Abstract
In vertebrates, mitochondrial DNA (mtDNA) transcription is initiated bidirectionally from closely spaced promoters, HSP and LSP, within the D-loop regulatory region. Early studies demonstrated that mtDNA transcription requires mitochondrial RNA polymerase and Tfam, a DNA binding stimulatory factor that is required for mtDNA maintenance. Recently, mitochondrial transcription specificity factors (TFB1M and TFB2M), which markedly enhance mtDNA transcription in the presence of Tfam and mitochondrial RNA polymerase, have been identified in mammalian cells. Here, we establish that the expression of human TFB1M and TFB2M promoters is governed by nuclear respiratory factors (NRF-1 and NRF-2), key transcription factors implicated in mitochondrial biogenesis. In addition, we show that NRF recognition sites within both TFB promoters are required for maximal trans activation by the PGC-1 family coactivators, PGC-1α and PRC. The physiological induction of these coactivators has been associated with the integration of NRFs and other transcription factors in a program of mitochondrial biogenesis. Finally, we demonstrate that the TFB genes are up-regulated along with Tfam and either PGC-1α or PRC in cellular systems where mitochondrial biogenesis is induced. Moreover, ectopic expression of PGC-1α is sufficient to induce the coordinate expression of all three nucleus-encoded mitochondrial transcription factors along with nuclear and mitochondrial respiratory subunits. These results support the conclusion that the coordinate regulation of nucleus-encoded mitochondrial transcription factors by NRFs and PGC-1 family coactivators is essential to the control of mitochondrial biogenesis.
The biogenesis of mitochondria requires the expression of a large number of genes encoded by both nuclear and mitochondrial genetic systems (10). However, because the protein coding capacity of mitochondrial DNA (mtDNA) is limited to 13 respiratory subunits, nuclear genes must provide the vast majority of products required for mitochondrial oxidative functions and biosynthetic capacity. In addition, nuclear genes must play a predominant role in controlling mitochondrial transcription, translation, and DNA replication.
Understanding the transcription and replication of mtDNA has been a major focus (6, 29). The majority of evidence points to a mechanism of bidirectional replication where the replication origins for the two strands, termed heavy (H) and light (L) based on their buoyant densities, are displaced by about two-thirds of the genome. The D-loop regulatory region contains bidirectional promoters, HSP and LSP, for transcribing H and L strands as well as the H-strand replication origin (OH). The activities of both HSP and LSP require a 15-nucleotide conserved sequence motif that defines the core promoter. In addition, the two promoters share an upstream enhancer that serves as the recognition site for Tfam (previously called mtTF-1 and mtTFA), a high-mobility-group (HMG) box protein that stimulates transcription through specific binding to the upstream enhancers.
In Saccharomyces cerevisiae, transcription is directed by a 145-kDa core polymerase and a 43-kDa specificity factor, also known as sc-mtTFB. The polymerase and specificity factor transiently interact, and both are required for specific transcription initiation in vitro (22). A vertebrate polymerase and a specificity factor have been characterized biochemically in Xenopus laevis (4), and a cDNA that encodes human mitochondrial RNA polymerase has been isolated (31). Most recently, two isoforms of a human mitochondrial transcription specificity factor, termed TFB1M and TFB2M (also known as h-mtTFB), have also been identified (9, 20). Although TFB1M has about 1/10 the transcriptional activity of TFB2M, both proteins work together with Tfam and mitochondrial RNA polymerase to direct proper initiation from HSP and LSP. Like sc-mtTFB, both TFBs are also related to rRNA methyltransferases and TFB1M can bind S-adenosylmethionine and methylate mitochondrial 12S rRNA (20, 28). Interestingly, TFB1M can also contact the carboxy-terminal domain of Tfam (21). The region of contact between TFB1M and Tfam is essential for transcriptional activation and corresponds to a 29-amino-acid domain that was previously identified as a Tfam activation domain.
A number of recent studies have contributed insights into the pathways regulating mitochondrial biogenesis in mammalian systems. The evidence supports a model whereby regulated coactivators communicate physiological signals to specific transcription factor targets. These events result in the activation of genes required for mitochondrial biogenesis and respiratory function (16, 26). Two transcription factors, nuclear respiratory factor 1 (NRF-1) and NRF-2, act on the majority of nuclear genes encoding subunits of the respiratory complexes. They are also involved in the expression of mitochondrial transcription and replication factors (Tfam and mitochondrial RNA processing RNA), heme biosynthetic enzymes, and other proteins required for respiratory function. Recently, consensus NRF-2 recognition sites have been observed in TFB1M and TFB2M promoters from both humans and mice (9, 20, 25), suggesting that NRFs may be important regulators of these genes as well.
In addition to these transcription factors, a transcriptional coactivator, designated PGC-1α, can induce mitochondrial biogenesis by interacting with NRF-1 (37), PPARα (32), and possibly other nuclear factors. PGC-1α is markedly up-regulated in brown fat during adaptive thermogenesis and can induce mitochondrial biogenesis when expressed ectopically in cultured cells or in transgenic mice (18, 37). A second coactivator, designated PGC-1-related coactivator (PRC), has several structural features in common with PGC-1, including an activation domain, an LXXLL coactivator signature, and an RNA recognition motif (1). Unlike PGC-1α, PRC is not significantly induced during adaptive thermogenesis but is induced upon cell proliferation and down-regulated when cells exit the cell cycle upon contact inhibition or withdrawal of serum. Both PGC-1α and PRC can trans activate NRF-1 target genes that are necessary for the biogenesis of mitochondria and the expression of a functional respiratory chain (1, 37). Both coactivators interact with NRF-1 in vitro and in vivo, and a dominant-negative allele of NRF-1 interferes with the mitochondrial proliferation by PGC-1α. Thus, the functional interplay between these factors appears to define a major regulatory pathway for the biogenesis of mitochondria.
It is of considerable interest to determine how the mtDNA transcriptional apparatus is controlled in the biogenesis of vertebrate mitochondria. The recent discovery of TFB1M and TFB2M raises the question of whether these factors are subject to regulatory pathways involving PGC-1 family coactivators and the NRFs. The coordinate control of the TFBs and Tfam would implicate a common set of nuclear factors in integrating the transcription and replication of mtDNA with a program of mitochondrial biogenesis. The present study is directed at elucidating the involvement of this pathway in TFB expression.
MATERIALS AND METHODS
Plasmids.
The hTFB1M promoter (accession number AL139101) was isolated by PCR amplification of HeLa DNA with sense (5′ AAAAAAGGTACCAGCATCTGCAGAGCGGCGGTTCT 3′) and antisense (5′ AAAAAAAAGCTTCCAACCCTACCTCACCCAGGACCT 3′) primers that yield a PCR product containing Acc65I (5′ end) and HindIII (3′ end) restriction sites to facilitate cloning into the luciferase reporter plasmid pGL3Basic (Promega). After verification of the amplified product by sequencing, the Acc65I-HindIII fragment was cloned first into pGEM-T and then into pGL3Basic to generate pGL3Basic/hTFB1Mwt. Similarly, the hTFB2M promoter (accession number AL356583) was isolated by PCR amplification of HeLa DNA with sense (5′ AAAAAAGGTACCTGTTTCCAGCCCCACTCGGCGACAT 3′) and antisense (5′ AAAAAAAAGCTTTTCTGGCGTCCGGGCCAGGTCAAG 3′) primers with the incorporated Acc65I (5′ end) and HindIII (3′ end) restriction sites. After verification of the PCR product by sequencing, the hTFB2M promoter was cloned into the Acc65I-HindIII sites of pGL3Basic to generate pGL3Basic/hTFB2Mwt.
A series of 5′ deletions of the hTFB1M promoter was generated by PCR amplification of pGEM-T/hTFB1Mwt with the use of a nested set of 5′ deletion primers containing the Acc65I restriction site and an antisense primer derived from the T7 promoter. The primers denoted by the deletion endpoints with the Acc65I cloning site underlined are as follows: −378, GACAGGTACCTAGAACGTTAAAG; −316, CGGTACCACCTCTCAGAGCAACT; −201, CGGTACCCCCCCGGCTCTCACA; −143, TCTCGCGGTACCACTTAGCGCAT; −117, CTCGGTACCCCGGGAATTTCCT; −71, AGGTACCACCAATGGGGCTGACT; −26, AGGTACCTCCCCTGCGCGTTTCT.
