genes responsible for extracellular adhesion, including those encoding clumping factor A (clfA) and an extracellular matrix protein (ssp), and genes required for anaerobic growth and antibiotic resistance are also repressed. Some of these genes are direct targets of MgrA, which harbors a single regulatory cysteine residue characterized by a range of oxidative modifications in cells (52, 53).
TABLE 4 .
List of genes significantly (≥3.5-fold) downregulated in the ΔcstR strain relative to the wild-type strain
| Locus tag (Newman) |
Locus tag (N315) |
Gene name |
Fold induction |
Function | Sulfide stress?a |
|---|---|---|---|---|---|
| NWMN_0022 | SA0022 | 5.3 | Hypothetical (5′-nucleotidase family) | − | |
| NWMN_0125 | SA0175 | 3.5 | Hypothetical | − | |
| NWMN_0162 | SA0218 | pflB | 21.7 | Formate acetyltransferase | − |
| NWMN_0163 | SA0219 | 7.2 | Hypothetical | − | |
| NWMN_0175 | SA0231 | 4.1 | Flavohemoprotein | − | |
| NWMN_0176 | SA0232 | ldh1 | 34.0 | l-Lactate dehydrogenase | − |
| NWMN_0247 | SA0293 | 6.0 | Formate/nitrite transporter family protein | − | |
| NWMN_0388 | SA0382 | ss1nm | 9.2 | Superantigen-like protein 1 | − |
| NWMN_0389 | SA0383 | ss2nm | 8.5 | Superantigen-like protein 2 | − |
| NWMN_0390 | SA0384 | ssl3nm | 22.6 | Superantigen-like protein 3 | − |
| NWMN_0391 | ssl4nm | 79.3 | Superantigen-like protein 4 | − | |
| NWMN_0392 | SA0386 | ss5nm | 9.2 | Superantigen-like protein 5 | − |
| NWMN_0393 | ssl6nm | 11.9 | Superantigen-like protein 6 | − | |
| NWMN_0394 | SA0387 | ssl7nm | 12.5 | Superantigen-like protein 7 | − |
| NWMN_0395 | SA0388 | ssl8nm | 5.9 | Superantigen-like protein 8 | − |
| NWMN_0396 | SA0389 | ssl9nm | 10.3 | Superantigen-like protein 9 | − |
| NWMN_0397 | SA0390 | ssl10nm | 6.4 | Superantigen-like protein 10 | − |
| NWMN_0426 | SA0420 | 3.6 | ABC transporter: amino acid (ATPase) | + | |
| NWMN_0436 | SA0430 | gltB | 3.9 | Glutamate synthase, large subunit | − |
| NWMN_0577 | SA0562 | adh1 | 24.5 | Alcohol dehydrogenase (eukaryote-like) | − |
| NWMN_0756 | SA0742 | clfA | 4.1 | Clumping factor A | − |
| NWMN_0758 | SA0744 | ssp | 6.9 | Extracellular matrix; plasma binding (cell wall) | − |
| NWMN_1067 | SA1001 | 4.8 | Formyl peptide receptor-like 1 inhibitory factorb | − | |
| NWMN_1075 | SA1009 | 8.1 | Superantigen-like protein (toxin c) | − | |
| NWMN_1076 | SA1010 | 10.6 | Superantigen-like protein | − | |
| NWMN_1077 | SA1011 | 11.8 | Superantigen-like protein | − | |
| NWMN_1082 | SA1016 | eta | 3.8 | Exfoliative toxin A | − |
| NWMN_1346 | SA1269 | 5.5 | Hypothetical (membrane efflux) | − | |
| NWMN_1347 | SA1270 | 7.2 | Amino acid permease | − | |
| NWMN_1348 | SA1271 | ilvA | 12.9 | Threonine dehydratase | − |
| NWMN_1349 | SA1272 | ald | 22.8 | Alanine dehydrogenase | − |
| NWMN_1749 | SA1674 | 3.9 | Glutamine transport | − | |
| NWMN_1850 | SA1728 | nadE | 3.8 | NAD synthetase (glutamine or ammonia dependent) | − |
| NWMN_1872 | SA1751 | map | 5.4 | MHC class II analog protein | − |
| NWMN_2268 | SA2156 | 12.7 | l-Lactate permease 2 | − | |
| NWMN_2301 | SA2189 | nirR | 10.3 | Nitrite reductase transcriptional regulatorc | − |
| NWMN_2375 | SA2266 | 5.0 | NAD short-chain dehydrogenase/reductase (SDR) | − | |
| NWMN_2377 | SA2268 | 4.6 | Hypothetical (63 residues) | − | |
| NWMN_2412 | SA2302 | 3.7 | ABC transporter (ATPase); lantibiotic | − | |
| NWMN_2448 | SA2336 | clpC | 5.0 | Clp protease C subunit (ATPase subunit) | − |
| NWMN_2465 | SA2352 | 5.2 | Hypothetical (NWMN_2365–2361 operon) | − | |
| NWMN_2473 | SA2360 | 3.8 | Hypothetical (76 residues) | − | |
| NWMN_2514 | SA2409 | nrdG | 4.5 | Ribonucleotide reductase, anaerobic (small subunit) | − |
| NWMN_2515 | SA2410 | nrdD | 5.9 | Ribonucleoside-triphosphate reductase, anaerobic | − |
See Table 2.
Secreted protein that specifically inhibits the activation of neutrophils and monocytes by binding to the formylated peptide receptor and the C5a receptor; blocks neutrophil migration toward the infection site, and hinders the establishment of the initial defense against the infection.
nirBG and narGHJ expression downregulated by approximately 2-fold (Table S1A).
Other genes that are downregulated in the ΔcstR strain are upregulated during anaerobic growth of wild-type cells (COL) (54), while a significant fraction of these genes are similarly repressed upon treatment of S. aureus Newman cells with antimicrobial peptides (55). These genes include those encoding the formate acetyltransferase (pflB) and the formate acetyltransferase-activating enzyme; a formate-nitrate transporter (NWMN_0247) and the nitrite reductase transcriptional regulator (nirR); and the anaerobic ribonucleotide reductase (nrdGD). In addition, genes encoding a candidate flavohemoglobin (NWMN_0175), NO-inducible l-lactate dehydrogenase (ldh1), and l-lactate permease 2, as well as alcohol dehydrogenase (adh1), and genes associated with amino acid metabolism are also significantly repressed in the ΔcstR strain.
Comparison of the ΔcstR strain and the wild-type strain results seen at times shortly following an acute-phase sulfide shock shows that their transcriptomic responses diverged considerably beyond the approximately 10 genes that were similarly upregulated or downregulated under both conditions (Fig. 2A and Tables 1 to 4). Sulfide-stressed cells differ strongly from the ΔcstR strain in the relative concentrations of low-molecular-weight thiol persulfides (organic RSS) (26), which are strongly elevated relative to the ΔcstR strain but reduced relative to the untreated wild-type strain (see below). This suggests the possibility that ambient RSS might directly impact gene expression through oxidative modification of one or more cysteine-containing global regulators, with genes required for infection, dissemination, adhesion, antibiotic resistance, and anaerobic growth largely repressed in the ΔcstR strain relative to the wild-type strain.
Transcriptomic profiling of the effects of an exogenous HNO donor, Angeli’s salt.
Emerging evidence suggests that many of the properties attributed to H2S as a signaling molecule may derive from a significant increase in the levels of organic and inorganic polysulfide species and, in some cases, of HNO (15). HNO reacts rapidly with LMW (and protein) thiols (10, 19). This leads to disulfide bond formation in the presence of resolving thiol with the release of hydroxylamine (10) and sulfinamides [RS(O)NH2], which are in turn slowly reduced by cellular thiols (56). Indeed, baker’s yeast encodes an enzyme that catalyzes the NADPH-dependent reduction of the S-nitrosoglutathione-derived glutathione sulfinamide to reduced glutathione, which accumulates in cells under conditions of NO· stress (57). We therefore tested the effects of HNO added to aerobically growing cells by the use of Angeli’s salt (AS), i.e., dinitrogen trioxide dianion (Na2N2O3), which undergoes cleavage to yield HNO and nitrite (NO2−) (Fig. 1C). In contrast to the minimal induction (Fig. S5) or absence of induction of the cstR-regulated genes resulting from the addition of an NO· donor or nitrite, respectively (29), we found that AS significantly induced cstA expression in a quantitative reverse transcription-PCR (qRT-PCR) experiment (Fig. 5A). Further, a ΔcstR strain complemented with a mutant CstR unable to sense persulfides cannot be induced under the same conditions, suggesting that HNO impacts CstR function. These cells also exhibit a dramatic growth defect relative to the wild-type strain stressed with HNO (Fig. 5B). We next tested if HNO is capable of reacting directly with CstR thiols, leading to disulfide bond formation (10), which would induce derepression of CstR-regulated genes (29). Treatment of reduced CstR with Angeli’s salt in vitro does indeed yield CstR characterized by an interprotomer disulfide bond between C31 and C60′ confirmed by both liquid chromatography-electrospray ionization-mass spectrometry (LC-ESI-MS) and LC-tandem MS (LC-MS/MS), with no cross-linked products obtained upon incubation with sodium nitrite (Fig. S6). This reveals that HNO is capable of impacting CstR-regulated transcription directly by forming a cross-linked CstR which has lower affinity for operator DNA (28).
