Abstract
Activation of canonical Wnt signaling inhibits brown adipogenesis of cultured cells by impeding induction of PPARγ and C/EBPα. Although enforced expression of these adipogenic transcription factors restores lipid accumulation and expression of FABP4 in Wnt-expressing cells, additional expression of PGC-1α is required for activation of uncoupling protein 1 (UCP1). Wnt10b blocks brown adipose tissue development and expression of UCP1 when expressed from the fatty acid binding protein 4 promoter, even when mice are administered a β3-agonist. In differentiated brown adipocytes, activation of Wnt signaling suppresses expression of UCP1 through repression of PGC-1α. Consistent with these in vitro observations, UCP1-Wnt10b transgenic mice, which express Wnt10b in interscapular tissue, lack functional brown adipose tissue. While interscapular tissue of UCP1-Wnt10b mice lacks expression of PGC-1α and UCP1, the presence of unilocular lipid droplets and expression of white adipocyte genes suggest conversion of brown adipose tissue to white. Reciprocal expression of Wnt10b with UCP1 and PGC-1α in interscapular tissue from cold-challenged or genetically obese mice provides further evidence for regulation of brown adipocyte metabolism by Wnt signaling. Taken together, these data suggest that activation of canonical Wnt signaling early in differentiation blocks brown adipogenesis, whereas activating Wnt signaling in mature brown adipocytes stimulates their conversion to white adipocytes.
Brown adipose tissue (BAT) releases energy in the form of heat and thus plays an important role in adaptive thermogenesis (reviewed in references 5, 20, and 34). In response to cold exposure or intake of excess calories, activation of the sympathetic nervous system stimulates the release of norepinephrine. This adrenergic stimulus causes brown adipocytes to mobilize stored triacylglycerol, and fatty acids are then metabolized in mitochondria. In most cell types, electrons from the resulting NADH and FADH2 pass through the respiratory chain to create an electrochemical proton gradient linked to ATP synthesis. However, in brown adipocytes this potential energy is uncoupled from ATP synthesis by the activity of uncoupling protein 1 (UCP1) and is lost as heat. While acute release of norepinephrine causes rapid activation of BAT thermogenesis, chronic exposure to adrenergic stimuli causes a dramatic expansion of BAT due, in part, to recruitment of new brown adipocytes (20).
Brown adipocytes are derived from mesenchymal stem cells, and development of BAT occurs late during embryonic life (23). Although the program of brown adipogenesis has not been as extensively studied as that of white, the process of differentiating into brown or white adipocytes appears to share some common features (46). In the paradigm for white adipocyte differentiation, transient expression of C/EBPβ and C/EBPδ in response to adipogenic stimuli induces expression of PPARγ and C/EBPα (12). These master transcription factors enforce each others' expression through a positive feedback loop, and working together, PPARγ and C/EBPα activate the genes involved in creating the complete adipocyte phenotype. In mice, genetic studies support a role for C/EBPβ, C/EBPδ, PPARγ, and C/EBPα in the development of BAT (2, 7, 51, 57).
While overexpression of adipogenic transcription factors in preadipocytes or mesenchymal precursors promotes accumulation of lipid and expression of adipocyte genes, these factors are not sufficient to stimulate the program of thermogenesis, including mitochondrial biogenesis and expression of UCP1. For full conversion of precursor cells to brown adipocytes, an additional requirement appears to be the expression and activity of PPARγ coactivator 1α (PGC-1α) (44). This protein coactivates members of the nuclear hormone receptor family, as well as other transcription factors, and plays an important regulatory role for energy metabolism in a number of cell types (32, 43). During adipogenesis of 3T3-F442A cells, expression of PGC-1α is sufficient to increase mitochondrial biogenesis and expression of UCP1 (44). PGC-1α is itself regulated by transcriptional coactivators, including SRC-1, TIF2, and p160 myb (15, 40). Evidence from rodents supports the notion that white adipocytes can be converted to multilocular mitochondria-rich adipocytes in response to adrenergic stimuli (22). A potential mechanism may involve PGC-1α as expression of this coactivator in human white adipocytes induces expression of brown adipocyte genes and stimulates oxidation of fatty acids (53). Other transcriptional regulators potentially involved in the transdifferentiation process include FOXC2 and the retinoblastoma protein (Rb) (8, 18).
Extracellular factors that regulate adipogenesis have been extensively studied in white adipocyte models, and lessons learned there may provide insight into brown adipogenesis (36). In addition, norepinephrine has been characterized as an important stimulator of brown adipocyte differentiation and metabolism, whereas inhibitory factors are less well-defined (5). Activation of canonical Wnt signaling blocks differentiation of white preadipocytes in vitro, and inhibition of endogenous Wnt signaling results in spontaneous adipogenesis (3, 48). While Wnt proteins do not appear to influence expression of C/EBPβ or C/EBPδ, they completely inhibit expression of PPARγ and C/EBPα, perhaps through a mechanism involving altered mitotic clonal expansion (47). Wnt10b is a good candidate for the endogenous inhibitory Wnt, since it is expressed in preadipocytes and stromal vascular cells, but not in adipocytes (3). In transgenic mice in which Wnt10b is expressed from the FABP4 promoter, Wnt10b inhibits accumulation of white adipose tissue (WAT), and transgenic mice are resistant to diet-induced obesity (33). In addition, these mice are not lipodystrophic but, instead, have improved glucose tolerance and insulin sensitivity. Expression of Wnt10b in interscapular tissue blocks development of BAT, and transgenic mice are unable to defend their core body temperature against a cold challenge; however, mechanisms whereby Wnt signaling inhibits brown adipocyte development were not reported (33).
In this report, we investigate the mechanisms whereby Wnt signaling regulates brown adipocyte differentiation and metabolism. Activation of Wnt signaling inhibits brown adipogenesis in two in vitro models. While enforced expression of C/EBPα or PPARγ rescues the adipogenic program, rescue of the thermogenic program requires expression of PPARγ and PGC-1α. Expression of Wnt10b from the FABP4 promoter profoundly inhibits development of BAT, and these transgenic mice are insensitive to adrenergic induction of UCP1 and thermogenesis. In addition to blocking development of brown adipocytes, Wnt signaling also influences metabolism after differentiation has occurred. For example, exposure of differentiated brown adipocytes to activators of Wnt signaling causes a decline in brown adipocyte genes, without influencing expression of common adipocyte markers. Suppression of PGC-1α appears to play an intermediary role in repression of UCP1 by Wnt. Consistent with these in vitro observations, expression of Wnt10b from the UCP1 promoter results in interscapular tissue with the histological and molecular characteristics of WAT, suggesting that Wnt signaling transdifferentiates brown adipocytes into white. Finally, reciprocal expression of PGC-1α and UCP1 with Wnt10b in cold-challenged or genetically obese mice suggests a role for Wnt signaling in the plasticity of white and brown adipocyte tissues in vivo.
MATERIALS AND METHODS
Cell culture.
Rb wild-type and Rb−/− mouse embryonic fibroblasts (MEFs) were prepared as described (18). Cells were maintained in Dulbecco's modified Eagle medium containing 25 mM glucose, 10% calf serum, 20 mM HEPES, and 100 U of penicillin-streptomycin per ml. Rb−/− MEFs were induced to differentiate 2 days after confluence (designated day 0) with 10% fetal calf serum, 1 μM dexamethasone, 0.5 mM methylisobutylxanthine, 5 μg of insulin per ml, and 90 nM darglitazone for the first 2 days. Cells were subsequently fed with medium containing 5 μg of insulin per ml and 90 nM darglitazone every other day.
HIB-1B cells (27) were generously provided by C. Ronald Kahn (Joslin Diabetes Center, Harvard University). These cells were grown in Dulbecco's modified Eagle medium containing 25 mM glucose, 20% fetal bovine serum, 20 mM HEPES, and 100 U of penicillin-streptomycin per ml. For differentiation, HIB-1B cells were grown to confluence in culture medium supplemented with 20 nM insulin and 1 nM triiodothyronine (differentiation medium). Confluent cells were incubated for 24 h in differentiation medium further supplemented with 0.5 mM methylisobutylxanthine, 0.5 μM dexamethasone, and 0.125 mM indomethacin (induction medium). Subsequently, cells were maintained in differentiation medium for 5 days. Cells were treated with 10 μM norepinephrine (Calbiochem) 4 h prior to lysis. LiCl (25 mM) or Chir99021 (3 μM in dimethyl sulfoxide; Chiron Corp.) were used to inhibit glycogen synthase kinase 3 (GSK3). All reagents were purchased from Sigma Chemical Co. unless otherwise designated.
Retroviral infection and constructs.
Genes were stably introduced into Rb−/− MEFs and HIB-1B cells by retroviral infection as described previously (14). Briefly, 10-cm plates of human embryonic kidney 293T cells were transiently transfected with 7.5 μg of retroviral expression vectors and the viral packaging vectors SV-E-MLV-env and SV-E-MLV. After precipitation of DNA, cells were shocked with 12.5% glycerol in phosphate-buffered saline (PBS). At 16 h after transfection, virus-containing medium was collected over three 12-h intervals and passed through a 0.45-μm-pore-size syringe filter. Filter-sterilized polybrene (hexadimethrine bromide; 8 μg/ml) was added to the virus-loaded medium. This medium was then applied to proliferating (∼40%) cells. After three rounds of infection, cells were trypsinized and replated in a medium supplemented with geneticin (Life Technologies, Inc.), hygromycin B (Invitrogen), or puromycin as selection antibiotics.