A series of 5′ deletions of the hTFB2M promoter was generated by PCR amplification of pGL3Basic/hTFB2Mwt with a nested set of 5′-end-deletion primers containing the Acc65I restriction site and a reverse primer (5′ CTTTATGTTTTTGGCGTCTTCC 3′) corresponding to GLprimer2 from Promega. All amplified 5′-deletion fragments from hTFB1M and hTFB2M promoters were cloned into Acc65I-HindIII sites of pGL3Basic plasmid and verified by sequencing. The primers denoted by the deletion endpoints with the Acc65I cloning site underlined are as follows: −272, TGGAGGAGGTACCTCTCGCCTTT; −80, ACTCAGGTACCTCGGGCGGCTGA; −26, CAGGTACCCTCCGCTGTTCGCAT; +1, GGTACCAGTGTTTACTTCCGCTT.
Site-directed mutagenesis of NRF-1, NRF-2, and Sp1 sites on both promoters was performed by PCR utilizing pGL3Basic/hTFB1M and two wild-type plasmids as templates. Pairs of internal overlapping oligonucleotides with the desired mutations along with flanking pGL3Basic primers (sense, 5′ ACTAGCAAAATAGGCTGTCC 3′; antisense, 5′ CTTTATGTTTTTGGCGTCTTCC 3′) were used to generate mutations (15). After verification of all site-directed mutations by sequencing, the mutant promoters were subcloned as Acc65I-HindIII fragments into pGL3Basic. Sense (S) and antisense (AS) mutagenesis primers with mutated nucleotides underlined are as follows: hTFB1M/NRF-1mut(S), GGACTTAGCGGAATTCCTCTCAGCAC; hTFB1M/NRF-1mut(AS), GTGCTGAGAGGAATTCCGCTAAGTCC; hTFB1M/NRF-2Amut(S), CAGCACGCCGAGATCTAGCTGTCCGCGG; hTFB1M/NRF-2Amut(AS), CCGCGGACAGCTAGATCTCGGCGTGCTG; hTFB1M/NRF-2Bmut(S), GCGGTCTTCGATATCGGTGGGATA; hTFB1M/NRF-2Bmut(AS), TATCCCACCGATATCGAAGACCGC; hTFB1M/Sp1Amut(S), TGTCCAGGCCTGCAGTCGTCCCGCC; hTFB1M/Sp1Amut(AS), GGCGGGACGACTGCAGGCCTGGACA; hTFB1M/Sp1Bmut(S), TCCCCGTAGCTCGAGCAGGAGAAGC; hTFB1M/Sp1Bmut(AS), GCTTCTCCTGCTCGAGCTACGGGGA; hTFB2M/NRF-1mut(S), CCGCTGTTCGACTGATCAGGCTCTAG; hTFB2M/NRF-1mut(AS), CTAGAGCCTGATCAGTCGAACAGCGG; hTFB2M/NRF-2Amut2(S), AGCCGAGGCTCAAGCCCAAGTGAGGGA; hTFB2M/NRF-2Amut2(AS), TCCCTCACTTGGGCTTGAGCCTCGGCT; hTFB2M/NRF-2Bmut2(S), GGGAGAAAAGCAACAAGGCTCCGCTG; hTFB2M/NRF-2Bmut2(AS), CAGCGGAGCCTTGTTGCTTTTCTCCC; hTFB2M/Sp1mut(S), CTCGCCTTTCGACAGCGTCTCCCTCTGC; hTFB2M/Sp1mut(AS), GCAGAGGGAGACGCTGTCGAAAGGCGAG.
Other plasmids used in transfections were δ-ALAS(−479)wt/pGL3, δ-ALAS(−479)m1m2/pGL3, FL PRC/pSV Sport (1), and PGC-1/pSV Sport (37).
Electromobility shift assays.
Nuclear extracts from HeLa cells were prepared as described previously (2). The following oligonucleotides (mutated nucleotides underlined) were employed in binding assays: TFB1M/NRF-1, GATCCGGACTTAGCGCATGCGCTCTCAGCA and GCCTGAATCGCGTACGCGAGAGTCGTTCGA; TFB1M/NRF-1mut, GATCCGGACTTAGCGGAATTCCTCTCAGCA and GCCTGAATCGCCTTAAGGAGAGTCGTTCGA; TFB2M/NRF-1, GATCCTCCGCTGTTCGCATGCGCAGGCTCTA and GAGGCGACAAGCGTACGCGTCCGAGATTCGA; TFB2M/NRF-1mut, GATCCTCCGCTGTTCGGAATTCCAGGCTCTA and GAGGCGACAAGCCTTAAGGTCCGAGATCGA; TFB1M/NRF-2, GATCCGCCGGGAATTTCCTGTCCGCGGTCATCGCTTCCGGTGGGA and GCGGCCCTTAAAGGACAGGCGCCAGTAGCGAAGGCCACCCTTCGA; TFB1M/NRF-2mut, GATCCGCCGTCTATTAGATGTCCGCGGTCATCGCTAGAGGTGGGA and GCGGCAGATAATCTACAGGCGCCAGTAGCGATCTCCACCCTTCGA; TFB2M/NRF-2, GATCCAGGCGGAAGCGGAAGTGAGGGAGAAAAGCAGGAAGGCTCA and GTCCGCCTTCGCCTTCACTCCCTCTTTTCGTCCTTCCGAGTTCGA; TFB2M/NRF-2mut, GATCCAGGCTCTAGCTCTAGTGAGGGAGAAAAGCATCTAGGCTCA and GTCCGAGATCGAGATCACTCCCTCTTTTCGTAGATCCGAGTTCGA; ratCO4/NRF-2, GATCCTTGCTCTTCCGGTGCGGGACCCGCTCTTCCGGTCGCGA and GAACGAGAAGGCCACGCCCAGGGCGAGAAGGCCAGCGCTTCGA; ratCO4/NRF-2mut, GATCCTTGCTCTAGAGGTGCGGGACCCGCTCTAGAGGTCGCGA and GAACGAGATCTCCACGCCCAGGGCGAGATCTCCAGCGCTTCGA; RC4(−172/−147), GATCCTGCTAGCCCGCATGCGCGCGCACCTTA and GACGATCGGGCGTACGCGCGCGTGGAATTCGA.
Annealed oligonucleotides were 3′ end labeled using Klenow enzyme and purified with the QIAquick nucleotide removal kit (Qiagen). Binding reaction mixtures (20 μl) contained either 5 μg of nuclear extract, 200 pg of recombinant NRF-1 (33), or various amounts of recombinant NRF-2α and NRF-2β1 (12) in 25 mM Tris-HCl (pH 7.9)-6.25 mM MgCl2-0.5 mM EDTA-10% (vol/vol) glycerol-1 mM dithiothreitol-5 μg of sonicated calf thymus DNA-∼0.15 pmol of labeled oligonucleotides. Specific and nonspecific unlabeled competitor oligonucleotides were added to the binding reaction mixtures in 50-fold molar excess prior to the addition of labeled oligonucleotides. Reaction mixtures were incubated at room temperature for 15 min, and DNA-protein complexes were resolved on a 5% polyacrylamide gel in Tris-borate-EDTA buffer at 300 V. Supershifting was carried out by addition of 1 μl of antiserum (either goat anti-NRF-1, rabbit anti-NRF-2α, rabbit anti-NRF-2β1, or preimmune serum) to the binding reaction mixtures 10 min after the components were mixed, followed by an additional 5 min of incubation before loading onto the gel. Gels were then dried and subjected to autoradiography.
Cell culture and transfections.
BALB/3T3 cells used in transfections were maintained in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% calf serum (HyClone) and 1% penicillin-streptomycin (Gibco). Nuclear extracts for electrophoretic mobility shift assay were prepared from HeLa S3 cells (American Type Culture Collection) grown in Ham's F-12 medium (CellGro). 3T3-L1 fibroblasts (American Type Culture Collection) were grown on 150-mm-diameter dishes in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% fetal bovine serum (HyClone) and 1% penicillin-streptomycin (Gibco). Fibroblasts were differentiated into adipocytes as described previously (36). Serum induction of quiescent BALB/3T3 cells was performed as described previously (14).
