Abstract
DNA replication can be a source of genetic instability. Given the tight connection between DNA replication and nucleosome assembly, we analyzed the effect of a partial depletion of histone H4 on genetic instability mediated by homologous recombination. A Saccharomyces cerevisiae strain was constructed in which the expression of histone H4 was driven by the regulated tet promoter. In agreement with defective nucleosome assembly, partial depletion of histone H4 led to subtle changes in plasmid superhelical density and chromatin sensitivity to micrococcal nuclease. Under these conditions, homologous recombination between ectopic DNA sequences was increased 20-fold above the wild-type levels. This hyperrecombination was not associated with either defective repair or transcription but with an accumulation of recombinogenic DNA lesions during the S and G2/M phases, as determined by an increase in the proportion of budded cells containing Rad52-yellow fluorescent protein foci. Consistently, partial depletion of histone H4 caused a delay during the S and G2/M phases. Our results suggest that histone deposition defects lead to the formation of recombinogenic DNA structures during replication that increase genomic instability.
Genetic instability leads to loss of cell fitness and is characteristic of cancer and a number of genetic diseases (31). This instability can be associated with errors during DNA replication. Thus, it is induced by either mutations affecting replication fork progression or replication inhibitors. In addition, genetic instability arises in S-phase checkpoint mutants in the absence of genotoxic agents, suggesting that DNA replication per se can be a source of spontaneous DNA rearrangements (28). Part of this instability involves DNA exchanges between homologous sequences, indicating that chromosomal rearrangements may result via homologous recombination from DNA lesions generated during replication. In accordance with this, homologous recombination has been shown to be required for the rescue of stalled replication forks in bacteria (13, 36, 50), and recombination intermediates have been physically detected during defective DNA replication in Saccharomyces cerevisiae (55, 69).
Newly replicated DNA is rapidly assembled into chromatin in a stepwise manner, with the initial deposition of the histone H3/H4 tetramers, followed by binding of two histone H2A/H2B dimers. Nucleosome assembly is catalyzed by histone chaperones and ATP-dependent chromatin remodeling complexes. Two different chaperones, conserved from S. cerevisiae to humans, have been involved in replication-dependent chromatin assembly: CAF-1 (chromatin assembly factor 1) and Asf1 (antisilencing function 1) (1, 37, 59). CAF-1 and Asf1 physically interact with each other, cooperate in chromatin assembly in vitro, and participate with redundant and specific roles in heterochromatin silencing, transcription, and DNA repair (29, 51, 60). In addition to CAF-1 and Asf1, the histone chaperone Spt6 has been shown to control chromatin structure in vivo and to participate in nucleosome assembly in vitro (9).
DNA replication and chromatin assembly are tightly coordinated. The coupling between DNA replication and chromatin assembly is mediated by the proliferating cell nuclear antigen, which interacts physically with the largest subunit of CAF-1 (52) and functionally with both CAF-1 and Asf1 in heterochromatin silencing and in vitro chromatin assembly (29, 51). In addition, the coordination of DNA replication and nucleosome assembly relies on the coregulation of the levels of DNA and histone synthesis. Thus, histone gene expression is cell cycle regulated to ensure the accumulation of histones during DNA replication: histone genes are actively transcribed in late G1/early S and repressed in early G1, G2, and M phases (22, 42, 56). Consistently, replication inhibitors and mutations that block DNA synthesis lead to histone gene repression from S. cerevisiae to human cells (34, 54), and repression of histone genes inhibits DNA synthesis in human cells (40). In addition, a Rad53-dependent surveillance mechanism that regulates histone levels during both normal cell cycle progression and in response to DNA damage in S. cerevisiae has been recently reported (21).
The absence of the Asf1 chromatin assembly factor in S. cerevisiae increases genomic instability and sister chromatid exchange, suggesting that defective chromatin assembly may generate recombinogenic DNA lesions during DNA replication (46). Nevertheless, the function of Asf1 is not restricted to DNA replication. Asf1 is required for histone deposition during nucleotide excision repair in vitro (38) and for the repair of double-strand breaks (DSBs) and DNA lesions caused by UV light, methyl-methane sulfonate (MMS), and hydroxyurea (HU) in vivo (30, 60). The activity of Asf1 in DNA repair seems to be mediated by its dynamic interaction with the checkpoint protein Rad53 (15, 23). In addition, Asf1 participates in the chromatin remodeling required for transcription regulation (2, 58).
To determine whether inefficient chromatin assembly during DNA replication leads to genetic instability mediated by homologous recombination, a yeast strain expressing histone H4 under the control of the regulated tet promoter was constructed. As expected from the requirement for high histone levels for proper nucleosome assembly during replication, partial depletion of histone H4 altered chromatin structure. Importantly, such a depletion led to an increase in the frequency of homologous recombination that was associated with an accumulation of budded cells containing Rad52-yellow fluorescent protein (YFP) foci and with a delay of the cell cycle during the S and G2/M phases. The increase in recombination was not associated with defective DNA repair or transcription. These results link replication-dependent chromatin assembly and the control of genetic instability mediated by homologous recombination.
MATERIALS AND METHODS
Yeast strains and growth conditions.
Yeast strains used in this study are listed in Table 1. Yeast cells were grown in synthetic complete (SC) medium as described previously (26). Doxycycline (DOX) was added to the medium at 0.25 or 5 μg/ml as indicated.
TABLE 1.
S. cerevisiae strains used in this studya
| Strain | Relevant genotype | Source |
|---|---|---|
| BY4741 | MATa | Euroscarf |
| Y07202 | MATatrp1Δ::kan | Euroscarf |
| Y03144 | MATahhf1Δ::kan | Euroscarf |
| Y15356 | MATα hhf2Δ::kan | Euroscarf |
| BYth2-1B | MATahhf2Δ::kan trp1Δ::kan | This work |
| Y11310 | MATα asf1Δ::kan | Euroscarf |
| Y02806 | MATarad1Δ::kan | Euroscarf |
| BY51 | MATarad51Δ::hyg | This work |
| BY59 | MATarad59Δ::hyg | This work |
| Y10540 | MATα rad52Δ::kan | Euroscarf |
| BYtetH4-10D | MATα hhf1Δ::kan hhf2Δ::kan(p413TARtetH4) | This work |
| BYtetH4-1C | MATahhf1Δ::kan hhf2Δ::kan trp1Δ::kan(p413TARtetH4) | This work |
| BY51tetH4-3C | MATahhf1Δ::kan hhf2Δ::kan rad51Δ::hyg(p413TARtetH4) | This work |
| BY59tetH4-2A | MATα hhf1Δ::kan hhf2Δ::kan rad59Δ::hyg(p413TARtetH4) | This work |
| BY52tetH4-2D | MATα hhf1Δ::kan hhf2Δ::kan rad52Δ::kan trp1Δ::kan(p413TARtetH4) | This work |
All strains are isogenic to BY4741 (MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0). BY51, BY59, and Y0 are met15Δ0 LYS2, Y1 strains are MET15 lys2Δ0, and BYth2 and BYtet strains are MET15 LYS2.
Plasmids.