Plasmids containing PPARγ and PGC-1α were kindly provided by Bruce Spiegelman (Dana Farber Cancer Institute, Harvard University). Other retroviral expression vectors were created as follows. C/EBPα from pcDNA3.1 was subcloned into the BamHI-HindIII sites of pBabepuro. In addition, PGC-1α was subcloned into the BglII-NotI sites of pLNCX2, and into the NruI-SalI sites of pBabepuro.
Oil Red-O staining.
Detection of neutral lipid with Oil Red-O was essentially as described previously (41). Briefly, plates were washed twice with PBS, fixed with 3.7% buffered formaldehyde, stained for 2 h at room temperature with Oil Red-O solution (0.5% Oil Red-O in 70% isopropyl alcohol), and then washed with H2O.
RNase protection analysis.
Total RNA was isolated by using RNA Stat60 (Tel-Test, Inc). Ten micrograms of total RNA was used for detection of UCP1, PPARγ, C/EBPα, FABP4, the Wnt10b transgene, and 36B4, and 15 μg of RNA was used for PGC-1α. Templates for each gene were amplified by reverse-transcription-PCR (RT-PCR) from brown adipose tissue RNA. PCR products for riboprobes were cloned into pCRII-Topo (Invitrogen, Inc.) and sequenced. After linearization, these constructs served as templates for in vitro transcription reactions using [α-32P]CTP (Perkin Elmer, Inc.) and T7 or Sp6 RNA polymerases (Promega, Inc.). RNase protection assays were performed as described previously (35). Briefly, labeled antisense riboprobe in 1× hybridization buffer was hybridized to RNA at 54°C for a 16-h incubation. RNase digestion with 150 mM RNase digestion buffer containing 80 μg of RNase A per μl and 250 U of RNase T1 (BD Biosciences) per μl was performed for 1 h at 37°C. Sodium dodecyl sulfate (SDS; 10%) and proteinase K (10 μg/μl; Roche Diagnostics) were added for 15 min at 37°C. The resulting protected fragments were isolated and electrophoresed through 5% polyacrylamide sequencing gels. The gels were then dried and subjected to autoradiography (Kodak Biomax MR) at −80°C. Quality of RNA was assessed by simultaneous analysis of 36B4 as an internal control.
Cell lysates and immunoblotting.
Cells were washed once with PBS and scraped in a lysis buffer containing 1% SDS and 60 mM Tris-Cl, pH 6.8. Lysates were boiled for 3 min, vortexed, and then boiled for an additional 7 min prior to storing at −21°C. Concentration of protein was estimated with the Bradford assay (Bio-Rad). Equal amounts of protein were used for immunoblot analysis, which was as described previously (30). Briefly, proteins were separated by electrophoresis in 10.5% or 15% polyacrylamide gels. Protein was transferred to a polyvinylidene difluoride membrane (Osmonics) and immunoblotted with the appropriate antibodies. Membranes were visualized by the bound immunoglobulin G-peroxidase by using Super Signal or Super Signal Ultra chemiluminescence substrates (Pierce). For the detection of C/EBPα, a polyclonal antibody generated against a synthetic polypeptide corresponding to amino acids 253 to 265 was used (31). Polyclonal FABP4 antibody was from David Bernlohr (University of Minnesota). PPARγ (Santa Cruz Biotechnology) and UCP1 (Alpha Diagnostic) antibodies were commercially available.
Creation of UCP1-Wnt10b transgenic mice.
A 3.1-kb fragment of mouse UCP1 promoter (generously provided by Leslie Kozak; Pennington Biomedical Research Center) was cloned upstream of the 1.2-kb Wnt10b open reading frame and a 1-kb fragment of the rabbit β-globin intron-poly(A) sequence (33). Transgenic mice that express Wnt10b under control of the UCP1 promoter were created on a congenic C57BL/6 background by the transgenic core facility at the University of Michigan. Genotyping was performed by PCR using transgene-specific primers. Tissue specificity was determined by RNase protection assay using a probe specific for the Wnt10b transgene. Creation and initial characterization of FABP4-Wnt10b mice are described elsewhere (33).
Characterization of UCP1-Wnt10b transgenic mice.
Animals were maintained on a C57BL/6 background. To assess development of BAT, interscapular tissue from transgenic and wild-type mice was dissected and fixed with 10% buffered formalin for at least 24 h. Tissue samples were embedded in paraffin, and 7-μm sections were stained with hematoxylin and eosin. For analysis of gene expression, interscapular tissues from wild-type and transgenic mice at 8 weeks of age were dissected. Isolated tissues were homogenized in RNA Stat60 for RNA or in a lysis buffer containing 1% SDS and 60 mM Tris-Cl, pH 6.8 for protein. Samples were further processed for RNase protection and immunoblotting assays as described above.
Animal studies.
To evaluate the effects of chronic exposure of mice to a β3-agonist, administration of CL316,243 was essentially as previously described (16). Briefly, wild-type and FABP4-Wnt10b mice (n = 3) were injected with saline or CL316,243 (1 mg/kg of body weight/day, intraperitoneally) daily for 21 days. Interscapular tissues were then isolated and processed to analyze changes in gene expression and histology as described above. To evaluate acute effects of CL316,243 on thermogenesis, wild-type and FABP4-Wnt10b mice (n = 4) were surgically implanted with precalibrated telemeters (model TA-F20; Data Sciences International, St. Paul, Minn.) under deep anesthesia. Mice were housed at 27°C with cycles of 12 h of light and 12 h of darkness for at least 2 weeks postsurgery. Core body temperatures for each animal were then recorded at an environmental temperature of 27°C. Saline was injected on day 1 at 9 am, and CL316,243 was injected on day 2 at 9 a.m., and core body temperatures were recorded simultaneously. Data acquisition and analysis were performed on ICELUS software (Mark J. Opp, University of Michigan).
To investigate the effects of cold exposure on gene expression, female C57BL/6 mice (n = 5) were singly housed and placed at 4°C with ad libitum access to both chow and water. After 24 h, mice were euthanized, and interscapular tissue was dissected and processed for RNA analysis. To evaluate gene expression in a genetic model of obesity, male ob/ob mice (n = 6) on a C57BL/6 genetic background were purchased from The Jackson Laboratory (Bar Harbor, Maine) and acclimatized for 2 weeks. After euthanasia, interscapular tissue was dissected and RNA was isolated with RNA Stat60.
Quantitative RT-PCR.
One microgram of total RNA was transcribed to cDNA by using TaqMan (Roche) according to the manufacturer's protocol. cDNA was synthesized by using the TaqMan system (Applied Biosystems) and random hexamer primers. Quantitative PCR was performed according to the manufacturer's protocol. SYBR green I was used to monitor amplification of DNA on the I-Cycler thermal cycler and IQ real-time PCR detection system (Bio-Rad). After amplification, melting curve analysis was performed as described in the manufacturer's protocol (Bio-Rad). Gene expression was normalized to 18S RNA. The oligonucleotide primers for amplification of PGC-1α (59), UCP1 (60), PPARγ (60), cytochrome c, long-chain acyl coenzyme A (CoA) dehydrogenase (50), Wnt10a (50), and Wnt10b (9) have been reported. Primers for the following genes were designed and validated for quantitative PCR: for C/EBPα, 5′-TGGACAAGAACAGCAACGAG-3′ (forward) and 5′-TCACTGGTCAACTCCAGCAC-3′ (reverse); for FABP4, 5′-TGGAAGCTTGTCTCCAGTGA-3′ (forward) and 5′-AATCCCCATTTACGCTGATG-3′ (reverse); for 18S, 5′-CGCTTCCTTACCTGGTTGAT-3′ (forward) and 5′-GAGCGACCAAAGGAACCATA-3′ (reverse); for mitochondrial transcription factor A (mtTFA), 5′-GCCTGGGGTCTTGTCTGTAT-3′ (forward) and 5′-TTTGGGTAGCTGTTCTGTGG-3′ (reverse); for ATP synthase (ATP5f1), 5′-CCGTGTCTGAAGAACG-3′ (forward) and 5′-GGCAAGGCGAGGCTGTC-3′ (reverse); for cytochrome c, 5′-TTCCTGCTTCGTGTGTTGTC-3′ (forward) and 5′-GATTGCAGAAGAGGTGACTGG-3′ (reverse).
RESULTS
Activation of canonical Wnt signaling blocks brown adipogenesis in vitro.
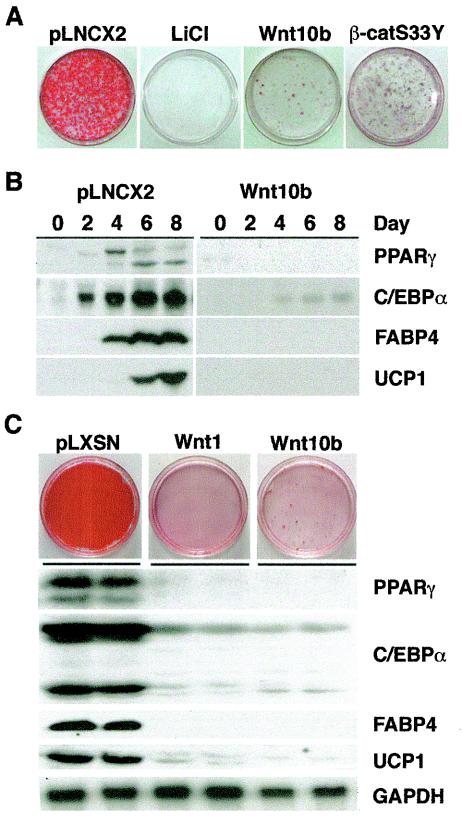
Wnt signaling influences the fate of many cell types. We along with others have demonstrated that Wnt signaling, potentially initiated by Wnt10b, inhibits differentiation of in vitro models of white adipogenesis (3, 26, 38, 48). To investigate whether Wnt signaling regulates brown adipogenesis, we tested whether activation of the canonical Wnt pathway blocks brown adipose conversion in two in vitro models, Rb−/− MEFs (18) and HIB-1B cells (Fig. 1) (27). To activate Wnt signaling, we treated cells with LiCl, which inhibits GSK3 (28), or we enforced expression of Wnt10b or a dominant-stable form of β-catenin (β-catS33Y). After induction of adipogenesis, control cells accumulated lipid droplets as assessed by Oil Red-O staining (Fig. 1A). In contrast, treatment with LiCl from day 0 to 4 of the differentiation protocol completely blocked lipid accumulation. As expected, inhibition of GSK3 with a more specific molecule, Chir99021 (3, 45, 56), also inhibited brown adipose conversion (data not shown). Finally, activation of canonical Wnt signaling by enforced expression of Wnt10b or β-catS33Y blocked accumulation of lipid (Fig. 1A).
