Abstract
We have generated a mouse model for hepatocellular carcinoma using somatic delivery of oncogene-bearing avian retroviral vectors to the liver cells of mice expressing the viral receptor TVA under the control of the albumin gene promoter (Alb-TVA mice). Viruses encoding mouse polyoma virus middle T antigen (PyMT) induced tumors, which can be visualized with magnetic resonance imaging, in 65% of TVA-positive animals. While these tumors can exceed 10 mm in diameter, they do not invade locally or metastasize to the lungs. Delivery of PyMT-expressing viruses to Alb-TVA mice lacking an intact p53 gene does not increase tumor incidence. However, the resulting tumors are poorly differentiated, invasive, and metastatic to the lungs. Gene expression microarrays identified over 100 genes that are differentially expressed between tumors found in p53 wild-type and p53 null mice. Some of these genes, such as cathepsin E and Igf2, have been previously implicated in tumor cell migration and invasion. Tumors induced in p53 null, TVA transgenic mice by PyMT mutants with changes in specific tyrosine residues fail to form metastases, indicating that metastasis is dependent on both the oncogene and the absence of p53.
Hepatocellular carcinoma (HCC) exerts a significant toll on human health worldwide, with more than 250,000 new cases annually and a 5-year survival rate of less than 5% (36). Current treatment modalities are largely unsuccessful: single-agent and combination chemotherapy regimens have proven to be ineffective in the treatment of HCC patients (24). Therefore, surgery remains the only curative option for HCC; however, for most patients this is not a viable option because of significant liver cirrhosis or invasive and metastatic disease that has spread to the lymph nodes, portal vein, or lungs (20).
HCC is strongly associated with chronic infection by hepatitis B virus (HBV) and hepatitis C virus (HCV): incidence of HCC correlates with HBV and HCV infection rates within a population, and the average age of onset is inversely correlated with the prevalence of HBV infection (4, 39, 45). Although the precise mechanisms by which HBV and HCV predispose to HCC are unclear, the current belief is that the liver damage and regeneration associated with chronic viral infection provide a context where oncogenic genetic alterations can occur and become fixed. The X protein encoded by HBV, through its regulation of calcium signaling that is critical for HBV replication, is thought to play an important role in this process (6). Other factors that predispose to the occurrence of HCC are prolonged exposure to aflatoxin B1, cirrhosis of any etiology, hemochromatosis, congenital hepatic fibrosis, and biliary atresia (22).
Alterations in the TP53 gene are frequently found in HCC. Further, the absence of TP53 gene alterations in hepatic adenomas suggests that this is a late event in tumor progression (7). Silencing of the INK4A locus has also been observed in a majority of HCC patients (7, 26, 28). In addition, several chromosome arms—including 1p, 4q, 6q, 9p, 13q, and 17p—are deleted in significant subsets of human HCC, suggesting the presence of important tumor suppressor genes at those loci (7). Interestingly, deregulation of the Wnt signaling pathway appears to be an important event in a subset of HCC tumors. Activating mutations in the positive effector β-catenin have been described, as have inactivating mutations in axin, a negative regulator of the Wnt signaling pathway (7). Furthermore, amplifications of the c-myc and CCND1 oncogenes, which are downstream targets of the Wnt signaling pathway, have been described in HCC patients (1, 32).
Mouse models for HCC have been generated by several investigators using traditional transgenic approaches and tetracycline-regulated transgene expression (5, 8, 9, 18, 27, 31, 42, 44, 48). These models have yielded important information about HCC but have two very important limitations. First, the transgene is expressed in all of the target cells, creating a field effect in which every hepatocyte within the liver has altered gene expression. Indeed, in some of the published models mice die shortly after birth with hepatic defects and atypical liver architecture (42). Second, it is very expensive and time consuming to cross these models to generate models with combinations of genetic lesions. This has prevented direct comparison of different mouse models for HCC. Clearly, a mouse model in which the effects of different oncogenes could be directly compared would represent a significant advance in the field. Recently, Harada et al. have described an HCC mouse model that bypasses some of these deficiencies by using adenovirus-mediated delivery of Cre recombinase to activate expression of constitutively active forms of the H-Ras and β-catenin oncoproteins (14).
We report here the generation of a mouse model for HCC through the targeted delivery of oncogene-bearing avian leukosis sarcoma virus subgroup A (ALSV-A)-based RCAS vectors (replication-competent ALSV retroviral vectors) into transgenic mice in which expression of the ALSV-A receptor TVA is regulated by the albumin gene promoter and enhancer sequences. Mammalian cells are normally resistant to infection by ALSV and ALSV-based vectors (49). However, expression of the ALSV-A receptor, TVA, renders mammalian cells susceptible to infection by ALSV-A and ALSV-A-based vectors (3, 51). Delivery of avian cells that produce RCAS viruses encoding mouse polyoma virus middle T antigen (PyMT) induces the formation of liver adenomas with high levels of penetrance. The induced tumors display increased proliferation, low apoptotic rates, induction of angiogenesis, and activation of downstream effectors of PyMT, including Akt and Erk (extracellular signal-regulated kinase). Delivery of RCAS-PyMT producer cells to TVA-expressing mice deficient for the tumor suppressor protein p53 induces progression to invasive carcinomas that metastasize to the lung. Introduction of phosphorylation site mutants of PyMT into TVA-positive, p53 null mice demonstrates that the activity of the oncogene is critical for the invasive and metastatic behavior of tumors in p53 null mice. Finally, utilizing gene expression microarrays, we have identified several potential mediators of the metastatic tumor behavior. Thus, we have generated a somatic model for HCC that can be utilized to dissect the molecular pathways involved in the initiation and progression of this malignancy.
MATERIALS AND METHODS
Transgenic mice and animal care.
The albumin-tv-a transgene was constructed with pBluescript SK(+) by fusing a 2.2-kb DNA fragment containing albumin gene promoter-enhancer sequences, consisting of 200 bp of proximal albumin gene promoter sequence fused to 2 kb of enhancer sequence located 8 kb upstream, to a 1.2-kb BamHI fragment containing the 800-bp tv-a cDNA fused to a simian virus 40 intron and polyadenylation sequences (gift of G. Fisher) (3, 38). The 3.4-kb transgene was isolated from vector sequences by digestion with NotI and XhoI, gel purified, and resuspended in Tris-EDTA for pronuclear injection. Albumin-tv-a animals were maintained on a CBA/CAJ × C57BL/6J mixed genetic background. p53 null mice with a Sv129 × C57BL/6 mixed genetic background have been previously described (17). Animals were kept in specific pathogen-free housing with abundant food and water under guidelines approved by the Memorial Sloan-Kettering Cancer Center Institutional Animal Care and Use Committee and Research Animal Resource Center.
Virus delivery.
The RCAS-green fluorescent protein (GFP), RCAS-PyMT, and RCAS-c-myc vectors have been previously described (12, 16, 35). DF1 chicken fibroblasts (15, 43) transfected with RCAS vectors were maintained in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum in humidified 37°C incubators under 5% CO2. Cells to be injected were harvested, washed once with phosphate-buffered saline (PBS), and resuspended in PBS at a final concentration of 2 × 105 cells/μl. A total of 5 μl of the cell suspension was delivered by injection into the liver parenchyma of 2- or 3-day old animals with Hamilton syringes attached to 26-gauge needles.
Tumor harvest and histology.