All plasmids used were centromeric. Plasmids pRS314-GL (44), pRS314-LNA, pRS314-LNAT (47), pRS316-SU, and pRS316-LYΔNS (18) contain the different recombination systems used in this study. p413TARtetH4 contains HHF2 under the control of the tet promoter. p413TARtetH4 was constructed by inserting the following fragments into the vector pRS413 (53): the XhoI-EcoRI fragment from pCM176 (8), containing the tetR′::VP16 activator, at the XhoI-EcoRI site; the SphI (blunt ended)-XhoI fragment from pCM242 (8), containing the tetR-SSN6 repressor, at the ApaI (blunt ended)-XhoI site; the XhoI (blunt ended)-BamHI fragment from pCM176, containing the ADHter::tetO7::CYC1TATA sequence, at the NotI (blunt ended)-BamHI site; and a BamHI fragment containing the HHF2 gene amplified by PCR with oligonucleotides 5′-GCATGGATCCCAATAAAATAATGTCCGGTA-3′ and 5′-TGAAGGATCCTTAGCCAGTGACTCAAAATT-3′, at the BamHI site. pWJ1344 is a centromeric plasmid containing the Rad52-YFP construct (R. Rothstein, Columbia University).
Recombination and DNA repair assays.
Spontaneous recombination frequencies in plasmids harboring direct and inverted repeat systems were obtained by fluctuation assays as the median value of six independent colonies as described previously (45). Yeast colonies were isolated in SC medium containing either 2% glucose or 2% galactose as indicated (44). MMS and HU sensitivities were determined by 10-fold serial dilutions plated onto SC medium containing either MMS or HU. Sensitivity to UV light was determined as previously described (20).
Flow cytometry.
Analysis of DNA content was performed by flow cytometry (12). Mid-log-phase cells were fixed in 70% ethanol and then incubated for 8 h in phosphate-buffered saline with 1 mg of RNase A/ml and stained for 30 min with 5 μg of propidium iodide/ml. Samples were sonicated to separate single cells and analyzed in a FACSCalibur flow cytometer (Becton Dickinson). Mid-log-phase BY4741 and BYtetH4l-1C cell growth with 5 μg of DOX/ml was arrested in G1 by twice adding α factor at 2 and 5 μg/ml at 1- and 2-h intervals, respectively. For synchronization of BYtetH4l-1C growth with 0.25 μg of DOX/ml, α factor at 5 μg/ml was added four times at intervals of 90 min. Then, cells were washed three times to remove the α factor and released in fresh medium for different times before DNA content analysis.
Chromatin analysis by MNase digestion.
Mapping of micrococcal nuclease (MNase) cleavage sites was performed as previously described (11). Briefly, spheroplasts were prepared from mid-log-phase cells and then treated with different amounts of MNase. MNase-cleaved genomic DNA was extracted and run in 0.8% agarose gels for bulk chromatin analysis. Nucleosome positioning along the endogenous GAL1 gene and the SU recombination system was performed by indirect end labeling: MNase-treated DNA was restricted with either EcoRI (GAL1) or ClaI (pRS316-SU), resolved in a 1.5% agarose gel, blotted onto a Quiabrane Nylon-Plus membrane (Quiagen), and probed with either the 196-bp fragment immediately downstream of the EcoRI site (GAL1) or the 221-bp fragment immediately downstream of the ClaI site (pRS316-SU).
Plasmid supercoiling analysis.
Total DNA from yeast cells transformed with pRS316-SU was isolated as described by Allers and Lichten (6) with the modifications of Wellinger et al. (63) in the absence of hexamine cobalt trichloride. DNA samples were run in 0.7% agarose gels containing either 4 or 8 μg of chloroquine/ml for 48 h at 2 V/cm to resolve plasmid topoisomers (10). These were probed with either a ClaI-EcoRV LEU2 (pRS316-SU) or an EcoRV-NdeI FLP1 (2μm circle) fragment and quantified in a Fuji FLA3000 reader.
RNA analysis.
Yeast RNA was extracted and analyzed by Northern hybridization as previously described (47). mRNA was probed with specific DNA fragments, quantified in a Fuji FLA3000 reader, and normalized with respect to the 25S rRNA value.
Western blot analysis.
Yeast protein extracts were prepared essentially as described previously (16), except that 20% trichloroacetic acid-treated cells were washed three times with acetone. Protein extracts were run on a sodium dodecyl sulfate-15% polyacrylamide gel, and histone H4 was detected with histone H4 polyclonal antibody (Abcam Ltd.) by standard Western analysis.
Detection of Rad52-YFP.
Rad52-YFP foci from mid-log-phase cells transformed with plasmid pWJ1344 were visualized with a Leyca TCS SL confocal microscope (excitation, 514 nm; emission, 535 nm) as described by Lisby et al. (33).
RESULTS
Construction of a yeast strain partially depleted of histone H4.
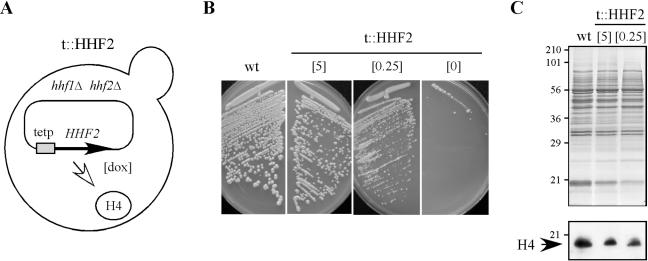
In order to understand the role of chromatin assembly in genetic instability, we constructed a yeast strain in which histone H4 could be partially depleted (Fig. 1A). This strain (t::HHF2) is a null mutant for the two copies of the histone H4 gene (hhf1Δ hhf2Δ) and contains a plasmid (p413TARtetH4) expressing HHF2 under the control of the bacterial tet promoter. This plasmid harbors copies of the tetR′-SSN6 and tetR-VP16 fusions, which positively regulate the tet promoter in response to DOX in a dose-dependent manner (8).
FIG. 1.
Construction of a yeast strain (t::HHF2) that can be gradually depleted of histone H4. (A) Scheme of the main genotype of strain t::HHF2. This strain is a null mutant for the two copies of the histone H4 gene (hhf1Δ hhf2Δ) that contains a plasmid (p413TARtetH4) expressing HHF2 under the control of the bacterial tet promoter, which responds to DOX in a dose-dependent manner. (B) Growth analysis of BYtetH4-10D at 5 (t::HHF2[5]) or 0.25 (t::HHF2[0.25]) μg of DOX/ml or in the absence of DOX and of BY4741 at 5 μg of DOX/ml (wild type [wt]). Cell growth of BY4741 was not affected by the presence or absence of DOX (data not shown). (C) Histone H4 content from strains described in panel B as determined by Western blot analysis. Total protein content was determined by Coomassie staining. The luminescence signals corresponding to histone H4 in t::HHF2[5] and t::HHF2[0.25] cells were 71 and 52% of the wild-type levels, respectively.
Since histone H4 is essential for cell viability, t::HHF2 cells do not grow in the absence of DOX (the tet promoter is repressed) and are extremely sick in 0.25 μg of DOX/ml (t::HHF2[0.25]), which leads to a reduction in the level of histone H4 and a doubling time of 5 h versus 2 h for the wild type (Fig. 1B and C). This growth defect was partially rescued with 5 μg of DOX/ml (t::HHF2[5]), which fully activates the tet promoter and results in cells with a slight reduction in the level of histone H4 and a doubling time of 3 h (Fig. 1B and C). Therefore, t::HHF2 cells are depleted partially of histone H4, which leads to growth defects that are more severe as the expression of histone H4 from the tet promoter is reduced.