FIG. 1.
Canonical Wnt signaling blocks brown adipogenesis in vitro. (A) Rb−/− MEFs were infected with control (pLNCX2), Wnt10b, or β-catS33Y retroviruses. Two days after confluence (day 0), cells were induced to undergo brown adipogenesis. In addition, cells infected with pLNCX2 were induced to differentiate in the presence of 25 mM LiCl on day 0 and day 2. At day 10, cells were stained with Oil Red-O to assess lipid accumulation. (B) Rb−/− MEFs infected with control (pLNCX2) or Wnt10b-expressing retroviruses were differentiated, and cells were lysed at days 0, 2, 4, 6, and 8. Lysates were analyzed for PPARγ, C/EBPα, FABP4, and UCP1 by immunoblotting. (C) HIB-1B cells were infected with control (pLNSX), Wnt1, or Wnt10b retroviruses. Cells were lysed 6 days after induction of brown adipogenesis and processed for immunoblotting (for PPARγ, C/EBPα, and FABP4) or RNase protection analyses (for UCP1 and GAPDH).
To determine molecular mechanisms whereby Wnt inhibits brown adipogenesis, expression of various adipocyte proteins was determined by immunoblot analysis (Fig. 1B). In Rb−/− MEFs, two adipogenic transcription factors, PPARγ and C/EBPα, as well as downstream genes, FABP4 and UCP1, were induced during adipogenesis. However, in cells expressing Wnt10b, induction of C/EBPα and PPARγ as well as FABP4 and UCP1 was greatly impaired. Similar results are observed in Rb−/− MEFs treated with LiCl (data not shown). In addition we observed that LiCl completely blocked induction of PGC-1α mRNA (data not shown). These observations in Rb−/− MEFs are supported by experiments in HIB-1B cells. In addition to inhibiting the accumulation of lipid (Fig. 1C), Wnt1 and Wnt10b inhibit induction of PPARγ, C/EBPα, and downstream genes such as FABP4 and UCP1, without influencing expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Taken together, these data demonstrate that Wnt signaling inhibits brown adipogenesis in vitro, and suggest that canonical Wnt signaling inhibits brown adipogenesis by interfering with induction of C/EBPα and PPARγ.
Wnt signaling inhibits adipogenesis through PPARγ and C/EBPα and expression of UCP1 through PGC-1α.
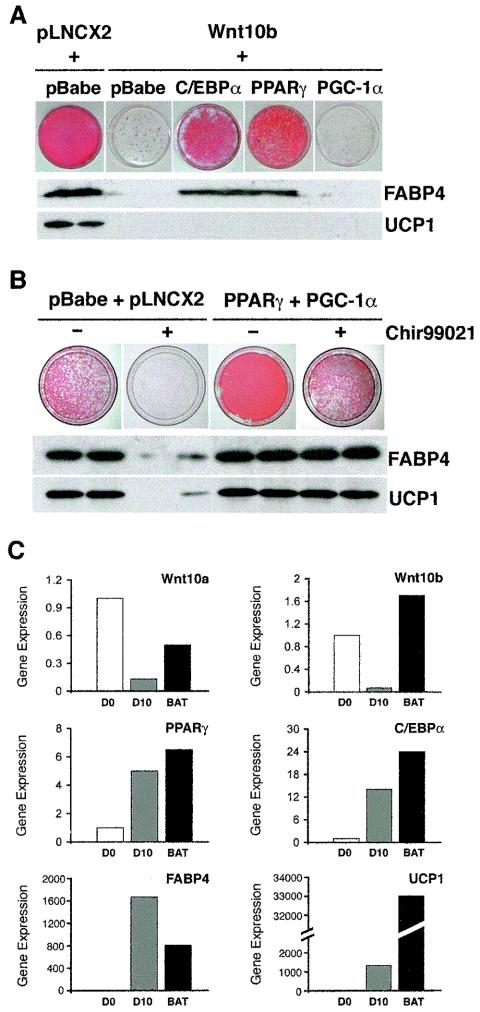
Wnt signaling blocks brown adipogenesis in two in vitro models by interfering with induction of C/EBPα, PPARγ and UCP1 (Fig. 1). To investigate further the molecular mechanisms whereby Wnt inhibits brown adipogenesis, we enforced expression of the adipogenic transcription factors PPARγ and C/EBPα. While expression of C/EBPα or PPARγ was sufficient to rescue accumulation of lipid and expression of FABP4 in Rb−/− MEFs expressing Wnt10b, these transcription factors did not stimulate expression of UCP1 (Fig. 2A). Consistent with these findings, C/EBPα and PPARγ also restored accumulation of lipid but not expression of UCP1 in HIB-1B cells (data not shown). In Rb−/− MEFs in which adipogenesis was blocked by Wnt10b, expression of PGC-1α alone was not sufficient to induce UCP1 (Fig. 2A). Thus, Wnt10b inhibits the adipogenic program through suppression of C/EBPα and PPARγ, but an additional mechanism appears to be required to constrain expression of UCP1.
FIG. 2.
Wnt signaling inhibits adipogenesis through PPARγ and C/EBPα and expression of UCP1 through PGC-1α. (A) Rb−/− MEFs were infected with pLNCX2 or Wnt10b retroviruses. After selection, both of these cell lines were infected with pBabe. Wnt10b cells were also infected with retroviruses containing C/EBPα, PPARγ, or PGC-1α. Two days after confluence (day 0), cells were induced to differentiate. At day 10, cells were stained with Oil Red-O to assess lipid accumulation. Cells were lysed at day 8 and subjected to immunoblotting for FABP4 and UCP1. (B) Rb−/− MEFs were sequentially infected with empty vectors (pBabe and pLNCX2) or with PPARγ and PGC-1α. Cells were induced to differentiate in the absence (−) or presence (+) of 3 μM Chir99021 on days 0 and 2. Cells were stained with Oil Red-O on day 10. Other cells were lysed at day 8 and subjected to immunoblot analysis for FABP4 and UCP1. (C) RNA was isolated from confluent Rb−/− MEFs (D0) and 10 days after induction of brown adipogenesis (D10). RNA was also isolated from BAT ofadult C57/BL6 mice. After reverse transcription, cDNAs were used as templates for quantitative RT-PCR, and expression of Wnt10a, Wnt10b, PPARγ, C/EBPα, FABP4, and UCP1 was evaluated. After normalizing expression to 18S RNA, expression is presented relative to that observed in undifferentiated Rb−/− MEFs.
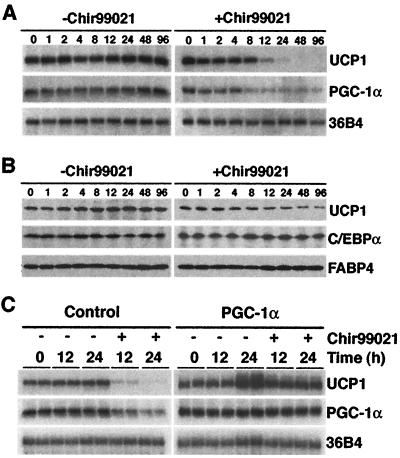
To determine if inhibition of the thermogenic program by Wnt signaling involves PGC-1α, we tested whether PPARγ and PGC-1α together can prevent the suppression of UCP1. For this experiment, PPARγ and PGC-1α were stably expressed in Rb−/− MEFs, and Chir99021 was used to activate canonical Wnt signaling. As expected, brown adipogenesis of Rb−/− MEFs was inhibited by Chir99021, as assessed by diminished lipid accumulation and expression of FABP4 and UCP1 (Fig. 2B). However, coexpression of PPARγ and PGC-1α largely repressed the effects of Wnt signaling on lipid accumulation and expression of FABP4 and UCP1. These results suggest that Wnt signaling blocks adipogenesis through repression of PPARγ and C/EBPα and inhibits UCP1 through suppression of PGC-1a.
To identify endogenous Wnt proteins that may act to repress brown adipogenesis, we used quantitative RT-PCR to evaluate patterns of expression during differentiation of Rb−/− MEFs. Wnt10a and Wnt10b are expressed in confluent Rb−/− MEFs, and their levels of expression decline during adipogenesis (Fig. 2C). Both of these Wnt proteins are also expressed in BAT. As expected, PPARγ and C/EBPα, as well as the downstream genes FABP4 and UCP1, are induced as Rb−/− MEFs differentiate into brown adipocytes (Fig. 2C). These factors are also highly expressed in BAT. Thus, candidates for activators of the endogenous inhibitory pathway are Wnt10a and Wnt10b.
Brown adipocyte recruitment and thermogenesis are blocked in FABP4-Wnt10b mice.