Animals were sacrificed with a lethal dose of CO2 per institutional guidelines. Tumors, liver tissue, and lungs were removed and either fixed in 10% buffered formalin overnight at room temperature or snap-frozen in liquid nitrogen. Fixed tissues were paraffin embedded, and 5-μm sections were placed on sialynated slides at Histoserv, Inc. (Gaithersburg, Md.).
Magnetic resonance imaging.
Mice were assessed individually by magnetic resonance imaging (MRI) for tumor establishment. Images were obtained on a 1.5T General Electric LX Echo Speed Signa scanner (General Electric Medical Systems, Milwaukee, Wis.) with homemade foil solenoid coils (inner diameter, 27 mm). The mice were anesthetized with isofluorane throughout image acquisition. Images were acquired with a two-dimensional spin-echo pulse sequence. Initially, the animal was positioned in the coil in a holder, and sagittal scout T1 weighted spin-echo images and axial scout T2 weighted spin-echo images were obtained to localize the region of interest. For tumor detection, high-quality T1 and T2 (spin-spin relaxation time) weighted fast spin-echo transverse images were obtained with in-plane resolution of 156 by 156 μm or 156 by 208 μm and 1.5-mm-thick slices.
Immunohistochemistry.
Paraffin sections were deparaffinized and rehydrated by passage through Clear-Rite 3 and a graded alcohol series. Endogenous peroxidase activity was inactivated by treatment with 3% hydrogen peroxide, after which antigen retrieval was performed utilizing heated citric buffer. Slides were blocked for 1 h and then incubated with a rabbit anti-Ki67 antibody (Novocastra) at a dilution of 1:1,000 for 1 h at room temperature. After being washed with PBS, slides were incubated with secondary antibody according to the manufacturer's instructions (Vector Labs, Burlingame, Calif.). Substrate incubation and color development were performed according to the manufacturer's instructions (Vector Labs).
In situ hybridization. Freshly generated paraffin sections were rehydrated as described above. After being blocked for 3 h in prehybridization solution (21), slides were hybridized to 4 × 106 cpm of 33P-labeled antisense RNA probes specific for PyMT. Washes were performed as previously described (50). Slides were then dipped in warm photographic emulsion and exposed for 2 weeks before development.
RNA isolation.
Frozen tissue samples were pulverized with a mortar and pestle, the ground tissue was lysed in Trizol reagent (Invitrogen), and RNA was extracted according to the manufacturer's instructions. The isolated RNA was purified over an RNeasy column (QIAGEN) according to the manufacturer's protocol. The RNA concentration was determined by a spectrophotometer, ethanol precipitated, and resuspended at a final concentration of 1 μg/μl.
Immunoblotting.
DF1 cells were collected in cell-scraping buffer, pelleted, and lysed in 10% sodium dodecyl sulfate by being boiled for 10 min. Chromatin was then sheared by passage through a 27-gauge needle. Protein concentration was determined with the BCA assay kit (Pierce, Rockford, Ill.), and equal amounts of protein were loaded per lane. Ground frozen tissue samples were lysed in 10% sodium dodecyl sulfate by being boiled as described above. Protein concentrations were determined as described above, and equal amounts of protein were loaded per lane. After being transferred to nitrocellulose membranes, primary antibodies were incubated overnight at 4°C in Tris-buffered saline-1% Tween 20 and the appropriate blocking reagent, according to the manufacturer's instructions. After being washed with Tris-buffered saline-0.1% Tween 20, blots were incubated with the appropriate secondary antibodies (Jackson Immunochemicals) at room temperature for 45 min. Chemiluminescence was performed with Super Signal reagent (Pierce). Primary antibodies used were as follows: rat anti-polyoma T antigens, 1:3,000 dilution (gift of Michelle Fluck); rabbit anti-phosphorylated AKT (anti-p-AKT), 1:1,000 dilution (Cell Signaling); rabbit anti-p-mitogen-activated protein kinase (anti-p-MAPK), 1:1,000 dilution (Cell Signaling); rabbit anti-Akt, 1:1,000 dilution (Cell Signaling); rabbit anti-MAPK, 1:1,000 dilution (Cell Signaling); mouse anti-α-tubulin, 1:2,000 dilution (Sigma).
Reverse transcription-PCR (RT-PCR).
A total of 0.5 μg of purified total RNA was utilized for RT-PCR. All RT-PCRs were performed with the Superscript One-Step RT-PCR kit (Invitrogen). For amplification of cathepsin E, reverse transcription was performed at 55°C for 30 min, followed by denaturing at 94°C for 2 min and amplification for 35 cycles with the following parameters: denaturing at 94°C for 30 s, annealing at 50°C for 30 s, and extension at 72°C for 1 min. Amplification conditions for insulin-like growth factor 2 (Igf2) and H19 were as above for cathepsin E, except the annealing temperature was 60°C for Igf2 and 58°C for H19. For β-actin, reactions were performed under identical conditions except the amplification was performed for 25 cycles. For tv-a, reactions were performed at an annealing temperature of 55°C for 35 cycles in the presence of 5% dimethyl sulfoxide.
Primers were as follows: tva5, 5′CTGCTGCCCGGTAACGTGACCGG 3′; tva31, 5′GCCCTGGGGAAGGTCCTGCCC 3′; cathepsin E F1, 5′ATAAGAGTTGCTTAAAGTCG 3′; cathepsin E B1, 5′GAGAAATGATTCCCTACCTC 3′; Igf2 F2, 5′TGGTCCCAGAGAGGTTTTAGGTGG 3′; Igf2 B2, 5′ACTTGCTCCCGCCTGATGTAAC 3′; H19 F1, 5′AGGGGGCCTGGTGAGAAGAA 3′; H19 B1, 5′ATGGGAATGGTGTGTCTGCA 3′; actin F1, 5′GGCTGTATTCCCCTCCATCG 3′; and actin B1, 5′AGATGGGCACAGTGTGGGTG 3′.
Gene expression array analysis.
The quality of RNA was ensured before labeling by analysis of 20 to 50 ng of each sample with the RNA 6000 NanoAssay and a Bioanalyzer 2100 (Agilent). Samples with a 28S/18S ribosomal peak ratio of 1.8 to 2.0 were considered suitable for labeling. For samples meeting this standard, 5 μg of total RNA was used for cDNA synthesis with an oligo(dT)-T7 primer and the SuperScript Double-Stranded cDNA Synthesis kit (Invitrogen). Synthesis, linear amplification, and labeling of cRNA were accomplished by transcription in vitro with the MessageAmp cRNA Kit (Ambion) and biotinylated nucleotides (Enzo Diagnostics). Ten micrograms of labeled and fragmented cRNA was then hybridized to the murine genome array U74A, version 2.0 (Affymetrix), which contains ∼12,000 oligonucleotide-based probe sets, at 45°C for 16 h. Automated washing and staining were performed with the Affymetrix Fluidics Station 400 according to the manufacturer's protocols, and probe intensities were measured with the argon laser confocal GeneArray scanner (Hewlett-Packard). Raw expression data were analyzed with Microarray Analysis 5.0 software (Affymetrix). Data were normalized to a target intensity of 500 to account for differences in global chip intensity. The data were then clustered both by genes and by samples. For gene clustering, the data were filtered with the P value from a Wilcoxon test between tumor and normal samples. The genes selected in this manner were then clustered with the angle correlation metric and a variant of the k means-clustering algorithm (46). For the clustering of samples, the genes were first filtered to include only those that scored present in at least four of the samples. The correlation metric was used with average linkage hierarchical clustering. To assess the robustness of the clustering results, a parametric bootstrapping procedure was done. The data was resampled and clustered 1,000 times, and a final tree was built by the consensus tree method (11). In addition, to find genes that discriminated between the tumor classes (p53 wild type versus p53 null) a variant of the t test was used. In this t test, the variance was regularized by a term that depended on the overall intensity of the samples (2).