Partial depletion of histone H4 affects chromatin assembly.
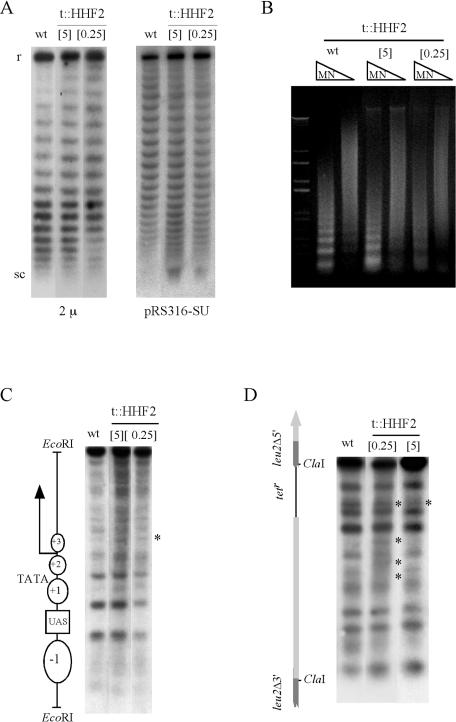
To analyze the effect of partial depletion of histone H4 on nucleosome assembly, plasmid supercoiling density and chromatin accessibility to MNase were determined. Since the assembly of one nucleosome introduces one negative superhelical turn in a circular DNA molecule (62, 65), a loss of plasmid superhelical density in cells partially depleted of histone H4 was expected. As can be seen in Fig. 2A, the endogenous 2μm plasmid in t::HHF2 cells displayed a shift in the distribution of topoisomers that indicated a loss of negative supercoiling, whereas the centromeric plasmid pRS314-SU displayed an increase in DNA supercoiling that was similar to that observed in the absence of the chromatin assembly factor Asf1 (data not shown; 46).
FIG. 2.
Analysis of plasmid superhelical density, chromatin accessibility, and nucleosome positioning in wild-type and t::HHF2 cells. (A) Topoisomer distribution of centromeric pRS316-SU and multicopy 2μm plasmids after electrophoresis in agarose gels containing 4 μg of chloroquine/ml. At this chloroquine concentration, negatively supercoiled topoisomers are resolved, consistent with a slower migration with 8 μg of chloroquine/ml (data not shown). r and sc indicate relaxed and negatively supercoiled plasmids, respectively. (B) MNase accessibility of bulk chromatin. MN, MNase I. (C and D) MNase digestion patterns of the endogenous GAL1 gene and the intervening sequence between the inverted leu2 repeats of the SU repeat system, respectively. A scheme of the analyzed sequences is shown on the left of each panel. Bands whose intensity is modified relative to the wild type are indicated with asterisks. Strains and other details are the same as for Fig. 1B.
Partial MNase digestion of bulk chromatin leads to the formation of a nucleosome ladder (41). As observed in Fig. 2B, this ladder was more diffuse in t::HHF2[0.25] cells, pointing to a general loss of nucleosome density. However, the depletion of histone H4 in t::HHF2[5] cells was not sufficient to induce this chromatin alteration.
For a more detailed molecular analysis of the effect of partial depletion of histone H4 on chromatin structure, we determined nucleosome positioning along two different DNA sequences: the endogenous GAL1 promoter and the LEU2 promoter of the SU recombination system. Figure 2C shows that nucleosomes remained well positioned along the regulatory region of GAL1 in both t::HHF2[0.25] and t::HHF2[5] cells. In the case of the SU recombination system, the LEU2 promoter region was more sensitive to MNase in t::HHF2[0.25] cells than in wild-type cells (Fig. 2D). As previously observed for bulk chromatin, most of these alterations were suppressed in t::HHF2[5] cells. Taken together, our results suggest that chromatin assembly is affected in t::HHF2 cells, leading to subtle changes in chromatin structure.
Partial depletion of histone H4 increases homologous recombination.
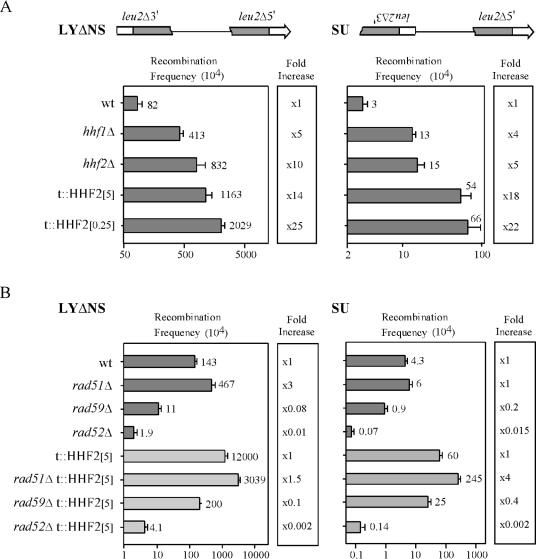
To determine the effect of partial depletion of histone H4 on homologous recombination, we used intramolecular recombination systems based on two direct repeats of a 0.6-kb internal fragment of the LEU2 gene (Fig. 3A, LYΔNS system). Recombination between these repeats leads to the deletion of the intervening sequence and one of the repeats, generating a wild-type copy of the LEU2 gene (45). The effect of partial depletion of histone H4 on inversions in the recombination system SU was also analyzed; the SU system differs from the LYΔNS system only in the orientation of the repeats (Fig. 3A). Recombination between these repeats leads to the inversion of the intervening sequence and the formation of a wild-type copy of the LEU2 gene (45). As shown in Fig. 3A, deletions and inversions increased 5- to 10-fold above wild-type levels in hhf1Δ and hhf2Δ single mutants, in which histone H4 RNA accumulated at 40 and 80% of the wild-type levels, respectively (data not shown). Notably, the frequency of deletions increased 25- and 14-fold in t::HHF2[0.25] and t::HHF2[5] cells, respectively, and the frequency of inversions increased 22- and 18-fold, respectively (Fig. 3A). Therefore, partial depletion of histone H4 causes genetic instability mediated by homologous recombination.
FIG. 3.
Recombination in cells partially depleted of histone H4. (A) Frequencies of Leu+ recombinants in BY4741 (wild type [wt]), Y03144 (hhf1Δ), and Y15356 (hhf2Δ) at 5 μg of DOX/ml and BYtetH4-10D at either 5 (t::HHF2[5]) or 0.25 (t::HHF2[0.25]) μg of DOX/ml. (B) Frequencies of Leu+ recombinants in BY4741 (wt), BY51 (rad51Δ), BY59 (rad59Δ), Y10540 (rad52Δ), BYtetH4-10D (t::HHF2[5]), BY51tetH4 (t::HHF2[5] rad51Δ), BY59tetH4 (t::HHF2[5] rad59Δ), and BY52tetH4 (t::HHF2[5] rad52Δ) at 5 μg of DOX/ml. All strains carried either the pRS316-LYΔNS or the pRS316-SU plasmid, harboring the direct repeat system LYΔNS or the inverted repeat system SU, respectively. A scheme of each recombination system is shown at the top of the panels. The frequencies of Leu+ recombinants of wild-type, hhf1Δ, hhf2Δ, rad51Δ, rad52Δ, and rad59Δ strains were not affected by the presence or absence of DOX (data not shown). The averages and standard deviations of three or four median frequencies obtained with two or three independent transformants are shown.