Measurement of the expression levels of cstA in the cst operon in cells treated with 0.5 mM methylamine hexamethylene methylamine NONOate (MAHMA NONOate) by qRT-PCR. cstA expression was seen 10 min or 30 min postexposure relative to unstressed cell results. Error bars represent standard deviations of results of triplicate biological experiments, with statistical significance established using a paired t test relative to untreated wild-type cell results (0 min) (*, P < 0.05; n.s., no significant change). Download FIG S5, EPS file, 0.6 MB (633.9KB, eps) .
Copyright © 2017 Peng et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
CstR reacts with AS to form cross-linked CstR characterized by an interprotomer disulfide bond between C31 and C60′. (A) CstR was mostly in reduced form. However, the cross-linked CstR was largely observed after the reaction with 20-fold molar excess AS (B) but not in the reaction with nitrite (C), another product of AS decomposition. (D) The formation of a disulfide bond between C31 and C60′ was confirmed by LC-MS/MS. The expected m/z of reduced CstR monomer is 9,641.2. Electron transfer dissociation (ETD) mode was used for the sequencing analysis. Download FIG S6, EPS file, 2.9 MB (2.9MB, eps) .
Copyright © 2017 Peng et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A transcriptomic analysis of all genes induced (Table 1) or repressed (Table 2; see Table S1A for a complete list) by AS reveals a significant genome-wide change in cellular transcription (Fig. 6). We focused our attention on those genes whose expression changed by approximately 1 standard deviation from the mean induction level (approximately 10-fold; Table 1) or the mean repression level (approximately 5-fold; Table 2). Beyond the cst operon, there was a subset of genes that were upregulated by both HNO and sulfide treatment relative to the results seen with untreated wild-type cells (Table 1 and Fig. 7). These genes included a gene that is part of the CymR regulon, aldA (NMWN_0113), which is among the most highly (56-fold) upregulated genes in the genome; cntKLM, encoding a broad-spectrum metallophore biosynthetic cluster (48) and among the genes most highly upregulated by sulfide treatment (Table 1); and NWMN_0815, a gene encoding a putative pyridine nucleotide (FAD) disulfide reductase (Fig. 7). These genes are also associated with the zinc limitation, CP-mediated transcriptomic response (Table 1) (Fig. 2), which suggests that zinc may well become limiting as a result of intracellular chelation of the metal upon AS treatment (58). Several other genes involved in the metal limitation response, e.g., rpmG2, rpsN2, cntAB, and NWMN_2483, are also detectably induced by HNO stress but to a level that is lower than that due to sulfide stress. Consistent with this, AS treatment leads to a detectable (approximately 3-fold) decrease in the Zn concentration and to a smaller (1.8-fold) (Fig. S4B) change in the Ni concentration in the growth medium, suggesting that HNO, like sulfide itself, may be capable of reducing the bioavailability of transition metals in cells.
FIG 6 .
RNAseq transcriptomic analysis of Staphylococcus aureus strain Newman cells treated with sulfide (red filled circles) versus Angeli’s salt (green filled circles). The fold change in expression for each locus tag is indicated (Tables 1 to 2; Table S1A). Gene names are indicated where known. Expression of those highlighted in red text was significantly induced or repressed under both experimental conditions; expression of those highlighted in light blue was induced during calprotectin (CP) and Angeli’s salt (nitroxyl) treatment.
FIG 7 .
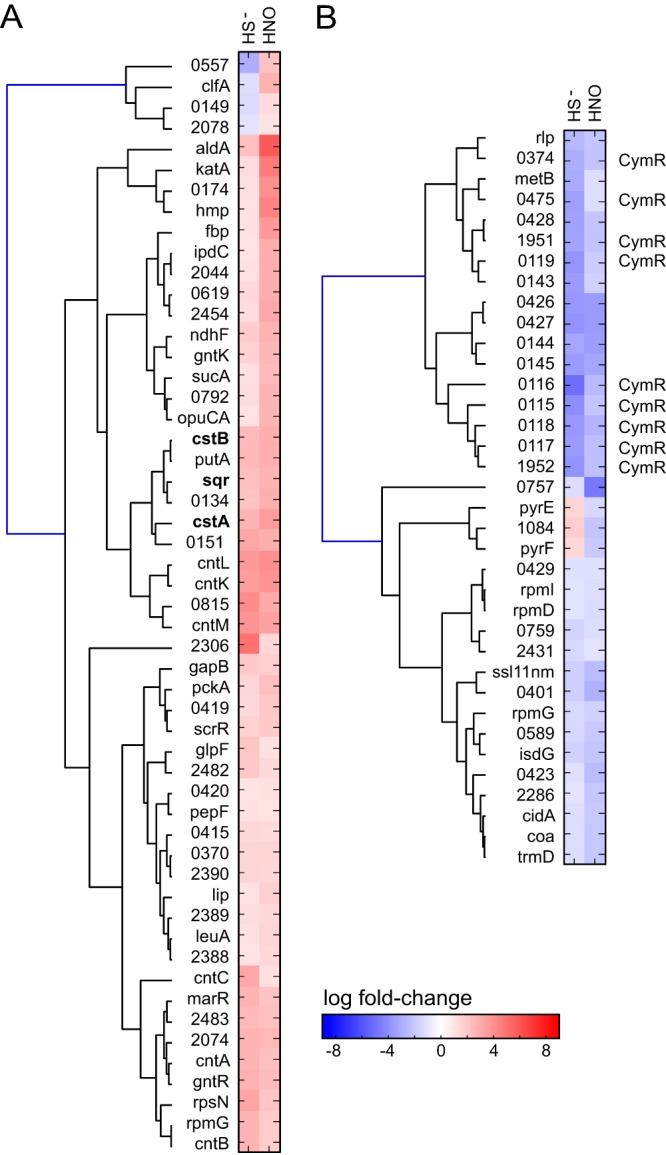
Clustering analysis of the RNAseq results based on pairwise comparisons of gene expression in sulfide (HS−)-treated versus AS (HNO)-treated S. aureus strain Newman cells. (A) Genes that were upregulated by HNO compared to the analogous change in HS−-treated cells. (B) Genes that were downregulated by HNO compared to the analogous change in HS−-treated cells. Genes of the cst operon are in boldface (panel A), while those genes previously identified as part of the CymR regulon (3) are marked "CymR" (panel B). Genes are clustered according to similarity in change in expression in each pair of experiments. Only the genes that were affected by both sulfide and AS stress are shown here (Tables 1 and 2, Fig. S9, and Table S1A for a compilation of all transcriptomic changes observed under these conditions).
The remaining genes in the far larger panel that are upregulated by AS treatment but unaltered by sulfide treatment show some overlap with respect to those genes induced by nitrite in nitrate-respiring, nitrite-stressed cells (59). Most notable is the massive upregulation of the copAZ operon, encoding a Cu(I)-transporting P-type ATPase effluxer and a Cu(I) chaperone, both under the transcriptional control of Cu(I)-sensing repressor CsoR (NWMN_1992) (28). CsoR is a paralog of CstR and is unresponsive to sulfide stress in cells (28). This suggests that HNO-mediated modification of LMW thiols, which maintain bioavailable Cu(I) at low levels, may result in the displacement of Cu(I) from these stores, which is then sensed by CsoR. Indeed, analysis of total cell-associated metal levels by inductively coupled plasma-mass spectrometry (ICP-MS) reveals a significant and specific increase in levels of cellular Cu (Fig. S7), consistent with this hypothesis. The remaining AS-affected genes represent an Fe overload response, the origin of which is probed below, coupled with a PerR-regulated ROS response and induction of the stress-associated CtsR regulon (clpB, clpC) and a DNA damage response (uvrBC) (Fig. S2). These transcriptomic changes, taken collectively, are consistent with an acute-phase combined ROS/RNS stress response induced by AS treatment (Fig. S2).
Analysis of cellular metal before and after treatment with 0.2 mM AS (A) and 0.2 mM sodium nitrite (B). ICP-MS was used to quantify the concentrations of transition metal ions in the untreated cell (gray bars) or in the cell treated with 0.2 mM AS or nitrite (light blue bars). (A) AS treatment did not lead to a significant change in the total levels of cellular transition metals, with the exception of Cu, whose level increased approximately 4-fold. (B) Nitrite treatment with the same concentration led to no significant change in all cellular metals. Download FIG S7, EPS file, 0.9 MB (944.2KB, eps) .
Copyright © 2017 Peng et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
AS also represses a number of virulence factors, including superantigen-like proteins; secreted von Willebrand factor-binding protein; proteins encoded by NWMN_1066 and NWMN_1068 to NWMN_1070, which are predicted to be fibrinogen binding, chemotaxis-inhibiting proteins; secretory antigen precursor SsaA; and immunoglobulin G-binding protein. We globally compared those genes whose expression is affected by AS to genes encoding other virulence factor regulons, including those encoded by sarA (60), sarZ (61), mgrA (62), sigB (63), and rot (64). We generally found no clear correlation of upregulation or downregulation between AS-responsive genes and those encoding these virulence regulons, with the exception of sigB, where many of the transcriptomic changes that were common under these two conditions occurred in the same direction (Fig. S8). SigB is an alternative sigma factor (σB) that controls the response to heat stress, oxidative stress, and antibiotic stress and appears to be linked to intracellular survival during chronic infection (65) while impacting the expression of genes encoding other virulence regulators, including sarA, sarS, and arlRS, a two-component system of autolysis (63). We note that most of the virulence factors repressed by AS treatment are not regulated by sulfide stress, suggesting that host NO·-derived RNS, e.g., HNO (Fig. 1B), likely have important regulatory roles in virulence expression distinct from that of sulfide.