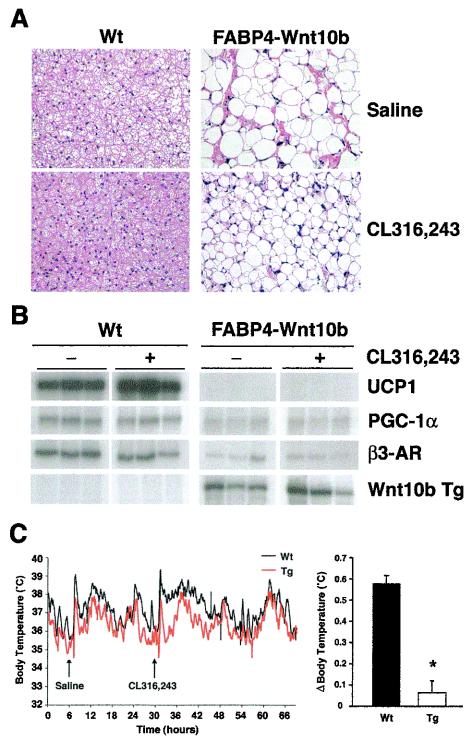
To determine the effect of Wnt signaling on the development of adipose tissues, our group developed transgenic mice in which Wnt10b is expressed in WAT and BAT from the FABP4 promoter (33). Our prior analyses indicated that Wnt10b impairs development of WAT and blocks development of BAT. Interscapular tissue of FABP4-Wnt10b mice has the visual appearance of WAT but lacks expression of white or brown adipocyte markers. The cold sensitivity of these mice provides compelling evidence that FABP4-Wnt10b mice lack functional BAT. To determine whether a strong adrenergic stimulus is sufficient to promote development of new brown adipocytes in these mice, we injected wild-type and FABP4-Wnt10b mice with CL316,243 daily for 21 days. In response to this β3-adrenergic agonist, lipid stores in interscapular BAT of wild-type mice became depleted (Fig. 3A), and expression of UCP1 was induced (Fig. 3B), consistent with a previous report (16). The amount of intracellular lipid was also decreased in FABP4-Wnt10b mice, but 21 days of CL316,243 treatment was not sufficient to increase expression of UCP1 in these mice (Fig. 3B). While long-term treatment of wild-type mice with CL316,243 caused lipid depletion in WAT and the appearance of a few multilocular adipocytes, loss of lipid and appearance of brown adipocytes were not observed in WAT of FABP4-Wnt10b mice (data not shown).
FIG. 3.
Brown adipocyte recruitment and thermogenesis is blocked in FABP4-Wnt10b mice. (A) Wild-type and FABP4-Wnt10b mice were administered saline or a β3-agonist (CL316,243) daily for 21 days. Interscapular tissue was then excised and fixed. Sections were stained with hematoxylin and eosin. Representative photomicrographs are presented. (B) RNA was purified from interscapular tissues (n = 3), and expression of UCP1, PGC-1α, β3-adrenergic receptor (β3-AR), and the Wnt10b transgene evaluated by a RNase protection assay. (C) Wild-type and FABP4-Wnt10b mice (n = 4) were implanted with telometers to measure core body temperature. Saline was injected on day 1 at 9 a.m., and CL316,243 was injected on day 2 at 9 a.m. and core body temperature was recorded (left panel). Average change in body temperature (Δ) was calculated over the 12 h following each injection for each animal. Body temperatures after saline injection were subtracted from that observed after CL316,243 (right panel). Wt, wild type; Tg, transgene.
To confirm the lack of functional BAT within FABP4-Wnt10b mice, we investigated thermogenesis in response to an acute injection of CL316,243. As expected, injection of CL316,243 in wild-type mice caused an increase in core body temperature that lasted more than 5 h (Fig. 3C). In contrast, injection of FABP4-Wnt10b mice with CL316,243 caused a spike in thermogenesis similar to that associated with the stress of injecting saline (Fig. 3C). When the thermal spike after injection of CL316,243 was normalized to that of saline, the increase in core body temperature in FABP4-Wnt10b mice was blunted compared to wild-type mice. Taken together, these data indicate that expression of Wnt10b from the FABP4 promoter blocks the ability of CL316,243 to recruit new brown adipocytes and to stimulate thermogenesis.
Wnt signaling specifically inhibits PGC-1α and UCP1 in differentiated brown adipocytes.
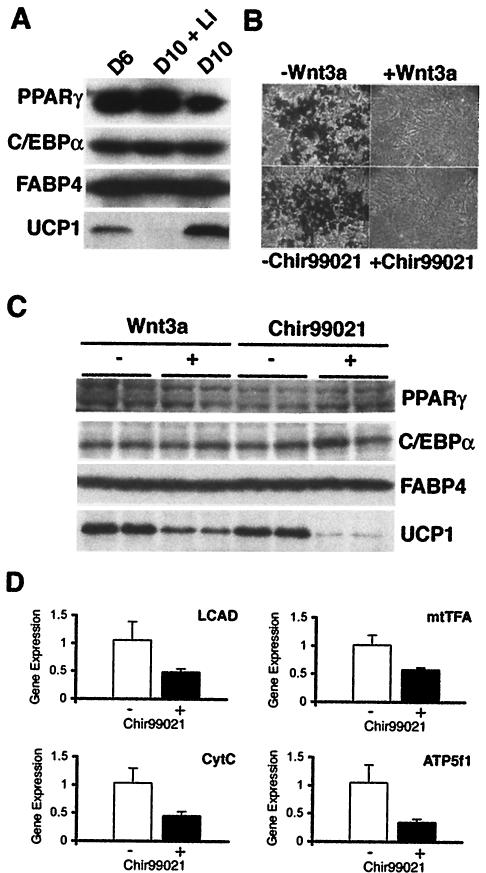
Our results that C/EBPα and PPARγ rescue lipid accumulation but not UCP1 expression prompted us to evaluate whether Wnt signaling also inhibits expression of UCP1 in differentiated brown adipocytes. For this experiment, differentiated Rb−/− MEFs were treated with the GSK3 inhibitor, LiCl, from day 6 to day 10 after induction of differentiation. Expression of various adipogenic genes was determined by immunoblot analysis at days 6 and 10 (Fig. 4A). Treatment of brown adipocytes with LiCl from days 6 to 10 did not influence expression of PPARγ, C/EBPα, or FABP4; however, LiCl dramatically decreased expression of UCP1. Similar regulation is observed at the mRNA level, with the expression of UCP1 transcript inhibited by LiCl, without effects on adipogenic markers such as C/EBPα, PPARγ, or FABP4 (data not shown).
FIG. 4.
Wnt signaling inhibits expression of UCP1 in differentiated Rb−/− MEFs. (A) LiCl (25 mM) or vehicle was added to media of Rb−/− MEFs from day 6s to 10 after induction of brown adipogenesis. Lysates were collected from cells on day 6 and on day 10. Expression of PPARγ, C/EBPα, FABP4, and UCP1 proteins was evaluated by immunoblot analyses. (B) Rb−/− MEFs were induced to differentiate in the presence of 20 ng of Wnt3a per ml, 3 μM Chir99021, or appropriate vehicles from days 0 to 4. At day 10, cells were stained with Oil-Red O and visualized by phase-contrast microscopy. (C and D) Brown adipocytes were treated with 20 ng of Wnt3a per ml, 3 μM Chir99021, or vehicle 8 and 10 days after induction of differentiation. On day12, cells were lysed, and expression of PPARγ, C/EBPα, FABP4, and UCP1 was determined by immunoblot analyses. Expression of mtTFA, cytochrome c (CytC), ATP synthase (ATP5f1), and long-chain acyl CoA dehydrogenase (LCAD) was analyzed by quantitative PCR.
Although inhibition of GSK3 by LiCl activates canonical Wnt signaling, the central role that GSK3 plays in other signaling pathways complicates the interpretation of this experiment. Thus, we used recombinant Wnt3a as an additional tool to activate Wnt signaling in differentiated Rb−/− MEFs (Fig. 4B and C). To confirm that activity of Wnt3a is similar to that of Wnt10b and Wnt1 (Fig. 1), 20 ng of purified Wnt3a per ml was added to Rb−/− MEFs from days 0 to 4 of the differentiation protocol. As expected, Wnt3a inhibits adipogenesis and blocks lipid accumulation to a similar extent as ectopic expression of Wnt10b or Wnt1 or addition of 3 μM Chir99021 (Fig. 4B). Furthermore, the addition of recombinant Wnt3a to differentiated brown adipocytes specifically suppressed expression of UCP1 without affecting expression of PPARγ, C/EBPα, or FABP4. Similar results were observed with Chir99021 (Fig. 4C). Suppression of nuclear- and mitochondrial-encoded genes involved in fatty acid oxidation (long-chain acyl CoA dehydrogenase), transcription and replication (mtTFA), and respiration (cytochrome c and ATP synthase) by Chir99021 (Fig. 4D) suggests that Wnt signaling in brown adipocytes represses both mitochondrial biogenesis and metabolism.
PGC-1α mediates the inhibition of UCP1 by Wnt signaling.