To determine the genetic basis of the recombination events induced by partial depletion of histone H4, we analyzed the effects of rad52Δ, rad59Δ, and rad51Δ mutations (Fig. 3B), whose genetic consequences on recombination are known to depend on the type of event. Thus, deletions and inversions require Rad52 and Rad59, but they can occur efficiently in the absence of Rad51 (5, 7, 24, 48). As observed with wild-type cells, both deletions and inversions were partially dependent on Rad59 and completely dependent on Rad52 in t::HHF2 cells. However, the level of inversions stimulated by histone H4 depletion in rad51Δ cells was fourfold above the level in RAD51 cells. This indicates that homologous recombination in the absence of Rad51 is more efficient under conditions of histone H4 depletion. This is in agreement with the increase in recombination in rad51Δ cells carrying either a single copy of the histone H2A and H2B loci or the spt6-140 mutation (35) and the requirement for Rad51 in DSB-induced recombination in the silenced mating-type loci (57).
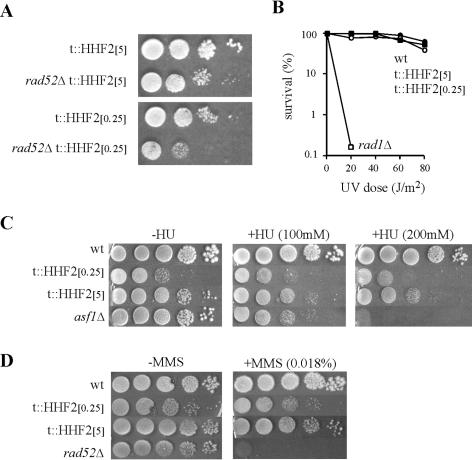
Our results suggest that recombinational repair may be a highly relevant process for the viability of cells partially depleted of histone H4. Consistent with this conclusion rad52Δ t::HHF2 cells grew poorly in 5 μg of DOX/ml and were extremely sick in 0.25 μg of DOX/ml (Fig. 4A).
FIG. 4.
DNA repair in cells partially depleted of histone H4. (A) Growth analysis of BYtetH4-10D (t::HHF2) and BY52tetH4 (t::HHF2 rad52Δ) at either 5 (t::HHF2[5]) or 0.25 (t::HHF2[0.25]) μg of DOX/ml. (B, C, and D) UV light, HU, and MMS sensitivities of the strains indicated in the legend to Fig. 1, respectively. rad1Δ, asf1Δ, and rad52Δ strains are controls for UV light-, HU-, and MMS-induced DNA repair, respectively, and were analyzed at both 5 (t::HHF2[5]) and 0.25 (t::HHF2[0.25]) μg of DOX/ml. Since UV light, HU, and MMS sensitivities of wild-type, rad52Δ, asf1Δ, and rad1Δ strains were not affected by the presence or absence of DOX (data not shown), only the data for 5 μg of DOX/ml are shown for these strains.
Hyperrecombination induced by partial depletion of histone H4 is not associated with defective repair or transcription.
Since the absence of the Asf1 and CAF1 chromatin assembly factors leads to defects in DNA repair and transcription (2, 17, 30, 58, 60), it was important to know whether partial depletion of histone H4 caused similar defects that could be associated with the increase in recombination. Thus, we analyzed the capacity of t::HHF2 cells to repair DNA lesions induced by either the alkylating agent MMS, the replication inhibitor HU, or UV light, which are subject to different DNA repair mechanisms (64). As can be seen in Fig. 4, t::HHF2 cell viability was not reduced by MMS or UV light and showed only a slight decrease at high levels of HU, as determined by CFU. Therefore, hyperrecombination in t::HHF2 cells seems not to be due to the recombinational processing of unrepaired DNA lesions.
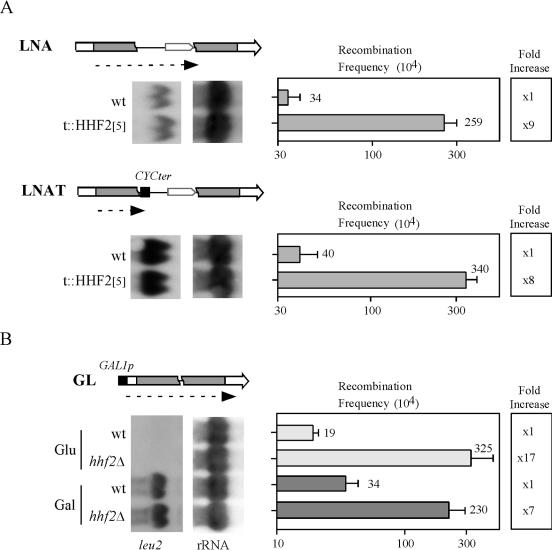
Histone H4 depletion affects global transcription regulation but not the expression levels of recombination proteins (66). However, since transcription induces homologous recombination (4), we wondered whether recombination induced by histone H4 depletion could be mediated by transcription. To analyze this possibility, three recombination systems based on the same direct repeats as those in the LYΔNS system were used (Fig. 5). The first two systems differ in the presence (LNAT) or absence (LNA) of a terminator just downstream of the first leu2 repeat, which impedes transcription elongation progress through the intervening sequence located between the repeats. The third system (GL) is under the control of the GAL1 promoter; thus, transcription could be turned on and off with galactose or glucose, respectively. Recombination was analyzed in t::HHF2[5] cells for the LNA and LNAT systems because recombination levels were similar at 5 and 0.25 μg of DOX/ml (Fig. 3) and in hhf2Δ cells for the GL system because the GAL1 promoter was derepressed in t::HHF2 cells (data not shown). As can be seen in Fig. 5, hyperrecombination in t::HHF2[5] and hhf2Δ cells was independent of the transcriptional state of the DNA sequence (compare the LNA and GL [galactose] systems versus the LNAT and GL [glucose] systems). Therefore, hyperrecombination mediated by partial depletion of histone H4 is not dependent on transcription.
FIG. 5.
Effect of transcription in recombination induced by partial depletion of histone H4. (A) leu2 RNA levels and frequencies of Leu+ recombinants of strains Y07202 (wild type [wt]) and BYtetH4-10D (t::HHF2[5]) transformed with pRS314-LNA or pRS314-LNAT and grown in 5 μg of DOX/ml. (B) leu2 RNA levels and frequencies of Leu+ recombinants of strains Y07202 (wt) and BYth2-1B (hhf2Δ) transformed with pRS314-GL and grown in either glucose- or galactose-containing medium, in which the GAL1 promoter of the GL system is either repressed or activated, respectively. Schemes of the direct repeat systems are shown. Dashed arrows indicate the transcripts produced by the recombination systems. As a probe, the ClaI-EcoRV LEU2 fragment was used. The averages and standard deviations of two to four median frequencies obtained with two to four independent transformants are shown.