Analysis of all transcriptomic (RNAseq) changes in the sulfide-stressed (HS−) and AS-treated cells compared to the regulons of selected virulence factors. Genes associated with the sarA (A) (60), sarZ (B) (61), mgrA (C) (62), sigB (D) (63), and rot (E) (64) regulons are shown. The cutoff is 2-fold change. Gene names are shown (where known) on the right. Note that only genes that are significantly regulated by either sulfide or AS stress or both sulfide and AS stress and the indicated virulence factor regulon are shown in each panel. With the exception of the sarZ and mgrA experiments, the transcriptomic profiling experiments were carried out using post-exponential-phase or stationary-phase cultures, which are different from the growth conditions used in our RNAseq experiments. (A) sarA: the 48 genes shown represent the 40% of the transcriptome that is affected by sarA, corresponding to 12% of upregulated and 2% of downregulated genes that change in the same direction. (B) sarZ: the 33 genes shown represent the 57% of the transcriptome that is affected by sarZ, corresponding to 0% of upregulated and 21% of downregulated genes that change in the same direction. (C) mgrA: the 53 genes shown represent the 15% of the transcriptome that is affected by mgrA, corresponding to 5% of upregulated and 2% of downregulated genes that change in the same direction. (D) sigB: the 136 genes shown represent the 54% of the transcriptome that is affected by sigB, corresponding to 56% of upregulated and 32% of downregulated genes that change in the same direction. (E) The 47 genes shown represent the 32% of the transcriptome that is affected by rot, corresponding to 19% of upregulated and 12% of downregulated genes that change in the same direction. Download FIG S8, PDF file, 0.6 MB (674.2KB, pdf) .
Copyright © 2017 Peng et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RSS profiling in sulfide-stressed versus Angeli’s salt-stressed cells.
We previously employed a fluorescence-based analytical method to detect and quantify LMW monobromobimane (mBBr)-derivatized sulfur-containing metabolites in cells (29). Here, we extended this method to incorporate ratiometric (32S/34S) tandem mass spectrometry, in which the concentrations of all organic thiols and of per- and polysulfides relative to those seen with an internal standard can be detected in a single experiment. We quantified bacillithiol persulfide (BSSH), cysteine persulfide (CysSSH), and coenzyme A persulfide (CoASSH) in the wild-type and ΔcstR S. aureus strains and in sulfide- or AS-stressed wild-type S. aureus. The ΔcstR strain showed slightly lower levels of LMW persulfides (Fig. 5C), consistent with an RSS clearance function of the cst operon-encoded enzymes. As predicted from the transcriptomics experiments, both sulfide stress (Fig. 5C) and AS treatment (Fig. 5D) caused an increase in cellular levels of LMW thiol persulfides, while nitrite, one of the AS decomposition products (Fig. 1B and C), did not (Fig. 5E). These results directly implicate HNO or a downstream reaction product(s) in this cellular increase in RSS (Fig. 1B). Thus, HNO may directly react with CstR thiols to induce transcriptional derepression of the cst operon (Fig. S6) or, alternatively, may induce higher levels of cellular LMW persulfides which are in turn sensed by CstR.
To further explore this, we reasoned that HNO could directly mediate an increase in cellular concentrations of RSS via at least two possible mechanisms. One is a mechanism by which HNO upregulates the expression of a sulfide biogenesis pathway(s), e.g., one involving CBS (CysM) and CSE (MetB). An alternative possibility is that AS-derived HNO disassembles Fe-S clusters directly or reacts with molecular oxygen to create ONOO– (21, 66), which is known to be capable of destabilizing Fe-S clusters in proteins (Fig. 1C) (24). Both would lead to an increase in endogenous sulfide as well as chelatable Fe(II) levels in the cell. Human CBS binds heme via coordination by Cys52 and His65 in an N-terminal heme-binding domain, and RNS are capable of regulating CBS activity by changing the oxidation state of the heme (67). However, this domain is not conserved in S. aureus CysM (alignment not shown). Consistent with this, RSS levels are in fact transiently elevated in a ΔcysM ΔmetB strain by AS treatment, albeit to an extent lower than that seen in the wild-type strain (Fig. 5F).
To measure the effect of molecular oxygen on this AS (HNO) stress-induced increase in RSS, we carried out this experiment under microaerophilic conditions, where oxygen levels are significantly decreased by growing static cultures in tightly capped tubes, with 5 mM nitrate as the electron acceptor of anaerobic respiration. AS-inducible increases in RSS were observed in these cultures (Fig. 5G) that were nearly equal to those seen with aerobically grown cells (Fig. 5D). Cells grown aerobically and treated with a burst of ONOO– also induced organic RSS but to a level that was lower overall than that seen with AS treatment (Fig. 5H). This result may have been related to the vanishingly short lifetime of ONOO– under these conditions. We propose that the presence of HNO or a downstream product(s) (Fig. 1B) leads to the disassembly of Fe-S clusters and to a corresponding increase in endogenous sulfide and RSS levels, with the oxidant ONOO– also capable of this chemistry (21, 66). In support of this idea, although the total cell-associated Fe level did not change (Fig. S7), our transcriptomic analysis strongly suggests an increase in the level of cytoplasmic chelatable iron, as exemplified by the upregulation of genes encoding the Fe-S cluster biogenesis pathway (sufC, sufD, sufS, NIFU, and sufB), the iron-storage proteins ferritin (ftn) and Dps (dps), and the iron uptake repressor (fur; NWMN_1406), which could chelate cellular Fe and further repress Fe import. Regardless of the precise mechanism involved, these data, taken collectively, suggest that HNO, as a principal product of H2S/NO· interplay (15, 68), directly impacts endogenous sulfur speciation and global gene expression in S. aureus.
Conclusions.
In this report, we show that the cellular transcriptomic response of the major human pathogen S. aureus to the effects of exogenous sulfide exhibits some parallels to the cellular response to Angeli’s salt, a commonly used HNO donor. Under these conditions, HNO appears to signal partly through perturbations in sulfur speciation in cells, as anticipated by much of the small-molecule chemistry that has been reported for this primary product of RNS/RSS interplay (15, 68). The origin of both effects derives in part from a significant increase in levels of cellular RSS (Fig. 5). The RSS-regulated transcriptomic response is opposite that induced by oxidants and suggests the possibility that bacterial cells can manage intracellular RSS as a means to provide protection against irreversible oxidation by oxidative stressors, as has been previously established in mammalian cells (25, 69). In contrast, decreases in levels of ambient RSS induced by overexpression of the cst-encoded sulfide oxidation system in a ΔcstR strain repressed virulence gene expression, i.e., expression of genes required for infection, dissemination, and adhesion to cells.
These results support the use of cellular RSS as a readily deployable chemical strategy to impact the physiological state of a bacterial community (32) and otherwise to mediate an adaptive response to changes in host microenvironments mediated by ROS and RNS. Indeed, multiple reactive small-molecule stressors often have a synergistic effect on microbial killing (70) and it is unusual for a pathogen to encounter a single stressor at a site of infection. This deployment of RSS, however, must be tightly managed, as intracellular sulfide and HNO (or downstream reaction products) significantly reduce zinc bioavailability, while HNO potentially increases levels of cytoplasmic free Cu (Fig. S7) and free Fe, the latter possibly via destruction or inhibition of assembly of Fe-S clusters. It is interesting in this context that one of the cst operon-encoded proteins, CstA, is capable of stripping the active site persulfide from the major S. aureus cysteine desulfurase SufS in a persulfide transfer reaction (30). This might provide a means to divert sulfur flow from Fe-S protein biogenesis to cellular RSS as a protective mechanism to minimize exposure to oxidative damage mediated by free Fe. This is consistent with the strong upregulation of Fe- and PerR-regulated ROS-inducible genes, including those encoding catalase (katA), flavohemoglobin (hmp), a candidate peroxiredoxin (bcp), alkylhydroperoxidases (ahpD, ahpF), a candidate nitroreductase, and the iron-storage protein Dps (dps) (Table S1A; Table 1; Fig. 7). These studies have set the stage for further elucidation of H2S/NO cross talk and proteome S-sulfhydration-based pathways in aerobically versus anaerobically growing bacterial cells.
Clustering analysis that summarizes all transcriptomic (RNAseq) changes in the ΔcstR, sulfide-stressed (HS–), AS (HNO)-treated, and calprotectin (CP)-treated cells that showed significant pairwise changes in expression. This figure recapitulates the data presented in Fig. 2 and 7 in the main text but provides more detail. A listing of all transcriptomic changes is provided in Table S1A. Locus tags are indicated on the left, with gene names shown (where known) on the right. Additional description is provided on the far right; the absence of an entry indicates a hypothetical protein of unknown function. Download FIG S9, PDF file, 0.6 MB (585.4KB, pdf) .
Copyright © 2017 Peng et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
MATERIALS AND METHODS
Chemicals and reagents.
AS (82230) and sodium peroxynitrite (81565) were purchased from Cayman Chemical. Monobromobimane (M-20381) was purchased from Invitrogen. Sulfur-34 metal (SLM-1085-PK) used for Na234S synthesis was purchased from Cambridge Isotope Laboratories, Inc. Sodium sulfide (407410), sodium nitrite (237213), sodium nitrate (S5506), oxidized coenzyme A (C2643) used to synthesize the CoASSH standard, l-cystine (C8755) used to synthesize the CysSSH standard, and other reagents were purchased from Sigma-Aldrich.
