Abstract
Talin1 is an adaptor protein that conjugates integrins to the cytoskeleton and regulates integrins and focal adhesion signaling. Several studies have found that Talin1 is overexpressed in several tumor types and promotes tumor progression. However, the explicit role of Talin1 in hepatocellular carcinoma (HCC) progression is still unclear and its functional mechanism remains largely unknown. In this study, we showed a trend of gradually decreasing expression of Talin1 from normal liver tissues to hepatocirrhosis, liver hyperplasia, the corresponding adjacent non‐tumor, primary HCC, and eventually metastatic foci, indicating that Talin1 may correlate with HCC initiation to progression. Talin1 was significantly downregulated in HCC tissues compared with adjacent non‐tumor tissues and low Talin1 expression was associated with HCC progression and poor prognosis. Furthermore, Talin1 knockdown induced epithelial–mesenchymal transition and promoted migration and invasion in SK‐Hep‐1 cells and HepG2 cells. Mechanistically, we found that the ERK pathway was responsible for these promoting effects of Talin1 knockdown in HCC cells. The promoting effects of Talin1 knockdown on epithelial–mesenchymal transition, migration, and invasion were reversed by U0126, a specific ERK1/2 inhibitor. Taken together, our results suggested that Talin1 might serve as a tumor suppressor in HCC and a potential prognostic biomarker for HCC patients.
Keywords: Epithelial–mesenchymal transition, ERK1/2 pathway, hepatocellular carcinoma, prognostic biomarker, Talin1
Hepatocellular carcinoma is the second‐leading cause of cancer‐related deaths globally.1, 2 Fewer than 30% of HCC patients are diagnosed at an early stage when they can be treated by hepatic resection, local ablation, or liver transplantation.3, 4 In advanced cases, the prognosis is poor due to the high rates of recurrence and metastasis.5, 6, 7 Therefore, due attention should be paid to the exploration of molecular mechanisms of HCC and identification of biomarkers of its progression and prognosis.
Talin1 is a macromolecular (270‐kDa) adaptor protein that is highly enriched at adhesion complexes formed at the junctions between cells and their ECM, thereby conjugating cell adhesion molecules (such as integrins, vinculin, and FAK) to the actin cytoskeleton.8, 9, 10 In addition to this structural role, Talin1 plays an indispensable role in activating integrins. Once activated, integrins initiate the activation of FAK, which regulates numerous processes concerning cancer development, including cell survival, proliferation, migration, invasion, and metastasis.8, 11, 12
Recently, the effect of Talin1 in cancers has drawn increasing attention. Some studies have reported that overexpression of Talin1 promotes tumor progression through FAK signaling and correlates with poor prognosis.13, 14, 15, 16, 17 For example, the aberrant enrichment of Talin1 in prostate tumors is significantly associated with higher tumor grades and it promotes tumor invasion and bone metastasis.13, 14 In OSCC, upregulated Talin1 correlates with advanced OSCC with invasion to the adjacent tissues and reduces overall survival of OCSS patients.15 Moreover, Talin1 is found to be involved in resistance to antitumor drugs.16, 18 For instance, inhibition of Talin1 attenuates the resistance of glioblastoma multiforme to bevacizumab.16 In triple‐negative breast cancer, Talin1 loss‐of‐function notably increases the chemosensitivity of triple‐negative breast cancer cells to docetaxel.18 As for HCC, a study based on a small sample size discovered a significant downregulation of Talin1 in HCC tissues compared with the adjacent non‐tumor tissues and indicated that the low Talin1 expression is largely associated with incomplete tumor capsule and portal vein tumor thrombus.19 However, another study found that MHCC‐97L cells, a well‐established HCC cell line, showed reduced invasion and migration abilities following Talin1 knockdown,20 which is contradictory to the former study indicating the tumor suppressor role of Talin1 in HCC. Hence, the explicit role of Talin1 in HCC progression is still disputed. In addition, its functional mechanism also remains largely unknown. For this reason, we carried out the following research.
In this study, the expression of Talin1 in a large number of HCC specimens and a tissue microarray of HCC progression was detected by IHC. In addition, we undertook migration assays, Matrigel invasion assays, cell counting assays, EdU assays, and Western blot and immunofluorescence analyses to further explore the functions of Talin1 in HCC and the underlying mechanism, using HCC cells with Talin1 knockdown by siRNAs.
Materials and Methods
Tissue microarray construction and samples
A tissue microarray with two normal liver tissues, seven hepatocirrhosis tissues, two liver hyperplasia tissues, three matched normal liver tissues with far‐end/non‐tumor/primary HCC tissues, 16 paired non‐tumor tissues and primary HCC tissues, seven metastatic foci (metastatic hepatic carcinoma to left index finger, the upper of right femur, distal stomach, right hemicolon, cervical lymph node, right lower lung, and the top of the brain), and one positive lymph node was purchased from Chaoxing Biotechnology (diameter, 2.0 mm, HLiv‐HCC060CD‐02; Shanxi, China). Fresh HCC tissues and adjacent non‐tumor tissues for qRT‐PCR, 36 samples) and Western blot (12 samples) analyses were collected from 36 patients who initially underwent surgery and were histologically diagnosed with HCC between January 2014 and July 2015 at the Nanfang Hospital affiliated with the Southern Medical University (Guangzhou, China). For IHC, the formalin‐fixed, paraffin‐embedded tissues including 200 HCC tissues and 196 adjacent non‐tumor tissues from 200 HCC patients who initially underwent surgery between March 2005 and September 2010 were selected randomly from the archives of the Department of Pathology of the same hospital. None of the patients was pretreated with chemotherapy or radiotherapy before undergoing hepatectomy.
Ethical approval for the study was given by the Research Ethics Committee of Nanfang Hospital. Informed consent was obtained from each patient prior to participation in the study. The 200 HCC patients enrolled between March 2005 and September 2010 were followed up for 5 years. The clinical information of the 200 HCC patients is described in Table S1. Tumor stage was defined according to the Barcelona Clinic Liver Cancer staging system.
Cell lines
The human HCC cell lines SK‐Hep‐1, HepG2, SMMC‐7721, MHCC97H, and HCCLM3, as well as the normal liver cell line HL‐7702, were obtained from the Cell Bank of the Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). Cell lines were all cultured at 37°C in a humidified atmosphere containing 5% CO2 in DMEM supplemented with 10% FBS (Gibco, Grand Island, NY, USA).
Immunohistochemical staining
Immunohistochemical staining of paraffin‐embedded, 4‐μm tumor sections or a tissue microarray of HCC progression was carried out using the primary antibody Talin1 (#14168‐1‐AP; Proteintech, Chicago, IL, USA) and a Dako Envision System (Dako, Glostrup, Denmark) following the manufacturer's recommended protocol. The IHC‐stained tissue sections were individually and independently reviewed and scored by two pathologists who were blinded to the clinical data. The score standard for the intensity of staining was as follows: 0, negative; 1, weak; 2, medium; and 3, strong. The score standard for the extent of staining was scored as: 0, 0%; 1, 1–25%; 2, 26–50%; 3, 51–75%; and 4, 76–100%. The total scores ranging from 0 to 7 include the scores of intensity and extent. We defined total scores of 2 or lower as the low‐expression group (negative group), whereas total scores of 3 or higher were defined as the high‐expression (positive group). The results from the two pathologists were highly consistent.