Wnt blocks expression of UCP1 during brown adipogenesis through effects on PGC-1α (Fig. 2), raising the possibility that the effects of Wnt on UCP1 in differentiated brown adipocytes are mediated by a similar mechanism. To establish whether PGC-1α plays an intermediary role in the suppression of UCP1 by Wnt signaling, we first examined the time frame whereby Wnt signaling decreases expression of these genes. Differentiated Rb−/− MEFs were treated with Chir99021 for the indicated times prior to preparation of RNA and protein. Expression of UCP1 mRNA began to decline 12 h after treatment with Chir99021, with complete suppression observed by 48 h (Fig. 5A). Importantly, activation of Wnt signaling with Chir99021 also caused a decline in the level of PGC-1α mRNA, which preceded that of UCP1 (Fig. 5A). PGC-1α mRNA levels decreased between 4 and 8 h after treatment, and expression remained low for at least 96 h. Chir99021 also stimulated a decrease in UCP1 protein, without influencing expression of C/EBPα and FABP4 proteins (Fig. 5B). These data demonstrate that in differentiated brown adipocytes, activation of canonical Wnt signaling by Chir99021 specifically inhibits expression of brown adipocyte genes, UCP1 and PGC-1α, without altering expression of those genes shared with white adipocytes. Suppression of PGC-1α precedes that of UCP1, suggesting a regulatory role for PGC-1α in repression of UCP1 by Wnt signaling.
FIG. 5.
PGC-1α mediates the inhibition of UCP1 by Wnt signaling. Six days after induction of differentiation, canonical Wnt signaling was activated in brown adipocytes by treatment with 3 μM Chir99021 or vehicle. Cells were lysed 0, 1, 2, 4, 8, 12, 24, 48, and 96 h after treatment. UCP1, PGC-1α, and 36B4 mRNA levels were evaluated by RNase protection assays, and UCP1, C/EBPα, and FABP4 proteins were determined by immunoblot analyses. (C) Rb−/− MEFs were infected with control or PGC-1α retroviruses. After differentiation into brown adipocytes (day 6), cells were treated with vehicle or Chir99021 for the indicated times. Expression of UCP1, PGC-1α, and 36Β4 mRNAs was evaluated by RNase protection assays.
To establish whether the decrease in PGC-1α is required for suppression of UCP1 by canonical Wnt signaling, we enforced expression of PGC-1α in Rb−/− MEFs. Six days after induction of adipogenesis, brown adipocytes were treated with 3 μM Chir99021 for 0, 12, or 24 h. RNA was then purified, and expression of PGC-1α and UCP1 was determined by an RNase protection assay (Fig. 5C). As expected, activation of canonical Wnt signaling with Chir99021 suppressed both UCP1 and PGC-1α in Rb−/− brown adipocytes. Ectopic expression of PGC-1α caused a slight increase in levels of total PGC-1α and UCP1 mRNAs. Furthermore, expression of PGC-1α blocked the suppression of UCP1 by Chir99021, indicating that PGC-1α plays an intermediary role in the suppression of UCP1 by Wnt signaling.
Expression of Wnt10b from the UCP1 promoter converts brown adipocytes into white.
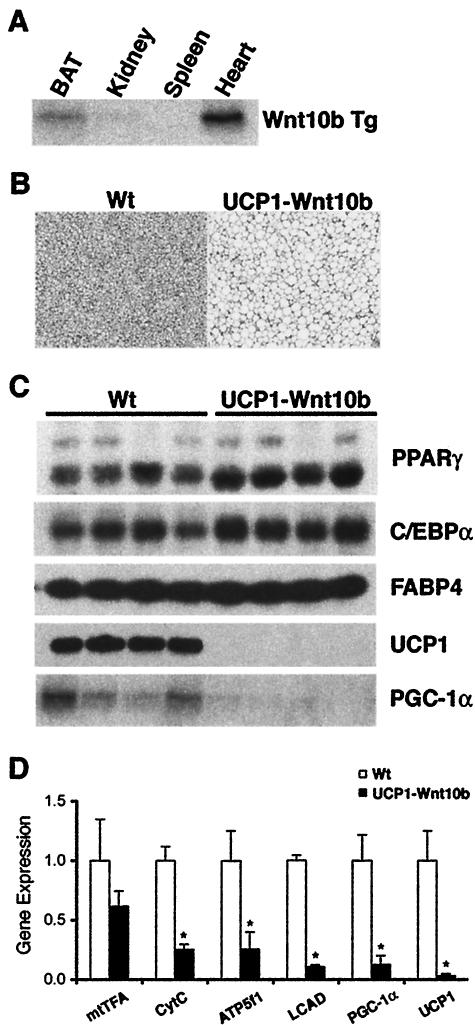
Based upon these in vitro findings in which Wnt signaling specifically inhibits expression of brown adipocyte genes, we explored whether Wnt10b could stimulate conversion of brown adipose into white. We developed a line of transgenic mice in which Wnt10b is expressed from ∼3.1 kb of the UCP1 promoter. In the founder line examined most extensively, the transgene is expressed more highly in interscapular tissue than in other tissues (Fig. 6A), with the exception of heart. No obvious differences in heart development or function were observed in these mice. Expression of the transgene is not observed in heart from a second founder line, which shows an intermediate BAT phenotype. Interscapular tissue from wild-type and UCP1-Wnt10b mice at 8 weeks of age was dissected for histological analyses (Fig. 6B). Wild-type mice have interscapular BAT characterized by cells with multilocular lipid droplets and a dark brown color due to high mitochondrial content. In contrast, interscapular tissue from UCP1-Wnt10b mice has a visual appearance reminiscent of WAT, with cells containing unilocular lipid droplets (Fig. 6B). A similar phenotype is observed as early as 72 h after birth (data not shown).
FIG. 6.
Wnt signaling blocks expression of UCP1 and PGC-1α in interscapular tissue of UCP1-Wnt10b mice without altering expression of white adipocyte genes. (A) RNA was isolated from the indicated tissues and expression of the Wnt10b transgene (Tg) was evaluated by an RNase protection assay. (B) Interscapular tissues from wild-type and UCP1-Wnt10b transgenic mice were isolated at 8 weeks of age, fixed with 10% buffered formalin, and embedded in paraffin. Tissue samples were sectioned and stained with hematoxylin and eosin. Representative photomicrographs are presented. (C) Interscapular tissues from wild-type mice (Wt) and UCP1-Wnt10b mice were dissected (n = 4). Expression levels of PPARγ, C/EBPα, FABP4, and UCP1 proteins were determined by immunoblot analysis, and expression of PGC-1α mRNA was evaluated by an RNase protection assay. (D) Expression of mtTFA, cytochrome c (CytC), ATP synthase (ATP5f1), long-chain acyl CoA dehydrogenase (LCAD), PGC-1α, UCP1, and 18S was analyzed by quantitative PCR. Expression is shown relative to wild-type mice and is expressed as mean ± standard deviation. Differences between wild-type and UCP1-Wnt10b mice were evaluated by a Student's t test. An asterisk indicates statistical significance (P < 0.05). Wt, wild type.
To characterize the effects of Wnt10b at the molecular level, interscapular tissues were dissected, and various adipocyte markers were analyzed. As expected from our histological findings, expression of UCP1 and PGC-1α proteins was greatly decreased in UCP1-Wnt10b mice (Fig. 6C). However, Wnt10b did not alter expression of the adipocyte markers PPARγ, C/EBPα, and FABP4 (Fig. 6C). Although expression of mRNAs for nuclear respiratory factor 1 (data not shown) and mtTFA were not significantly different in UCP1-Wnt10b interscapular tissue, the decline observed in levels of cytochrome c, ATP synthase (ATP5f1), long-chain acyl CoA dehydrogenase, and UCP1 mRNAs indicates that Wnt signaling suppresses mitochondrial biogenesis and metabolism in these mice. These effects are likely to be mediated by the repression of PGC-1α (Fig. 6C). Reduced expression of genes involved in mitochondrial biogenesis and metabolism, without alteration of white adipocyte gene expression, suggests that exposure of brown adipocytes to Wnt signaling in vitro (Fig. 4 and 5) and in vivo (Fig. 6) converts brown adipose into white.
Reciprocal regulation of brown adipocyte and Wnt signaling genes in response to cold challenge or genetic obesity.
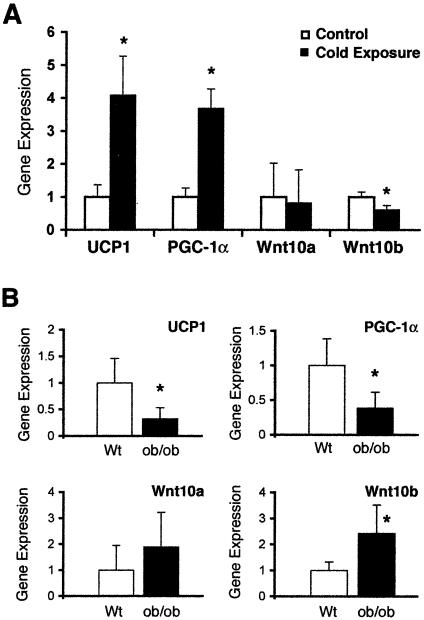
When mice are placed in a cold environment, adrenergic stimulation increases thermogenesis of BAT to help maintain their core body temperature. To determine whether expression of Wnt10b and other Wnt signaling molecules are regulated during this adaptive thermogenesis, we placed individually housed mice (n = 5) at 4°C for 24 h. As expected (44), expression of UCP1 and PGC-1α mRNAs in interscapular BAT increased by about fourfold in mice exposed to the cold temperature (Fig. 7A). Although differences could not be detected with Wnt10a, the cold challenge stimulated a 40% decline in Wnt10b expression. These data are consistent with alterations in Wnt10b and with Wnt signaling components' playing a regulatory role in expression of PGC-1α and UCP1 and, thus, adaptive thermogenesis.
FIG. 7.