S. aureus RNAseq and qRT-PCR experiments.
Sample collection, RNA extraction, and the procedure for qRT-PCR experiments were described previously (29). The primers used in qRT-PCR experiments are listed in Table S1B in the supplemental material. Sequencing reads were trimmed using Trimmomatic (version 0.33 [71]) with the following parameters: ILLUMINACLIP:adapter.fa:2:20:6 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:20 MINLEN:35. The trimmed reads were mapped onto the Staphylococcus aureus subsp. aureus strain Newman genome using bowtie2 (version 2.1 with default parameters) (72). Read counts for genes and intergenic intervals were calculated using a custom perl script. The resulting gene/interval counts were used to conduct differential expression analysis using the program DESeq2 algorithm (73) with default parameters. For transcriptome sequencing (RNAseq) analysis of calprotectin (CP)-treated cultures, bacteria were cultured overnight in Chelex-treated RPMI medium plus 1% Casamino Acids supplemented with 1 mM MgCl2, 100 μM CaCl2, and 1 μM FeSO4. These samples were then back-diluted 1 to 100 into growth medium containing 38% tryptic soy broth (TSB)–72% calprotectin buffer (20 mM Tris [pH 7.5], 100 mM NaCl, 3 mM CaCl2, 10 mM β-mercaptoethanol), supplemented with 1 µM Zn and 1 µM Mn. The bacteria were grown to an exponential-phase optical density at 600 nm (OD600) of 0.25 to 0.4 in the presence and absence of 960 µg/ml CP. Samples were harvested and processed as previously described (29), with the exception that RNA was isolated using the method reported by Collins et al. (74). The mean level of induction of all genes that showed an increase in expression of ≥2.0-fold at an adjusted P value of ≤0.05 in the CP-treated cells was 20-fold (±59-fold), with a median genome-wide level of induction of 4.4-fold (330 genes). The mean level of expression for all genes that were downregulated as a result of CP treatment was 7.4-fold (±8.0-fold), with a median level of genome-wide repression of 4.3-fold (289 genes). The results of all RNAseq experiments have been deposited in the GEO database (see below).
CstR reaction with AS.
The purification of CstR was carried out as described previously (29). A 200-µl volume of 25 µM apo- and reduced CstR was incubated anaerobically with a 20-fold molar excess over levels of thiols of sodium nitrite or Angeli’s salt in 25 mM Tris (pH 8.0)–500 mM NaCl–5 mM EDTA at room temperature. After 30 min, 100 μl of the reaction mixture was transferred to a vial, the vial was tightly capped, and the contents of the vial were injected in LC-ESI-MS experiments to determine if cross-linked CstR was formed, as previously described (29). The remainder of each sample was alkylated with iodoacetamide, digested by trypsin, and sequenced to further confirm the formation of a disulfide bond.
Bacterial growth curves and cell culture.
The wild-type S. aureus Newman strain and the ΔcstR CstRC31A/C60A strain were described previously (29). The ΔcysM ΔmetB strain was constructed by allelic exchange (75). The fragment upstream of cysM was amplified using primers 5′-TTGAGCCTCGGAACCGGTACCAACATTAGATGGCGCCTTAG-3′ and 5′-TCCTAGCTTAGCTAGCAATTAAATCATAAGTAATCATAGATGC-3′; a spectinomycin resistance cassette was amplified from pSPC using primers 5′-GCTAGCTAAGCTAGGATCGAATCCC-3′ and 5′-GCTAGCCTAATTGAGAGAAGTTTCTATAGAATTTTTC-3′; and the fragment downstream of metB was amplified using primers 5′-TCTCAATTAGGCTAGCCAAGCACTAGATACTTTATAAATAATAGC-3′ and 5′-ACAGCTATGACATAGTCACGAATTCAAACACCTCTTTAACAGTTC-3′. These fragments were assembled with pKOR1 linearized by digestion with EcoRI and KpnI using NEB Gibson assembly and were integrated into the S. aureus genome. The genetic lesion was then transduced into a clean S. aureus Newman background using bacteriophage φ85, and transductants were selected with spectinomycin at 1,000 mg/liter. All bacterial strains were grown overnight in TSB with 10 μg/ml chloramphenicol. Cells were pelleted and resuspended in Hussain-Hastings-White modified (HHWm) minimal media (76) supplemented with 50 μg/ml chloramphenicol and 0.5 mM thiosulfate as the sole sulfur source. For growth curve analyses, cultures were initiated at an OD600 of 0.007 with or without 0.2 mM AS stress added to the growth medium. All aerobically grown cultures were grown at 37°C with shaking (200 rpm), with the OD600 measured every hour from h 2 to h 10.
Quantitation of cellular LMW thiol persulfides.
Overnight S. aureus cells grown in TSB were diluted to an OD600 of 0.02 in HHWm minimal medium (76) supplemented with 0.5 mM thiosulfate as the sole sulfur source and grown aerobically. When these cultures reached an OD600 of 0.2, 0.2 mM disodium sulfide, AS, sodium nitrite, or sodium peroxynitrite was added. For microaerophilic conditions, 5 mM sodium nitrate was added as an electron acceptor to 50-ml tubes that were capped tightly without shaking. The tubes were opened for addition of AS and were gently inverted for mixing. Samples (5 ml plus 1 ml for protein quantification) were collected before (t = 0 min) and after addition of the stressor at the indicated times following addition of stressors and were centrifuged at 3,000 rpm for 10 min. The resulting pellets were washed with ice-cold phosphate-buffered saline (PBS), pelleted again by centrifugation (16,100 rpm for 5 min), and stored frozen at −80°C until use. Thawed cell pellets were resuspended in 100 μl monobromobimane (mBBr) labeling solution containing 20 mM Tris-HBr (pH 8.0), 50% acetonitrile, and 1 mM mBBr and subjected to three freeze-thaw cycles in liquid nitrogen in the dark in screw-cap tubes (77). Cell debris was removed by centrifugation, and the supernatant was transferred to a tube containing 100 μl 15 mM methanesulfonic acid (MA) to quench the labeling reaction (31). Finally, particulates were removed via passage through a 0.2-μm-pore-size centrifugal filter unit prior to injection into a liquid chromatograph mass spectrometry (LC-MS) system for quantitation of LMW thiol persulfides as follows.
Samples (10 μl) were injected into a Triart C18 column (YMC, Inc.) (50 by 2.0 mm inner diameter) and subjected to chromatography on a Waters Acquity Ultra Performance Liquid Chromatography (UPLC) I-class system, using a methanol-based gradient system (for solvent A, 10% methanol and 0.25% acetic acid, pH 3.0; for solvent B, 90% methanol and 0.25% acetic acid, pH 3.0) with the elution protocol at 25°C and a flow rate of 0.2 ml/min as follows: at 0 to 3 min, 0% B isocratic; at 3 to 7 min, 0% to 25% B, linear gradient; at 7 to 9 min, 25% B isocratic; at 9 to 12 min, 25% to 75% B, linear gradient; at 12 to 14 min, 75% to 100% B, linear gradient; at 14 to 14.5 min, 100% B isocratic, followed by reequilibration to 0% B. Quantitation of LMW thiols and persulfides was carried out with a Waters Synapt G2S mass spectrometer by spiking in a specific amount of authentic 34S-containing LMW persulfide standards synthesized with Na234S in place of Na232S (31) to achieve a typical 34S/32S persulfide ratio of approximately 0.2 to 1.0. To quantify the relative change in the BSH/BSSB ratio, the peak area of mBBr-derivatized BSH and the peak area of underivatized BSSB in each sample were obtained and normalized to that of a known concentration (peak area) of mBBr-labeled N-acetyl-cysteine (NAC), with the same concentration added to each sample. The change of the ratio was obtained by dividing the ratio seen under stress conditions by the ratio for the unstressed sample, with standard propagation of errors. Analysis of peak areas was performed in Masslynx (v 4.1) software, and the data were normalized to protein concentrations measured using a Bradford assay with bovine serum albumin (BSA) as the standard, as previously described (26). Data shown represent means and standard deviations of results from three biological replicates.
Synthesis of isotope-labeled internal standards.
Na234S was synthesized using a published protocol (78). Bacillithiol persulfide (BS34SH), cysteine persulfide (CysS34SH), and coenzyme A persulfide (CoAS34SH) were synthesized by reacting the appropriate oxidized disulfide (RSSR) with Na2S to obtain an equimolar mixture of the thiol RS− and the persulfide RSS– (16). Bacillithiol disulfide (BSSB; 5 mM), generously provided by M. Kiethly (Vanderbilt University), CoA disulfide (5 mM), or l-cystine (2.5 mM) was reacted anaerobically with a 5-fold molar excess of Na234S–300 mM degassed phosphate buffer (pH 7.4) at 30°C for 30 min. The concentration of persulfide in the final mixture was quantified by a cold cyanolysis assay (30), and the persulfide was diluted to 0.1 mM in 20 mM Tris-HBr (pH 8.0)–50% acetonitrile–2 mM mBBr and labeled as described above. Standard samples were used without further purification.
Transition metal measurements.