Small interfering RNA transfection
Talin1 was silenced by siRNAs synthesized by Qiagen (Valencia, CA, USA). The sequences are listed in Table S2. Transfection of siRNA in SK‐Hep‐1 and HepG2 cells was carried out by the Lipofectamine RNAiMAX Transfection Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's recommended protocol. Briefly, HCC cells were seeded into a 6‐well plate and then cultured under complete medium conditions. After 12 h, each well was transiently transfected with siTalin1 (10 μM) or scramble sequences with 5 μL Lipofectamine RNAiMAX. The cells were incubated for 48 h and ultimately harvested for real‐time PCR and Western blotting to detect the efficiency of interference.
RNA extraction and qRT‐PCR
Total RNA from HCC tissues, corresponding adjacent non‐tumor tissues, and SK‐Hep‐1 and HepG2 cells was isolated with TRIzol reagent (Invitrogen), according to the manufacturer's instructions. The qRT‐PCR was carried out using RT primers and a SYBR Green PCR kit purchased from Takara Biotechnology (Tokyo, Japan). The amplification of Talin1 and β‐actin was carried out using the following primers: Talin1 sense, 5′‐TGTAGAGGAGCACGAGACGC‐3′ and anti‐sense, 5′‐AAGGAGACAGGGTGGGAGC‐3′; β‐actin sense, 5′‐TCAAGATCATTGCTCCTCCTGA‐3′ and anti‐sense, 5′‐CTCGTCATACTCCTGCTTGCTG‐3′.
Western blot analysis
SK‐Hep‐1 and HepG2 cells were lysed in RIPA buffer supplemented with protease inhibitor and phosphoesterase inhibitor (Roche, Basel, Switzerland). Total protein was quantified by the Bradford method (Bio‐Rad Laboratories, Hercules, CA, USA). For Western blot analysis, equal amounts of protein (30–50 μg) were separated by 6–12% SDS‐PAGE and blotted onto PVDF membranes (Bio‐Rad Laboratories). Next, the membranes were incubated with 5% BSA for 40 min at room temperature and then with primary antibodies at 4°C overnight, followed by incubation with the matched rabbit or mouse secondary antibodies conjugated to HRP (Abcam, Cambridge, MA, USA). Proteins were detected using enhanced chemiluminescence (Thermo Fisher Scientific, Rockford, IL, USA). The primary antibodies are listed in Table S3.
Immunofluorescence and FITC‐phalloidin staining
SK‐Hep‐1 cells were stained by immunofluorescence on cover slips. Briefly, SK‐Hep‐1 cells were fixed in 4% paraformaldehyde for 15 min and permeabilized with 0.25% Triton X‐100 for 7 min, then incubated with 5% BSA for 40 min. The cells were incubated with primary antibodies at 4°C overnight and then for 1 h with rhodamine‐conjugated goat antibodies against rabbit IgG. In addition, the nuclei were counterstained with DAPI (Abcam). The cover slips were imaged by a confocal laser‐scanning microscope (FV1000; Olympus, Center Valley, PA, USA). For phalloidin‐FITC staining, cells on cover slips were fixed in 4% paraformaldehyde for 15 min, incubated with 1% BSA for 20 min, and then 5 μg/mL phalloidin‐FITC (Sigma‐Aldrich, St. Louis, MO, USA) for 1 h at room temperature.
Migration and invasion and cell proliferation assays
The Transwell migration and invasion and cell proliferation assays are described in the Data S1 section.
Statistical analyses
All statistical analyses were carried out using spss 21.0 software (IBM, Armonk, NY, USA). Analyses of the relationship between Talin1 expression and clinicopathological features were undertaken using Pearson's χ2‐test. Overall survival and disease‐free survival of HCC patients were evaluated using Kaplan–Meier and log–rank tests. The effects of variables on overall survival were assessed using univariate and multivariate Cox's proportional hazard regression model. Student's t‐test was used to determine significance of data from qRT‐PCR experiments, Transwell migration and invasion assays, and EdU assays. Multiway classification analysis of variance text was used to determine significance of data from cell counting kit assays. A two‐tailed P < 0.05 was considered to be statistically significant.
Results
Talin1 is significantly downregulated in HCC tissues
To investigate the underlying role of Talin1 in HCC, we first detected the protein expression levels of Talin1 on a tissue microarray of HCC progression by IHC. As shown in Figure 1(a), the positive expression rate of Talin1 was 100% (5/5) in normal liver, 83.3% (5/6) in hepatocirrhosis, 100% (2/2) in liver hyperplasia, 83.3% (15/18) in the corresponding adjacent non‐tumor tissues, 42.1% (8/19) in primary HCC tissues, and 14.3% (1/7) in metastatic foci. These results showed a trend of gradually decreasing expression of Talin1 from normal liver to HCC metastasis, indicating that Talin1 may correlate with HCC initiation to progression. Compared with non‐tumor tissues, Talin1 was significantly downregulated in primary HCC tissues (P < 0.05) and metastatic foci (P < 0.01) (Fig. 1b). Representative images of IHC staining are shown in Figure 1(b). Next, we detected the mRNA and protein expression of Talin1 in 36 paired tumor and non‐tumor liver samples using qRT‐PCR and Western blot analysis. The results showed that, compared with non‐tumor tissues, Talin1 was significantly downregulated in tumor tissues (Fig. 1c–e, P < 0.001).
Figure 1.
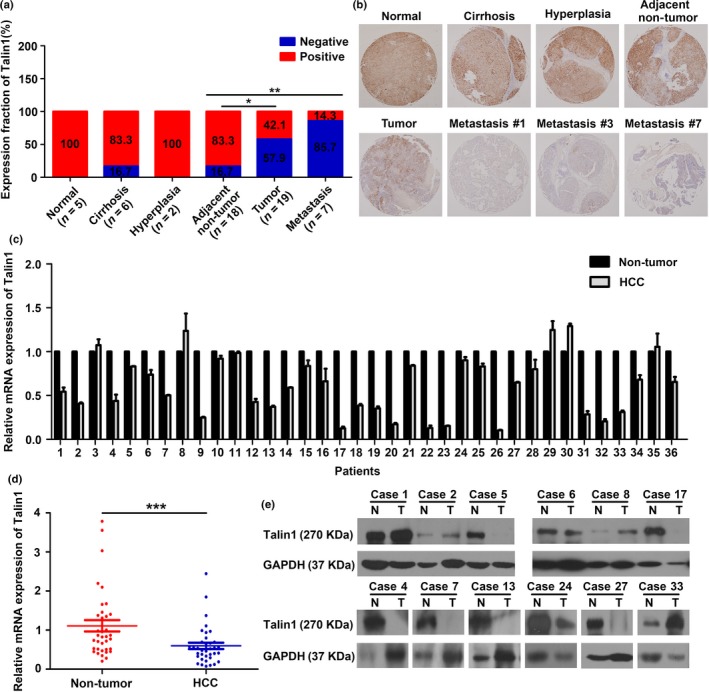
Talin1 is significantly down‐regulated in HCC tissues. (a) Talin1 expression in indicated tissues was determined by IHC assays (*P < 0.05, **P < 0.01). (b) Representative images of Talin1 expression from normal liver tissues, cirrhosis tissues, hyperplasia tissues, corresponding adjacent non‐tumor tissues, primary tumor tissues, and metastasis #1 (left index finger), metastasis #3 (distal stomach), and metastasis #7 (the top of the brain) by IHC assays. (c) qRT‐PCR analysis of relative Talin1 mRNA level in 36 HCC tissues and the corresponding non‐tumor liver tissues after normalization for the endogenous control (β‐actin). (d) Quantification of Talin1 mRNA expression in HCC tissues and non‐tumor tissues (***P < 0.001). (e) Western blot analysis of Talin1 expression in HCC tissues (T) and corresponding adjacent non‐tumor tissues (N) (n = 12).