Reciprocal regulation of brown adipocyte and Wnt signaling genes in response to cold challenge or genetic obesity. (A) C57BL/6 mice were singly housed and maintained at room temperature (Control; n = 5) or at 4°C for 24 h (Cold exposure; n = 5). After euthanasia, interscapular tissues were dissected and RNA was prepared. Expression of UCP1, PGC-1α, Wnt10a, and Wnt10b was evaluated by quantitative PCR. Relative levels of RNA are shown graphically as the mean + standard deviation. Differences between treatments were evaluated by a Student's t test. An asterisk indicates statistical significance (P < 0.05). (B) RNA from interscapular tissue of wild-type (Wt) and ob/ob mice was isolated. Expression of UCP1, PGC-1α, Wnt10a, and Wnt10b mRNAs was evaluated by quantitative PCR, and relative expression is presented as the mean ± standard deviation. Differences between wild-type and ob/ob mice were evaluated by a Student's t test. An asterisk indicates statistical significance (P < 0.05).
To establish whether reciprocal regulation of UCP1 and Wnt10b also occurs in physiological states where thermogenesis is suppressed, we explored leptin-deficient mice (ob/ob) as a model. Consistent with previous reports (10, 25), the interscapular tissue of ob/ob mice has characteristics of WAT, with a high proportion of unilocular fat cells and reduced expression of UCP1 and PGC-1α (Fig. 7B). We report here that expression of Wnt10b in interscapular tissue of ob/ob mice is increased by about 2.5-fold (Fig. 7B), and although not statistically significant, the twofold increase in Wnt10a expression suggests that additional Wnt signaling components may also be regulated (Fig. 7B). The inverse relationship between Wnt10b and expression of PGC-1α and UCP1 in interscapular tissue of ob/ob mice provides further correlative evidence that Wnt signaling inhibits metabolism of brown adipocytes.
DISCUSSION
We have shown previously that activation of Wnt signaling inhibits differentiation of preadipocytes in vitro and development of white adipocytes in vivo (3, 26, 33, 48). We also demonstrated that expression of Wnt10b from the FABP4 promoter results in a lack of functional BAT (33). In the present study, we use cell culture and transgenic models to investigate mechanisms whereby Wnt signaling regulates differentiation and metabolism of brown adipocytes. Activation of canonical Wnt signaling in Rb−/− MEFs and HIB-1B cells inhibits accumulation of lipid and blocks expression of adipocyte genes, including the brown adipocyte marker, UCP1 (Fig. 1). Enforced expression of PPARγ or C/EBPα in Wnt10b-expressing Rb−/− MEFs rescues the adipogenic program but not the thermogenic program (i.e., expression of UCP1). To overcome the suppression of UCP1 by Wnt signaling requires expression of PPARγ and the important metabolic coactivator PGC-1α (Fig. 2). Endogenous signaling in brown adipocytes is likely mediated by Wnt10a and/or Wnt10b. Both of these factors are expressed in preadipocytes and BAT but not within differentiated brown adipocytes (Fig. 2). The work of Tseng and coworkers supports this idea as they observed an inverse relationship between expression of Wnt10a and the ability of various insulin receptor substrate-deficient cells to differentiate into brown adipocytes (54).
Inhibition of brown adipocyte differentiation is also observed in transgenic mice where Wnt10b is overexpressed from the FABP4 promoter. We demonstrated previously that interscapular tissue from FABP4-Wnt10b mice lacks expression of brown adipocyte markers, including UCP1, PGC-1α, and β3-adrenergic receptors (33). Here we report that expression of UCP1 is undetectable in these mice, even when mice are chronically treated with a β3-agonist, possibly due to a paucity of β3-adrenergic receptors and/or continued active suppression of PGC-1α and UCP1 by Wnt signaling (Fig. 3). While adrenergic stimuli normally cause mobilization of lipid from BAT of wild-type mice (17, 21), interscapular tissue from FABP4-Wnt10b mice is not depleted to the same extent. Furthermore, the thermogenic spike that usually occurs subsequent to injection of CL316,243 is lacking in FABP4-Wnt10b mice, providing further support for the lack of functional BAT (Fig. 3). Thus, Wnt signaling profoundly inhibits development of brown adipocytes in vitro and in vivo.
Lipid filled cells within interscapular tissue are often observed when the function of BAT is inhibited (1, 13, 52). However, interscapular tissue which lacks expression of brown and most white adipocyte genes has only been observed previously when adipogenesis was inhibited by overexpression of a nuclear form of sterol regulatory element-binding protein 1c (49). While the mechanism whereby lipid accumulates in cells without expression of adipocyte markers is unknown, undetectable levels of fatty acid synthase suggest that de novo synthesis of fatty acids is an unlikely cause. However, low levels of lipoprotein lipase in FABP4-Wnt10b mice suggest uptake of lipids from extracellular sources as a possible mechanism (data not shown).
Interestingly, Wnt signaling also influences brown adipocyte metabolism after differentiation has occurred. For instance, activation of canonical Wnt signaling in differentiated brown adipocytes represses expression of UCP1 and genes involved in mitochondrial biogenesis and metabolism, without altering genes typically associated with white adipocytes (Fig. 4). Consistent with the intermediary role of PGC-1α (44), suppression of UCP1 mRNA is preceded by a decline in PGC-1α, and enforced expression of PGC-1α blocks the ability of Wnt signaling to inhibit UCP1 (Fig. 5). These in vitro results suggest that PGC-1α plays a central role in suppression of UCP1 by Wnt signaling. Both mtTFA and long-chain acyl CoA dehydrogenase are regulated by PGC-1α (53, 58), suggesting a role for this transcriptional coactivator in mediating the effects of Wnt10b on these and other downstream genes. the rapid effects of Wnt signaling on suppression of PGC-1α mRNA could be due to altered RNA stability, it is likely that Wnt regulates transcription of the PGC-1α gene. Suppression of PGC-1α transcription might be direct, perhaps through a β-catenin-LEF-1 complex (24), or Wnt signaling could interfere with transcriptional effects of ATF-2, CREB, or FOXO1, all of which have been reported to transactivate the PGC-1α promoter (6, 11, 19). Alternatively, Wnt10b may inhibit upstream regulators such as protein kinase A or p38 mitogen-activated protein kinase, two signaling molecules known to regulate PGC-1α expression and activity (6, 42). Similar levels of UCP1 in PGC-1α−/− mice (32) suggest that repression of UCP1 by Wnt signaling may involve multiple mechanisms.
Although expression of Wnt10b from the FABP4 promoter blocks development of BAT, expression of Wnt10b from the UCP1 promoter appears to cause transdifferentiation of brown adipose into white. While interscapular tissue of UCP1-Wnt10b mice has exceedingly low expression of PGC-1α, UCP1, and genes involved in mitochondrial biogenesis and metabolism, this tissue has unilocular adipocytes and normal expression of white adipocyte genes such as PPARγ, C/EBPα, and FABP4 (Fig. 6). This phenotype is reminiscent of that observed in UCP1−/− and A-ZIP mice (13, 37). Interscapular tissues from these mice have cells with unilocular lipid droplets but comparable expression of various adipocyte markers. Our work in cultured cells suggests that the effects of Wnt10b on brown adipocyte differentiation in FABP4-Wnt10b mice and transdifferentiation in UCP1-Wnt10b mice are due to temporal differences in transgene activation.
Human infants have large depots of BAT to help defend against cold exposure and to maintain energy homeostasis; however, by adulthood, defined BAT depots are lost, and brown adipocytes are instead found interspersed within WAT. While UCP1 is not normally expressed at high levels within human adipose tissue, this important thermogenic protein can be induced in adults under certain pathological conditions (4). Although speculative, it should nonetheless be noted that increased Wnt signaling is a potential mechanism whereby formation of new brown adipocytes is blocked and existing brown adipocytes may be converted into white adipocytes. In rodents, BAT has a crucial role in nonshivering thermogenesis throughout life. Reciprocal expression of Wnt10b with UCP1 and PGC-1α in interscapular tissue from cold-challenged or genetically obese mice supports the notion that Wnt signaling may regulate expression of PGC-1α and UCP1 under conditions of plasticity between WAT and BAT (Fig. 7).
Expression and activity of PGC-1α play important roles in regulating cellular respiration in numerous tissues, and PGC-1α may influence insulin sensitivity in liver and muscle (29, 39). We demonstrate here that Wnt signaling blocks expression of PGC-1α in brown adipocytes. Furthermore, inhibition of PGC-1α by canonical Wnt signaling plays an important intermediary role in the suppression of UCP1 and appears to be sufficient to transform brown adipocytes into white. Consistent with this notion, mice in which FOXC2 is overexpressed (8) or in which 4E-BP1 is disrupted (55) have increased PGC-1α protein, and this is correlated with expression of UCP1 in traditional WAT depots and increased metabolic rate. Furthermore, enforced expression of PGC-1α in white adipocytes is sufficient to cause conversion to a brown adipocyte phenotype, with expression of UCP1 and increased fatty acid oxidation (53). If Wnt signaling regulates PGC-1α in other cell types, this may be a mechanism whereby Wnt proteins influence cellular metabolism.
Acknowledgments
This work was supported by grants from the National Institutes of Health to O.A.M. (grants DK51563 and DK62876). Other support was from the University of Michigan Center for Integrative Genomics, Diabetes Research and Training Center, Nathan Shock Mutant and Transgenic Rodent Core, the Jack Lapides Fellowship, and a mentor-based postdoctoral fellowship from the American Diabetes Association.
We thank Mark Ribick and Randy Kaufman for sharing unpublished qPCR primers for C/EBPα, FABP4, and 18S rRNA. Chir99021 was the generous gift of Allan S. Wagman at Chiron Corporation, Emeryville, Calif.