Hyperrecombination induced by partial depletion of histone H4 is related to defective DNA replication.
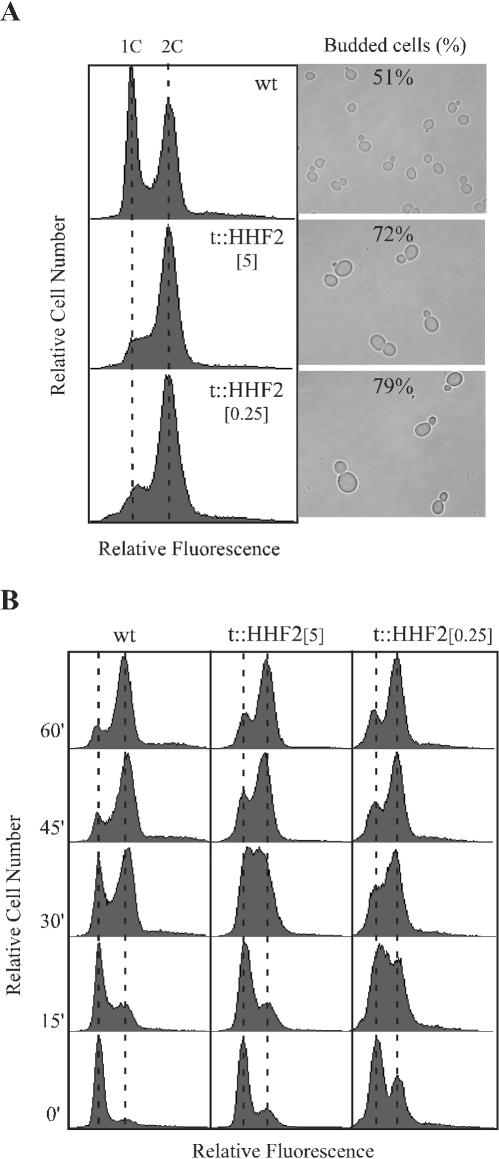
Given the tight connection between nucleosome assembly and DNA synthesis, we wondered whether hyperrecombination in t::HHF2 cells was linked to DNA replication defects. Thus, we would expect that growth retardation in t::HHF2 cells was due to a delay during the S and G2/M phases of the cell cycle. In accordance with this, DNA content analysis of asynchronous cultures by flow cytometry showed an accumulation of t::HHF2 cells during the S and G2/M phases with both 0.25 and 5 μg of DOX/ml (Fig. 6A). Also, t::HHF2 cells accumulated as budded cells and showed a heterogeneous increase in cell size (Fig. 6A). To confirm the delay during S phase, cell cycle progression of α-factor-synchronized cultures was monitored by flow cytometry. As shown in Fig. 6B (time zero), a fraction of t::HHF2 cells could not be arrested in G1 with α factor, particularly in 0.25 μg of DOX/ml. It is possible that this fraction corresponds to those t::HHF2 cells accumulated in G2/M that do not to progress into G1 and consequently do not respond to α factor. Alternatively, t::HHF2 cells could be less sensitive to α factor. Indeed, t::HHF2[0.25] cells required five times more α factor for synchronization than wild-type cells. This reduced sensitivity could also explain the apparent early entrance of t::HHF2[0.25] cells into S phase upon α-factor release (Fig. 6B, time 15′). Nevertheless, it is clear that most wild-type cells reached G2 while t::HHF2 cells accumulated in S phase at 30 min upon release from α-factor arrest. Therefore, cells partially depleted of histone H4 accumulate in S and G2/M phases, in agreement with replication-associated cell cycle defects.
FIG. 6.
Cell cycle analysis of cells partially depleted of histone H4. (A) DNA content and cell morphology of asynchronous cultures of the strains and at the conditions indicated in the legend to Fig. 1. Similar profiles were obtained with BYtetH4l-1C (data not shown). (B) Cell cycle progression of BYtetH4l-1C at either 5 (t::HHF2[5]) or 0.25 (t::HHF2[0.25]) μg of DOX/ml and of BY4741 at 5 μg of DOX/ml (wild type [wt]). DNA content of wild-type and t::HHF2 cells was determined by flow cytometry at the indicated times upon release from α-factor arrest in G1.
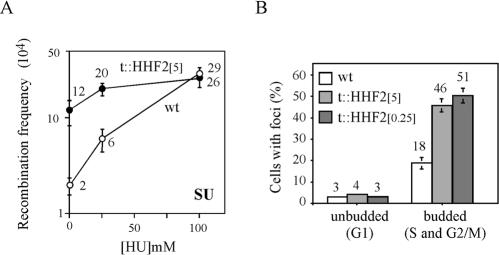
To determine the importance of DNA replication, recombination in t::HHF2 cells was measured in the presence of HU, an inhibitor of nucleotide reductase, which pauses replication by limiting deoxynucleoside triphosphate pools (68). HU-mediated impairment of DNA replication has been shown to lead to single- and double-strand DNA breaks and an increase in homologous recombination (19, 49). Importantly, HU-induced DNA damage is repaired in t::HHF2 cells (Fig. 4C). As shown in Fig. 7A, HU treatment increased recombination up to 15-fold in wild-type cells and only 2-fold in t::HHF2[5] cells. In addition, recombination frequencies were the same in wild-type and t::HHF2[5] cells at 100 mM HU. The lack of an additive effect of histone H4 depletion and HU on recombination is consistent with the idea that replication is the major process affected in histone H4-depleted cells. In this regard, the spt16-197 point mutation of SPT16, an essential chromatin remodeling gene involved in replication (25), does not affect growth of t::HHF2 cells at either 5 or 0.25 μg of DOX/ml (data not shown). These results are consistent with the idea that the recombination events induced by partial depletion of histone H4 may by associated with replication defects.
FIG. 7.
(A) Effect of HU on recombination induced by partial depletion of histone H4. Frequencies of Leu+ recombinants of the strains indicated in the legend to Fig. 1 grown with 5 μg of DOX/ml and either 0, 25, or 100 mM HU. (B) Accumulation of Rad52-YFP foci in t::HHF2 cells. Rad52-YFP foci were visualized by fluorescence microscopy in asynchronous cultures of BYtetH4-10D transformed with pWJ1344 and grown in either 5 (t::HHF2[5]) or 0.25 (t::HHF2[0.25]) μg of DOX/ml and of BY4741 transformed with pWJ1344 and grown in 5 μg of DOX/ml (wild type [wt]). The percentages of unbudded (G1) and budded (S and G2/M) wild-type and t::HHF2 cells containing Rad52-YFP foci are shown. The total numbers of analyzed cells were 50 for unbudded cells and 150 for budded cells. The averages and standard deviations of two independent experiments are plotted.
Partial depletion of histone H4 leads to an accumulation of Rad52 foci during DNA replication.
To determine whether recombinogenic DNA lesions were accumulated during replication as a consequence of the depletion of histone H4, the formation of Rad52-YFP foci was analyzed. These foci have previously been shown to appear both spontaneously and in response to DSBs during S and G2/M phases, providing a specific link between DNA replication and homologous recombination (32, 33). As previously published, Rad52-YFP foci were detected in 3% of unbudded (G1) and 18% of budded (S and G2/M) wild-type cells (Fig. 7B) (33). In t::HHF2 cells the Rad52-YFP foci appeared at wild-type levels (4%) in unbudded cells but increased to 46 and 51% in budded cells in the presence of 5 and 0.25 μg of DOX/ml, respectively (Fig. 7B). Therefore, partial depletion of histone H4 leads to an accumulation of DNA lesions during DNA replication that are repaired by homologous recombination.