The expression of Talin1 was also detected in 200 HCC tissues and 196 adjacent non‐tumor tissues by IHC. As shown in Figure 2(a), Talin1 was mainly expressed in the cytoplasm of liver tissues. Furthermore, 67.5% of HCC tissues showed low Talin1 expression, whereas only 7.2% of adjacent non‐tumor tissues showed low Talin1 expression (Fig. 2b, P < 0.001). The images of HE staining matched with the images of IHC staining in Figures 1(a) and 2(a) are shown in Figure S1. Collectively, these findings strongly suggest that Talin1 is significantly downregulated in HCC.
Figure 2.
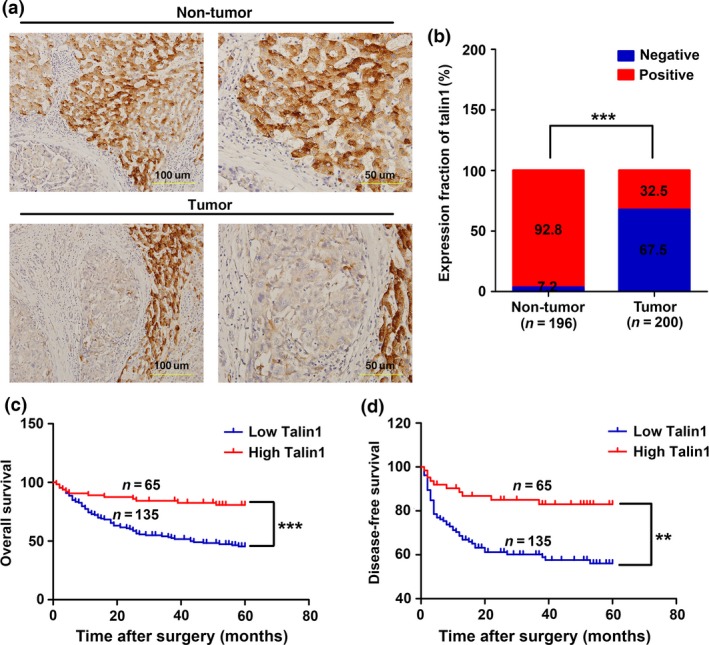
Low Talin1 expression correlates with poor prognosis of hepatocellular carcinoma (HCC) patients. (a) Representative images of Talin1 expression from non‐tumor tissues and HCC tissues by immunohistochemical assays. Scale bar = 50 μm (right panels), 100 μm (left panels). (b) Talin1 expression in non‐tumor tissues (n = 196) and HCC tissues (n = 200) was determined by immunohistochemistry. ***P < 0.001. (c,d) Kaplan–Meier and log–rank test analyses of overall survival (c) and disease‐free survival (d) in 200 HCC patients based on Talin1 expression level of HCC tissues. **P < 0.01; ***P < 0.001.
Low Talin1 expression significantly correlates with HCC progression and poor prognosis
To determine whether the expression levels of Talin1 are related to HCC progression, we analyzed the association between Talin1 expression and clinicopathological characteristics in 200 HCC patients. As shown in Table 1, Talin1 downregulation significantly correlated with younger age (P = 0.032), larger tumor size (P = 0.006), higher α‐fetoprotein levels (P = 0.003), and tumor recurrence (P < 0.001). Furthermore, Kaplan–Meier and log–rank test analyses revealed that patients with low Talin1 expression had worse overall survival (P < 0.001) and disease‐free survival rates (P = 0.001) than patients with high Talin1 expression (Fig. 2c,d). In addition, the multivariate analysis showed that Talin1 expression (P = 0.010), portal vein tumor thrombus (P < 0.001), and gender (P = 0.013) were independent prognostic factors for overall survival in HCC patients (Table 2).
Table 1.
Clinicopathologic correlations of Talin1 expression in 200 patients with hepatocellular carcinoma
| Characteristic | Talin1 expression | P‐value | |
|---|---|---|---|
| Low (n = 135) | High (n = 65) | ||
| Age, years | |||
| ≤55 | 82 | 29 | 0.032a |
| >55 | 53 | 36 | |
| Gender | |||
| Male | 111 | 57 | 0.323 |
| Female | 24 | 8 | |
| Liver cirrhosis | |||
| No | 36 | 13 | 0.305 |
| Yes | 99 | 52 | |
| AFP, ng/mL | |||
| ≤20 | 38 | 32 | 0.003a |
| >20 | 97 | 33 | |
| Tumor size, cm | |||
| ≤5 | 47 | 36 | 0.006a |
| >5 | 88 | 29 | |
| Tumor number | |||
| Single | 101 | 47 | 0.705 |
| Multiple | 34 | 18 | |
| Portal vein thrombosis | |||
| No | 108 | 58 | 0.104 |
| Yes | 27 | 7 | |
| Distant metastasis | |||
| No | 122 | 60 | 0.654 |
| Yes | 13 | 5 | |
| Pathological grade | |||
| I–II | 102 | 55 | 0.144 |
| III–IV | 33 | 10 | |
| BCLC stage | |||
| 0+A | 82 | 38 | 0.758 |
| B+C | 53 | 27 | |
| Recurrence | |||
| No | 83 | 56 | 0.000a |
| Yes | 52 | 9 | |
P < 0.05 considered statistically significant. AFP, α‐fetoprotein; BCLC, Barcelona Clinic Liver Cancer.
Table 2.
Univariate and multivariate analyses of overall survival in 200 patients with hepatocellular carcinoma by Cox regression analysis
| Variable | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| Hazard ratio | 95% CI | P‐value | Hazard ratio | 95% CI | P‐value | |
| Age, years: >50 vs ≤50 | 0.946 | 0.617–1.449 | 0.797 | |||
| Cirrhosis: yes versus no | 0.963 | 0.589–1.576 | 0.882 | |||
| Tumor number: multiple versus single | 1.254 | 0.787–1.998 | 0.340 | |||
| BCLC stage: B+C versus 0+A | 1.120 | 0.729–1.719 | 0.605 | |||
| Pathological grade: III–IV versus I–II | 1.274 | 0.779–2.083 | 0.335 | |||
| Metastasis: yes versus no | 1.620 | 0.837–3.134 | 0.152 | |||
| Gender: male versus female | 0.589 | 0.353–0.981 | 0.042a | 0.512 | 0.302–0.871 | 0.013a |
| Tumor size, cm: >5 vs ≤5 | 1.938 | 1.228–3.060 | 0.005a | 1.426 | 0.880–2.311 | 0.150 |
| AFP, ng/mL: >25 vs ≤25 | 2.025 | 1.237–3.314 | 0.005a | 1.592 | 0.958–2.648 | 0.073 |
| Portal vein tumor thrombus: yes versus no | 3.596 | 2.255–5.734 | 0.000a | 2.899 | 1.772–4.743 | 0.000a |
| Relapse: yes versus no | 1.913 | 1.247–2.933 | 0.003a | 1.513 | 0.962–2.380 | 0.073 |
| Talin1 expression: high versus low | 0.301 | 0.167–0.544 | 0.000a | 0.444 | 0.238–0.826 | 0.010a |
P < 0.05 considered statistically significant. AFP, α‐fetoprotein; BCLC, Barcelona Clinic Liver Cancer; CI, confidence interval.