RESULTS
Generation of albumin-tv-a transgenic mice.
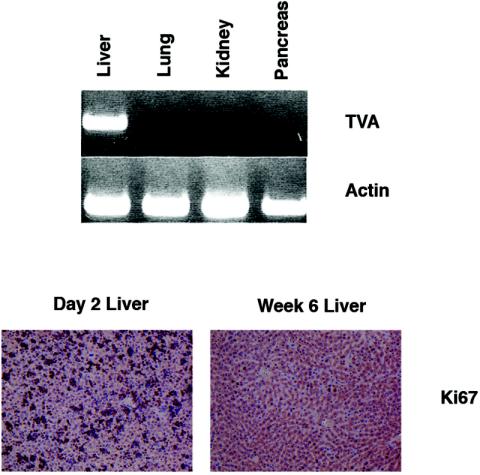
We sought to develop a new mouse model for HCC through the somatic introduction of oncogenes into hepatocytes. We therefore generated transgenic animals in which the receptor for ALSV-A, TVA, was produced specifically within the liver. This approach has been successfully utilized in previously described mouse models (25, 34). We fused promoter and enhancer sequences from the albumin gene, previously demonstrated to direct liver-specific expression (38), upstream of a cDNA encoding the glycosylphosphatidylinositol-anchored isoform of TVA (3, 51). Attempts to identify the presence of the receptor in liver tissue sections from albumin-tv-a transgenic mice by immunostaining failed (data not shown). We have, however, detected TVA by immunostaining in transgenic mice designed to express the receptor in other tissues (25, 34). Despite the absence of visible staining in the liver, the tv-a transcript could be readily identified by RT-PCR within the liver but not in other organs tested, including the lung (Fig. 1, top). Three independent transgenic lines showed liver-specific expression of tv-a by RT-PCR. One of these lines, line B, was subsequently used for tumor induction studies.
FIG. 1.
Characterization of albumin-tv-a transgenic mice. (Top) Ethidium bromide-stained agarose gel showing the presence of TVA mRNA in selected tissues in albumin-tv-a transgenic mice, as detected by RT-PCR. Amplification of β-actin mRNA is shown as a control in the lower panel. (Bottom) Identification of proliferating cells in the liver of 2-day-old mice (left) and 6-week-old mice (right) by immunohistochemical staining for the proliferation-associated marker Ki67. Original magnification, ×100.
RCAS vectors are replication-competent ALSV retroviral vectors. Like other oncoretroviruses, RCAS viruses require division of target cells for integration of the reverse-transcribed viral genome into the host genome (30, 40). We therefore assayed the proliferation status of liver cells in newborn mice and 6-week-old adults. Staining for the proliferation marker Ki67 demonstrated high rates of cell division in the livers of newborn mice, with significantly lower rates in adult livers (Fig. 1, bottom).
RCAS-PyMT induces liver tumors with high penetrance.
To attempt to induce liver tumors, we injected DF-1 chicken fibroblasts producing RCAS viruses encoding PyMT into the livers of 2- or 3-day-old albumin-tv-a transgenic animals, when the organ is easily visualized beneath the skin. A pilot experiment was performed on a litter of three tv-a-positive mice. Sacrifice of the litter at 3 months of age identified a single tumor-bearing animal. Given these findings, we subsequently injected other litters with RCAS-PyMT and sacrificed the mice at either 4 or 6 months of age to identify the presence of tumors. Of the animals analyzed in this fashion, 8 of 11 mice sacrificed at 4 months of age, and 9 of 15 mice sacrificed at 6 months of age displayed grossly visible liver tumors (Table 1). Further, tumor formation was dependent on the presence of TVA, as none of 11 tv-a-negative littermates injected with RCAS-PyMT developed tumors. In addition, only 1 of 16 tv-a-positive animals injected with RCAS-GFP and aged for 13 months developed a liver tumor (Table 1). This tumor may have occurred as a result of insertional mutagenesis, although we have not assayed for the presence of integrated proviral DNA.
TABLE 1.
Tumor induction by RCAS-PyMT in albumin-tv-a transgenic micea
| Albumin-tv-a transgene | p53 status | Tumor incidence | Metastasis |
|---|---|---|---|
| − | +/+ | 0/11 | N/A |
| + | +/+ | 17/26 | 1/17 |
| + | +/− | 14/38 | 1/14 |
| + | −/− | 18/43 | 6/16 |
Incidence of grossly visible tumors in albumin-tv-a transgenic mice detected after sacrifice. p53+/+ and p53+/− mice were sacrificed at either 4 or 6 months of age. p53−/− mice were sacrificed at either 4 months of age or upon the detection of lymphomas or sarcomas, whichever came first. N/A, not applicable.
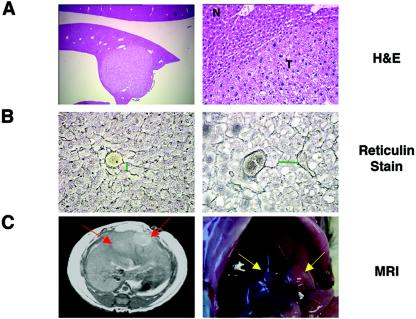
Examination of tissue sections obtained from the tumors demonstrated well-circumscribed lesions composed of enlarged hepatocytes with clear, vacuolated cytoplasm and moderate nuclear atypia (Fig. 2A). The clear cytoplasm is believed to contain high levels of fat, which was removed during fixation and paraffin embedding, as the tumors were negative after periodic acid Schiff staining, which detects glycogen (data not shown). Consistent with this hypothesis, gene expression arrays demonstrated increased transcripts from several adipocyte-specific genes in RCAS-PyMT-induced tumors relative to normal liver tissue (see the supplemental data). In addition to the morphological changes in the nucleus and cytoplasm, the tumors displayed abnormal tissue architecture, as determined by a reticulin stain for the basement membrane. While normal hepatocyte plates are a single cell thick, the plates of the induced liver tumors were two to three cells thick (Fig. 2B).
FIG. 2.
RCAS-PyMT induces liver tumors in albumin-tv-a transgenic mice. (A) Hematoxylin and eosin (H&E)-stained sections demonstrating the presence of liver lesions in a 6-month-old albumin-tv-a transgenic animal injected with RCAS-PyMT. Magnification, ×25 (left), ×100 (right). N, normal liver tissue; T, tumor. (B) Determination of hepatocyte plate size by reticulin staining for basement membrane in normal adult liver tissue (left) and tumor tissue (right) sections. Green bars indicate widths of hepatocyte plates. Original magnification, ×200. (C) Detection of liver tumors by MRI. (Left) T2 weighted MRI image demonstrating the presence of liver tumors (red arrows); (right) a photograph of the liver of the animal after euthanasia 24 h later. Arrows indicate the location of the tumors.