Aliquots of growth medium with or without the addition of 0.2 mM sulfide or 0.2 mM AS were taken, centrifuged to remove any precipitates, diluted 10-fold in 2.5% HNO3, and analyzed using a PerkinElmer Elan II DRC ICP-MS system essentially as described in our previous work (79). To analyze the total cell-associated metal content, 5-ml cultures (OD600 of 0.2) were pelleted, the pellet was resuspended in 400 μl 30% nitric acid, and metal concentrations were determined as described above for the growth medium, with normalization to the amount of protein in each sample (in nanomoles per milligram of protein) as previously described (80). Metal concentrations were determined from a standard curve of 1 to 30 ppb metal stock solutions.
Accession number(s).
The results of all RNAseq experiments have been deposited in the GEO database under GenBank accession number GSE99432.
ACKNOWLEDGMENTS
We thank the late Richard Armstrong and the Vanderbilt Center for Chemical Biology for the gift of bacillithiol disulfide (supported by NIH R01 GM030910), Yifan Zhang for the gift of CstR, Yixiang Zhang and Jon Trinidad for analysis of cross-linked CstR species by mass spectrometry, Jiefei Wang for ICP-MS analysis, and Julia E. Martin for detailed comments on the manuscript.
We gratefully acknowledge support by NIH grants R01 GM097225 and R35 GM118157 (to D.P.G.), R01 AI073843 and R01 AI069233 (to E.P.S.), F32 AI122516 and T32 HL094296 (to L.D.P.), and K22 AI104805 and R01 AI118880 (to T.E.K.-F.) from the U.S. National Institutes of Health.
We declare that we have no competing financial interests.
REFERENCES
- 1.Wang T, Leyh TS. 2012. Three-stage assembly of the cysteine synthase complex from Escherichia coli. J Biol Chem 287:4360–4367. doi: 10.1074/jbc.M111.288423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ayala-Castro C, Saini A, Outten FW. 2008. Fe-S cluster assembly pathways in bacteria. Microbiol Mol Biol Rev 72:110–125, table of contents. doi: 10.1128/MMBR.00034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soutourina O, Poupel O, Coppée JY, Danchin A, Msadek T, Martin-Verstraete I. 2009. CymR, the master regulator of cysteine metabolism in Staphylococcus aureus, controls host sulphur source utilization and plays a role in biofilm formation. Mol Microbiol 73:194–211. doi: 10.1111/j.1365-2958.2009.06760.x. [DOI] [PubMed] [Google Scholar]
- 4.Tanous C, Soutourina O, Raynal B, Hullo MF, Mervelet P, Gilles AM, Noirot P, Danchin A, England P, Martin-Verstraete I. 2008. The CymR regulator in complex with the enzyme CysK controls cysteine metabolism in Bacillus subtilis. J Biol Chem 283:35551–35560. doi: 10.1074/jbc.M805951200. [DOI] [PubMed] [Google Scholar]
- 5.Ji Q, Zhang L, Sun F, Deng X, Liang H, Bae T, He C. 2012. Staphylococcus aureus CymR is a new thiol-based oxidation-sensing regulator of stress resistance and oxidative response. J Biol Chem 287:21102–21109. doi: 10.1074/jbc.M112.359737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gadalla MM, Snyder SH. 2010. Hydrogen-sulfide as a gasotransmitter. J Neurochem 113:14–26. doi: 10.1111/j.1471-4159.2010.06580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mustafa AK, Gadalla MM, Sen N, Kim S, Mu W, Gazi SK, Barrow RK, Yang G, Wang R, Snyder SH. 2009. H2S signals through protein S-sulfhydration. Sci Signal 2:ra72. doi: 10.1126/scisignal.2000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yong QC, Hu LF, Wang S, Huang D, Bian JS. 2010. Hydrogen-sulfide interacts with nitric oxide in the heart: possible involvement of nitroxyl. Cardiovasc Res 88:482–491. doi: 10.1093/cvr/cvq248. [DOI] [PubMed] [Google Scholar]
- 9.Yong QC, Cheong JL, Hua F, Deng LW, Khoo YM, Lee HS, Perry A, Wood M, Whiteman M, Bian JS. 2011. Regulation of heart function by endogenous gaseous mediators—crosstalk between nitric oxide and hydrogen sulfide. Antioxid Redox Signal 14:2081–2091. doi: 10.1089/ars.2010.3572. [DOI] [PubMed] [Google Scholar]
- 10.Eberhardt M, Dux M, Namer B, Miljkovic J, Cordasic N, Will C, Kichko TI, de la Roche J, Fischer M, Suárez SA, Bikiel D, Dorsch K, Leffler A, Babes A, Lampert A, Lennerz JK, Jacobi J, Martí MA, Doctorovich F, Högestätt ED, Zygmunt PM, Ivanovic-Burmazovic I, Messlinger K, Reeh P, Filipovic MR. 2014. H2S and NO cooperatively regulate vascular tone by activating a neuroendocrine HNO-TRPA1-CGRP signalling pathway. Nat Commun 5:4381. doi: 10.1038/ncomms5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shatalin K, Shatalina E, Mironov A, Nudler E. 2011. H2S: a universal defense against antibiotics in bacteria. Science 334:986–990. doi: 10.1126/science.1209855. [DOI] [PubMed] [Google Scholar]
- 12.Gusarov I, Shatalin K, Starodubtseva M, Nudler E. 2009. Endogenous nitric oxide protects bacteria against a wide spectrum of antibiotics. Science 325:1380–1384. doi: 10.1126/science.1175439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adak S, Aulak KS, Stuehr DJ. 2002. Direct evidence for nitric oxide production by a nitric-oxide synthase-like protein from Bacillus subtilis. J Biol Chem 277:16167–16171. doi: 10.1074/jbc.M201136200. [DOI] [PubMed] [Google Scholar]
- 14.Gusarov I, Nudler E. 2005. NO-mediated cytoprotection: instant adaptation to oxidative stress in bacteria. Proc Natl Acad Sci U S A 102:13855–13860. doi: 10.1073/pnas.0504307102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cortese-Krott MM, Kuhnle GG, Dyson A, Fernandez BO, Grman M, DuMond JF, Barrow MP, McLeod G, Nakagawa H, Ondrias K, Nagy P, King SB, Saavedra JE, Keefer LK, Singer M, Kelm M, Butler AR, Feelisch M. 2015. Key bioactive reaction products of the NO/H2S interaction are S/N-hybrid species, polysulfides, and nitroxyl. Proc Natl Acad Sci U S A 112:E4651–E4660. doi: 10.1073/pnas.1509277112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ida T, Sawa T, Ihara H, Tsuchiya Y, Watanabe Y, Kumagai Y, Suematsu M, Motohashi H, Fujii S, Matsunaga T, Yamamoto M, Ono K, Devarie-Baez NO, Xian M, Fukuto JM, Akaike T. 2014. Reactive cysteine persulfides and S-polythiolation regulate oxidative stress and redox signaling. Proc Natl Acad Sci U S A 111:7606–7611. doi: 10.1073/pnas.1321232111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivanovic-Burmazovic I. 2017. HNO generation from NO·, nitrite, inorganic or organic nitrosyls, and crosstalk with H2S, p 68–104. In Doctorovich F, Farmer PJ, Marti MA (ed), The chemistry and biology of nitroxyl (HNO). Elsevier, Amsterdam, Netherlands. [Google Scholar]
- 18.Fehling C, Friedrichs G. 2011. Dimerization of HNO in aqueous solution: an interplay of solvation effects, fast acid-base equilibria, and intramolecular hydrogen bonding? J Am Chem Soc 133:17912–17922. doi: 10.1021/ja2075949. [DOI] [PubMed] [Google Scholar]
- 19.Keceli G, Moore CD, Labonte JW, Toscano JP. 2013. NMR detection and study of hydrolysis of HNO-derived sulfinamides. Biochemistry 52:7387–7396. doi: 10.1021/bi401110f. [DOI] [PubMed] [Google Scholar]
- 20.Filipovic MR. 2017. HNO-thiol relationship, p 105–126. In Doctorovich F, Farmer PJ, Marti MA (ed), The chemistry and biology of nitroxyl (HNO). Elsevior, Amsterdam, Netherlands. [Google Scholar]
- 21.Smulik R, Dębski D, Zielonka J, Michałowski B, Adamus J, Marcinek A, Kalyanaraman B, Sikora A. 2014. Nitroxyl (HNO) reacts with molecular oxygen and forms peroxynitrite at physiological pH. Biological implications. J Biol Chem 289:35570–35581. doi: 10.1074/jbc.M114.597740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldstein S, Czapski G. 1995. The reaction of NO. with O2.- and HO2.: a pulse radiolysis study. Free Radic Biol Med 19:505–510. doi: 10.1016/0891-5849(95)00034-U. [DOI] [PubMed] [Google Scholar]
- 23.Hamer M, Morales Vasquez MA, Doctorovich F. 2017. HNO: redox chemistry and interactions with small inorganic molecules, p 1–9. In Doctorovich F, Farmer PJ, Marti MA (ed), The chemistry and biology of nitroxyl (HNO). Elsevier, Amsterdam, Netherlands. [Google Scholar]
- 24.Castro LA, Robalinho RL, Cayota A, Meneghini R, Radi R. 1998. Nitric oxide and peroxynitrite-dependent aconitase inactivation and iron-regulatory protein-1 activation in mammalian fibroblasts. Arch Biochem Biophys 359:215–224. doi: 10.1006/abbi.1998.0898. [DOI] [PubMed] [Google Scholar]