Knockdown of Talin1 induces EMT and promotes migration and invasion in HCC cells
To explore the exact biological function of Talin1 in HCC, we examined the expression of Talin1 in five HCC cell lines and a normal liver cell line by Western blot analysis. As shown in Figure 3(a), higher Talin1 levels were found in the normal liver cell line (HL‐7702) and HCC cell lines with low malignancy (SK‐Hep‐1 and HepG2) rather than in the more aggressive HCC cell lines (SMCC‐7721, MHCC97H, and HCCLM3). Hence, SK‐Hep‐1 and HepG2 cells were selected for Talin1 knockdown by using siRNAs (Figs. 3b,S2). Interestingly, a dramatic morphological change emerged in Talin1‐knockdown cells, with polygonal or oval cells becoming spindle‐like fibroblastic cells, which is regarded as a main characteristic of EMT (Fig. 3c). At the molecular level, the typical EMT phenotype, including a decrease of the epithelial marker E‐cadherin and an increase of mesenchymal markers N‐cadherin and vimentin, was also observed in Talin1‐knockdown cells (Fig. 3d). In addition, we found a significantly positive relationship between Talin1 and E‐cadherin proteins by Western blot analysis in 12 paired tumor and non‐tumor liver samples (Figs 1e,S3,3e, r = 0.622, P = 0.031). Moreover, increased β‐catenin expression and its nuclear accumulation that contribute to induction of the EMT in tumor cells21, 22, 23 were observed in Talin1‐knockdown cells (Fig. 3d,f). Taken together, these findings indicated that Talin1 could modulate the phenotypic shift of EMT/mesenchymal–epithelial transition in HCC cells.
Figure 3.
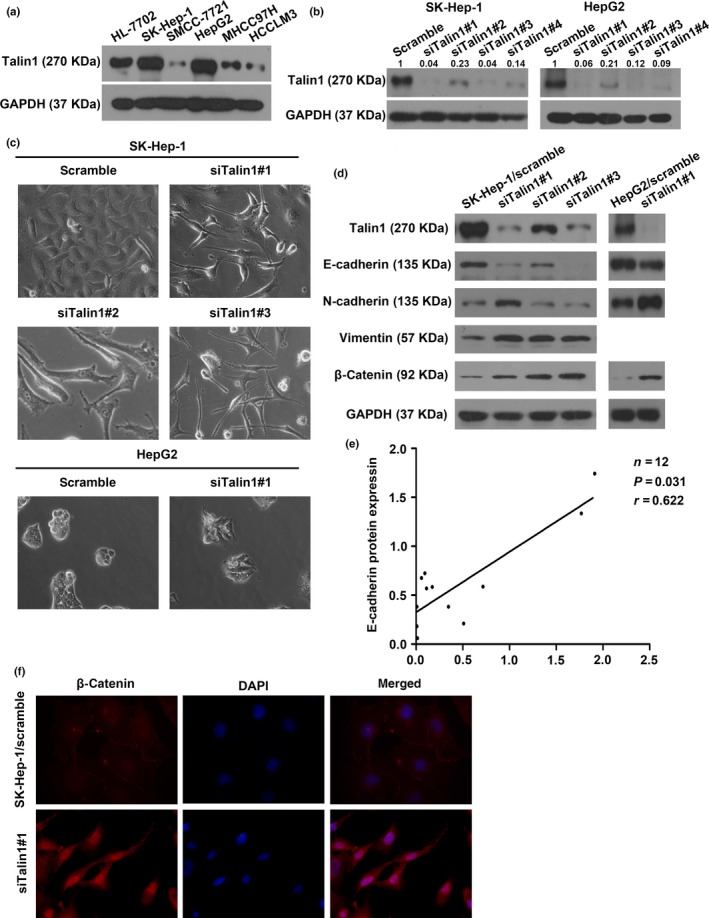
Knockdown of Talin1 induces epithelial–mesenchymal transition in hepatocellular carcinoma (HCC) cells. (a) Talin1 expression was assessed in indicated HCC cell lines by Western blot analysis. (b) Western blot analysis showed knockdown of Talin1 protein in SK‐Hep‐1 and HepG2 cells transfected with Talin1‐specific siRNA#1, #2, #3, or #4. Talin1 was quantified as integrated density to represent the amount of protein. Numerical values represented the quantified data as normalized to the scramble group. (c) SK‐Hep‐1 and HepG2 cells transfected with scrambled or Talin1‐specific siRNA are shown by phase contrast microscopy. Original magnification, ×400. (d) Western blot analysis for the expression of Talin1, E‐cadherin, N‐cadherin, vimentin, and β‐catenin in Talin1‐silenced SK‐Hep‐1 and HepG2 cells. (e) Correlation between Talin1 and E‐cadherin protein expression was detected in 12 pairs of HCC tissues and corresponding adjacent non‐tumor tissues by Western blot analysis. Spearman's rank correlation, r = 0.622; P = 0.031. (f) Immunofluorescence staining showed that knockdown of Talin1 altered the subcellular redistribution of β‐catenin. Blue, DAPI; red, β‐catenin antibody.
Epithelial–mesenchymal transition is thought to be a key process contributing to migration and invasion of tumor cells.24, 25, 26 Therefore, as expected, migration and Matrigel invasion assays showed that Talin1‐knockdown cells exhibited a significant increase of migration and invasion capacities compared with the corresponding control cells (Fig. 4a,b). In addition, given that Talin1 can act as an adaptor protein binding to the actin cytoskeleton, we explored whether silencing Talin1 in HCC tumors was beneficial for actin cytoskeletal reorganization. Staining with FITC‐phalloidin showed that silencing of Talin1 in SK‐Hep‐1 cells promoted the growth of lamellipodia (Fig. 4c), a membrane protrusion at the leading edge of the cell that is powered by actin polymerization and is critical for driving cell migration.27, 28 Collectively, our data suggested that Talin1 plays a negative role in HCC cell migration and invasion.
Figure 4.