Since tumor-bearing animals did not show signs of disease before sacrifice, we attempted to identify animals with tumors noninvasively by subjecting a small cohort of animals to MRI at 2- to 3-week intervals. In this way, we observed small tumors, less than 5 mm in diameter, in two of four animals at 5 months of age; the presence of the tumors was confirmed by pathological examination 24 h later (Fig. 2C).
Characterization of PyMT-induced tumors.
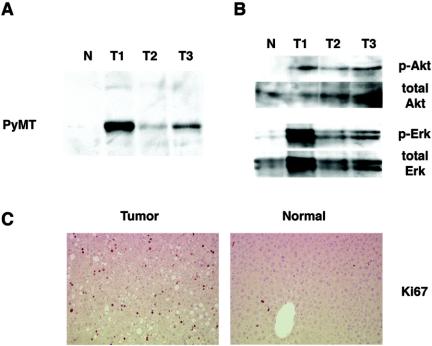
To confirm that the tumors in mice exposed to RCAS-PyMT arose from RCAS-infected cells, we first determined whether the identified tumors expressed PyMT. Protein lysates from tumors and normal liver tissue were analyzed by immunoblotting with a rat serum raised against mouse polyoma virus T antigens. PyMT was readily detected in all tumor samples but not in normal liver tissue from an uninfected transgenic mouse (Fig. 3A). We then assayed the activation state of the Akt and Erk kinases, which are known to act downstream of PyMT-mediated signaling pathways (13). Western blot analysis showed that both Akt and Erk are more highly phosphorylated, and presumably more active, in tumors than in normal tissue (Fig. 3B).
FIG. 3.
Characterization of induced liver tumors. (A) Detection of PyMT by immunoblotting with anti-PyMT serum of protein lysates from normal adult liver tissue (N) and three representative tumors (T1 to T3). (B) Increased p-Akt/total Akt, and p-MAPK/total MAPK ratios in RCAS-PyMT-induced liver tumors in comparison to age-matched adult liver tissue detected by immunoblotting. (C) Enhanced proliferation in induced liver tumor (left) in comparison to adjacent normal tissue (right) as determined by immunohistochemical staining for Ki67.
We next measured cell proliferation and apoptosis in the liver tumors. The PyMT-induced liver tumors displayed an elevated fraction of cells in S phase, compared to surrounding normal tissue, as determined by Ki67 staining (Fig. 3C). In addition, the tumors did not display increased levels of apoptosis by terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling staining, consistent with the antiapoptotic effects of Akt-mediated signaling downstream of the PyMT oncoprotein (data not shown).
The loss of the TP53 tumor suppressor gene is a common event in HCC. We therefore sought to determine whether loss of this tumor suppressor was required for tumor formation. We analyzed genomic DNA from three tumors induced in albumin-tv-a mice for the presence of the p53 locus by PCR. All tumors were positive for the presence of p53. We next amplified cDNA from four tumors corresponding to nucleotides 719 to 1150 of the p53 coding sequence, which contains several mutation hotspots found in human tumors, and sequenced the PCR products. No mutations were identified in any of the tumors. Thus, inactivation of p53 does not appear to be required for tumor formation.
The p19ARF tumor suppressor mediates oncoprotein signaling to p53 (53). We therefore determined whether this tumor suppressor was inactivated during tumor formation. We performed RT-PCR on RNA isolated from four tumor samples and four normal liver tissue samples. All tumors displayed elevated p16 and p19 mRNA levels compared to normal liver tissue, indicating that the Ink4a locus is intact and not silenced by gene methylation (data not shown).
The absence of p53 induces lung metastasis.
The TP53 tumor suppressor is frequently inactivated in human HCC. We therefore sought to determine the effect of p53 deficiency on tumor formation. Albumin-tv-a transgenic mice were bred to animals null for p53 as a result of gene targeting to generate cohorts of transgenic mice that were either heterozygous or null for p53 (17). Newborn mice from these crosses were injected with RCAS-PyMT producer cells and monitored for tumor formation. Of 38 TVA-positive, p53 heterozygous animals injected with RCAS-PyMT, 14 were found to have tumors after sacrifice at either 4 or 6 months of age (Table 1). Similarly, tumor incidence in the injected p53 null littermates was not increased, with 18 of 43 animals found to have tumors after sacrifice (Table 1). These data suggest that the absence of p53 does not promote tumor initiation in this model. It should be noted, however, that some of the p53 null animals were sacrificed as early as 9 weeks of age due to the presence of thymic lymphomas, sarcomas, or hemangiosarcomas and were maintained for no longer than 4 months. The reduced life span of these mice may have reduced tumor occurrence or detection of liver tumors.
Although the absence of p53 appeared to have no effect on tumor initiation, it greatly influenced tumor behavior. While only 1 of 17 p53 wild-type and 1 of 14 p53 heterozygous tumor-bearing mice developed lung metastases, 6 of 16 tumor-bearing p53 null mice analyzed displayed lung metastases (Table 1). Furthermore, in p53 null animals bearing primary tumors larger than 6 mm in diameter, lung metastases were observed in six of seven animals. By contrast, only one of six p53 wild-type mice bearing tumors larger than 6 mm in diameter had lung metastases. On histologic examination, the tumors in p53 null mice were shown to contain regions that resembled the tumors induced in p53 wild-type mice. However, the tumors from p53 null animals also contained regions with highly disorganized and undifferentiated cells (Fig. 4A). Such regions were not seen in tumors identified in p53 wild-type or p53 heterozygous animals, including those that exceeded 6 mm in diameter. Histologically, the pulmonary lesions resembled the undifferentiated regions identified within the primary tumors, suggesting that the metastatic cells were derived from these regions (Fig. 4B).
FIG. 4.
RCAS-PyMT-induced tumors in p53 null mice. (A) H&E-stained tissue sections demonstrating the histologic features of p53-deficient liver tumors. Three regions of a single tumor are shown. (B) Histologic features of lung metastasis identified in the same animal. Note that the metastasis most closely resembles region 2 of the primary tumor. (C) Detection of PyMT RNA in the lung metastasis by in situ hybridization with 33P-labeled antisense PyMT RNA.
To confirm that the lung lesions were metastases and not other tumors arising from uninfected cells within the p53-deficient animals, we performed in situ hybridizations to detect PyMT mRNA. The lung metastases but not the surrounding normal lung tissue showed evidence of hybridization of cellular RNA to radiolabeled antisense PyMT RNA probes (Fig. 4C). Furthermore, the hepatic transcription factor hepatocyte nuclear factor 1 was detected by immunostaining in the nuclei of cells within the lung lesions but not in surrounding lung tissue (data not shown).
p53 null and p53 wild-type tumors display different gene expression profiles.
The difference in metastatic potential displayed by tumors in p53 null and wild-type mice raised the possibility that these effects may be mediated by differences in gene expression. We therefore determined the gene expression profiles of a subset of tumors from p53 wild-type and p53 null mice, as well as normal liver tissue obtained from p53 wild-type and p53 null adult animals, by Affymetrix oligonucleotide arrays. Unsupervised hierarchical clustering of the expression array data demonstrated that the tumor samples were readily distinguished from the normal tissue samples (Fig. 5A). We used the Wilcoxon rank sum test to find genes that differentiated between normal and tumor samples. Over 500 genes were identified that differentiated between normal and tumor samples with a P value of ≤0.00067 (see Table S1 in the supplemental material). Among the genes more highly expressed in tumors was the proliferation marker Ki67, reflecting the increased proliferation of the tumor cells compared to normal cells (Fig. 2C), and JunB, a member of the AP-1 transcription factor family. We also identified elevated expression of another AP-1 family member, JunD, by immunohistochemical staining (data not shown). Interestingly, several proapoptotic genes, including Bax and 24p3 (lipocalin), were elevated in the tumors compared to normal tissues. However, increased mRNA levels were detected for the antiapoptotic gene TIAP (survivin), suggesting a possible mechanism for the absence of increased apoptosis in the tumors.