Transient transfections were performed by calcium phosphate precipitation as described previously (1). Cells were plated at a density of 2,600 to 6,200 cells per cm2 in six-well plates, washed twice at 5 h posttransfection with Dulbecco's phosphate-buffered saline (Gibco), and grown for an additional 40 h in fresh medium. Cell extracts were prepared, and luciferase assays were performed with PharMingen reagents. Values were normalized to β-galactosidase activity measured spectrophotometrically by using the β-galactosidase enzyme assay system (Promega).
The recombinant adenoviral plasmids Ad-PGC-1 and Ad-GFP were a gift from D. P. Kelly (Washington University). The Ad-PGC-1 plasmid contains, in tandem, the green fluorescent protein (GFP) gene and the myc-tagged PGC-1 cDNA, whereas the control Ad-GFP plasmid contains only the GFP gene (18). These plasmids were linearized and transfected into the packaging cell line 293 with Lipofectamine (Invitrogen), and the resulting viral stock was amplified to high titer. C2C12 mouse myoblasts were infected, and the infection efficiency was determined by GFP expression 24 h after infection. RNA was isolated with Trizol (Invitrogen) at 72 h postinfection and subjected to real-time PCR analysis.
Real-time quantitative reverse transcription-PCR.
For real-time PCR expression analysis, cells were washed in phosphate-buffered saline and total RNA was extracted using Trizol reagent (Invitrogen). RNA samples were then DNase treated with a DNA-Free kit (Ambion) and analyzed for quality with an Agilent 2100 Bioanalyzer. Treated RNA (500 ng to 1 μg) was reverse transcribed in 20- to 30-μl reaction mixtures with random hexamer primers and the TaqMan reverse transcription reagents kit (Applied Biosystems) according to the manufacturer's protocol. Target sequences were selected to span an exon-intron junction, except for COXII, which has no introns. Gene-specific primer-probe mixtures for each amplicon were manufactured by Applied Biosystems. Aliquots of the reverse-transcribed products were combined with TaqMan Universal PCR Master Mix (Applied Biosystems) and an appropriate primer-probe mix in 20-μl reaction mixtures. PCR amplifications were carried out in 384-well plates with the ABI Prism 7900HT sequence detection system, and the results were analyzed with a Relative Quantification Study program with SDS 2.1 software (Applied Biosystems). Samples were analyzed in triplicate, and mRNA quantities were normalized against 18S RNA (primer-probe mix from Applied Biosystems). Reactions were carried out using the following conditions: an initial step of 2 min at 50°C and 10 min at 95°C, followed by 45 cycles of 15 s at 95°C and 1 min at 60°C. Relative gene expression levels were determined by the comparative Ct method.
ChIP.
Chromatin immunoprecipitations (ChIPs) were performed using a modification of an established method (30). HeLa cells, infected for 48 h with recombinant adenovirus expressing GFP or PGC-1, were fixed with 1% formaldehyde for 10 min at 25°C, lysed, and subjected to sonication. Soluble chromatin was first precleared with a salmon sperm DNA-protein A agarose slurry for 3 h at 4°C and then immunoprecipitated at 4°C overnight with rabbit anti-NRF-1 (11), anti-NRF-2α, or anti-dynein light chain (13) antibodies or without the addition of antibodies. Immune complexes were collected by incubation with salmon sperm DNA-protein A agarose slurry for 1 h at 4°C, the beads were washed extensively, and the chromatin immune complexes were eluted. After de-cross-linking, DNA was purified using the QiaQuick PCR purification kit (Qiagen) and subjected to PCR with primer sets specific for the human TFB1M promoter (forward, 5′ CCTAGTCCACCCGGCTCT 3′; reverse, 5′ GAGGAACCTGCGAGACCTAA 3′), TFB2M promoter (forward, 5′ ACGGTCCACTCACAATCCTC 3′; reverse, 5′ CCCACGTGGAACATTTTCTG 3′), and a control fragment from the human β-actin gene (forward, 5′ GTCATCACCATTGGCAATGAG 3′; reverse, 5′ CGTCATACTCCTGCTTGCTG 3′). The regions of amplification included the NRF-1 and NRF-2α binding sites of both the TFB1M and the TFB2M promoters, whereas the region amplified in the β-actin gene does not bind the factors. Linearity of the PCR amplifications was established experimentally (TFB1M and TFB2M, 30 cycles; β-actin, 26 cycles), and PCR products were resolved on a 2% agarose gel and visualized by ethidium bromide staining.
RESULTS
Authentic recognition sites for NRF-1 and NRF-2 reside in the human TFB promoters.
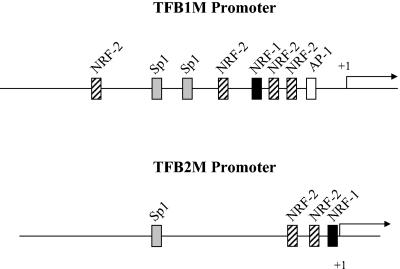
NRFs have been implicated in the expression of both Tfam and mitochondrial RNA processing RNA, key constituents of the mitochondrial transcription and replication machinery. To explore whether TFB expression is also governed by NRFs, we cloned the human TFB promoters (Fig. 1). A search for transcription factor binding sites revealed relatively simple promoter structures consisting of recognition sites for NRF-1, NRF-2, and Sp1. In addition, the TFB1M promoter has a consensus AP-1 site. Both promoters have tandem NRF-2 sites and an NRF-1 site in close proximity to the transcription start site. In addition, the TFB1M promoter has two upstream Sp1 sites flanked by NRF-2 sites, whereas the TFB2M promoter has a single upstream Sp1 site. A similar configuration of NRF and Sp1 sites has been observed in many well-characterized genes that are essential to the expression and function of the respiratory chain.
FIG. 1.
Schematic representation of hTFB1M and hTFB2M promoters. The hTFB1M promoter was identified by aligning the CGI-75 cDNA (accession number NP_057104) corresponding to hTFB1M (20) with the human genomic sequence from chromosome 6 encoding this gene (accession number AL139101). The hTFB2M promoter was identified by aligning the human cDNA (accession number NM_022366) with the human genomic clone from chromosome 1 (accession number AL356583). The cis-acting elements, denoted by labeled rectangular boxes, were initially identified by the Match internet search tool and by visual inspection of the sequence.
As shown in Fig. 2A, the TFB1M and TFB2M NRF-1 sites are near-perfect matches to the NRF-1 consensus and contain all of the essential nucleotides that are invariant in a large number of functional NRF-1 recognition sites (26). Similarly, the TFB NRF-2 sites contain the GGAA motifs that are characteristic of the binding sites for ETS family transcription factors, of which NRF-2 is a member (Fig. 2B). As originally observed in the COXIV promoter, these sites are often tandemly arranged in respiratory genes to promote cooperative high-affinity binding of NRF-2 (12, 34). Like the COXIV promoter, both TFB promoters contain tandem NRF-2 sites that are separated by 16 nucleotides (Fig. 2B). This conservation of spacing suggests that cooperative binding of NRF-2 to these promoters may occur as well.
FIG. 2.
Comparison of hTFB NRF-1 and NRF-2 sites. (A) NRF-1 sites from hTFB1M and hTFB2M are compared to the consensus. Numbers below the consensus represent the percent representation of the nucleotide at that position in over 20 functional NRF-1 sites. Highly invariant nucleotides are in boldface. (B) Comparison of tandemly arranged NRF-2 sites in the TFB promoters to those present in the COXIV promoter in which they were first identified. The GGAA core motifs are boxed. Numbers adjacent to the sequences denote the nucleotide positions relative to the mRNA 5′ ends.
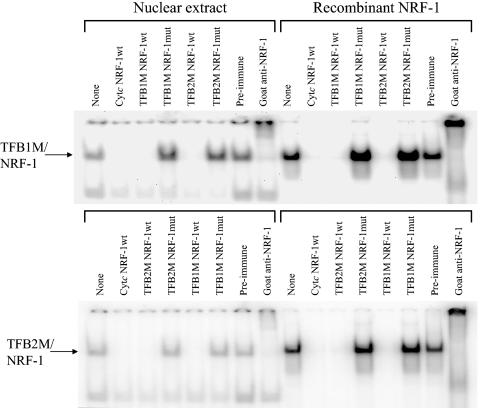
Both HeLa cell nuclear extract and purified recombinant NRF-1 were compared for their ability to form specific DNA-protein complexes with the TFB NRF-1 sites. As shown in Fig. 3, DNA-protein complexes were formed using radiolabeled TFB1M and TFB2M NRF-1 oligomers. The complexes formed using crude nuclear extract or recombinant NRF-1 displayed identical electrophoretic migrations and were competed away by an excess of an unlabeled oligomer containing an authentic cytochrome c (Cyt c) NRF-1 recognition sequence. The TFB-NRF-1 complexes were also supershifted upon inclusion of goat anti-NRF-1 serum in the binding reaction mixtures, confirming that NRF-1 is present in the complex. Complexes formed with either TFB site were also competitively displaced by excess oligomers containing either site but not with those synthetic oligomers where essential guanine nucleotide contacts were mutated. No shifted complexes were observed in the absence of extract or recombinant protein (data not shown). These results establish that the TFB NRF-1 sites are indistinguishable from those present in other respiratory gene promoters.