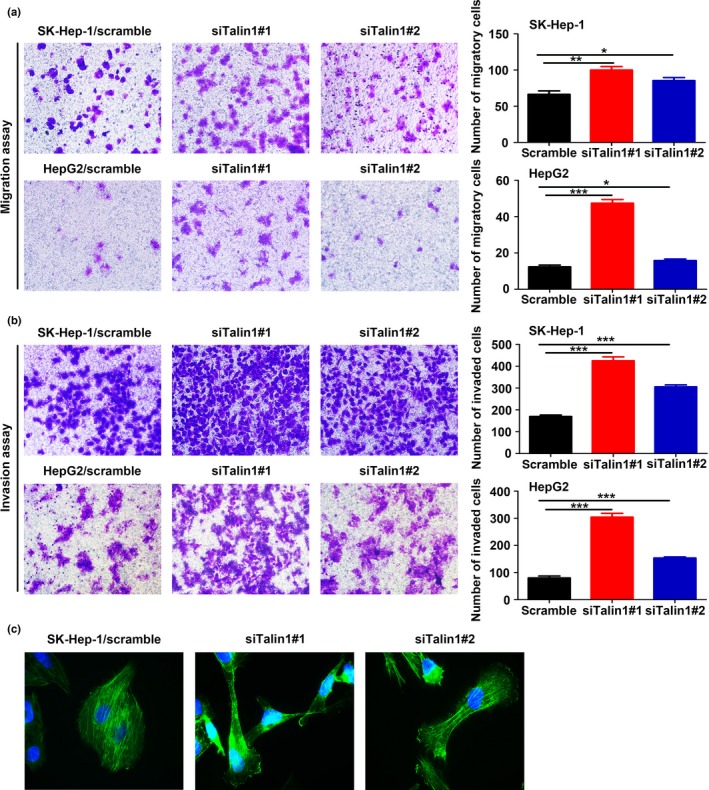
Knockdown of Talin1 promotes hepatocellular carcinoma cell migration and invasion. (a,b) Representative images of Transwell migration assays and matrix‐coated Transwell invasion assays in SK‐Hep‐1 and HepG2 cells from indicated groups (left panels). Bar charts represent the average number of migratory cells and invaded cells from five random microscopic fields. *P < 0.05; **P < 0.01; ***P < 0.001. (c) FITC‐phalloidin staining determined the effect of Talin1 knockdown on the growth of lamellipodia in SK‐Hep‐1 cells.
In addition, Talin1‐knockdown cells showed a significant decrease of proliferation ability compared with scramble cells (Fig. S4a,b). The expression of cell cycle‐related protein cyclinD1 was clearly downregulated in siTalin1 cells (Fig. S4c). However, the expression of apoptosis‐related proteins, including cleaved caspase3, cleaved poly ADP‐ribose polymerase, Bcl‐2, and Bax, showed no distinct changes in siTalin1 cells, indicating that Talin1 had no effect on HCC cell apoptosis (Fig. S4d).
Extracellular signal‐regulated kinase1/2 pathway is responsible for the Talin1‐mediated EMT process in HCC cells
It is well known that Talin1 plays an indispensable role in activating integrins. Once activated, integrins initiate the activation of FAK.8 It has also been recently reported that the activation of FAK facilitates EMT of HCC cells.29, 30, 31 In this study, as expected, the phosphorylation of FAK was notably inactivated in Talin1‐knockdown cells, indicating that Talin1‐mediated EMT is FAK‐independent (Fig. 5a). Hence, to clarify the molecular basis of the promoting effects of Talin1 knockdown on HCC cell EMT, we assessed the activation of several signaling pathways, including the transforming growth factor‐β, Smad2/3, phosphatidylinositol 3‐kinase, AKT, Rho family, and ERK1/2 pathways, which have been known to be involved in the induction of EMT.26, 32, 33 Among the screened pathways, phosphorylated ERK1/2 was significantly elevated in Talin1‐knockdown cells, whereas phosphorylated Smad2/3, phosphorylated AKT, RhoA, and Rac1 showed no such changes (Fig. 5b,c). Furthermore, Western blot assays showed that inhibition of ERK1/2 activation by U0126 in Talin1‐knockdown cells restored the expression of epithelial marker E‐cadherin and abolished the expression of mesenchymal markers N‐cadherin and vimentin, as well as β‐catenin (Fig. 5d,e).
Figure 5.
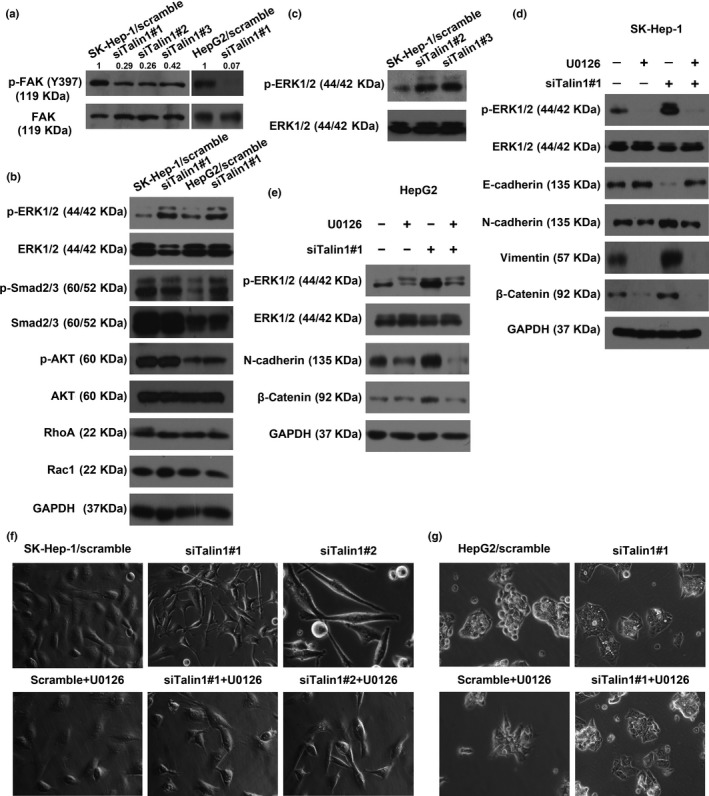
The ERK1/2 pathway is involved in Talin1 knockdown‐induced epithelial–mesenchymal transition effects in hepatocellular carcinoma (HCC) cells. (a) Expression of phosphorylated focal adhesion kinase (p‐FAK) (Y397) and total FAK in Talin1‐silenced SK‐Hep‐1 and HepG2 cells was determined using Western blot analysis. p‐FAK (Y397) was quantified as integrated density to represent the amount of protein. Numerical values represent the quantified data as normalized to the scramble group. (b) Western blot analysis for the expression of p‐ERK1/2, total ERK1/2, p‐Smad2/3, total Smad2/3, phosphorylated protein kinase B (p‐AKT), total AKT, RhoA, and Rac1 in Talin1‐silenced HCC cells. (c). Western blot analysis for the expression of p‐ERK1/2 and total ERK1/2 in SK‐Hep‐1 cells transfected with scrambled or Talin1‐specific siRNA#2 or #3. (d,e) Western blot analysis detected the expression of p‐ERK1/2, total ERK1/2, E‐cadherin, N‐cadherin, vimentin, and β‐catenin in SK‐Hep‐1 and HepG2 HCC cells transfected with scrambled or Talin1‐specific siRNA#1 and then treated with or without U0126 (10 μM) for 48 h. (f,g) Phase contrast microscopy detected the morphology of SK‐Hep‐1 cells and HepG2 cells transfected with scrambled or Talin1‐specific siRNA and then treated with or without U0126 (10 μM) for 48 h.
The typical morphological change of EMT induced by silencing Talin1 in HCC cells was clearly reversed by U0126 (a specific inhibitor of ERK1/2) (Fig. 5f,g). In addition, the enhanced migration and invasion capacities caused by silencing Talin1 in SK‐Hep‐1 cells (Fig. 6a,b) and HepG2 cells (Fig. 6c,d) were significantly suppressed by U0126. Collectively, these results suggested that inhibition of Talin1 could induce EMT and promote migration and invasion in HCC cells partly through the ERK1/2 pathway.