- 25.Gao XH, Krokowski D, Guan BJ, Bederman I, Majumder M, Parisien M, Diatchenko L, Kabil O, Willard B, Banerjee R, Wang B, Bebek G, Evans CR, Fox PL, Gerson SL, Hoppel CL, Liu M, Arvan P, Hatzoglou M. 2015. Quantitative H2S−mediated protein sulfhydration reveals metabolic reprogramming during the integrated stress response. Elife 4:e10067. doi: 10.7554/eLife.10067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen J, Peng H, Zhang Y, Trinidad JC, Giedroc DP. 2016. Staphylococcus aureus sqr encodes a type II sulfide:quinone oxidoreductase and impacts reactive sulfur speciation in cells. Biochemistry 55:6524–6534. doi: 10.1021/acs.biochem.6b00714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wedmann R, Onderka C, Wei S, Szijártó IA, Miljkovic JL, Mitrovic A, Lange M, Savitsky S, Yadav PK, Torregrossa R, Harrer EG, Harrer T, Ishii I, Gollasch M, Wood ME, Galardon E, Xian M, Whiteman M, Banerjee R, Filipovic MR. 2016. Improved tag-switch method reveals that thioredoxin acts as depersulfidase and controls the intracellular levels of protein persulfidation. Chem Sci 7:3414–3426. doi: 10.1039/c5sc04818d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grossoehme N, Kehl-Fie TE, Ma Z, Adams KW, Cowart DM, Scott RA, Skaar EP, Giedroc DP. 2011. Control of copper resistance and inorganic sulfur metabolism by paralogous regulators in Staphylococcus aureus. J Biol Chem 286:13522–13531. doi: 10.1074/jbc.M111.220012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luebke JL, Shen J, Bruce KE, Kehl-Fie TE, Peng H, Skaar EP, Giedroc DP. 2014. The CsoR-like sulfurtransferase repressor (CstR) is a persulfide sensor in Staphylococcus aureus. Mol Microbiol 94:1343–1360. doi: 10.1111/mmi.12835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Higgins KA, Peng H, Luebke JL, Chang FM, Giedroc DP. 2015. Conformational analysis and chemical reactivity of the multidomain sulfurtransferase, Staphylococcus aureus CstA. Biochemistry 54:2385–2398. doi: 10.1021/acs.biochem.5b00056. [DOI] [PubMed] [Google Scholar]
- 31.Shen J, Keithly ME, Armstrong RN, Higgins KA, Edmonds KA, Giedroc DP. 2015. Staphylococcus aureus CstB is a novel multidomain persulfide dioxygenase-sulfurtransferase involved in hydrogen sulfide detoxification. Biochemistry 54:4542–4554. doi: 10.1021/acs.biochem.5b00584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guimarães BG, Barbosa RL, Soprano AS, Campos BM, de Souza TA, Tonoli CC, Leme AF, Murakami MT, Benedetti CE. 2011. Plant pathogenic bacteria utilize biofilm growth-associated repressor (BigR), a novel winged-helix redox switch, to control hydrogen sulfide detoxification under hypoxia. J Biol Chem 286:26148–26157. doi: 10.1074/jbc.M111.234039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shimizu T, Shen J, Fang M, Zhang Y, Hori K, Trinidad JC, Bauer CE, Giedroc DP, Masuda S. 2017. Sulfide-responsive transcriptional repressor SqrR functions as a master regulator of sulfide-dependent photosynthesis. Proc Natl Acad Sci U S A 114:2355–2360. doi: 10.1073/pnas.1614133114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amatore C, Arbault S, Ducrocq C, Hu S, Tapsoba I. 2007. Angeli’s salt (Na2N2O3) is a precursor of HNO and NO: a voltammetric study of the reactive intermediates released by Angeli’s salt decomposition. ChemMedChem 2:898–903. doi: 10.1002/cmdc.200700016. [DOI] [PubMed] [Google Scholar]
- 35.Trotonda MP, Tamber S, Memmi G, Cheung AL. 2008. MgrA represses biofilm formation in Staphylococcus aureus. Infect Immun 76:5645–5654. doi: 10.1128/IAI.00735-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gopal T, Nagarajan V, Elasri MO. 2015. SATRAT: Staphylococcus aureus transcript regulatory network analysis tool. PeerJ 3:e717. doi: 10.7717/peerj.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nobre LS, Saraiva LM. 2013. Effect of combined oxidative and nitrosative stresses on Staphylococcus aureus transcriptome. Appl Microbiol Biotechnol 97:2563–2573. doi: 10.1007/s00253-013-4730-3. [DOI] [PubMed] [Google Scholar]
- 38.Morrissey JA, Cockayne A, Brummell K, Williams P. 2004. The staphylococcal ferritins are differentially regulated in response to iron and manganese and via PerR and Fur. Infect Immun 72:972–979. doi: 10.1128/IAI.72.2.972-979.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hood MI, Mortensen BL, Moore JL, Zhang Y, Kehl-Fie TE, Sugitani N, Chazin WJ, Caprioli RM, Skaar EP. 2012. Identification of an Acinetobacter baumannii zinc acquisition system that facilitates resistance to calprotectin-mediated zinc sequestration. PLoS Pathog 8:e1003068. doi: 10.1371/journal.ppat.1003068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh VK, Xiong A, Usgaard TR, Chakrabarti S, Deora R, Misra TK, Jayaswal RK. 1999. ZntR is an autoregulatory protein and negatively regulates the chromosomal zinc resistance operon znt of Staphylococcus aureus. Mol Microbiol 33:200–207. doi: 10.1046/j.1365-2958.1999.01466.x. [DOI] [PubMed] [Google Scholar]
- 41.Kuroda M, Hayashi H, Ohta T. 1999. Chromosome-determined zinc-responsible operon czr in Staphylococcus aureus strain 912. Microbiol Immunol 43:115–125. doi: 10.1111/j.1348-0421.1999.tb02382.x. [DOI] [PubMed] [Google Scholar]
- 42.Eicken C, Pennella MA, Chen X, Koshlap KM, VanZile ML, Sacchettini JC, Giedroc DP. 2003. A metal-ligand-mediated intersubunit allosteric switch in related SmtB/ArsR zinc sensor proteins. J Mol Biol 333:683–695. doi: 10.1016/j.jmb.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 43.Kehl-Fie TE, Zhang Y, Moore JL, Farrand AJ, Hood MI, Rathi S, Chazin WJ, Caprioli RM, Skaar EP. 2013. MntABC and MntH contribute to systemic Staphylococcus aureus infection by competing with calprotectin for nutrient manganese. Infect Immun 81:3395–3405. doi: 10.1128/IAI.00420-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haas CE, Rodionov DA, Kropat J, Malasarn D, Merchant SS, de Crécy-Lagard V. 2009. A subset of the diverse COG0523 family of putative metal chaperones is linked to zinc homeostasis in all kingdoms of life. BMC Genomics 10:470. doi: 10.1186/1471-2164-10-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nairn BL, Lonergan ZR, Wang J, Braymer JJ, Zhang Y, Calcutt MW, Lisher JP, Gilston BA, Chazin WJ, de Crécy-Lagard V, Giedroc DP, Skaar EP. 2016. The response of Acinetobacter baumannii to zinc starvation. Cell Host Microbe 19:826–836. doi: 10.1016/j.chom.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Panina EM, Mironov AA, Gelfand MS. 2003. Comparative genomics of bacterial zinc regulons: enhanced ion transport, pathogenesis, and rearrangement of ribosomal proteins. Proc Natl Acad Sci U S A 100:9912–9917. doi: 10.1073/pnas.1733691100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Remy L, Carrière M, Derré-Bobillot A, Martini C, Sanguinetti M, Borezée-Durant E. 2013. The Staphylococcus aureus Opp1 ABC transporter imports nickel and cobalt in zinc-depleted conditions and contributes to virulence. Mol Microbiol 87:730–743. doi: 10.1111/mmi.12126. [DOI] [PubMed] [Google Scholar]
- 48.Ghssein G, Brutesco C, Ouerdane L, Izaute A, Foljcik C, Wang S, Hajjar C, Lobinski R, Lemaire D, Richaud P, Voulhoux R, Espaillet A, Cava F, Pignol D, Borezee-Durant E, Arnoux P. 2016. Biosynthesis of a broad-spectrum nicotianamine-like metallophore in Staphylococcus aureus. Science 352:1105–1109. doi: 10.1126/science.aaf1018. [DOI] [PubMed] [Google Scholar]
- 49.Luther GW, Rickard DT, Theberge S, Olroyd A. 1996. Determination of metal (bi)sulfide stability constants of Mn2+, Fe2+, CO2+, Ni2+, Cu2+, And Zn2+ by voltammetric methods. Environ Sci Technol 30:671–679. doi: 10.1021/es950417i. [DOI] [Google Scholar]
- 50.Shepard W, Soutourina O, Courtois E, England P, Haouz A, Martin-Verstraete I. 2011. Insights into the Rrf2 repressor family—the structure of CymR, the global cysteine regulator of Bacillus subtilis. FEBS J 278:2689–2701. doi: 10.1111/j.1742-4658.2011.08195.x. [DOI] [PubMed] [Google Scholar]
- 51.Nakano MM, Kominos-Marvell W, Sane B, Nader YM, Barendt SM, Jones MB, Zuber P. 2014. spxA2, encoding a regulator of stress resistance in Bacillus anthracis, is controlled by SaiR, a new member of the Rrf2 protein family. Mol Microbiol 94:815–827. doi: 10.1111/mmi.12798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen PR, Bae T, Williams WA, Duguid EM, Rice PA, Schneewind O, He C. 2006. An oxidation-sensing mechanism is used by the global regulator MgrA in Staphylococcus aureus. Nat Chem Biol 2:591–595. doi: 10.1038/nchembio820. [DOI] [PubMed] [Google Scholar]
- 53.Sun F, Ding Y, Ji Q, Liang Z, Deng X, Wong CC, Yi C, Zhang L, Xie S, Alvarez S, Hicks LM, Luo C, Jiang H, Lan L, He C. 2012. Protein cysteine phosphorylation of SarA/MgrA family transcriptional regulators mediates bacterial virulence and antibiotic resistance. Proc Natl Acad Sci U S A 109:15461–15466. doi: 10.1073/pnas.1205952109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fuchs S, Pané-Farré J, Kohler C, Hecker M, Engelmann S. 2007. Anaerobic gene expression in Staphylococcus aureus. J Bacteriol 189:4275–4289. doi: 10.1128/JB.00081-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pietiäinen M, François P, Hyyryläinen HL, Tangomo M, Sass V, Sahl HG, Schrenzel J, Kontinen VP. 2009. Transcriptome analysis of the responses of Staphylococcus aureus to antimicrobial peptides and characterization of the roles of vraDE and vraSR in antimicrobial resistance. BMC Genomics 10:429. doi: 10.1186/1471-2164-10-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Keceli G, Toscano JP. 2012. Reactivity of nitroxyl-derived sulfinamides. Biochemistry 51:4206–4216. doi: 10.1021/bi300015u. [DOI] [PubMed] [Google Scholar]