FIG. 3.
Specific binding of NRF-1 to recognition sites within the hTFB1M and hTFB2M promoters. Radiolabeled synthetic oligonucleotides containing either the TFB1M or TFB2M NRF-1 site were bound to either crude nuclear extracts or purified recombinant NRF-1, and the complexes (indicated by the arrows) were separated by gel electrophoresis. The unlabeled competitor oligonucleotide or the supershifting antiserum added to the binding reaction is indicated above each lane.
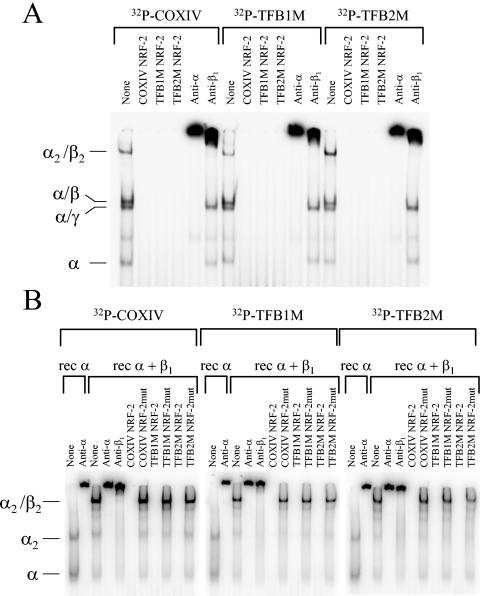
Similar DNA binding experiments were conducted by comparing radiolabeled oligomers containing the tandem NFR-2 sites from the COXIV, TFB1M, and TFB2M promoter regions. As shown in Fig. 4A, identically migrating DNA-protein complexes were formed with all three synthetic NRF-2 oligomers by using a crude heparin agarose chromatography fraction prepared from a HeLa cell nuclear extract. Fractionation of nuclear extracts on heparin agarose is necessary for detection of the NRF-2 DNA binding activity present in crude nuclear extracts (34). The complexes included a rapidly migrating complex containing the DNA binding α subunit, a doublet of intermediate migration containing the α/γ and α/β heterodimers, and a slowly migrating heterotetrameric α2/β2 complex (Fig. 4A). All of the complexes were competitively displaced by an excess of unlabeled COXIV oligomer containing authentic NRF-2 sites as well as by TFB1M and TFB2M NRF-2 oligomers. In addition, all of the complexes were supershifted by inclusion of rabbit anti-NRF-2α serum in binding reaction mixtures because they all contain the α subunit. In contrast, only those complexes containing the β subunit (α/β and α2/β2) were supershifted by inclusion of rabbit anti-NRF-2β1 serum.
FIG. 4.
Specific binding of NRF-2 to recognition sites within the hTFB1M and hTFB2M promoters. (A) Radiolabeled synthetic oligonucleotides containing COXIV, hTFB1M, or hTFB2M recognition sites were bound to an aliquot of partially purified nuclear extract that was eluted from heparin agarose at 0.25 M NaCl. The indicated DNA-protein complexes were separated by gel electrophoresis. The unlabeled competitor oligonucleotide or the supershifting antiserum added to the binding reactions is indicated above each lane. (B) The same radiolabeled oligonucleotides as in panel A were bound to either recombinant NRF-2α (rec α) or a mixture of recombinant NRF-2α and NRF-2β1 (rec α + β1). The indicated DNA-protein complexes were separated by gel electrophoresis. The unlabeled competitor oligonucleotide or the supershifting antiserum added to the binding reactions is indicated above each lane.
These results were confirmed using purified recombinant α and β1 subunits. As shown in Fig. 4B, the migration of DNA-protein complexes formed with recombinant subunits was identical for all three radiolabeled oligomers, as was their ability to be supershifted by anti-NRF-2α and anti-NRF-2β1 sera. Likewise, the complexes formed between a mixture of recombinant α and β1 subunits and labeled COXIV, TFB1M, or TFB2M NRF-2 oligomers were competed away by an excess of unlabeled oligomer containing each site but not by those where the GGAA binding motif had been mutated. No shifted complexes were observed in the absence of extract or recombinant protein (data not shown). These results establish the authenticity of the TFB NRF-2 sites and their specificity of interaction with the transcription factor.
NRFs are major determinants of TFB promoter function.
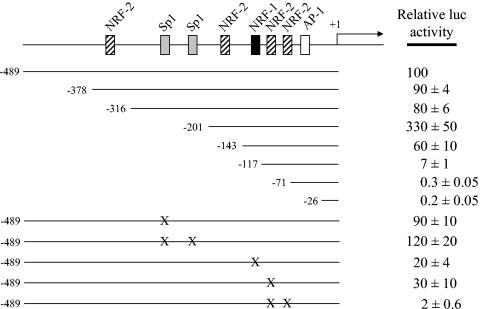
It was of interest to investigate the functional contribution of the NRF-1 and NRF-2 recognition sites within the context of the TFB1M and TFB2M promoters. To this end, a series of 5′ deletions and point mutations were constructed in luciferase reporter plasmids, and their effects on promoter function were assayed by gene transfection. The 5′ deletions were designed to progressively remove the sequences identified as transcription factor binding sites. As depicted in Fig. 5, deletion of the TFB1M promoter to −316, removing the NRF-2 site most distal to the transcription start site, had little effect on activity whereas deletion to −201, removing the tandem Sp1 sites, increased activity severalfold. Competition mobility shift and supershift assays demonstrated that these sites actually bind Sp1 (data not shown). The results suggest that the Sp1 sites function as negative elements within the context of this promoter. Progressive removal of a second NRF-2 site by deletion to −143 reduced activity to 60% of wild-type level but less than 20% of the −201 deletion level, indicating that this site exerts a strong positive effect on the truncated −201 promoter. Further deletion to −117, removing the NRF-1 site, resulted in an additional ninefold reduction in activity to a level approximately 14-fold below that of the wild type. Removal of the tandem NRF-2 sites by deletion to −71 markedly reduced activity to minimally detectable levels with no significant further reduction observed by removal of the AP-1 site by deletion to −26. These results demonstrate that the NRF sites between −201 and −71 constitute major determinants of promoter function.
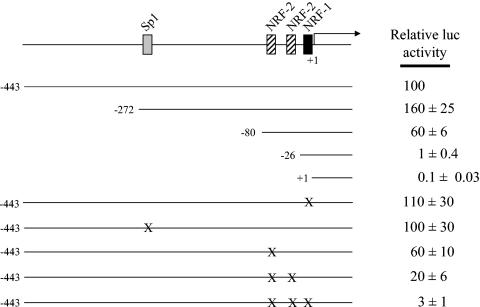
FIG. 5.
Mutational analysis of the hTFB1M promoter region. A series of 5′ deletions designed to progressively remove putative cis-acting elements was analyzed by gene transfection. The normalized luciferase activity obtained from the promoter fragment containing 489 nucleotides of 5′-flanking DNA (−489) was designated as 100%. Point mutations represented by X were introduced into the −489 promoter as described under Materials and Methods. The activities of all of the mutated promoters were expressed as a percentage of that obtained from the −489 promoter. Numbers represent the average ± standard error for three separate determinations.
The results were confirmed by altering some of the key elements by the introduction of point mutations. In contrast to the Sp1 deletion, point mutations in the Sp1 sites increased activity by only 20%, suggesting that upstream elements exert a compensatory positive effect upon removal of these sites. Again, competition mobility shift and supershift assays demonstrated that the wild-type but not the mutant sites bound Sp1 (data not shown). Point mutations in the NRF-1 site exerted a large negative effect on promoter activity, confirming its importance for promoter function. Interestingly, mutation of one of the tandem NRF-2 sites reduced activity severalfold whereas mutation of both reduced activity about 50-fold. This apparent cooperativity most likely results from the high-affinity binding of the NRF-2 heterotetramer to these tandem sites.