Figure 6.
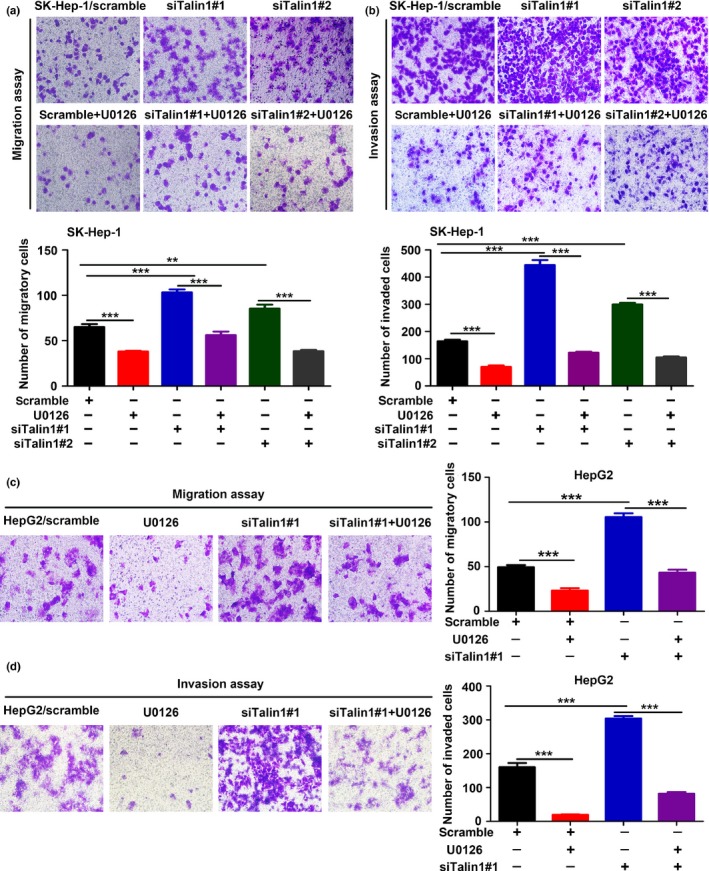
The ERK1/2 pathway is involved in Talin1 knockdown‐induced migration and invasion effects in hepatocellular carcinoma cells. (a,b) Transwell migration and matrix‐coated Transwell invasion assays were carried out to evaluate the migratory and invasive abilities of SK‐Hep‐1 cells transfected with scrambled or Talin1‐specific siRNA#1 or #2 and then treated with or without U0126 (10 μM) for 48 h. Top panels, representative images of Transwell migration (a) and matrix‐coated Transwell invasion (b) assays from indicated groups. Bottom panels, bar charts representing the average number of migratory (a) and invaded (b) cells from five random microscopic fields. **P < 0.01; ***P < 0.001. (c,d) Transwell migration and matrix‐coated Transwell invasion assays were carried out to evaluate the migratory and invasive abilities of HepG2 cells transfected with scrambled or Talin1‐specific siRNA#1 and then treated with or without U0126 (10 μM) for 24 h. Left panels, representative images of Transwell migration (c) and matrix‐coated Transwell invasion (d) assays from indicated groups. Right panels, bar charts representing the average number of migratory (c) and invaded (d) cells from five random microscopic fields. ***P < 0.001.
Discussion
In this study, we showed a trend of gradually decreasing expression of Talin1 from normal liver tissues to hepatocirrhosis, liver hyperplasia, the corresponding adjacent non‐tumor, primary HCC, and eventually metastatic foci. Furthermore, low Talin1 expression is significantly associated with tumor recurrence and poor prognosis in HCC patients. More importantly, we provided novel data to show that Talin1 inhibition can act as a positive regulator of EMT, migration, and invasion in HCC cells through activation of the ERK1/2 pathway.
Kanamori et al.34 reported that Talin1 is significantly upregulated in HCC nodules and its overexpression correlates with portal vein invasion and tumor recurrence. In our study, Talin1 was significantly downregulated in HCC tissues and its downregulation was correlated with larger tumor size, higher α‐fetoprotein levels, and tumor recurrence. This may be explained by environmental and dietary differences. Moreover, we found that HCC patients with low Talin1 expression had worse survival than those with high Talin1 expression, and Talin1 expression was an independent prognostic factor for OS in HCC patients. Therefore, our study revealed a tumor suppressor role of Talin1 in HCC. Furthermore, although Talin1 is also overexpressed in several tumors and there is strong evidence linking it to oncogenic progress,13, 14, 15, 16, 17 our findings indicated that Talin1 might play distinct roles in tumor progression in a tumor type‐dependent manner.
Fang et al.20 have recently described that, after transfection with Talin1 shRNA, MHCC‐97L cells, with originally low invasion capability, show even more decreased migration and invasion abilities. The team also previously showed that the expression of Talin1 in MHCC‐97L cells is the highest among other HCC cell lines and higher levels of Talin1 expression are associated with reduced migration and invasion capabilities in these cell lines.35 The findings of these two studies are contradictory and untenable. In our study, the finding, drawing from analysis of a considerably large number of clinical data, that low Talin1 expression significantly correlated with HCC recurrence, and the results that Talin1 inhibition induced EMT and the growth of lamellipodia in HCC cells, encouraged us to infer that decreased Talin1 expression might induce migration and invasion of HCC cells. As expected, we clearly showed that HCC cells with Talin1 knockdown exhibited strong migratory and highly invasive potential.
Furthermore, although Talin1 expression triggers the EMT and increases migration and invasion in several malignancies,13, 14, 15, 16, 17 our findings also suggested that Talin1 could regulate these processes either positively or negatively in a tumor cell type‐dependent manner. However, further studies should design shRNA aimed at Talin1 for exploring the role of Talin1 in HCC metastasis. Published work has shown that the capacity of tumor cells to migrate and invade is a key property of the metastatic phenotype; our finding that Talin1 was remarkably downregulated in HCC metastatic foci indicated that Talin1 might have a powerful antimetastatic impact on HCC.
Focal adhesion kinase has been shown by accumulating evidence to be crucial for tumor EMT, migration, and invasion.12, 36 Interestingly, while the HCC cells with Talin1 inhibition showed evident inactivation of FAK, Talin1 knockdown‐induced EMT, migration, and invasion remained unaffected, indicating that there are alternative molecular mechanisms engaged in these functions of Talin1. By screening several pathways and applying pathway inhibitor, we found that the ERK1/2 pathway is indispensable for Talin1 knockdown‐mediated EMT, migration, and invasion in HCC cells. It also provides a novel perspective to explore the functional mechanisms of Talin1 in other malignancies. Nevertheless, uncovering the mechanisms by which Talin1 regulates the activation of ERK1/2 in HCC cells requires further investigation. One previous study has shown that Calpain2 physically forms a complex with p42ERK/MAPK,37 which can mediate activation of the protease Calpain2.38, 39 Previous studies have also reported that Talin1 contains Calpain2‐cleavage sites40, 41 and Calpain2 activity is required for Talin1 proteolysis.42, 43 Therefore, it is possible that Calpain2, ERK1/2, and Talin1 could physically form a complex in HCC cells. Thus, Talin1 could reduce its phosphorylation by interacting with ERK1/2, which in turn could lead to a decrease of Talin1 proteolysis caused by Calpain2. In addition, the ERK1/2 pathway could be activated through protein kinase C,44 which could directly bind to Talin1.8 Therefore, it is also possible that Talin1 indirectly regulates the activity of the ERK1/2 pathway through suppressing activation of protein kinase C in HCC cells.