DISCUSSION
In this study we show that partial depletion of histone H4 affects chromatin assembly and causes genetic instability by increasing the frequency of homologous recombination between ectopic DNA sequences. This increase in recombination is associated with an accumulation of Rad52-YFP foci and a cell cycle delay during the S and G2/M phases. Our results suggest that defective chromatin assembly mediated by partial depletion of histone H4 impairs DNA synthesis and leads to the formation of recombinogenic structures.
To impair nucleosome deposition during DNA replication, a yeast strain (t::HHF2) in which the expression of histone H4 is under the control of the regulated tet promoter was constructed. This strain can be gradually depleted of histone H4. Our results suggest that chromatin assembly is affected in this strain. Thus, partial depletion of histone H4 leads to subtle changes in plasmid superhelical density and chromatin accessibility to MNase, particularly at 0.25 μg of DOX/ml. Also, t::HHF2 cells are delayed in the S and G2/M phases and accumulate as budded cells with a heterogeneous increase in cell size. Our results are reminiscent of those obtained for cells completely depleted of histone H4, which show dramatic changes in plasmid topology and chromatin accessibility and stop growing at the S and G2/M phases of the cell cycle (27). It is worth noting that chromatin assembly factor mutants display phenotypes similar to those of t::HHF2 cells. Thus, yeast mutants lacking the chromatin assembly factor Asf1 accumulate as large cells in S and G2/M phases (60) and display similar alterations in chromatin accessibility to MNase (46). In addition, the superhelical density of the centromeric plasmid pRS316-SU is increased in t::HHF2 and asf1Δ cells (Fig. 2A; 46), whereas the supercoiling of the endogenous 2μm plasmid is decreased in t::HHF2 cells (Fig. 2A) and increased in asf1Δ cells (data not shown; 3). Similar observations have been reported for human cells, in which HIRA-mediated histone repression and expression of a dominant-negative mutation of the chromatin assembly factor p150CAF-1 cause chromatin hypersensitivity and replication defects (40, 67). In this sense, it is worth noting that histone depletion causes S-phase arrest in human cells (40) but S-phase delay and G2/M arrest in yeast cells as determined by flow cytometry (Fig. 6B; 27).
The most relevant observation of this study is that partial depletion of histone H4 increases recombination between ectopic DNA sequences up to 20-fold above wild-type levels. These recombination events are RAD52 dependent, as expected for events occurring by homologous recombination mechanisms. The importance of recombinational repair in cells depleted partially of histone H4 is supported by the severe growth defects of t::HHF2 rad52Δ cells. Notably, recombination was 5- to 10-fold above wild-type levels in hhf1Δ and hhf2Δ single mutants, in which histone H4 RNA levels were reduced to 80% of that of the wild type. Our results strongly suggest that DNA stability is sensitive to slight reductions in the pool of histone H4. In accordance with this idea, deletion of HHF1 has been shown to result in telomeric sequence instability, manifested by changes in their length and sequence organization (61). The effect on recombination is less evident for a deletion of one of the two genes coding for histones H2A and H2B (35). Altogether, these results are consistent with the central role of the H3/H4 tetramer in nucleosome assembly (59).
The increase in recombination and its association with an accumulation of Rad52-YFP foci in t::HHF2 cells are consistent with a general role of proper chromatin assembly in preventing genetic instability. This is strongly supported by our recent observation that yeast cells lacking the chromatin assembly factor Asf1 also display an increase in recombination and an accumulation of Rad52-YFP foci (46). In contrast to cells lacking Asf1, histone H4 depletion does not impair DNA repair, and consequently, this rules out the possibility that the increase in recombination could be due to the channeling of unrepaired DNA lesions into homologous recombination. The influence of chromatin assembly in genome integrity is also supported by the observations that a thermosensitive spt6-140 mutation leads to hyperrecombination (35) and that a chromatin assembly factor p150CAF-1 dominant-negative mutant accumulates DNA breaks and induces S-phase arrest in human cells (67). In the same vein, the absence of the yeast chromatin assembly factors CAF-1 and Asf1 leads to gross chromosomal rearrangements (GCRs) (39). These rearrangements are Rad52 and homology independent. Indeed, the frequency of GCRs is synergistically increased in rad52Δ mutants, indicating that homologous recombination partially suppresses GCRs (39). The results of the present study provide evidence that the defects in chromatin assembly also increase genetic instability mediated by homologous recombination.
The tight connection between chromatin assembly and DNA synthesis provides a framework to explain the genetic instability reported here. Defective chromatin assembly could impair DNA synthesis, thus leading to the accumulation of recombinogenic structures. Consistent with this interpretation, we have shown that t::HHF2 cells are delayed during S phase and accumulate in G2/M. Also, the observation that hyperrecombination in t::HHF2 cells is not further increased by the presence of the replication inhibitor HU suggests that it results from lesions generated during DNA replication. Indeed, t::HHF2 cells accumulate Rad52-YFP foci (Fig. 7), which have been shown to appear spontaneously and in response to DSBs during the S and G2/M phases (33). Our results are in accordance with the observation that the lethality associated with total depletion of histone H4 occurs in S phase (27). Altogether, these results strengthen the idea that homologous recombination has an important role in the control of the accuracy of DNA replication. Presumably, defective nucleosome assembly increases the basal rate of replication-mediated DNA lesions and, in turn, the requirement of the recombination machinery for the repair of these lesions. Partial depletion of histone H4 provides a unique tool to study the role of proper chromatin assembly in genome integrity during replication.
We do not yet know the nature of the recombinogenic structures generated in t::HHF2 cells. DSBs have been extensively shown to induce homologous recombination (43), and it has recently been reported that phosphorylation of histone H2A (P-H2A), which occurs in response to DSBs (14), increases in yeast asf1Δ mutants (46) and in human cells defective in the chromatin assembly factor p150CAF-1 (67). We have not been able to detect P-H2A in t::HHF2 cells (data not shown). Whether DSBs, replication fork blocks, or single-stranded DNA gaps or tails are the primary structures leading to recombination as a consequence of impaired nucleosome assembly remains to be determined.
Acknowledgments
We thank R. Rothstein for providing the Rad52-YFP construct, G. Vicent for the anti-H4 antibodies, M. Nieto for excellent technical assistance, M. Fidalgo for help with confocal microscopy and flow cytometry, R. E. Wellinger and M. C. Muñoz-Centeno for critical reading of the manuscript, and Diane Haun for style supervision.
The research was funded by the Spanish Ministry of Science and Technology (BMC2000-0439 and SAF2003-00204 grants).