A similar analysis was performed on the TFB2M promoter (Fig. 6). Deletion to −272 exerted a small positive effect on activity whereas deletion to −80, removing the single Sp1 site, modestly reduced activity to 60% of wild-type level. As with the TFB1M promoter, the largest negative effects on activity occurred upon deletion of the NRF-1 and NRF-2 sites that are proximal to the transcription initiation site. Removal of the NRF-2 sites by deletion to −26 reduced activity to 1% of wild-type levels while further deletion of the NRF-1 site essentially eliminated any residual activity. Surprisingly, point mutations in the NRF-1 site alone had no effect on promoter activity, again suggesting that compensatory effects occur in the context of the full promoter. In addition, although mutation of the tandem NRF-2 sites reduced activity significantly, the synergism observed in the TFB1M promoter was not observed. However, combined mutation of all three NRF sites reduced activity over 30-fold to near-background levels. These results are indicative of strong interactions between the NRF-1 and NRF-2 sites within the TFB2M promoter context.
FIG. 6.
Mutational analysis of the hTFB2M promoter region. A series of 5′ deletions designed to progressively remove putative cis-acting elements was analyzed by gene transfection. The normalized luciferase activity obtained from the promoter fragment containing 443 nucleotides of 5′-flanking DNA (−443) was designated as 100%. Point mutations represented by X were introduced into the −443 promoter as described under Materials and Methods. The activities of all of the mutated promoters were expressed as a percentage of that obtained from the −443 promoter. Numbers represent the average ± standard error for three separate determinations.
NRF-1 and -2 binding sites in the TFB promoters are targets for trans activation by the PGC-1 family coactivators.
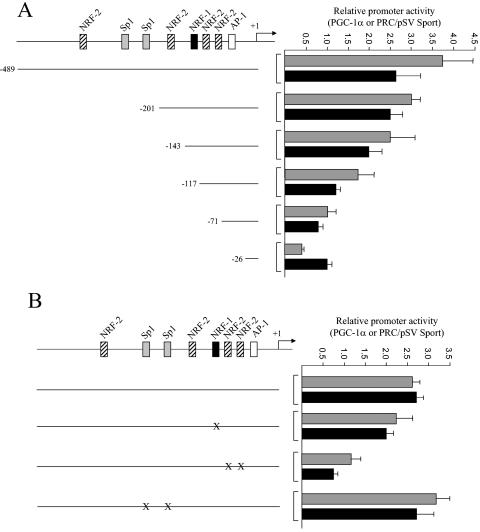
Regulation of mitochondrial biogenesis occurs through the induction of PGC-1α coactivator and its subsequent interaction with NRF-1 and other transcription factors (37). Both PGC-1α and PRC act through NRF-1 to induce NRF-1 target genes (1). trans-activation experiments were performed in order to investigate the potential role of these coactivators in TFB gene expression. As shown in Fig. 7 and 8, both TFB promoters are trans activated to an equivalent degree by PGC-1α and PRC. To identify the cis elements that are required for the enhancement of promoter activity by the coactivators, each of the TFB promoter deletions was tested in the assay. It is clear that the full-length TFB1M promoter and the −201 deletion are similarly trans activated by both PGC-1α and PRC (Fig. 7A). Further deletion beyond −201 removes the NRF sites that contribute most to promoter function, and by deletion to −71, promoter activation is reduced to uninduced levels.
FIG. 7.
trans activation of the hTFB1M promoter and its mutated derivatives by PGC-1α and PRC. (A) The hTFB1M promoter deletions shown in Fig. 5 were assayed for trans activation by PGC-1α (gray bars) and PRC (black bars) by cotransfection with vectors expressing each coactivator. Values are the average fold activation for three separate determinations ± standard error measured relative to the pSVSport negative control. (B) Same as panel A except that the indicated point mutations were assayed for trans activation by the coactivators.
FIG. 8.
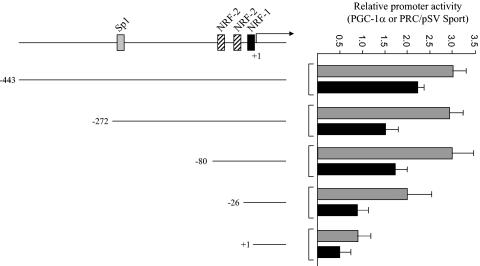
trans activation of the hTFB2M promoter and its mutated derivatives by PGC-1α and PRC. The hTFB2M promoter deletions shown in Fig. 6 were assayed for trans activation by PGC-1α (gray bars) and PRC (black bars) by cotransfection with vectors expressing each coactivator. Values are the average fold activation for three separate determinations ± standard error measured relative to the pSVSport negative control.
The requirement for the TFB1M NRF sites for promoter activation by the coactivators was confirmed by point mutation. As shown in Fig. 7B, elimination of the Sp1 sites alone had no effect on promoter induction by either coactivator, whereas mutation of the major NRF-1 and NRF-2 sites reduced or eliminated trans activation by both coactivators. Similar experiments were conducted on the TFB2M promoter region. The results show that the −80 promoter, containing only the NRF sites, can support near-maximal levels of trans activation by PGC-1α or PRC (Fig. 8). Deletions removing these sites reduce or abolish promoter activation. The results support the conclusion that the NRF sites most proximal to the transcription start sites in both TFB promoters are the major targets for coactivator trans activation.
TFB expression coincides with enhanced mitochondrial biogenesis.
Nothing is known about whether the transcriptional control of TFB expression is associated with that of other genes implicated in the regulation of mitochondrial biogenesis and/or function. A series of real-time PCR assays was therefore developed to compare, quantitatively, the expression of a number of key genes involved in the process. The collection includes representatives from several classes of genes. The first consists of NRF target genes whose products function in the mitochondria, as represented by mitochondrial transcription factors (Tfam, TFB1M, and TFB2M) as well as respiratory proteins (Cyt c and COXIV). A second class consists of the nuclear regulatory factors (NRF-1, PGC-1α, and PRC) that regulate nuclear genes involved in mitochondrial respiratory function. A third class includes a mitochondrially encoded respiratory subunit (COXII).
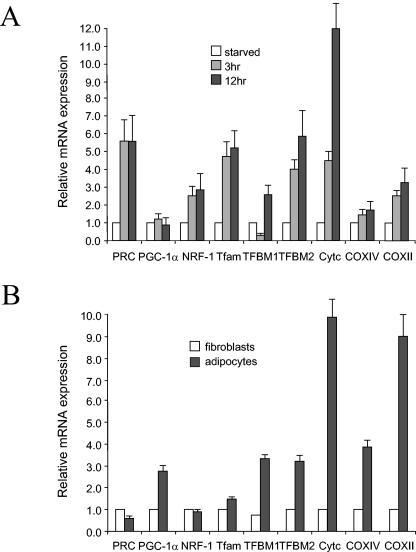
The expression of these genes was monitored in cellular systems where Cyt c, a mitochondrial marker, is known to be up-regulated. In the first, quiescent fibroblasts were induced to proliferate in response to stimulation by serum growth factors. As shown previously (14) and in Fig. 9A, this results in a rapid induction of both PRC and Cyt c mRNAs. This induction is accompanied by increased mitochondrial respiratory activity. Notably, both TFB1M and TFB2M mRNAs are induced under these conditions, suggesting that their up-regulation is an integral part of the program leading to cell growth. Interestingly, TFB1M mRNA is initially down-regulated severalfold before it is induced, a pattern that may be mediated by the negatively acting Sp1 sites that reside in the promoter. The induction of these factors is not part of a generalized transcriptional response because PGC-1α and COXIV are not induced beyond the level of the internal rRNA control. However, the mitochondrially encoded COXII mRNA is induced in parallel with the factors (Tfam and the TFBs) required for its transcription. Thus, all three NRF target genes encoding mitochondrial transcription factors are up-regulated in preparation for cell division.
FIG. 9.