In addition, we found that Talin1 inhibition significantly reduced proliferation of HCC cells, consistent with another study in which Talin1‐knockdown MHCC‐97L cells were significantly more blocked in G0/G1 phase.20 Some genes also have analogous characteristics, possessing diverse and complex biological functions in regulating tumor progression through distinct pathways.45, 46, 47 For example, SPARC suppresses glioblastoma cell growth by interfering with growth factor–growth factor receptor interactions, while promoting cell migration and invasion by activating integrin‐regulated kinases including integrin‐linked kinase and FAK, and signaling.45 Previous studies have shown that Talin1 expression promotes proliferation of glioblastoma multiforme cells16 and ovarian serous carcinoma cells17 through FAK signaling. Our study found that Talin1 knockdown distinctly reduced activation of FAK in HCC cells. Thus, it is possible that FAK signaling plays an important role in Talin1‐mediated proliferation in HCC cells.
In summary, our study illustrated that Talin1 acts as a tumor suppressor to inhibit HCC cell EMT, migration, and invasion by decreasing activation of the ERK1/2 pathway and can be considered a potential prognostic biomarker for HCC patients.
Disclosure Statement
The authors have no conflict of interest.
Abbreviations
- AKT
protein kinase B
- EMT
epithelial–mesenchymal transition
- FAK
focal adhesion kinase
- HCC
hepatocellular carcinoma
- IHC
immunohistochemistry
- OSCC
oral squamous cell carcinoma
- qRT‐PCR
quantitative RT‐PCR
Supporting information
Data S1. Supplementary methods.
Fig. S1. Images of HE staining matched with the images of immunohistochemical staining in Figures 1(a) and 2(a).
Fig. S2. Quantitative RT‐PCR analysis showing knockdown of Talin1 mRNA in SK‐Hep‐1 and HepG2 cells transfected with Talin1‐specific siRNA#1, #2, #3, or #4. ***P < 0.001.
Fig. S3. Western blot analysis for the expression of E‐cadherin in 12 pairs of hepatocellular carcinoma tissues (T) and corresponding adjacent non‐tumor liver tissues (N), also used in Figure 1(b).
Fig. S4. Knockdown of Talin1 suppresses hepatocellular carcinoma cell proliferation.
Table S1. Clinicopathologic characteristics of 200 patients with hepatocellular carcinoma.
Table S2. Small interfering RNA sequences used in this study.
Table S3. Antibodies used in this study.
Acknowledgments
This work was supported by the National Nature Science Foundation of China (Grant Nos. 81401180, 81372283, and 91540111).
Cancer Sci 108 (2017) 1157–1168
Funding Information
National Nature Science Foundation of China.
References
- 1. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin 2011; 61: 69–90. [DOI] [PubMed] [Google Scholar]
- 2. Kim HY, Park J‐W. Clinical trials of combined molecular targeted therapy and locoregional therapy in hepatocellular carcinoma: past, present, and future. Liver Cancer 2014; 3: 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chu KK, Cheung TT. Update in management of hepatocellular carcinoma in Eastern population. World J Hepatol 2015; 7: 1562–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kabbach G, Assi HA, Bolotin G, Schuster M, Lee HJ, Tadros M. Hepatobiliary tumors: update on diagnosis and management. J Clin Transl Hepatol 2015; 3: 169–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shao YY, Shau WY, Chan SY, Lu LC, Hsu CH, Cheng AL. Treatment efficacy differences of sorafenib for advanced hepatocellular carcinoma: a meta‐analysis of randomized clinical trials. Oncology 2015; 88: 345–52. [DOI] [PubMed] [Google Scholar]
- 6. Kitano K, Murayama T, Sakamoto M et al Outcome and survival analysis of pulmonary metastasectomy for hepatocellular carcinoma. Eur J Cardiothorac Surg 2012; 41: 376–82. [DOI] [PubMed] [Google Scholar]
- 7. Li T, Fan J, Qin LX et al Risk factors, prognosis, and management of early and late intrahepatic recurrence after resection of primary clear cell carcinoma of the liver. Ann Surg Oncol 2011; 18: 1955–63. [DOI] [PubMed] [Google Scholar]
- 8. Das M, Subbayya Ithychanda S, Qin J, Plow EF. Mechanisms of talin‐dependent integrin signaling and crosstalk. Biochim Biophys Acta 2014; 1838: 579–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Calderwood DA, Ginsberg MH. Talin forges the links between integrins and actin. Nat Cell Biol 2003; 5: 694–7. [DOI] [PubMed] [Google Scholar]
- 10. Hu X, Jing C, Xu X et al Cooperative vinculin binding to talin mapped by time‐resolved super resolution microscopy. Nano Lett 2016; 16: 4062–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nader GP, Ezratty EJ, Gundersen GG. FAK, talin and PIPKIgamma regulate endocytosed integrin activation to polarize focal adhesion assembly. Nat Cell Biol 2016; 18: 491–503. [DOI] [PubMed] [Google Scholar]
- 12. Tai YL, Chen LC, Shen TL. Emerging roles of focal adhesion kinase in cancer. Biomed Res Int 2015; 2015: 690690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sakamoto S, McCann RO, Dhir R, Kyprianou N. Talin1 promotes tumor invasion and metastasis via focal adhesion signaling and anoikis resistance. Cancer Res 2010; 70: 1885–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jin JK, Tien PC, Cheng CJ et al Talin1 phosphorylation activates beta1 integrins: a novel mechanism to promote prostate cancer bone metastasis. Oncogene 2015; 34: 1811–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lai MT, Hua CH, Tsai MH et al Talin‐1 overexpression defines high risk for aggressive oral squamous cell carcinoma and promotes cancer metastasis. J Pathol 2011; 224: 367–76. [DOI] [PubMed] [Google Scholar]
- 16. Kang W, Kim SH, Cho HJ et al Talin1 targeting potentiates anti‐angiogenic therapy by attenuating invasion and stem‐like features of glioblastoma multiforme. Oncotarget 2015; 6: 27239–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tang H, Yao L, Tao X et al miR‐9 functions as a tumor suppressor in ovarian serous carcinoma by targeting TLN1. Int J Mol Med 2013; 32: 381–8. [DOI] [PubMed] [Google Scholar]
- 18. Singel SM, Cornelius C, Batten K et al A targeted RNAi screen of the breast cancer genome identifies KIF14 and TLN1 as genes that modulate docetaxel chemosensitivity in triple‐negative breast cancer. Clin Cancer Res 2013; 19: 2061–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang JL, Qian YB, Zhu LX, Xiong QR. Talin1, a valuable marker for diagnosis and prognostic assessment of human hepatocelluar carcinomas. Asian Pac J Cancer Prev 2011; 12: 3265–9. [PubMed] [Google Scholar]