FIG. 5.
p53 null and p53 wild-type tumors have different gene expression profiles. (A) Hierarchial clustering of normal liver tissue and tumor samples using 532 genes that most strongly discriminate between the two classes. The sample marked by an asterisk (*) shows data from a small p53 null tumor that clusters with normal tissue from p53 null livers, indicating possible contamination with normal tissue. The genes present in the cluster are identified in Table S1 in the supplemental data. (B) Analysis of clustering of p53 null and p53 wild-type tumors using 100 genes that most strongly discriminate between the two tumor groups. The genes present in the cluster are identified in Table S2 in the supplemental data. (C) Confirmation of differential expression by RT-PCR. RT-PCR assays demonstrating elevated expression of cathepsin E, Igf2, and H19 genes in p53 null tumors relative to p53 wild-type tumors.
Among the genes with lower mRNA levels in the tumors were genes associated with differentiated liver cells such as hydroxysteroid dehydrogenase and liver-specific arginase. Further, in comparison to normal liver tissue, the tumors had lower levels of biliary glycoproteins 1 and 2, suggesting that the tumors were indeed derived from the transformation of hepatocytes and not from cells in the bile ducts.
The gene expression studies further demonstrated that the tumors with the ability to metastasize have an expression profile that is distinct from that of the nonmetastatic tumors induced in p53 wild-type mice. To identify genes that differentiated between these two groups, a variant t test was used (see Materials and Methods) which corrects the variance with a term that depends on the intensity of the samples. (There were not enough samples in each group to use the Wilcoxon test.) By means of the variance-corrected t test, 105 genes were identified as highly differentially expressed between the two tumor types (Fig. 5B; see Table S2 in the supplemental material). The greatest fold-change in expression was found for the cathepsin E gene, which encodes a protein previously shown to colocalize to the invasive edge of gastric tumors (29). RT-PCR of RNA extracted from tumors from p53 null and p53 wild-type animals confirmed the strongly elevated expression of this gene in the p53 null tumors (Fig. 5C). Several other genes of potential significance were also differentially expressed between the two tumor groups. These differences include an increase in Igf2 gene mRNA, and decreased RNA carrying the insulin-like growth factor binding protein 2 gene (Igfbp2), a negative regulator of Igf2 signaling. Both Igf2 and Igfbp2 have been previously implicated in tumor cell migration and invasion (37, 41, 52). We confirmed by RT-PCR the differential expression of Igf2 and H19, a gene located adjacent to Igf2 and shown to be elevated in p53 null tumors compared to tumors induced in p53 wild-type mice (Fig. 5C). Interestingly, differences in the mRNA levels for known p53 target genes such as p21 and mdm2 were not detected by the gene array studies. Thus, gene expression profiling of liver tumors induced in this model system identifies a set of changes that are associated with, and might be responsible for, invasion and metastasis.
PyMT phosphorylation mutants fail to induce lung metastasis.
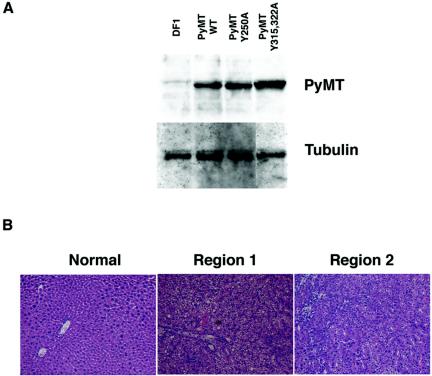
The above findings demonstrated that PyMT can induce liver tumors with metastatic potential in the absence of p53. We therefore sought to identify the signaling pathways induced by PyMT that might be crucial to this process. PyMT is activated by binding to the c-Src protein, followed by Src-mediated phosphorylation of several critical tyrosine residues. We therefore generated RCAS vectors encoding PyMT proteins with mutations at tyrosine residues previously demonstrated to play critical roles in cellular transformation in other systems (33). One mutant, RCAS-PyMT Y250A, expresses a protein with a tyrosine-to-alanine change at residue 250 that eliminates binding of ShcA to PyMT and prevents activation of Ras-mediated signaling pathways downstream of PyMT (13). Another mutant, RCAS-PyMT Y315 322A, expresses a protein with tyrosine-to-alanine changes at residues 315 and 322 which have been demonstrated to be critical for activation of phosphatidylinositol 3 (PI3)-kinase-mediated signaling pathways by PyMT (13). Production of the mutant PyMT proteins with the expected size was verified by immunoblotting of extracts from DF1 cells infected with RCAS vectors encoding the PyMT mutants (Fig. 6A).
FIG. 6.
Phosphorylation site mutants of PyMT fail to induce lung metastasis. (A) Western blot demonstrating the expression of wild-type and mutant PyMT proteins in DF-1 fibroblasts (top). α-Tubulin is shown as a loading control (bottom). (B) Histology of tumors induced by RCAS-PyMT Y315 322A in albumin-tv-a p53 null mice. H&E-stained tissue sections from normal liver tissue (left) and two regions of a single tumor (middle and right). Note the retention of hepatocyte cords in region 2 of the tumor (right).
To determine the ability of the PyMT mutant proteins to induce liver tumors, we injected litters from albumin-tv-a p53−/− × albumin-tv-a p53+/− matings. Introduction of RCAS-PyMT Y250A failed to induce tumors in either p53 null animals or their heterozygous littermates (Table 2), implying that activation of Ras signaling via ShcA is a crucial event in the induction of hepatic tumors by PyMT. In contrast, RCAS-PyMT Y315 322A induced liver tumors in both p53 heterozygous and p53 null animals, albeit at a reduced frequency relative to that of wild-type PyMT (Table 2). However, none of the RCAS-PyMT Y315 322A-injected p53 null tumor-bearing mice had lung metastases, including three mice bearing tumors of >6 mm in diameter. In contrast, as mentioned previously, six of seven TVA-positive, p53 null mice with wild-type PyMT-induced tumors of >6 mm in diameter had lung metastases. Consistent with the absence of metastases, the tumors induced by the PyMT phosphorylation mutant in albumin-tv-a p53 null mice were well circumscribed, displayed histology similar to that of the tumors induced by PyMT in p53 wild-type and heterozygous backgrounds (Fig. 6B, middle), and occasionally contained well-differentiated regions where normal hepatocyte plate architecture remained (Fig. 6B, right). Notably absent in these tumors were the undifferentiated regions seen in tumors induced by wild-type PyMT in p53 null animals. These findings suggest that metastatic potential in this mouse model is dependent on both the absence of p53 and the strength of signaling from the initiating oncoprotein.
TABLE 2.