Expression of hTFB genes compared to that of a collection of regulatory and structural genes involved in mitochondrial biogenesis. (A) Gene expression was monitored during serum stimulation of quiescent BALB/3T3 fibroblasts by quantitative real-time PCR. The battery of genes examined represented nuclear regulatory factors (PRC, PGC-1α, and NRF-1), mitochondrial transcription and replication factors (Tfam, TFB1M, and TFB2M), and nucleus (Cyt c and COXIV)- and mitochondrion (COXII)-encoded respiratory subunits. Relative steady-state mRNA levels were normalized to 18S rRNA as an internal control. (B) Relative mRNA expression was monitored during differentiation of fibroblasts to adipocytes for the same battery of genes as in panel A. Steady-state mRNA levels were normalized to rRNA and represent the average of at least three separate determinations with error bars denoting ± standard error.
In a second system, 3T3-L1 cells were allowed to differentiate into adipocytes, a process that is accompanied by enhanced mitochondrial biogenesis and a large induction of Cyt c (36). As shown in Fig. 9B, there is a striking increase in the expression of both nucleus (Cyt c and COXIV)- and mitochondrion (COXII)-encoded respiratory subunit mRNAs upon differentiation. This is accompanied by the coordinate induction of Tfam, TFB1M, and TFB2M. Again, the induction of these factors does not result from a generalized transcriptional response because the expression of PRC and NRF-1 remains unchanged. PGC-1α is induced upon differentiation whereas PRC is not, suggesting that PGC-1α is the coactivator that drives mitochondrial biogenesis during differentiation.
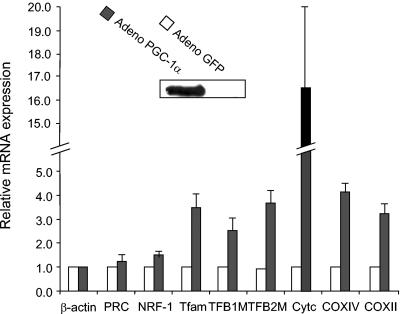
Finally, to determine whether increasing the expression of a PGC-1 family coactivator is sufficient to induce TFB expression, C2C12 cells were infected with an adenovirus that drives the overproduction of PGC-1α. Expression of PGC-1α from this virus has been shown to up-regulate the expression of respiratory subunit mRNAs and mitochondrial biogenesis in cardiac myocytes (18). Although C2C12 cells normally do not express PGC-1α, its expression is easily detected in infected cells (Fig. 10). Under these conditions, Cyt c and both nuclear and mitochondrial COX subunit mRNAs are markedly induced relative to the GFP control. The induction of respiratory subunit mRNAs is accompanied by coordinate increases in transcripts encoding Tfam and the TFBs. The expression of β-actin is not induced and serves as a normalizing control. PRC and NRF-1 mRNAs are increased only modestly. These results are consistent with the notion that PGC-1α is limiting for expression of NRF target genes and that modulating its expression is sufficient to up-regulate endogenous genes encoding components of the mitochondrial transcriptional machinery.
FIG. 10.
Expression of hTFB genes in response to ectopic expression of PGC-1α. Expression of mRNAs was measured in response to production of PGC-1α from an adenovirus vector in C2C12 myoblasts compared to a GFP-producing control. The inset panel shows the expression of PGC-1α protein by immunoblotting. Steady-state mRNA levels determined 72 h postinfection were normalized to that of β-actin and represent the average of three separate determinations with error bars denoting ± standard error.
In vivo occupancy of TFB1M and TFB2M promoters by nuclear factors.
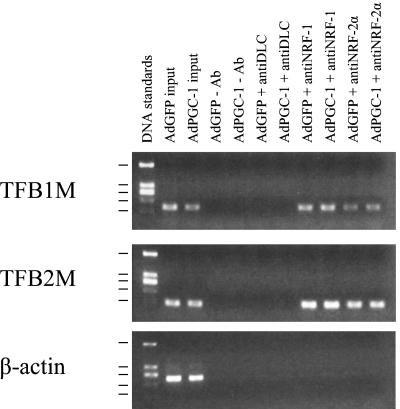
ChIPs were performed to determine whether the NRFs are actually bound to the TFB promoters in vivo. These experiments were carried out in human cells because the human promoters bind both factors in vitro. It is apparent from the results in Fig. 11 that both NRF-1 and NRF-2α are bound specifically to both hTFB1M and hTFB2M promoters in vivo as well. The immunoprecipitated promoter fragments were not detected in the absence of antibody or when an unrelated anti-dynein light chain antibody was used. In addition, no NRF-1- or NRF-2α-dependent product was obtained using primers specific for a region in the β-actin gene (Fig. 11), which lacks recognition sites for these factors, or when the analysis was performed using primers specific for the TFB 5′-flanking regions that are upstream from the NRF recognition sites (data not shown). No difference in NRF occupancy of these promoters was observed in the presence of overexpressed PGC-1α. This suggests that the mechanism for PGC-1α activation of these promoters does not involve a major increase in the recruitment of either NRF to the promoter. In preliminary experiments, we failed to detect PGC-1α at either promoter in cells overexpressing the protein from an adenovirus vector compared to a GFP-expressing control (data not shown).
FIG. 11.
NRF-1 and NRF-2α occupancy of TFB1M and TFB2M promoters in vivo detected by ChIP. HeLa cells infected with Ad-GFP- or Ad-PGC-1-expressing virus were subjected to ChIP assays as described in Materials and Methods. Input lanes show the PCR product derived from chromatin prior to immunoprecipitation. Antibodies used for immunoprecipitation are indicated above each lane. Precipitated DNA was analyzed by semiquantitative PCR with primer sets specific for the TFB1M or TFB2M promoter or the control β-actin fragment as indicated. Sizes of the DNA standards indicated at the left are, from top to bottom, 1,207, 540, 400 (doublet), 275, and 166 bp.
DISCUSSION
Major advances have been made in characterizing factors required for mitochondrial transcription. Early on, Tfam was identified as a vertebrate factor that binds a specific sequence motif within HSP and LSP and stimulates transcription in the presence of mitochondrial RNA polymerase. Tfam is related to yeast ABF2p, and both proteins are required for mtDNA maintenance in their respective organisms (8, 17). Although the existence of a vertebrate homologue of yeast sc-mtTFB was supported by studies with Xenopus laevis, its molecular cloning was elusive (3, 4). However, the recent cloning and characterization of the human and mouse TFB transcription specificity factors support the idea that mitochondrial transcription, in organisms as divergent as yeast and humans, occurs through the concerted action of a small number of proteins. Both TFBs work through a single mitochondrial RNA polymerase to stimulate transcription in the presence of Tfam. Although the TFBs bind DNA in a sequence-independent manner, neither DNA binding nor the RNA methyltransferase activities of these factors are required for transcriptional activation (21). TFB1M is thought to facilitate promoter-specific recognition by binding the carboxy-terminal transcriptional activation domain of Tfam. This represents a significant departure from the yeast system, where ABF2p does not have an activation domain and does not stimulate transcription (7). Currently there is no explanation as to why there are two TFB isoforms or why TFB1M is markedly less active than TFB2M in transcriptional stimulation.
Despite these advances, little is known about the regulatory pathways that govern mitochondrial transcriptional expression. Previous studies demonstrated that the human Tfam promoter is subject to regulation by NRFs, suggesting that these factors might be involved in integrating the expression of respiratory subunits with components of the mitochondrial transcriptional machinery (35). Here, we establish that the human nuclear genes encoding TFB1M and TFB2M are also subject to regulation by NRF-1 and NRF-2. This reinforces the idea that common transcriptional regulators link the expression of key mitochondrial transcription factors to that of respiratory chain subunits. The arrangement of the consensus NRF-1 and NRF-2 sites present in both of the human TFB promoters is similar to that found in many nuclear genes required for mitochondrial respiratory function (16). Of particular note is the conservation of spacing between tandem NRF-2 sites between the TFB and COXIV promoters. This is optimal for promoting high-affinity binding of the heterotetrameric NRF-2 complex (12). The TFB NRF sites can bind NRFs, either from crude extracts or as purified recombinant proteins, in a manner that is indistinguishable from previously authenticated sites present in the Cyt c and COXIV genes. In addition, mutagenesis experiments establish that the TFB NRF sites are major determinants of promoter function. These in vitro results are corroborated by the ChIP assays, which clearly show occupancy of both TFB promoters by NRF-1 and NRF-2α in vivo.