- 20. Fang KP, Dai W, Ren YH, Xu YC, Zhang SM, Qian YB. Both Talin‐1 and Talin‐2 correlate with malignancy potential of the human hepatocellular carcinoma MHCC‐97 L cell. BMC Cancer 2016; 16: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mimeault M, Batra SK. Altered gene products involved in the malignant reprogramming of cancer stem/progenitor cells and multitargeted therapies. Mol Aspects Med 2014; 39: 3–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nejak‐Bowen KN, Monga SP. Beta‐catenin signaling, liver regeneration and hepatocellular cancer: sorting the good from the bad. Semin Cancer Biol 2011; 21: 44–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu L, Dong Z, Liang J et al As an independent prognostic factor, FAT10 promotes hepatitis B virus‐related hepatocellular carcinoma progression via Akt/GSK3beta pathway. Oncogene 2014; 33: 909–20. [DOI] [PubMed] [Google Scholar]
- 24. Qin Q, Xu Y, He T, Qin C, Xu J. Normal and disease‐related biological functions of Twist1 and underlying molecular mechanisms. Cell Res 2012; 22: 90–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Frame MC, Inman GJ. NCAM is at the heart of reciprocal regulation of E‐cadherin‐ and integrin‐mediated adhesions via signaling modulation. Dev Cell 2008; 15: 494–6. [DOI] [PubMed] [Google Scholar]
- 26. Giannelli G, Koudelkova P, Dituri F, Mikulits W. Role of epithelial to mesenchymal transition in hepatocellular carcinoma. J Hepatol 2016; 65: 798–808. [DOI] [PubMed] [Google Scholar]
- 27. Skau CT, Waterman CM. Specification of architecture and function of actin structures by actin nucleation factors. Annu Rev Biophys 2015; 44: 285–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Krause M, Gautreau A. Steering cell migration: lamellipodium dynamics and the regulation of directional persistence. Nat Rev Mol Cell Biol 2014; 15: 577–90. [DOI] [PubMed] [Google Scholar]
- 29. Tao YM, Huang JL, Zeng S et al BTB/POZ domain‐containing protein 7: epithelial‐mesenchymal transition promoter and prognostic biomarker of hepatocellular carcinoma. Hepatology 2013; 57: 2326–37. [DOI] [PubMed] [Google Scholar]
- 30. Zhang PF, Li KS, Shen YH et al Galectin‐1 induces hepatocellular carcinoma EMT and sorafenib resistance by activating FAK/PI3K/AKT signaling. Cell Death Dis 2016; 7: e2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen JS, Li HS, Huang JQ et al MicroRNA‐379‐5p inhibits tumor invasion and metastasis by targeting FAK/AKT signaling in hepatocellular carcinoma. Cancer Lett 2016; 375: 73–83. [DOI] [PubMed] [Google Scholar]
- 32. Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial‐mesenchymal transition. Nat Rev Mol Cell Biol 2014; 15: 178–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xia H, Chen J, Shi M et al EDIL3 is a novel regulator of epithelial‐mesenchymal transition controlling early recurrence of hepatocellular carcinoma. J Hepatol 2015; 63: 863–73. [DOI] [PubMed] [Google Scholar]
- 34. Kanamori H, Kawakami T, Effendi K et al Identification by differential tissue proteome analysis of talin‐1 as a novel molecular marker of progression of hepatocellular carcinoma. Oncology 2011; 80: 406–15. [DOI] [PubMed] [Google Scholar]
- 35. Fang KP, Zhang JL, Ren YH, Qian YB. Talin‐1 correlates with reduced invasion and migration in human hepatocellular carcinoma cells. Asian Pac J Cancer Prev 2014; 15: 2655–61. [DOI] [PubMed] [Google Scholar]
- 36. Avizienyte E, Frame MC. Src and FAK signalling controls adhesion fate and the epithelial‐to‐mesenchymal transition. Curr Opin Cell Biol 2005; 17: 542–7. [DOI] [PubMed] [Google Scholar]
- 37. Carragher NO, Westhoff MA, Fincham VJ, Schaller MD, Frame MC. A novel role for FAK as a protease‐targeting adaptor protein: regulation by p42 ERK and Src. Curr Biol 2003; 13: 1442–50. [DOI] [PubMed] [Google Scholar]
- 38. Glading A, Chang P, Lauffenburger DA, Wells A. Epidermal growth factor receptor activation of calpain is required for fibroblast motility and occurs via an ERK/MAP kinase signaling pathway. J Biol Chem 2000; 275: 2390–8. [DOI] [PubMed] [Google Scholar]
- 39. Glading A, Lauffenburger DA, Wells A. Cutting to the chase: calpain proteases in cell motility. Trends Cell Biol 2002; 12: 46–54. [DOI] [PubMed] [Google Scholar]
- 40. Bate N, Gingras AR, Bachir A et al Talin contains a C‐terminal calpain2 cleavage site important in focal adhesion dynamics. PLoS ONE 2012; 7: e34461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Franco SJ, Rodgers MA, Perrin BJ et al Calpain‐mediated proteolysis of talin regulates adhesion dynamics. Nat Cell Biol 2004; 6: 977–83. [DOI] [PubMed] [Google Scholar]
- 42. Dourdin N, Bhatt AK, Dutt P et al Reduced cell migration and disruption of the actin cytoskeleton in calpain‐deficient embryonic fibroblasts. J Biol Chem 2001; 276: 48382–8. [DOI] [PubMed] [Google Scholar]
- 43. Franco S, Perrin B, Huttenlocher A. Isoform specific function of calpain 2 in regulating membrane protrusion. Exp Cell Res 2004; 299: 179–87. [DOI] [PubMed] [Google Scholar]
- 44. Naor Z. Signaling by G‐protein‐coupled receptor (GPCR): studies on the GnRH receptor. Front Neuroendocrinol 2009; 30: 10–29. [DOI] [PubMed] [Google Scholar]
- 45. Thomas SL, Alam R, Lemke N, Schultz LR, Gutierrez JA, Rempel SA. PTEN augments SPARC suppression of proliferation and inhibits SPARC‐induced migration by suppressing SHC‐RAF‐ERK and AKT signaling. Neuro Oncol 2010; 12: 941–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Murthy SR, Dupart E, Al‐Sweel N, Chen A, Cawley NX, Loh YP. Carboxypeptidase E promotes cancer cell survival, but inhibits migration and invasion. Cancer Lett 2013; 341: 204–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li J, Weinberg MS, Zerbini L, Prince S. The oncogenic TBX3 is a downstream target and mediator of the TGF‐beta1 signaling pathway. Mol Biol Cell 2013; 24: 3569–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Supplementary methods.
Fig. S1. Images of HE staining matched with the images of immunohistochemical staining in Figures 1(a) and 2(a).
Fig. S2. Quantitative RT‐PCR analysis showing knockdown of Talin1 mRNA in SK‐Hep‐1 and HepG2 cells transfected with Talin1‐specific siRNA#1, #2, #3, or #4. ***P < 0.001.
Fig. S3. Western blot analysis for the expression of E‐cadherin in 12 pairs of hepatocellular carcinoma tissues (T) and corresponding adjacent non‐tumor liver tissues (N), also used in Figure 1(b).
Fig. S4. Knockdown of Talin1 suppresses hepatocellular carcinoma cell proliferation.
Table S1. Clinicopathologic characteristics of 200 patients with hepatocellular carcinoma.
Table S2. Small interfering RNA sequences used in this study.
Table S3. Antibodies used in this study.