Tumor induction by RCAS viruses encoding PyMT phosphorylation site mutantsa
| RCAS virus(es) |
p53 heterozygote
|
p53 null
|
||
|---|---|---|---|---|
| Tumor | Metastasis | Tumor | Metastasis | |
| PyMTb | 14/38 | 1/14 | 18/43 | 6/16 |
| PyMT Y250A | 0/32 | N/A | 0/17 | N/A |
| PyMT Y315A, -Y322A | 2/24 | 0/2 | 6/26 | 0/6 |
Incidence of grossly visible tumors, detected after sacrifice at 4 months of age or after the detection of lymphomas or sarcomas, in albumin-tv-a p53−/− mice injected with indicated viruses. N/A, not applicable.
Data for RCAS-PyMT are taken from Table 1.
DISCUSSION
Despite the devastating impact of HCC on human health, there are very few mouse models for this disease. We describe here a novel somatic mouse model for HCC generated using the RCAS-TVA gene delivery system. This mouse model resembles human sporadic HCC through the somatic activation of oncogenic pathways, the emergence of tumors from a single transformed cell, and the clonal nature of the resulting tumors. The tumors induced by RCAS-PyMT in albumin-tv-a mice, by histology and their well-circumscribed appearance, resemble human hepatocellular adenomas (23). We also found that we can detect developing tumors noninvasively by MRI. This finding will allow us to follow the course of disease in living animals, to screen animals for serum markers that correlate with disease induced by various genetic changes, and to monitor responses to therapy.
While the tumors induced in albumin-tv-a transgenic mice with an otherwise normal genetic background can grow to sizes exceeding 1 cm in diameter, they rarely metastasize to the lung or other organs. This suggests that additional genetic alterations are required to produce an invasive and metastatic phenotype. In agreement with this hypothesis, we find that while p53 nullizygosity does not enhance tumor initiation, the resulting tumors are more aggressive and have a greater capacity for invasion and metastasis to the lung. This increased aggressiveness is not simply due to increased proliferation or decreased apoptosis, as these occur at rates similar to those of p53 wild-type tumors (data not shown). Despite our use of mice with mixed genetic backgrounds, the phenotypic changes seen in the p53 null background are unlikely to be the result of strain background changes, as tumors induced in p53 heterozygous littermates are nonmetastatic and display the histologic features of tumors induced in p53 wild-type animals.
Interestingly, we find that inactivation of one or both alleles encoding the p53 tumor suppressor protein does not enhance the appearance of liver tumors. These findings are consistent with those reported by Balmain and colleagues a decade ago that demonstrated that loss of p53 did not promote tumor initiation in a skin carcinogenesis model (19). They are also consistent with findings in human hepatic adenomas and HCC that suggest that mutation of TP53 is a late event in HCC development (7). In agreement with this, we find that tumors induced in p53 wild-type mice retain p53 and do not lose expression of the p19Arf tumor suppressor protein, which mediates oncoprotein stimulation of p53 activity (53).
The differences in tumor behavior may be mediated through changes at multiple levels, including transcription regulation and mRNA stability. To determine the contribution of changes in mRNA levels, we used Affymetrix oligonucleotide arrays to explore the gene expression profiles of tumors identified in tv-a transgenic mice in p53 null and p53 wild-type backgrounds. Statistical analysis of the gene expression data identified over 100 genes that were differentially expressed between the two groups and which therefore represent potential mediators of the metastatic phenotype of the p53 null tumors. Consistent with this hypothesis, among the genes shown to be differentially expressed are the cathepsin E, Igf2, and Igfbp2 genes. These three genes encode proteins that have been demonstrated to be involved in tumor cell invasion and metastasis in various experimental systems, and their expression correlates with the invasive capacity of human tumors in vivo (29, 37, 41, 52). Whether these molecules play important roles in tumor invasion in human HCC is yet to be determined and provides an avenue for further exploration. Interestingly, our gene expression studies did not identify any known p53 transcriptional targets, such as p21CIP1, as molecules that differentiate between tumors induced in p53 wild-type and p53 null animals.
Our gene expression arrays also identified more than 500 genes whose mRNA levels distinguish RCAS-PyMT-induced liver tumors from normal liver tissue, independent of p53 gene status. Several of these genes or their family members, such as JunB and Rbl2, have been identified as misexpressed in human HCC or are critical mediators of tumor initiation in animal models of HCC (7, 10). These findings validate our model and suggest that there are common critical signaling pathways required for HCC tumor formation independent of the identity of the initiating oncogenic lesion. Interestingly, we identified elevated expression of multiple members of the AP-1 transcription factor family in the PyMT-induced tumors, irrespective of p53 gene status. These data are consistent with findings in other HCC models, suggesting a critical role for AP-1-mediated transcription in the initiation of liver tumorigenesis (10). However, the identity of the AP-1 transcription targets involved in HCC initiation remains to be elucidated. Importantly, because it is not necessary to generate a new transgenic line for each oncogene to be evaluated, the hypothesis that there are common pathways to HCC initiation can be readily tested in our model system through the introduction of other oncogene-bearing RCAS viruses—including those encoding c-Myc, β-catenin, and cyclin D1—which have all been implicated in the pathogenesis of the human disease (7).
The induction of tumors by RCAS-PyMT in genetically normal mice suggests that the activation of Ras- and PI3-kinase-mediated signaling pathways downstream of PyMT is sufficient for the initiation of tumorigenesis in the liver. Moreover, activation of these pathways is required as reduced signaling through either of these pathways impairs tumor initiation, consistent with findings reported in other model systems (33). Interestingly, impairment of signaling through these pathways inhibits the formation of lung metastases in a p53-deficient genetic background. Thus, the development of lung metastasis is dependent on both the absence of p53 function and signaling through Ras- and PI3-kinase-dependent signaling pathways.
In the experiments described here, we illustrated the utility of somatic delivery of oncogenes to the liver using the potent mouse PyMT oncoprotein. However, the model is also amenable to testing the requirements for the induction of liver tumors by the c-Myc, cyclin D1, and β-catenin oncoproteins, all of which are either overexpressed or activated in a significant fraction of human HCC. Indeed, we have analyzed the ability of c-Myc to induce liver tumors when delivered in RCAS vectors and have not observed liver tumors even in the absence of p53 (B. C. Lewis and H. E. Varmus, unpublished results). These findings suggest that additional mutations are required for initiation of liver tumorigenesis by c-Myc. The additional mutations may include expression of the HBV X protein or silencing of the INK4A locus, both highly prevalent events in HCC (7). These findings are consistent with earlier transgenic mouse models that demonstrated that c-Myc required the presence of other oncogenic lesions to induce liver tumors (31, 47). Further, the requirements for tumor induction by c-Myc can be compared to those for β-catenin and cyclin D1, using a single transgenic mouse line, as can the molecular characteristics of the induced tumors. Thus, the tumor model described here should prove to be a valuable tool in the elucidation of the molecular features of HCC development.
Supplementary Material
Acknowledgments
We thank the MSKCC transgenic core for the generation of albumin-tv-a mice; Michelle Fluck for reagents; Mary Barrett, Jennifer Doherty, Ya-Wen Chen, Meena Tanwar, and Irina Linkov for technical assistance; Cornelia Matei and Mihaela Lupu for assistance with MRI; Agnes Viale, Juan Li and the MSKCC Genomics Core Lab for assistance with the microarray experiments; the MSKCC Molecular Cytology Core for assistance with in situ hybridization; and Kirsten Hubbard and members of the Varmus lab for discussion and critical reading of the manuscript.