REFERENCES
- 1.Adams, C. R., and R. T. Kamakaka. 1999. Chromatin assembly: biochemical identities and genetic redundancy. Curr. Opin. Genet. Dev. 9:185-190. [DOI] [PubMed] [Google Scholar]
- 2.Adkins, M. W., S. R. Howar, and J. K. Tyler. 2004. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol. Cell 14:657-666. [DOI] [PubMed] [Google Scholar]
- 3.Adkins, M. W., and J. K. Tyler. 2004. The histone chaperone Asf1p mediates chromatin disassembly in vivo. J. Biol. Chem. 279:52069-52074. [DOI] [PubMed] [Google Scholar]
- 4.Aguilera, A. 2002. The connection between transcription and genomic instability. EMBO J. 21:195-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aguilera, A. 1995. Genetic evidence for different RAD52-dependent intrachromosomal recombination pathways in Saccharomyces cerevisiae. Curr. Genet. 27:298-305. [DOI] [PubMed] [Google Scholar]
- 6.Allers, T., and M. Lichten. 2000. A method for preparing genomic DNA that restrains branch migration of Holliday junctions. Nucleic Acids Res. 28:e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bai, Y., and L. S. Symington. 1996. A Rad52 homolog is required for RAD51-independent mitotic recombination in Saccharomyces cerevisiae. Genes Dev. 10:2025-2037. [DOI] [PubMed] [Google Scholar]
- 8.Belli, G., E. Gari, L. Piedrafita, M. Aldea, and E. Herrero. 1998. An activator/repressor dual system allows tight tetracycline-regulated gene expression in budding yeast. Nucleic Acids Res. 26:942-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bortvin, A., and F. Winston. 1996. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science 272:1473-1476. [DOI] [PubMed] [Google Scholar]
- 10.Brill, S. J., and R. Sternglanz. 1988. Transcription-dependent DNA supercoiling in yeast DNA topoisomerase mutants. Cell 54:403-411. [DOI] [PubMed] [Google Scholar]
- 11.Chavez, S., R. Candau, M. Truss, and M. Beato. 1995. Constitutive repression and nuclear factor I-dependent hormone activation of the mouse mammary tumor virus promoter in Saccharomyces cerevisiae. Mol. Cell. Biol. 15:6987-6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corliss, D. A., and W. E. White, Jr. 1981. Fluorescence of yeast vitally stained with ethidium bromide and propidium iodide. J. Histochem. Cytochem. 29:45-48. [DOI] [PubMed] [Google Scholar]
- 13.Courcelle, J., J. R. Donaldson, K. H. Chow, and C. T. Courcelle. 2003. DNA damage-induced replication fork regression and processing in Escherichia coli. Science 299:1064-1067. [DOI] [PubMed] [Google Scholar]
- 14.Downs, J. A., N. F. Lowndes, and S. P. Jackson. 2000. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature 408:1001-1004. [DOI] [PubMed] [Google Scholar]
- 15.Emili, A., D. M. Schieltz, J. R. Yates, 3rd, and L. H. Hartwell. 2001. Dynamic interaction of DNA damage checkpoint protein Rad53 with chromatin assembly factor Asf1. Mol. Cell 7:13-20. [DOI] [PubMed] [Google Scholar]
- 16.Foiani, M., F. Marini, D. Gamba, G. Lucchini, and P. Plevani. 1994. The B subunit of the DNA polymerase alpha-primase complex in Saccharomyces cerevisiae executes an essential function at the initial stage of DNA replication. Mol. Cell. Biol. 14:923-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaillard, P. H., E. M. Martini, P. D. Kaufman, B. Stillman, E. Moustacchi, and G. Almouzni. 1996. Chromatin assembly coupled to DNA repair: a new role for chromatin assembly factor I. Cell 86:887-896. [DOI] [PubMed] [Google Scholar]
- 18.Gallardo, M., R. Luna, H. Erdjument-Bromage, P. Tempst, and A. Aguilera. 2003. Nab2p and the Thp1p-Sac3p complex functionally interact at the interface between transcription and mRNA metabolism. J. Biol. Chem. 278:24225-24232. [DOI] [PubMed] [Google Scholar]
- 19.Galli, A., and R. H. Schiestl. 1996. Hydroxyurea induces recombination in dividing but not in G1 or G2 cell cycle arrested yeast cells. Mutat. Res. 354:69-75. [DOI] [PubMed] [Google Scholar]
- 20.Gonzalez-Barrera., F. Prado, R. Verhage, J. Brouwer, and A. Aguilera. 2002. Defective nucleotide excision repair in yeast hpr1 and tho2 mutants. Nucleic Acids Res. 30:2193-2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gunjan, A., and A. Verreault. 2003. A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell 115:537-549. [DOI] [PubMed] [Google Scholar]
- 22.Holstege, F. C., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 23.Hu, F., A. A. Alcasabas, and S. J. Elledge. 2001. Asf1 links Rad53 to control of chromatin assembly. Genes Dev. 15:1061-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jablonovich, Z., B. Liefshitz, R. Steinlauf, and M. Kupiec. 1999. Characterization of the role played by the RAD59 gene of Saccharomyces cerevisiae in ectopic recombination. Curr. Genet. 36:13-20. [DOI] [PubMed] [Google Scholar]
- 25.John, S., L. Howe, S. T. Tafrov, P. A. Grant, R. Sternglanz, and J. L. Workman. 2000. The something about silencing protein, Sas3, is the catalytic subunit of NuA3, a yTAF(II)30-containing HAT complex that interacts with the Spt16 subunit of the yeast CP (Cdc68/Pob3)-FACT complex. Genes Dev. 14:1196-1208. [PMC free article] [PubMed] [Google Scholar]
- 26.Kaiser, C., M. Michaelis, and A. Mitchell. 1994. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Kim, U. J., M. Han, P. Kayne, and M. Grunstein. 1988. Effects of histone H4 depletion on the cell cycle and transcription of Saccharomyces cerevisiae. EMBO J. 7:2211-2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolodner, R. D., C. D. Putnam, and K. Myung. 2002. Maintenance of genome stability in Saccharomyces cerevisiae. Science 297:552-557. [DOI] [PubMed] [Google Scholar]
- 29.Krawitz, D. C., T. Kama, and P. D. Kaufman. 2002. Chromatin assembly factor I mutants defective for PCNA binding require Asf1/Hir proteins for silencing. Mol. Cell. Biol. 22:614-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le, S., C. Davis, J. B. Konopka, and R. Sternglanz. 1997. Two new S-phase-specific genes from Saccharomyces cerevisiae. Yeast 13:1029-1042. [DOI] [PubMed] [Google Scholar]
- 31.Lengauer, C., K. W. Kinzler, and B. Vogelstein. 1998. Genetic instabilities in human cancers. Nature 396:643-649. [DOI] [PubMed] [Google Scholar]
- 32.Lisby, M., U. H. Mortensen, and R. Rothstein. 2003. Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nat. Cell Biol. 5:572-577. [DOI] [PubMed] [Google Scholar]
- 33.Lisby, M., R. Rothstein, and U. H. Mortensen. 2001. Rad52 forms DNA repair and recombination centers during S phase. Proc. Natl. Acad. Sci. USA 98:8276-8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lycan, D. E., M. A. Osley, and L. M. Hereford. 1987. Role of transcriptional and posttranscriptional regulation in expression of histone genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 7:614-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malagon, F., and A. Aguilera. 2001. Yeast spt6-140 mutation, affecting chromatin and transcription, preferentially increases recombination in which Rad51p-mediated strand exchange is dispensable. Genetics 158:597-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McGlynn, P., and R. G. Lloyd. 2000. Modulation of RNA polymerase by (p)ppGpp reveals a RecG-dependent mechanism for replication fork progression. Cell 101:35-45. [DOI] [PubMed] [Google Scholar]