REFERENCES
- 1.Bachman, E. S., H. Dhillon, C. Y. Zhang, S. Cinti, A. C. Bianco, B. K. Kobilka, and B. B. Lowell. 2002. betaAR signaling required for diet-induced thermogenesis and obesity resistance. Science 297:843-845. [DOI] [PubMed] [Google Scholar]
- 2.Barak, Y., M. C. Nelson, E. S. Ong, Y. Z. Jones, P. Ruiz-Lozano, C. K. R., A. Koder, and R. M. Evans. 1999. PPARγ is required for placental, cardiac, and adipose tissue development. Mol. Cell 4:585-595. [DOI] [PubMed] [Google Scholar]
- 3.Bennett, C. N., S. E. Ross, K. A. Longo, L. Bajnok, N. Hemati, K. W. Johnson, S. D. Harrison, and O. A. MacDougald. 2002. Regulation of Wnt signaling during adipogenesis. J. Biol. Chem. 277:30998-31004. [DOI] [PubMed] [Google Scholar]
- 4.Bouillaud, F., F. Villarroya, E. Hentz, S. Raimbault, A. M. Cassard, and D. Ricquier. 1988. Detection of brown adipose tissue uncoupling protein mRNA in adult patients by a human genomic probe. Clin. Sci. (London) 75:21-27. [DOI] [PubMed] [Google Scholar]
- 5.Cannon, B., and J. Nedergaard. 2004. Brown adipose tissue: function and physiological significance. Physiol. Rev. 84:277-359. [DOI] [PubMed] [Google Scholar]
- 6.Cao, W., K. W. Daniel, J. Robidoux, P. Puigserver, A. V. Medvedev, X. Bai, L. M. Floering, B. M. Spiegelman, and S. Collins. 2004. p38 mitogen-activated protein kinase is the central regulator of cyclic AMP-dependent transcription of the brown fat uncoupling protein 1 gene. Mol. Cell. Biol. 24:3057-3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carmona, M. C., R. Iglesias, M. J. Obregon, G. J. Darlington, F. Villarroya, and M. Giralt. 2002. Mitochondrial biogenesis and thyroid status maturation in brown fat require CCAAT/enhancer-binding protein alpha. J. Biol. Chem. 277:21489-21498. [DOI] [PubMed] [Google Scholar]
- 8.Cederberg, A., L. M. Gronning, B. Ahren, K. Tasken, P. Carlsson, and S. Enerback. 2001. FOXC2 is a winged helix gene that counteracts obesity, hypertriglyceridemia, and diet-induced insulin resistance. Cell 106:563-573. [DOI] [PubMed] [Google Scholar]
- 9.Chavey, C., B. Mari, M. N. Monthouel, S. Bonnafous, P. Anglard, E. Van Obberghen, and S. Tartare-Deckert. 2003. Matrix metalloproteinases are differentially expressed in adipose tissue during obesity and modulate adipocyte differentiation. J. Biol. Chem. 278:11888-11896. [DOI] [PubMed] [Google Scholar]
- 10.Commins, S. P., P. M. Watson, M. A. Padgett, A. Dudley, G. Argyropoulos, and T. W. Gettys. 1999. Induction of uncoupling protein expression in brown and white adipose tissue by leptin. Endocrinology 140:292-300. [DOI] [PubMed] [Google Scholar]
- 11.Daitoku, H., K. Yamagata, H. Matsuzaki, M. Hatta, and A. Fukamizu. 2003. Regulation of PGC-1 promoter activity by protein kinase B and the forkhead transcription factor FKHR. Diabetes 52:642-649. [DOI] [PubMed] [Google Scholar]
- 12.Darlington, G. J., S. E. Ross, and O. A. MacDougald. 1998. The role of C/EBP genes in adipocyte differentiation. J. Biol. Chem. 273:30057-30060. [DOI] [PubMed] [Google Scholar]
- 13.Enerback, S., A. Jacobsson, E. M. Simpson, C. Guerra, H. Yamashita, M. E. Harper, and L. P. Kozak. 1997. Mice lacking mitochondrial uncoupling protein are cold-sensitive but not obese. Nature 387:90-94. [DOI] [PubMed] [Google Scholar]
- 14.Erickson, R. L., N. Hemati, S. E. Ross, and O. A. MacDougald. 2001. p300 coactivates the adipogenic transcription factor CCAAT/enhancer-binding protein alpha. J. Biol. Chem. 276:16348-16355. [DOI] [PubMed] [Google Scholar]
- 15.Fan, M., J. Rhee, J. St.-Pierre, C. Handschin, P. Puigserver, J. Lin, S. Jaeger, H. Erdjument-Bromage, P. Tempst, and B. M. Spiegelman. 2004. Suppression of mitochondrial respiration through recruitment of p160 myb binding protein to PGC-1alpha: modulation by p38 MAPK. Genes Dev. 18:278-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gavrilova, O., B. Marcus-Samuels, and M. L. Reitman. 2000. Lack of responses to a beta3-adrenergic agonist in lipoatrophic A-ZIP/F-1 mice. Diabetes 49:1910-1916. [DOI] [PubMed] [Google Scholar]
- 17.Ghorbani, M., T. H. Claus, and J. Himms-Hagen. 1997. Hypertrophy of brown adipocytes in brown and white adipose tissues and reversal of diet-induced obesity in rats treated with a beta3-adrenoceptor agonist. Biochem. Pharmacol. 54:121-131. [DOI] [PubMed] [Google Scholar]
- 18.Hansen, J. B., C. Jorgensen, R. K. Petersen, P. Hallenborg, R. De Matteis, H. A. Boye, N. Petrovic, S. Enerback, J. Nedergaard, S. Cinti, H. te Riele, and K. Kristiansen. 2004. Retinoblastoma protein functions as a molecular switch determining white versus brown adipocyte differentiation. Proc. Natl. Acad. Sci. USA 101:4112-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herzig, S., F. Long, U. S. Jhala, S. Hedrick, R. Quinn, A. Bauer, D. Rudolph, G. Schutz, C. Yoon, P. Puigserver, B. Spiegelman, and M. Montminy. 2001. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature 413:179-183. [DOI] [PubMed] [Google Scholar]
- 20.Himms-Hagen, J. 2001. Does brown adipose tissue (BAT) have a role in the physiology or treatment of human obesity? Rev. Endocr. Metab. Disord. 2:395-401. [DOI] [PubMed] [Google Scholar]
- 21.Himms-Hagen, J., J. Cui, E. Danforth, Jr., D. J. Taatjes, S. S. Lang, B. L. Waters, and T. H. Claus. 1994. Effect of CL-316,243, a thermogenic beta 3-agonist, on energy balance and brown and white adipose tissues in rats. Am. J. Physiol. 266:R1371-R1382. [DOI] [PubMed] [Google Scholar]
- 22.Himms-Hagen, J., A. Melnyk, M. C. Zingaretti, E. Ceresi, G. Barbatelli, and S. Cinti. 2000. Multilocular fat cells in WAT of CL-316243-treated rats derive directly from white adipocytes. Am. J. Physiol. Cell Physiol. 279:C670-C681. [DOI] [PubMed] [Google Scholar]
- 23.Houstek, J., K. Vizek, S. Pavelka, J. Kopecky, E. Krejcova, J. Hermanska, and M. Cermakova. 1993. Type II iodothyronine 5′-deiodinase and uncoupling protein in brown adipose tissue of human newborns. J. Clin. Endocrinol. Metab. 77:382-387. [DOI] [PubMed] [Google Scholar]
- 24.Jamora, C., R. DasGupta, P. Kocieniewski, and E. Fuchs. 2003. Links between signal transduction, transcription and adhesion in epithelial bud development. Nature 422:317-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kakuma, T., Z. W. Wang, W. Pan, R. H. Unger, and Y. T. Zhou. 2000. Role of leptin in peroxisome proliferator-activated receptor gamma coactivator-1 expression. Endocrinology 141:4576-4582. [DOI] [PubMed] [Google Scholar]
- 26.Kennell, J. A., E. E. O'Leary, B. M. Gummow, G. D. Hammer, and O. A. MacDougald. 2003. T-cell factor 4N (TCF-4N), a novel isoform of mouse TCF-4, synergizes with beta-catenin to coactivate C/EBPalpha and steroidogenic factor 1 transcription factors. Mol. Cell. Biol. 23:5366-5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klaus, S., L. Choy, O. Champigny, A. M. Cassard-Doulcier, S. Ross, B. Spiegelman, and D. Ricquier. 1994. Characterization of the novel brown adipocyte cell line HIB 1B. Adrenergic pathways involved in regulation of uncoupling protein gene expression. J. Cell Sci. 107:313-319. [DOI] [PubMed] [Google Scholar]
- 28.Klein, P. S., and D. A. Melton. 1996. A molecular mechanism for the effect of lithium on development. Proc. Natl. Acad. Sci. USA 93:8455-8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koo, S. H., H. Satoh, S. Herzig, C. H. Lee, S. Hedrick, R. Kulkarni, R. M. Evans, J. Olefsky, and M. Montminy. 2004. PGC-1 promotes insulin resistance in liver through PPAR-alpha-dependent induction of TRB-3. Nat. Med. 10:530-534. [DOI] [PubMed] [Google Scholar]
- 30.Lin, F.-T., and M. D. Lane. 1992. Antisense CCAAT/enhancer-binding protein RNA suppresses coordinate gene expression and triglyceride accumulation during differentiation of 3T3-L1 preadipocytes. Genes Dev. 6:533-544. [DOI] [PubMed] [Google Scholar]