This work was supported by NIH grants R24CA83084, CA08748, and CA94060 (J.A.K.). B.C.L. was supported by a Helen Hay Whitney Foundation postdoctoral fellowship and a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abou-Elella, A., T. Gramlich, C. Fritsch, and T. Gansler. 1996. c-myc amplification in hepatocellular carcinoma predicts unfavorable prognosis. Mod. Pathol. 9:95-98. [PubMed] [Google Scholar]
- 2.Baldi, P., and A. D. Long. 2001. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics 17:509-519. [DOI] [PubMed] [Google Scholar]
- 3.Bates, P., J. A. Young, and H. E. Varmus. 1993. A receptor for subgroup A Rous sarcoma virus is related to the low density lipoprotein receptor. Cell 74:1043-1051. [DOI] [PubMed] [Google Scholar]
- 4.Beasley, R. P. 1988. Hepatitis B virus. The major etiology of hepatocellular carcinoma. Cancer 61:1942-1956. [DOI] [PubMed] [Google Scholar]
- 5.Beer, S., A. Zetterberg, R. A. Ihrie, R. A. McTaggart, Q. Yang, N. Bradon, C. Arvanitis, L. D. Attardi, S. Feng, B. Ruebner, R. D. Cardiff, and D. W. Felsher. 2004. Developmental context determines latency of MYC-induced tumorigenesis. PLoS Biol. 2:e332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouchard, M. J., L. H. Wang, and R. J. Schneider. 2001. Calcium signaling by HBx protein in hepatitis B virus DNA replication. Science 294:2376-2378. [DOI] [PubMed] [Google Scholar]
- 7.Buendia, M. A. 2000. Genetics of hepatocellular carcinoma. Semin. Cancer Biol. 10:185-200. [DOI] [PubMed] [Google Scholar]
- 8.Deane, N. G., H. Lee, J. Hamaamen, A. Ruley, M. K. Washington, B. LaFleur, S. S. Thorgeirsson, R. Price, and R. D. Beauchamp. 2004. Enhanced tumor formation in cyclin D1 × transforming growth factor β1 double transgenic mice with characterization by magnetic resonance imaging. Cancer Res. 64:1315-1322. [DOI] [PubMed] [Google Scholar]
- 9.Deane, N. G., M. A. Parker, R. Aramandla, L. Diehl, W. J. Lee, M. K. Washington, L. B. Nanney, Y. Shyr, and R. D. Beauchamp. 2001. Hepatocellular carcinoma results from chronic cyclin D1 overexpression in transgenic mice. Cancer Res. 61:5389-5395. [PubMed] [Google Scholar]
- 10.Eferl, R., R. Ricci, L. Kenner, R. Zenz, J. P. David, M. Rath, and E. F. Wagner. 2003. Liver tumor development. c-Jun antagonizes the proapoptotic activity of p53. Cell 112:181-192. [DOI] [PubMed] [Google Scholar]
- 11.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 12.Fults, D., C. Pedone, C. Dai, and E. C. Holland. 2002. MYC expression promotes the proliferation of neural progenitor cells in culture and in vivo. Neoplasia 4:32-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gottlieb, K. A., and L. P. Villarreal. 2001. Natural biology of polyomavirus middle T antigen. Microbiol. Mol. Biol. Rev. 65:288-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harada, N., H. Oshima, M. Katoh, Y. Tamai, M. Oshima, and M. M. Taketo. 2004. Hepatocarcinogenesis in mice with beta-catenin and Ha-ras gene mutations. Cancer Res. 64:48-54. [DOI] [PubMed] [Google Scholar]
- 15.Himly, M., D. N. Foster, I. Bottoli, J. S. Iacovoni, and P. K. Vogt. 1998. The DF-1 chicken fibroblast cell line: transformation induced by diverse oncogenes and cell death resulting from infection by avian leukosis viruses. Virology 248:295-304. [DOI] [PubMed] [Google Scholar]
- 16.Holland, E. C., Y. Li, J. Celestino, C. Dai, L. Schaefer, R. A. Sawaya, and G. N. Fuller. 2000. Astrocytes give rise to oligodendrogliomas and astrocytomas after gene transfer of polyoma virus middle T antigen in vivo. Am. J. Pathol. 157:1031-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacks, T., L. Remington, B. O. Williams, E. M. Schmitt, S. Halachmi, R. T. Bronson, and R. A. Weinberg. 1994. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 4:1-7. [DOI] [PubMed] [Google Scholar]
- 18.Jhappan, C., C. Stahle, R. N. Harkins, N. Fausto, G. H. Smith, and G. T. Merlino. 1990. TGF alpha overexpression in transgenic mice induces liver neoplasia and abnormal development of the mammary gland and pancreas. Cell 61:1137-1146. [DOI] [PubMed] [Google Scholar]
- 19.Kemp, C. J., L. A. Donehower, A. Bradley, and A. Balmain. 1993. Reduction of p53 gene dosage does not increase initiation or promotion but enhances malignant progression of chemically induced skin tumors. Cell 74:813-822. [DOI] [PubMed] [Google Scholar]
- 20.Lau, J. W. Y., and C. K. Leow. 1999. Surgical management, p. 147-172. In A. S.-Y. Leong, C. T. Liew, J. W. Y. Lau, and P. J. Johnson (ed.), Hepatocellular carcinoma: diagnosis, investigation and management. Arnold, London, England.
- 21.Lehnert, S. A., and R. J. Akhurst. 1988. Embryonic expression pattern of TGF beta type-1 RNA suggests both paracrine and autocrine mechanisms of action. Development 104:263-273. [DOI] [PubMed] [Google Scholar]
- 22.Leong, A. S.-Y. 1999. Epidemiology, risk factors, etiology, premalignant lesions and carcinogenesis, p. 1-18. In A. S.-Y. Leong, C. T. Liew, J. W. Y. Lau, and P. J. Johnson (ed.), Hepatocellular carcinoma: diagnosis, investigation and management. Arnold, London, England.
- 23.Leong, A. S.-Y., and C. T. Liew. 1999. Needle biopsy diagnosis of hepatocellular carcinoma, p. 107-129. In A. S.-Y. Leong, C. T. Liew, J. W. Y. Lau, and P. J. Johnson (ed.), Hepatocellular carcinoma: diagnosis, investigation and management. Arnold, London, England.
- 24.Leung, T. W. T. 1999. Non-surgical management, p. 173-191. In A. S.-Y. Leong, C. T. Liew, J. W. Y. Lau, and P. J. Johnson (ed.), Hepatocellular carcinoma: diagnosis, investigation and management. Arnold, London, England.