- 57.Anand P, Hausladen A, Wang YJ, Zhang GF, Stomberski C, Brunengraber H, Hess DT, Stamler JS. 2014. Identification of S-nitroso-CoA reductases that regulate protein S-nitrosylation. Proc Natl Acad Sci U S A 111:18572–18577. doi: 10.1073/pnas.1417816112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ma Z, Chandrangsu P, Helmann TC, Romsang A, Gaballa A, Helmann JD. 2014. Bacillithiol is a major buffer of the labile zinc pool in Bacillus subtilis. Mol Microbiol 94:756–770. doi: 10.1111/mmi.12794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schlag S, Nerz C, Birkenstock TA, Altenberend F, Götz F. 2007. Inhibition of staphylococcal biofilm formation by nitrite. J Bacteriol 189:7911–7919. doi: 10.1128/JB.00598-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dunman PM, Murphy E, Haney S, Palacios D, Tucker-Kellogg G, Wu S, Brown EL, Zagursky RJ, Shlaes D, Projan SJ. 2001. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J Bacteriol 183:7341–7353. doi: 10.1128/JB.183.24.7341-7353.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen PR, Nishida S, Poor CB, Cheng A, Bae T, Kuechenmeister L, Dunman PM, Missiakas D, He C. 2009. A new oxidative sensing and regulation pathway mediated by the MgrA homologue SarZ in Staphylococcus aureus. Mol Microbiol 71:198–211. doi: 10.1111/j.1365-2958.2008.06518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Luong TT, Dunman PM, Murphy E, Projan SJ, Lee CY. 2006. Transcription profiling of the mgrA regulon in Staphylococcus aureus. J Bacteriol 188:1899–1910. doi: 10.1128/JB.188.5.1899-1910.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bischoff M, Dunman P, Kormanec J, Macapagal D, Murphy E, Mounts W, Berger-Bächi B, Projan S. 2004. Microarray-based analysis of the Staphylococcus aureus sigmaB regulon. J Bacteriol 186:4085–4099. doi: 10.1128/JB.186.13.4085-4099.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saïd-Salim B, Dunman PM, McAleese FM, Macapagal D, Murphy E, McNamara PJ, Arvidson S, Foster TJ, Projan SJ, Kreiswirth BN. 2003. Global regulation of Staphylococcus aureus genes by Rot. J Bacteriol 185:610–619. doi: 10.1128/JB.185.2.610-619.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tuchscherr L, Bischoff M, Lattar SM, Noto Llana M, Pförtner H, Niemann S, Geraci J, Van de Vyver H, Fraunholz MJ, Cheung AL, Herrmann M, Völker U, Sordelli DO, Peters G, Löffler B. 2015. Sigma factor SigB Is crucial to mediate Staphylococcus aureus adaptation during chronic infections. PLoS Pathog 11:e1004870. doi: 10.1371/journal.ppat.1004870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shafirovich V, Lymar SV. 2002. Nitroxyl and its anion in aqueous solutions: spin states, protic equilibria, and reactivities toward oxygen and nitric oxide. Proc Natl Acad Sci U S A 99:7340–7345. doi: 10.1073/pnas.112202099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gherasim C, Yadav PK, Kabil O, Niu WN, Banerjee R. 2014. Nitrite reductase activity and inhibition of H2S biogenesis by human cystathionine β-synthase. PLoS One 9:e85544. doi: 10.1371/journal.pone.0085544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Filipovic MR, Eberhardt M, Prokopovic V, Mijuskovic A, Orescanin-Dusic Z, Reeh P, Ivanovic-Burmazovic I. 2013. Beyond H2S and NO interplay: hydrogen sulfide and nitroprusside react directly to give nitroxyl (HNO). A new pharmacological source of HNO. J Med Chem 56:1499–1508. doi: 10.1021/jm3012036. [DOI] [PubMed] [Google Scholar]
- 69.Sen N, Paul BD, Gadalla MM, Mustafa AK, Sen T, Xu R, Kim S, Snyder SH. 2012. Hydrogen-sulfide-linked sulfhydration of NF-kappaB mediates its antiapoptotic actions. Mol Cell 45:13–24. doi: 10.1016/j.molcel.2011.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pacelli R, Wink DA, Cook JA, Krishna MC, Degraff W, Friedman N, Tsokos M, Samuni A, Mitchell JB. 1995. Nitric-oxide potentiates hydrogen peroxide-induced killing of Escherichia coli. J Exp Med 182:1469–1479. doi: 10.1084/jem.182.5.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Collins JA, Irnov I, Baker S, Winkler WC. 2007. Mechanism of mRNA destabilization by the glmS ribozyme. Genes Dev 21:3356–3368. doi: 10.1101/gad.1605307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bae T, Schneewind O. 2006. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55:58–63. doi: 10.1016/j.plasmid.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 76.Toledo-Arana A, Merino N, Vergara-Irigaray M, Débarbouillé M, Penadés JR, Lasa I. 2005. Staphylococcus aureus develops an alternative, ica-independent biofilm in the absence of the arlRS two-component system. J Bacteriol 187:5318–5329. doi: 10.1128/JB.187.15.5318-5329.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shen X, Kolluru GK, Yuan S, Kevil CG. 2015. Measurement of H2S in vivo and in vitro by the monobromobimane method. Methods Enzymol 554:31–45. doi: 10.1016/bs.mie.2014.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Seela JL, Knapp MJ, Kolack KS, Chang HR, Huffman JC, Hendrickson DN, Christou G. 1998. Structural and magnetochemical properties of mono-, di-, and trinuclear manganese(III) dithiolate complexes. Inorg Chem 37:516–525. doi: 10.1021/ic970587r. [DOI] [PubMed] [Google Scholar]
- 79.Jacobsen FE, Kazmierczak KM, Lisher JP, Winkler ME, Giedroc DP. 2011. Interplay between manganese and zinc homeostasis in the human pathogen Streptococcus pneumoniae. Metallomics 3:38–41. doi: 10.1039/C0MT00050G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Martin JE, Lisher JP, Winkler ME, Giedroc DP. 2017. Perturbation of manganese metabolism disrupts cell division in Streptococcus pneumoniae. Mol Microbiol 104:334–348. doi: 10.1111/mmi.13630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mishanina TV, Libiad M, Banerjee R. 2015. Biogenesis of reactive sulfur species for signaling by hydrogen sulfide oxidation pathways. Nat Chem Biol 11:457–464. doi: 10.1038/nchembio.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cuevasanta E, Zeida A, Carballal S, Wedmann R, Morzan UN, Trujillo M, Radi R, Estrin DA, Filipovic MR, Alvarez B. 2015. Insights into the mechanism of the reaction between hydrogen sulfide and peroxynitrite. Free Radic Biol Med 80:93–100. doi: 10.1016/j.freeradbiomed.2014.12.017. [DOI] [PubMed] [Google Scholar]
- 83.Wedmann R, Zahl A, Shubina TE, Dürr M, Heinemann FW, Bugenhagen BE, Burger P, Ivanovic-Burmazovic I, Filipovic MR. 2015. Does perthionitrite (SSNO–) account for sustained bioactivity of NO? A (bio)chemical characterization. Inorg Chem 54:9367–9380. doi: 10.1021/acs.inorgchem.5b00831. [DOI] [PubMed] [Google Scholar]
- 84.Cuevasanta E, Lange M, Bonanata J, Coitiño EL, Ferrer-Sueta G, Filipovic MR, Alvarez B. 2015. Reaction of hydrogen sulfide with disulfide and sulfenic acid to form the strongly nucleophilic persulfide. J Biol Chem 290:26866–26880. doi: 10.1074/jbc.M115.672816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Suarez SA, Neuman NI, Muñoz M, Álvarez L, Bikiel DE, Brondino CD, Ivanović-Burmazović I, Miljkovic JLJ, Filipovic MR, Martí MA, Doctorovich F. 2015. Nitric oxide is reduced to HNO by proton-coupled nucleophilic attack by ascorbate, tyrosine, and other alcohols. A new route to HNO in biological media? J Am Chem Soc 137:4720–4727. doi: 10.1021/ja512343w. [DOI] [PubMed] [Google Scholar]
- 86.Filipovic MR, Miljkovic JLJ, Nauser T, Royzen M, Klos K, Shubina T, Koppenol WH, Lippard SJ, Ivanović-Burmazović I. 2012. Chemical characterization of the smallest S-nitrosothiol, HSNO; cellular cross-talk of H2S and S-nitrosothiols. J Am Chem Soc 134:12016–12027. doi: 10.1021/ja3009693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Doctorovich F, Bikiel DE, Pellegrino J, Suárez SA, Martí MA. 2014. Reactions of HNO with metal porphyrins: underscoring the biological relevance of HNO. Acc Chem Res 47:2907–2916. doi: 10.1021/ar500153c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
qRT-PCR validation of the RNAseq results. (A) Subset of the genes (expressed as S. aureus strain Newman locus tags, NWMN_wxyz) induced (compare to Table 1) or repressed (Table 2) upon treatment with 0.2 mM Na2S at 10 min posttreatment relative to unstressed cells. (B) Subset of genes induced (compare to Table 3) and repressed (Table 4) in the unstressed, early log-phase ΔcstR strain relative to the isogenic wild-type strain cultured under the same conditions. (C) Subset of the genes induced (compare to Table 1) and repressed (Table 2) upon treatment with 0.2 mM AS at 10 min posttreatment relative to unstressed cells. Download FIG S1, EPS file, 1.1 MB (1.1MB, eps) .