It is notable that the NRF-1 sites do not appear to be conserved in the rodent Tfam or TFB promoters whereas the NRF-2 and Sp1 sites are present in both humans and rodents (5, 25). This is atypical in that NRF-1 sites in respiratory subunit promoters are usually conserved between mammalian species (33). Either NRF-1 is not involved in expression of the rodent Tfam and TFB promoters, or it participates in expression through protein-protein contacts rather than high-affinity protein-DNA contacts. It is of interest in this context that NRF-1 can trans activate the rat Tfam promoter and can bind a nonconsensus GC-rich element near the transcription start site (5). The Sp1 consensus sites appear to be the only other cis-acting elements that affect promoter activity. Interestingly, the TFB1M Sp1 sites function as negative elements within the promoter context whereas the TFB2M Sp1 site exerts a positive effect on transcription. Such differences in the action of Sp1 have been documented in respiratory gene promoters. In general, Sp1 exerts a positive effect on many COX promoters but is also known to act negatively in the ANT2 (19) and ATP synthase β-subunit (38) promoters. The physiological significance of these findings remains to be explored.
A current model for mitochondrial biogenesis involves the coordination of nuclear transcription factors through interactions with regulated coactivators of the PGC-1 family (16, 26, 27). The docking of NRFs and other transcription factors with PGC-1α and its relatives may induce a conformational change in these coactivators. This allows the assembly of additional cofactors containing histone-modifying enzymatic activities that promote gene expression. These assemble by interaction with a transcriptional activation domain that is conserved among family members (23). An important characteristic of the PGC-1 family coactivators is that their expression is regulated physiologically (24). PGC-1α, the founding member of the group, is induced during adaptive thermogenesis through a cyclic AMP-dependent pathway that is activated by β-adrenergic stimulation. PGC-1α is also up-regulated by cyclic AMP during fasting where it promotes the expression of gluconeogenic enzymes. PGC-1β, a putative homologue of PGC-1α, is similarly up-regulated in fasted liver but not during thermogenesis in brown fat. Likewise, PRC is not regulated during thermogenesis but is induced by serum growth factors and down-regulated upon withdrawal of serum or contact inhibition (1). Despite these differences in their responses to regulatory signals, all three family members can bind NRF-1 and promote the trans activation of NRF-1 target genes.
The data presented here place TFB1M and TFB2M among the growing list of genes that can be trans activated by PGC-1 family coactivators. It is clear that the trans activation of both TFB promoters by PGC-1α and PRC maps to their NRF-1 and NRF-2 recognition sites. As previously observed for the Cyt c and Tfam promoters, the Sp1 sites do not appear to be direct or indirect targets for these coactivators (1, 37). Thus, as observed for respiratory chain components, it is likely that physiological control of TFB expression is exerted through NRF-1, NRF-2, and the PGC-1 family coactivators.
The mRNA expression results are consistent with the argument that many of the regulatory factors that have been implicated in mitochondrial biogenesis are coordinately regulated. Previous work established that Cyt c mRNA is markedly induced upon serum stimulation of quiescent fibroblasts and that this induction is preceded by the rapid increase in PRC mRNA. This result is confirmed here using a real-time PCR assay to measure these transcripts quantitatively. Under these conditions TFB1M and TFB2M mRNAs are also induced, suggesting that these factors are up-regulated as part of the program of cell growth. The initial reduction of TFB1M mRNA expression at 3 h of serum treatment was also verified by RNase protection (data not shown). Under certain conditions it may be desirable to reduce TFB1M expression relative to that of TFB2M because TFB2M is the more potent of the two in driving mitochondrial transcription. It is notable that the mitochondrial COXII transcript is also induced. Induction of mitochondrial transcripts is consistent with the model since their expression requires the action of Tfam and one or both TFBs.
Even more dramatic results were obtained using an adipocyte differentiation model. Previous work demonstrated that Cyt c protein levels increase approximately 20-fold upon differentiation of 3T3-L1 fibroblasts to adipocytes (36). This was accompanied by increases in a number of other mitochondrial proteins and by elevated oxygen consumption, suggesting that mitochondrial biogenesis itself is enhanced. Here, we show large increases in nucleus- (Cyt c, COXIV) and mitochondrially (COXII) encoded respiratory subunit mRNAs including a 10-fold induction of Cyt c. Both TFBs along with Tfam mRNAs are also markedly induced upon differentiation, indicating that coordinate increases in these mitochondrial transcriptional regulators are part of the program of mitochondrial biogenesis in this system. Of the nuclear regulatory factors examined, only PGC-1α is induced. This contrasts with the serum growth response, where PRC mRNA is induced and PGC-1α is not. On this basis it is tempting to speculate that PRC may function as a growth regulatory factor whereas PGC-1α is more involved in differentiation as observed here and in brown fat.
We have not yet been successful in overexpressing PRC. However, the overexpression of PGC-1α from an adenovirus vector increased the expression of respiratory subunit mRNAs along with those for Tfam and the TFBs. This demonstrates a causal relationship between increased PGC-1α expression and the induction of the TFBs as part of a program of mitochondrial biogenesis. This result is consistent with the results of others showing that elevated PGC-1α is sufficient to induce mitochondrial biogenesis (37). It remains to be determined whether modifications of the activators or coactivators play a role in controlling target gene expression. For example, the phosphorylation of NRF-1 that occurs in response to serum growth factors (11, 14) may enhance its ability to utilize a particular coactivator. Nevertheless, the results show that varying the expression of a PGC-1 family coactivator is sufficient to coordinately induce all of the known factors required for the transcription of mtDNA. This contributes to the integration of mitochondrial transcription and replication with the expression of the respiratory apparatus.
Questions remain concerning the mechanism by which the coactivators induce gene expression. One possibility is that the PGC-1 family coactivators induce the expression of NRFs. Overexpression of PGC-1α in mouse myoblasts and myotubules has been associated with a dramatic increase in NRF-1 and NRF-2α mRNA levels (37). Here, we observe a small but significant increase in NRF-1 transcript levels in response to serum stimulation but little or no increase upon adipocyte differentiation or upon PGC-1α overexpression in mouse myoblasts. The reasons for the discrepancy with previous findings are unknown. However, the latter results are consistent with the ChIP experiments, which show no major increase in NRF-1 or NRF-2α occupancy of either promoter in response to PGC-1α overexpression. An alternative but not mutually exclusive notion is that the coactivators work through direct interaction with their cognate transcription factors. Although we did not detect PGC-1α in an NRF complex at the TFB promoters by ChIP assay, there is good evidence that both PGC-1α and PRC form a functional complex with NRF-1 both in vivo and in vitro (1, 37). By contrast, we detected no direct interaction of either PGC-1α or PRC with NRF-2α or NRF-2β (unpublished observations), suggesting that if such an interaction exists it may be mediated by a third party. The negative ChIP result does not allow us to rule out the possibility that PGC-1α is present in a promoter-associated complex. PGC-1α is known to interact with many transcriptional components including transcription factors, other coactivators, and RNA splicing factors. Thus, it is possible that, in the cross-linked complex, the epitopes recognized by the available antibody are inaccessible. Future studies should resolve these important mechanistic questions.
Acknowledgments
We thank Daniel P. Kelly of Washington University School of Medicine for the PGC-1α-expressing adenovirus vector and antibodies.
This work was supported by United States Public Health Service grant GM32525-22.