- 25.Lewis, B. C., D. S. Klimstra, and H. E. Varmus. 2003. The c-myc and PyMT oncogenes induce different tumor types in a somatic mouse model for pancreatic cancer. Genes Dev. 17:3127-3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liew, C. T., H. M. Li, K. W. Lo, C. K. Leow, J. Y. Chan, L. Y. Hin, W. Y. Lau, P. B. Lai, B. K. Lim, J. Huang, W. T. Leung, S. Wu, and J. C. Lee. 1999. High frequency of p16INK4A gene alterations in hepatocellular carcinoma. Oncogene 18:789-795. [DOI] [PubMed] [Google Scholar]
- 27.Manickan, E., J. Satoi, T. C. Wang, and T. J. Liang. 2001. Conditional liver-specific expression of simian virus 40 T antigen leads to regulatable development of hepatic neoplasm in transgenic mice. J. Biol. Chem. 276:13989-13994. [DOI] [PubMed] [Google Scholar]
- 28.Matsuda, Y., T. Ichida, J. Matsuzawa, K. Sugimura, and H. Asakura. 1999. p16(INK4) is inactivated by extensive CpG methylation in human hepatocellular carcinoma. Gastroenterology 116:394-400. [DOI] [PubMed] [Google Scholar]
- 29.Matsuo, K., I. Kobayashi, T. Tsukuba, T. Kiyoshima, Y. Ishibashi, A. Miyoshi, K. Yamamoto, and H. Sakai. 1996. Immunohistochemical localization of cathepsins D and E in human gastric cancer: a possible correlation with local invasive and metastatic activities of carcinoma cells. Hum. Pathol. 27:184-190. [DOI] [PubMed] [Google Scholar]
- 30.Miller, D. G., M. A. Adam, and A. D. Miller. 1990. Gene transfer by retrovirus vectors occurs only in cells that are actively replicating at the time of infection. Mol. Cell. Biol. 10:4239-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murakami, H., N. D. Sanderson, P. Nagy, P. A. Marino, G. Merlino, and S. S. Thorgeirsson. 1993. Transgenic mouse model for synergistic effects of nuclear oncogenes and growth factors in tumorigenesis: interaction of c-myc and transforming growth factor alpha in hepatic oncogenesis. Cancer Res. 53:1719-1723. [PubMed] [Google Scholar]
- 32.Nishida, N., Y. Fukuda, T. Komeda, R. Kita, T. Sando, M. Furukawa, M. Amenomori, I. Shibagaki, K. Nakao, M. Ikenaga, et al. 1994. Amplification and overexpression of the cyclin D1 gene in aggressive human hepatocellular carcinoma. Cancer Res. 54:3107-3110. [PubMed] [Google Scholar]
- 33.Ong, S. H., S. Dilworth, I. Hauck-Schmalenberger, T. Pawson, and F. Kiefer. 2001. ShcA and Grb2 mediate polyoma middle T antigen-induced endothelial transformation and Gab1 tyrosine phosphorylation. EMBO J. 20:6327-6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orsulic, S. 2002. An RCAS-TVA-based approach to designer mouse models. Mamm. Genome 13:543-547. [DOI] [PubMed] [Google Scholar]
- 35.Orsulic, S., Y. Li, R. A. Soslow, L. A. Vitale-Cross, S. J. Gutkind, and H. E. Varmus. 2002. Induction of ovarian cancer by defined multiple genetic changes in a mouse model system. Cancer Cell 1:53-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parkin, D. M. 2001. Global cancer statistics in the year 2000. Lancet Oncol. 2:533-543. [DOI] [PubMed] [Google Scholar]
- 37.Pereira, J. J., T. Meyer, S. E. Docherty, H. H. Reid, J. Marshall, E. W. Thompson, J. Rossjohn, and J. T. Price. 2004. Bimolecular interaction of insulin-like growth factor (IGF) binding protein-2 with αβ3 negatively modulates IGF-I-mediated migration and tumor growth. Cancer Res. 64:977-984. [DOI] [PubMed] [Google Scholar]
- 38.Pinkert, C. A., D. M. Ornitz, R. L. Brinster, and R. D. Palmiter. 1987. An albumin enhancer located 10 kb upstream functions along with its promoter to direct efficient, liver-specific expression in transgenic mice. Genes Dev. 1:268-276. [DOI] [PubMed] [Google Scholar]
- 39.Robinson, W. S. 1994. Molecular events in the pathogenesis of hepadnavirus-associated hepatocellular carcinoma. Annu. Rev. Med. 45:297-323. [DOI] [PubMed] [Google Scholar]
- 40.Roe, T., T. C. Reynolds, G. Yu, and P. O. Brown. 1993. Integration of murine leukemia virus DNA depends on mitosis. EMBO J. 12:2099-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sachdev, D., J. S. Hartell, A. V. Lee, X. Zhang, and D. Yee. 2004. A dominant negative type I insulin-like growth factor receptor inhibits metastasis of human cancer cells. J. Biol. Chem. 279:5017-5024. [DOI] [PubMed] [Google Scholar]
- 42.Sandgren, E. P., C. J. Quaife, C. A. Pinkert, R. D. Palmiter, and R. L. Brinster. 1989. Oncogene-induced liver neoplasia in transgenic mice. Oncogene 4:715-724. [PubMed] [Google Scholar]
- 43.Schaefer-Klein, J., I. Givol, E. V. Barsov, J. M. Whitcomb, M. VanBrocklin, D. N. Foster, M. J. Federspiel, and S. H. Hughes. 1998. The EV-O-derived cell line DF-1 supports the efficient replication of avian leukosis-sarcoma viruses and vectors. Virology 248:305-311. [DOI] [PubMed] [Google Scholar]
- 44.Shachaf, C. M., A. M. Kopelman, C. Arvanitis, A. Karlsson, S. Beer, S. Mandl, M. H. Bachmann, A. D. Borowsky, B. Ruebner, R. D. Cardiff, Q. Yang, J. M. Bishop, C. H. Contag, and D. W. Felsher. 2004. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature 431:1112-1117. [DOI] [PubMed] [Google Scholar]
- 45.Simonetti, R. G., M. Cottone, A. Craxi, L. Pagliaro, M. Rapicetta, P. Chionne, and A. Costantino. 1989. Prevalence of antibodies to hepatitis C virus in hepatocellular carcinoma. Lancet 2:1338. [DOI] [PubMed] [Google Scholar]
- 46.Soukas, A., P. Cohen, N. D. Socci, and J. M. Friedman. 2000. Leptin-specific patterns of gene expression in white adipose tissue. Genes Dev. 14:963-980. [PMC free article] [PubMed] [Google Scholar]
- 47.Terradillos, O., O. Billet, C. A. Renard, R. Levy, T. Molina, P. Briand, and M. A. Buendia. 1997. The hepatitis B virus X gene potentiates c-myc-induced liver oncogenesis in transgenic mice. Oncogene 14:395-404. [DOI] [PubMed] [Google Scholar]
- 48.Wang, R., L. D. Ferrell, S. Faouzi, J. J. Maher, and J. M. Bishop. 2001. Activation of the Met receptor by cell attachment induces and sustains hepatocellular carcinomas in transgenic mice. J. Cell Biol. 153:1023-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weiss, R. 1982. Experimental biology and assay of RNA tumor viruses, p. 209-260. In R. Weiss, N. Teich, H. Varmus, and J. Coffin (ed.), RNA tumor viruses, vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 50.Wilkinson, D. G., J. A. Bailes, and A. P. McMahon. 1987. Expression of the proto-oncogene int-1 is restricted to specific neural cells in the developing mouse embryo. Cell 50:79-88. [DOI] [PubMed] [Google Scholar]
- 51.Young, J. A., P. Bates, and H. E. Varmus. 1993. Isolation of a chicken gene that confers susceptibility to infection by subgroup A avian leukosis and sarcoma viruses. J. Virol. 67:1811-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang, D., A. A. Samani, and P. Brodt. 2003. The role of the IGF-I receptor in the regulation of matrix metalloproteinases, tumor invasion and metastasis. Horm. Metab. Res. 35:802-808. [DOI] [PubMed] [Google Scholar]
- 53.Zindy, F., C. M. Eischen, D. H. Randle, T. Kamijo, J. L. Cleveland, C. J. Sherr, and M. F. Roussel. 1998. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 12:2424-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.