- 31.Lin, F.-T., O. A. MacDougald, A. M. Diehl, and M. D. Lane. 1993. A 30 kilodalton alternative translation product of the CCAAT/enhancer binding protein α message: transcriptional activator lacking antimitotic activity. Proc. Natl. Acad. Sci. USA 90:9606-9610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin, J., P. H. Wu, P. T. Tarr, K. S. Lindenberg, J. St-Pierre, C. Y. Zhang, V. K. Mootha, S. Jager, C. R. Vianna, R. M. Reznick, L. Cui, M. Manieri, M. X. Donovan, Z. Wu, M. P. Cooper, M. C. Fan, L. M. Rohas, A. M. Zavacki, S. Cinti, G. I. Shulman, B. B. Lowell, D. Krainc, and B. M. Spiegelman. 2004. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1α null mice. Cell 119:121-135. [DOI] [PubMed] [Google Scholar]
- 33.Longo, K. A., W. S. Wright, S. Kang, I. Gerin, S. H. Chiang, P. C. Lucas, M. R. Opp, and O. A. MacDougald. 2004. Wnt10b inhibits development of white and brown adipose tissues. J. Biol. Chem. 279:35503-35509. [DOI] [PubMed] [Google Scholar]
- 34.Lowell, B. B., and B. M. Spiegelman. 2000. Towards a molecular understanding of adaptive thermogenesis. Nature 404:652-660. [DOI] [PubMed] [Google Scholar]
- 35.MacDougald, O. A., C.-S. Hwang, H. Fan, and M. D. Lane. 1995. Regulated expression of the obese gene product (leptin) in white adipose tissue and 3T3-L1 adipocytes. Proc. Natl. Acad. Sci. USA 92:9034-9037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.MacDougald, O. A., and S. Mandrup. 2002. Adipogenesis: forces that tip the scales. Trends Endocrinol. Metab. 13:5-11. [DOI] [PubMed] [Google Scholar]
- 37.Moitra, J., M. M. Mason, M. Olive, D. Krylov, O. Gavrilova, B. Marcus-Samuels, L. Feigenbaum, E. Lee, T. Aoyama, M. Eckhaus, M. L. Reitman, and C. Vinson. 1998. Life without white fat: a transgenic mouse. Genes Dev. 12:3168-3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moldes, M., Y. Zuo, R. F. Morrison, D. Silva, B. H. Park, J. Liu, and S. R. Farmer. 2003. Peroxisome-proliferator-activated receptor gamma suppresses Wnt/beta-catenin signalling during adipogenesis. Biochem. J. 376:607-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mootha, V. K., C. Handschin, D. Arlow, X. Xie, J. St. Pierre, S. Sihag, W. Yang, D. Altshuler, P. Puigserver, N. Patterson, P. J. Willy, I. G. Schulman, R. A. Heyman, E. S. Lander, and B. M. Spiegelman. 2004. Errα and Gabpa/b specify PGC-1α-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc. Natl. Acad. Sci. USA 101:6570-6575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Picard, F., M. Gehin, J. Annicotte, S. Rocchi, M. F. Champy, B. W. O'Malley, P. Chambon, and J. Auwerx. 2002. SRC-1 and TIF2 control energy balance between white and brown adipose tissues. Cell 111:931-941. [DOI] [PubMed] [Google Scholar]
- 41.Preece, A. 1972. A manual for histologic technicians. Little Brown, Boston, Mass.
- 42.Puigserver, P., J. Rhee, J. Lin, Z. Wu, J. C. Yoon, C. Y. Zhang, S. Krauss, V. K. Mootha, B. B. Lowell, and B. M. Spiegelman. 2001. Cytokine stimulation of energy expenditure through p38 MAP kinase activation of PPARγ coactivator-1. Mol. Cell 8:971-982. [DOI] [PubMed] [Google Scholar]
- 43.Puigserver, P., and B. M. Spiegelman. 2003. Peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1 alpha): transcriptional coactivator and metabolic regulator. Endocr. Rev. 24:78-90. [DOI] [PubMed] [Google Scholar]
- 44.Puigserver, P., Z. Wu, C. W. Park, R. Graves, M. Wright, and B. M. Spiegelman. 1998. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92:829-839. [DOI] [PubMed] [Google Scholar]
- 45.Ring, D. B., K. W. Johnson, E. J. Henriksen, J. M. Nuss, D. Goff, T. R. Kinnick, S. T. Ma, J. W. Reeder, I. Samuels, T. Slabiak, A. S. Wagman, M. E. Hammond, and S. D. Harrison. 2003. Selective glycogen synthase kinase 3 inhibitors potentiate insulin activation of glucose transport and utilization in vitro and in vivo. Diabetes 52:588-595. [DOI] [PubMed] [Google Scholar]
- 46.Rosen, E. D., C. J. Walkey, P. Puigserver, and B. M. Spiegelman. 2000. Transcriptional regulation of adipogenesis. Genes Dev. 14:1293-1307. [PubMed] [Google Scholar]
- 47.Ross, S. E., R. L. Erickson, I. Gerin, P. M. DeRose, L. Bajnok, D. A. Longo, D. E. Misek, R. Kuick, S. M. Hanask, K. B. Atkins, S. Maehle, H. I. Nebb, L. Madsen, K. Kristiansen, and O. A. MacDougald. 2002. Microarray analyses during adipogenesis: understanding the effects of Wnt signaling on adipogenesis and the roles of liver X receptor α in adipocyte metabolism. Mol. Cell. Biol. 22:5989-5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ross, S. E., N. Hemati, K. A. Longo, C. N. Bennett, P. C. Lucas, R. L. Erickson, and O. A. MacDougald. 2000. Inhibition of adipogenesis by Wnt signaling. Science 289:950-953. [DOI] [PubMed] [Google Scholar]
- 49.Shimomura, I., R. E. Hammer, J. A. Richardson, S. Ikemoto, Y. Bashmakov, J. L. Goldstein, and M. S. Brown. 1998. Insulin resistance and diabetes mellitus in transgenic mice expressing nuclear SREBP-1c in adipose tissue: model for congenital generalized lipodystrophy. Genes Dev. 12:3182-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tanaka, T., J. Yamamoto, S. Iwasaki, H. Asaba, H. Hamura, Y. Ikeda, M. Watanabe, K. Magoori, R. X. Ioka, K. Tachibana, Y. Watanabe, Y. Uchiyama, K. Sumi, H. Iguchi, S. Ito, T. Doi, T. Hamakubo, M. Naito, J. Auwerx, M. Yanagisawa, T. Kodama, and J. Sakai. 2003. Activation of peroxisome proliferator-activated receptor delta induces fatty acid beta-oxidation in skeletal muscle and attenuates metabolic syndrome. Proc. Natl. Acad. Sci. USA 100:15924-15929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tanaka, T., N. Yoshida, T. Kishimoto, and S. Akira. 1997. Defective adipocyte differentiation in mice lacking the C/EBPβ and/or C/EBPδ gene. EMBO J. 16:7432-7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thomas, S. A., and R. D. Palmiter. 1997. Thermoregulatory and metabolic phenotypes of mice lacking noradrenaline and adrenaline. Nature 387:94-97. [DOI] [PubMed] [Google Scholar]
- 53.Tiraby, C., G. Tavernier, C. Lefort, D. Larrouy, F. Bouillaud, D. Ricquier, and D. Langin. 2003. Acquirement of brown fat cell features by human white adipocytes. J. Biol. Chem. 278:33370-33376. [DOI] [PubMed] [Google Scholar]
- 54.Tseng, Y. H., K. M. Kriauciunas, E. Kokkotou, and C. R. Kahn. 2004. Differential roles of insulin receptor substrates in brown adipocyte differentiation. Mol. Cell. Biol. 24:1918-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsukiyama-Kohara, K., F. Poulin, M. Kohara, C. T. DeMaria, A. Cheng, Z. Wu, A. C. Gingras, A. Katsume, M. Elchebly, B. M. Spiegelman, M. E. Harper, M. L. Tremblay, and N. Sonenberg. 2001. Adipose tissue reduction in mice lacking the translational inhibitor 4E-BP1. Nat. Med. 7:1128-1132. [DOI] [PubMed] [Google Scholar]
- 56.Wagman, A. S., K. W. Johnson, and D. E. Bussiere. 2004. Discovery and development of GSK3 inhibitors for the treatment of type 2 diabetes. Curr. Pharm. Des. 10:1105-1137. [DOI] [PubMed] [Google Scholar]
- 57.Wang, N. D., M. J. Finegold, A. Bradley, C. N. Ou, S. V. Abdelsayed, M. D. Wilde, L. R. Taylor, D. R. Wilson, and G. J. Darlington. 1995. Impaired energy homeostasis in C/EBPα knockout mice. Science 269:1108-1112. [DOI] [PubMed] [Google Scholar]
- 58.Wu, Z., P. Puigserver, U. Andersson, C. Zhang, G. Adelmant, V. Mootha, A. Troy, S. Cinti, B. Lowell, R. C. Scarpulla, and B. M. Spiegelman. 1999. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 98:115-124. [DOI] [PubMed] [Google Scholar]
- 59.Yu, X. X., J. L. Barger, B. B. Boyer, M. D. Brand, G. Pan, and S. H. Adams. 2000. Impact of endotoxin on UCP homolog mRNA abundance, thermoregulation, and mitochondrial proton leak kinetics. Am. J. Physiol. Endocrinol. Metab. 279:E433-E446. [DOI] [PubMed] [Google Scholar]
- 60.Yu, X. X., D. A. Lewin, W. Forrest, and S. H. Adams. 2002. Cold elicits the simultaneous induction of fatty acid synthesis and beta-oxidation in murine brown adipose tissue: prediction from differential gene expression and confirmation in vivo. FASEB J. 16:155-168. [DOI] [PubMed] [Google Scholar]