REFERENCES
- 1.Andersson, U., and R. C. Scarpulla. 2001. PGC-l-related coactivator, a novel, serum-inducible coactivator of nuclear respiratory factor 1-dependent transcription in mammalian cells. Mol. Cell. Biol. 21:3738-3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews, N. C., and D. V. Faller. 1991. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 19:2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antoshechkin, I., and D. F. Bogenhagen. 1995. Distinct roles for two purified factors in transcription of Xenopus mitochondrial DNA. Mol. Cell. Biol. 15:7032-7042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogenhagen, D. F. 1996. Interaction of mtTFB and mtRNA polymerase at core promoters for transcription of Xenopus laevis mtDNA. J. Biol. Chem. 271:12036-12041. [PubMed] [Google Scholar]
- 5.Choi, Y. S., H. K. Lee, and Y. K. Pak. 2002. Characterization of the 5′-flanking region of the rat gene for mitochondrial transcription factor A (Tfam). Biochim. Biophys. Acta Gene Struct. Expr. 1574:200-204. [DOI] [PubMed] [Google Scholar]
- 6.Clayton, D. A. 2003. Mitochondrial DNA replication: what we know. IUBMB Life 55:213-217. [DOI] [PubMed] [Google Scholar]
- 7.Dairaghi, D. J., G. S. Shadel, and D. A. Clayton. 1995. Addition of a 29 residue carboxyl-terminal tail converts a simple HMG box-containing protein into a transcriptional activator. J. Mol. Biol. 249:11-28. [DOI] [PubMed] [Google Scholar]
- 8.Diffley, J. F., and B. Stillman. 1991. A close relative of the nuclear, chromosomal high-mobility group protein HMG1 in yeast mitochondria. Proc. Natl. Acad. Sci. USA 88:7864-7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falkenberg, M., M. Gaspari, A. Rantanen, A. Trifunovic, N.-G. Larsson, and C. M. Gustafsson. 2002. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat. Genet. 31:289-294. [DOI] [PubMed] [Google Scholar]
- 10.Garesse, R., and C. G. Vallejo. 2001. Animal mitochondrial biogenesis and function: a regulatory cross-talk between two genomes. Gene 263:1-16. [DOI] [PubMed] [Google Scholar]
- 11.Gugneja, S., and R. C. Scarpulla. 1997. Serine phosphorylation within a concise amino-terminal domain in nuclear respiratory factor 1 enhances DNA binding. J. Biol. Chem. 272:18732-18739. [DOI] [PubMed] [Google Scholar]
- 12.Gugneja, S., J. V. Virbasius, and R. C. Scarpulla. 1995. Four structurally distinct, non-DNA-binding subunits of human nuclear respiratory factor 2 share a conserved transcriptional activation domain. Mol. Cell. Biol. 15:102-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herzig, R. P., U. Andersson, and R. C. Scarpulla. 2000. Dynein light chain interacts with NRF-1 and EWG, structurally and functionally related transcription factors from humans and Drosophila. J. Cell Sci. 113:4263-4273. [DOI] [PubMed] [Google Scholar]
- 14.Herzig, R. P., S. Scacco, and R. C. Scarpulla. 2000. Sequential serum-dependent activation of CREB and NRF-1 leads to enhanced mitochondrial respiration through the induction of cytochrome c. J. Biol. Chem. 275:13134-13141. [DOI] [PubMed] [Google Scholar]
- 15.Higuchi, R. 1990. Recombinant PCR, p. 177-183. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols. A guide to methods and applications. Academic Press, San Diego, Calif.
- 16.Kelly, D. P., and R. C. Scarpulla. 2004. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 18:357-368. [DOI] [PubMed] [Google Scholar]
- 17.Larsson, N. G., J. M. Wang, H. Wilhelmsson, A. Oldfors, P. Rustin, M. Lewandoski, G. S. Barsh, and D. A. Clayton. 1998. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat. Genet. 18:231-236. [DOI] [PubMed] [Google Scholar]
- 18.Lehman, J. J., P. M. Barger, A. Kovacs, J. E. Saffitz, D. M. Medeiros, and D. P. Kelly. 2000. Peroxisome proliferator-activated receptor γ coactivator-1 promotes cardiac mitochondrial biogenesis. J. Clin. Investig. 106:847-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, R., Z. Hodny, K. Luciakova, P. Barath, and B. D. Nelson. 1996. Sp1 activates and inhibits transcription from separate elements in the proximal promoter of the human adenine nucleotide translocase 2 (ANT2) gene. J. Biol. Chem. 271:18925-18930. [DOI] [PubMed] [Google Scholar]
- 20.McCulloch, V., B. L. Seidel-Rogol, and G. S. Shadel. 2002. A human mitochondrial transcription factor is related to RNA adenine methyltransferases and binds S-adenosylmethionine. Mol. Cell. Biol. 22:1116-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCulloch, V., and G. S. Shadel. 2003. Human mitochondrial transcription factor B1 interacts with the C-terminal activation region of h-mtTFA and stimulates transcription independently of its RNA methyltransferase activity. Mol. Cell. Biol. 23:5816-5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poyton, R. O., and J. E. McEwen. 1996. Crosstalk between nuclear and mitochondrial genomes. Annu. Rev. Biochem. 65:563-607. [DOI] [PubMed] [Google Scholar]
- 23.Puigserver, P., C. Adelmant, Z. D. Wu, M. Fan, J. M. Xu, B. O'Malley, and B. M. Spiegelman. 1999. Activation of PPARγ coactivator-1 through transcription factor docking. Science 286:1368-1371. [DOI] [PubMed] [Google Scholar]
- 24.Puigserver, P., and B. M. Spiegelman. 2003. Peroxisome proliferator-activated receptor-gamma coactivator 1α (PGC-1α): transcriptional coactivator and metabolic regulator. Endocr. Rev. 24:78-90. [DOI] [PubMed] [Google Scholar]
- 25.Rantanen, A., M. Gaspari, M. Falkenberg, C. M. Gustafsson, and N.-G. Larsson. 2003. Characterization of the mouse genes for mitochondrial transcription factors B1 and B2. Mamm. Genome 14:1-6. [DOI] [PubMed] [Google Scholar]
- 26.Scarpulla, R. C. 2002. Nuclear activators and coactivators in mammalian mitochondrial biogenesis. Biochim. Biophys. Acta 1576:1-14. [DOI] [PubMed] [Google Scholar]
- 27.Scarpulla, R. C. 2002. Transcriptional activators and coactivators in the nuclear control of mitochondrial function in mammalian cells. Gene 286:81-89. [DOI] [PubMed] [Google Scholar]
- 28.Seidel-Rogol, B. L., V. McCulloch, and G. S. Shadel. 2003. Human mitochondrial transcription factor B1 methylates ribosomal RNA at a conserved stem-loop. Nat. Genet. 33:23-24. [DOI] [PubMed] [Google Scholar]
- 29.Shadel, G. S., and D. A. Clayton. 1997. Mitochondrial DNA maintenance in vertebrates. Annu. Rev. Biochem. 66:409-435. [DOI] [PubMed] [Google Scholar]
- 30.Takahashi, Y., J. B. Rayman, and B. D. Dynlacht. 2000. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 14:804-816. [PMC free article] [PubMed] [Google Scholar]
- 31.Tiranti, V., A. Savoia, F. Forti, M. F. D'Apolito, M. Centra, M. Racchi, and M. Zeviani. 1997. Identification of the gene encoding the human mitochondrial RNA polymerase (h-mtRPOL) by cyberscreening of the expressed sequence tags database. Hum. Mol. Genet. 6:615-625. [DOI] [PubMed] [Google Scholar]
- 32.Vega, R. B., J. M. Huss, and D. P. Kelly. 2000. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor α in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol. Cell. Biol. 20:1868-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Virbasius, C. A., J. V. Virbasius, and R. C. Scarpulla. 1993. NRF-1, an activator involved in nuclear-mitochondrial interactions, utilizes a new DNA-binding domain conserved in a family of developmental regulators. Genes Dev. 7:2431-2445. [DOI] [PubMed] [Google Scholar]
- 34.Virbasius, J. V., and R. C. Scarpulla. 1991. Transcriptional activation through ETS domain binding sites in the cytochrome c oxidase subunit IV gene. Mol. Cell. Biol. 11:5631-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Virbasius, J. V., and R. C. Scarpulla. 1994. Activation of the human mitochondrial transcription factor A gene by nuclear respiratory factors: a potential regulatory link between nuclear and mitochondrial gene expression in organelle biogenesis. Proc. Natl. Acad. Sci. USA 91:1309-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson-Fritch, L., A. Burkart, G. Bell, K. Mendelson, J. Leszyk, S. Nicoloro, M. Czech, and S. Corvera. 2003. Mitochondrial biogenesis and remodeling during adipogenesis and in response to the insulin sensitizer rosiglitazone. Mol. Cell. Biol. 23:1085-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu, Z., P. Puigserver, U. Andersson, C. Zhang, G. Adelmant, V. Mootha, A. Troy, S. Cinti, B. Lowell, R. C. Scarpulla, and B. M. Spiegelman. 1999. Mechanisms controlling mitochondrial biogenesis and function through the thermogenic coactivator PGC-1. Cell 98:115-124. [DOI] [PubMed] [Google Scholar]
- 38.Zaid, A., R. Li, K. Luciakova, P. Barath, S. Nery, and B. D. Nelson. 1999. On the role of the general transcription factor Sp1 in the activation and repression of diverse mammalian oxidative phosphorylation genes. J. Bioenerg. Biomembr. 31:129-135. [DOI] [PubMed] [Google Scholar]