- 37.Mello, J. A., and G. Almouzni. 2001. The ins and outs of nucleosome assembly. Curr. Opin. Genet. Dev. 11:136-141. [DOI] [PubMed] [Google Scholar]
- 38.Mello, J. A., H. H. Sillje, D. M. Roche, D. B. Kirschner, E. A. Nigg, and G. Almouzni. 2002. Human Asf1 and CAF-1 interact and synergize in a repair-coupled nucleosome assembly pathway. EMBO Rep. 3:329-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Myung, K., V. Pennaneach, E. S. Kats, and R. D. Kolodner. 2003. Saccharomyces cerevisiae chromatin-assembly factors that act during DNA replication function in the maintenance of genome stability. Proc. Natl. Acad. Sci. USA 100:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nelson, D. M., X. Ye, C. Hall, H. Santos, T. Ma, G. D. Kao, T. J. Yen, J. W. Harper, and P. D. Adams. 2002. Coupling of DNA synthesis and histone synthesis in S phase independent of cyclin/cdk2 activity. Mol. Cell. Biol. 22:7459-7472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Noll, M., and R. D. Kornberg. 1977. Action of micrococcal nuclease on chromatin and the location of histone H1. J. Mol. Biol. 109:393-404. [DOI] [PubMed] [Google Scholar]
- 42.Osley, M. A. 1991. The regulation of histone synthesis in the cell cycle. Annu. Rev. Biochem. 60:827-861. [DOI] [PubMed] [Google Scholar]
- 43.Paques, F., and J. E. Haber. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63:349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Piruat, J. I., and A. Aguilera. 1998. A novel yeast gene, THO2, is involved in RNA pol II transcription and provides new evidence for transcriptional elongation-associated recombination. EMBO J. 17:4859-4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prado, F., and A. Aguilera. 1995. Role of reciprocal exchange, one-ended invasion crossover and single-strand annealing on inverted and direct repeat recombination in yeast: different requirements for the RAD1, RAD10, and RAD52 genes. Genetics 139:109-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prado, F., F. Cortes-Ledesma, and A. Aguilera. 2004. The absence of the yeast chromatin assembly factor Asf1 increases genomic instability and sister chromatid exchange. EMBO Rep. 5:497-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prado, F., J. I. Piruat, and A. Aguilera. 1997. Recombination between DNA repeats in yeast hpr1Δ cells is linked to transcription elongation. EMBO J. 16:2826-2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rattray, A. J., and L. S. Symington. 1994. Use of a chromosomal inverted repeat to demonstrate that the RAD51 and RAD52 genes of Saccharomyces cerevisiae have different roles in mitotic recombination. Genetics 138:587-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saintigny, Y., F. Delacote, G. Vares, F. Petitot, S. Lambert, D. Averbeck, and B. S. Lopez. 2001. Characterization of homologous recombination induced by replication inhibition in mammalian cells. EMBO J. 20:3861-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seigneur, M., V. Bidnenko, S. D. Ehrlich, and B. Michel. 1998. RuvAB acts at arrested replication forks. Cell 95:419-430. [DOI] [PubMed] [Google Scholar]
- 51.Sharp, J. A., E. T. Fouts, D. C. Krawitz, and P. D. Kaufman. 2001. Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Curr. Biol. 11:463-473. [DOI] [PubMed] [Google Scholar]
- 52.Shibahara, K., and B. Stillman. 1999. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell 96:575-585. [DOI] [PubMed] [Google Scholar]
- 53.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sittman, D. B., R. A. Graves, and W. F. Marzluff. 1983. Histone mRNA concentrations are regulated at the level of transcription and mRNA degradation. Proc. Natl. Acad. Sci. USA 80:1849-1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sogo, J. M., M. Lopes, and M. Foiani. 2002. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297:599-602. [DOI] [PubMed] [Google Scholar]
- 56.Spellman, P. T., G. Sherlock, M. Q. Zhang, V. R. Iyer, K. Anders, M. B. Eisen, P. O. Brown, D. Botstein, and B. Futcher. 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9:3273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sugawara, N., E. L. Ivanov, J. Fishman-Lobell, B. L. Ray, X. Wu, and J. E. Haber. 1995. DNA structure-dependent requirements for yeast RAD genes in gene conversion. Nature 373:84-86. [DOI] [PubMed] [Google Scholar]
- 58.Sutton, A., J. Bucaria, M. A. Osley, and R. Sternglanz. 2001. Yeast Asf1 protein is required for cell cycle regulation of histone gene transcription. Genetics 158:587-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tyler, J. K. 2002. Chromatin assembly. Cooperation between histone chaperones and ATP-dependent nucleosome remodeling machines. Eur. J. Biochem. 269:2268-2274. [DOI] [PubMed] [Google Scholar]
- 60.Tyler, J. K., C. R. Adams, S. R. Chen, R. Kobayashi, R. T. Kamakaka, and J. T. Kadonaga. 1999. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature 402:555-560. [DOI] [PubMed] [Google Scholar]
- 61.Venditti, S., M. A. Vega-Palas, G. Di Stefano, and E. Di Mauro. 1999. Imbalance in dosage of the genes for the heterochromatin components Sir3p and histone H4 results in changes in the length and sequence organization of yeast telomeres. Mol. Gen. Genet. 262:367-377. [DOI] [PubMed] [Google Scholar]
- 62.Wang, J. C. 1982. The path of DNA in the nucleosome. Cell 29:724-726. [DOI] [PubMed] [Google Scholar]
- 63.Wellinger, R. E., A. Schar, and J. M. Sogo. 2003. Rad52-independent accumulation of joint circular minichromosomes during S phase in Saccharomyces cerevisiae. Mol. Cell. Biol. 23:6363-6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wood, R. D. 1996. DNA repair in eukaryotes. Annu. Rev. Biochem. 65:135-167. [DOI] [PubMed] [Google Scholar]
- 65.Worcel, A., S. Strogatz, and D. Riley. 1981. Structure of chromatin and the linking number of DNA. Proc. Natl. Acad. Sci. USA 78:1461-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wyrick, J. J., F. C. Holstege, E. G. Jennings, H. C. Causton, D. Shore, M. Grunstein, E. S. Lander, and R. A. Young. 1999. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature 402:418-421. [DOI] [PubMed] [Google Scholar]
- 67.Ye, X., A. A. Franco, H. Santos, D. M. Nelson, P. D. Kaufman, and P. D. Adams. 2003. Defective S phase chromatin assembly causes DNA damage, activation of the S phase checkpoint, and S phase arrest. Mol. Cell 11:341-351. [DOI] [PubMed] [Google Scholar]
- 68.Zhao, X., E. G. Muller, and R. Rothstein. 1998. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell 2:329-340. [DOI] [PubMed] [Google Scholar]
- 69.Zou, H., and R. Rothstein. 1997. Holliday junctions accumulate in replication mutants via a RecA homolog-independent mechanism. Cell 90:87-96. [DOI] [PubMed] [Google Scholar]