Copyright © 2017 Peng et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Complete transcriptomic data and sequences of qRT-PCR primers used in this study. (A) Excel file listing all genes whose expression is altered in sulfide-treated cells (sheets 1 to 2, corresponding to Fig. 4 and Tables 1 and 2, main text), in the ΔcstR strain (sheets 3 to 4, corresponding to Fig. 4 and Tables 3 and 4, main text), in AS-treated cells (sheets 5 to 6, corresponding to Fig. 6 and Tables 1 and 2, main text), and in CP-treated cells (sheets 7 to 8, corresponding to Fig. 3, main text) that meet the minimal acceptance criteria (2-fold change in expression; adjusted P value of ≤0.05). (B) A list of all the primers used in the qRT-PCR experiments. Download TABLE S1, XLSX file, 0.1 MB (103KB, xlsx) .
Copyright © 2017 Peng et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Analysis of all transcriptomic (RNAseq) changes in the sulfide-stressed (HS−) and AS-treated cells that showed significant changes in other stress-treated cells. These stresses included oxidative stress (ROS) (10 mM H2O2) (A), nitrosative stress (RNS) (150 μM GSNO [S-nitrosoglutathione]) (B), and combined ROS/RNS stress (10 mM H2O2 plus 150 μM GSNO) (C) (37). The cutoff is 2-fold change. Gene names are shown (where known) on the right. Note that only genes that are significantly regulated by either sulfide or AS stress or both sulfide and AS stress and under other stress conditions are shown. (A) The 90 genes shown represent the 45% of the transcriptome that is affected by acute-phase ROS, corresponding to 56% of upregulated and 18% of downregulated genes that change in the same direction. (B) The 43 genes shown represent the 41% of the transcriptome that is affected by GSNO, corresponding to 37% of upregulated and 19% of downregulated genes that change in the same direction. (C) The 123 genes shown represent the 43% of the transcriptome that is affected by the combined ROS/RNS stress, corresponding to 33% of upregulated and 36% of downregulated genes that change in the same direction. Download FIG S2, EPS file, 2.6 MB (2.6MB, eps) .
Copyright © 2017 Peng et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The ratio of reduced to oxidized bacillithiol (BSH/BSSB) in the sulfide-stressed (HS–) and HNO-treated cells compared to untreated cells. Sulfide treatment increased BSH/BSSB levels significantly, but the ratio remained unchanged under AS treatment. The experiments were done in triplicate, with standard deviations shown as error bars. Statistical significance relative to untreated cells was established by using a paired t test (***, P < 0.001; n.s., no significant change). Download FIG S3, EPS file, 0.6 MB (615.9KB, eps) .
Copyright © 2017 Peng et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Metal analysis of the HHWm growth medium before and after treatment with 0.2 mM NaHS (A) and 0.2 mM AS (B). ICP-MS was used to quantify the concentrations of transition metal ions in the HHWm growth medium (gray bars), HHWm plus chloramphenicol plus 0.2 mM thiosulfate (TS) as the sole sulfur source (red bars), or HHWm plus chloramphenicol plus 0.2 mM thiosulfate plus 0.2 mM NaHS or AS (light blue bars). Sulfide treatment led to a significant decrease in the level of bioavailable (soluble) Zn (15.2 µM to 1.2 µM), with correspondingly small changes in the levels of Cu (approximately 2-fold) and Ni (less than 2-fold). AS treatment led to a significant decrease in the level of bioavailable (soluble) Zn (18.2 µM to 5.9 µM), with a correspondingly small change in the level of Ni (1.8-fold). Download FIG S4, EPS file, 2.7 MB (2.8MB, eps) .
Copyright © 2017 Peng et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Measurement of the expression levels of cstA in the cst operon in cells treated with 0.5 mM methylamine hexamethylene methylamine NONOate (MAHMA NONOate) by qRT-PCR. cstA expression was seen 10 min or 30 min postexposure relative to unstressed cell results. Error bars represent standard deviations of results of triplicate biological experiments, with statistical significance established using a paired t test relative to untreated wild-type cell results (0 min) (*, P < 0.05; n.s., no significant change). Download FIG S5, EPS file, 0.6 MB (633.9KB, eps) .
Copyright © 2017 Peng et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
CstR reacts with AS to form cross-linked CstR characterized by an interprotomer disulfide bond between C31 and C60′. (A) CstR was mostly in reduced form. However, the cross-linked CstR was largely observed after the reaction with 20-fold molar excess AS (B) but not in the reaction with nitrite (C), another product of AS decomposition. (D) The formation of a disulfide bond between C31 and C60′ was confirmed by LC-MS/MS. The expected m/z of reduced CstR monomer is 9,641.2. Electron transfer dissociation (ETD) mode was used for the sequencing analysis. Download FIG S6, EPS file, 2.9 MB (2.9MB, eps) .
Copyright © 2017 Peng et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Analysis of cellular metal before and after treatment with 0.2 mM AS (A) and 0.2 mM sodium nitrite (B). ICP-MS was used to quantify the concentrations of transition metal ions in the untreated cell (gray bars) or in the cell treated with 0.2 mM AS or nitrite (light blue bars). (A) AS treatment did not lead to a significant change in the total levels of cellular transition metals, with the exception of Cu, whose level increased approximately 4-fold. (B) Nitrite treatment with the same concentration led to no significant change in all cellular metals. Download FIG S7, EPS file, 0.9 MB (944.2KB, eps) .
Copyright © 2017 Peng et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Analysis of all transcriptomic (RNAseq) changes in the sulfide-stressed (HS−) and AS-treated cells compared to the regulons of selected virulence factors. Genes associated with the sarA (A) (60), sarZ (B) (61), mgrA (C) (62), sigB (D) (63), and rot (E) (64) regulons are shown. The cutoff is 2-fold change. Gene names are shown (where known) on the right. Note that only genes that are significantly regulated by either sulfide or AS stress or both sulfide and AS stress and the indicated virulence factor regulon are shown in each panel. With the exception of the sarZ and mgrA experiments, the transcriptomic profiling experiments were carried out using post-exponential-phase or stationary-phase cultures, which are different from the growth conditions used in our RNAseq experiments. (A) sarA: the 48 genes shown represent the 40% of the transcriptome that is affected by sarA, corresponding to 12% of upregulated and 2% of downregulated genes that change in the same direction. (B) sarZ: the 33 genes shown represent the 57% of the transcriptome that is affected by sarZ, corresponding to 0% of upregulated and 21% of downregulated genes that change in the same direction. (C) mgrA: the 53 genes shown represent the 15% of the transcriptome that is affected by mgrA, corresponding to 5% of upregulated and 2% of downregulated genes that change in the same direction. (D) sigB: the 136 genes shown represent the 54% of the transcriptome that is affected by sigB, corresponding to 56% of upregulated and 32% of downregulated genes that change in the same direction. (E) The 47 genes shown represent the 32% of the transcriptome that is affected by rot, corresponding to 19% of upregulated and 12% of downregulated genes that change in the same direction. Download FIG S8, PDF file, 0.6 MB (674.2KB, pdf) .
Copyright © 2017 Peng et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Clustering analysis that summarizes all transcriptomic (RNAseq) changes in the ΔcstR, sulfide-stressed (HS–), AS (HNO)-treated, and calprotectin (CP)-treated cells that showed significant pairwise changes in expression. This figure recapitulates the data presented in Fig. 2 and 7 in the main text but provides more detail. A listing of all transcriptomic changes is provided in Table S1A. Locus tags are indicated on the left, with gene names shown (where known) on the right. Additional description is provided on the far right; the absence of an entry indicates a hypothetical protein of unknown function. Download FIG S9, PDF file, 0.6 MB (585.4KB, pdf) .
Copyright © 2017 Peng et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.