Abstract
The Notch signaling pathway controls several cell fate decisions during lymphocyte development, from T-cell lineage commitment to the peripheral differentiation of B and T lymphocytes. Deltex-1 is a RING finger ubiquitin ligase which is conserved from Drosophila to humans and has been proposed to be a regulator of Notch signaling. Its pattern of lymphoid expression as well as gain-of-function experiments suggest that Deltex-1 regulates both B-cell lineage and splenic marginal-zone B-cell commitment. Deltex-1 was also found to be highly expressed in germinal-center B cells. To investigate the physiological function of Deltex-1, we generated a mouse strain lacking the Deltex-1 RING finger domain, which is essential for its ubiquitin ligase activity. Deltex-1Δ/Δ mice were viable and fertile. A detailed histological analysis did not reveal any defects in major organs. T- and B-cell development was normal, as were humoral responses against T-dependent and T-independent antigens. These data indicate that the Deltex-1 ubiquitin ligase activity is dispensable for mouse development and immune function. Possible compensatory mechanisms, in particular those from a fourth Deltex gene identified during the course of this study, are also discussed.
Notch proteins are evolutionarily conserved transmembrane receptors which control cellular differentiation processes in nearly every organ (reviewed in references 2 and 20). Mammals have four homologues of the Drosophila melanogaster Notch gene. Upon interaction with one of their ligands, Notch receptors undergo a cascade of proteolytic cleavages, which ultimately releases the Notch intracellular domain (NICD) from the membrane (reviewed in reference 42). The NICD subsequently translocates to the nucleus and forms a complex with RBP-J, which upregulates the transcription of various targets. RBP-J is thus the central mediator of Notch signal transduction. However, an RBP-J-independent pathway has also been described (39; reviewed in reference 24), but its relevance in vivo for mammals remains to be addressed.
Notch-1 is essential for lymphoid development (reviewed in reference 37). Notch-1 activation in the thymus (14) commits common lymphocyte precursors to the T-cell lineage and prevents them from adopting a B-cell fate (36, 38, 48). In agreement with this function, the NICD can repress the activity of E2A (32, 34), a transcription factor that is required for early B-cell ontogeny. The Notch/RBP-J pathway also regulates later steps of T-cell differentiation, such as VDJβ rearrangement, β-selection (50), and αβ-γδ (46) and TH1-TH2 (1) lineage decisions. Gain-of-function experiments suggested that Notch-1 might similarly influence the CD4+/CD8+ lineage decision (40), but a conditional inactivation of RBP-J did not alter the CD4/CD8 ratio (46). Lastly, new data point to a potential function of the Notch pathway in the regulation of peripheral T-cell activation (7) and the development of regulatory T cells (reviewed in reference 29).
A role for the Notch pathway in B-cell development was recently discovered. A conditional deletion of either RBP-J (45) or Notch-2 (41) in the B-cell lineage results in the selective loss of splenic marginal-zone B cells. Moreover, an RBP-J deletion leads to a concomitant increase in the number of follicular B cells, while mice deficient in Mint (19), a suppressor of the Notch/RBP-J pathway, display the reciprocal phenotype. It was thus suggested that Notch-2 and RBP-J instruct splenic transitional B cells to adopt a marginal-zone B-cell versus follicular B-cell fate. Notch-2 has also been proposed to regulate the B1-B2 lineage decision (49), but conflicting data have been reported on this question (45).
E3 ubiquitin ligases are key regulators of Notch signaling which control the trafficking and stability of Notch receptors and ligands (21, 22). For example, Itchy/Suppressor of Deltex and c-Cbl are thought to drive the endocytosis of a membrane-anchored form of Notch to the lysosomal compartment (17, 28), while a ubiquitin ligase complex containing SEL-10 targets nuclear NICD to the proteasome (12, 33, 51).
Deltex is another ubiquitin ligase that binds Notch and modulates its signaling, but its precise function remains unclear. Deltex was initially identified in Drosophila as a positive regulator of the Notch pathway: deltex loss-of-function mutants display a phenotype similar to that caused by weak Notch alleles (11, 52), whereas deltex overexpression partially mimics Notch gain-of-function mutations (25). However, the overexpression of Deltex-1, one of the three mammalian homologues of the Drosophila deltex gene (18, 35), can either enhance (26) or antagonize (43) Notch/RBP-J signaling, depending on the cellular context. In addition, Deltex is thought to mediate RBP-J-independent Notch signals (34, 39).
Deltex proteins share three functional domains (18). Domain I mediates a physical interaction with Notch ankyrin repeats (25). Domain II consists of proline-rich sequences, which may serve as a docking site for an unknown WW- or SH3-containing protein (26). Domain III contains a highly conserved RING finger domain which mediates homo- and heterodimerization (27) and confers ubiquitin ligase activity in vitro (44).
Deltex-1 is expressed in a wide array of tissues, particularly in the central nervous system, testes, and endothelial cells (26, 30). Deltex-1 has been suggested to play a role in neurogenesis (18), myogenesis (34), and oligodendrocyte maturation and myelination (5, 15). Deltex-1 also displays a dynamic pattern of expression in thymocytes (6, 16) and peripheral T cells (31) and may thus play a role in T-cell development. On the basis of its ability to inhibit E2A in vitro (34), Deltex-1 was initially thought to mediate Notch signaling during T-cell commitment. However, Izon et al. reported opposite effects of Deltex-1 on E2A and showed that the enforced expression of Deltex-1 in hematopoietic stem cells results in a phenotype that mirrors that caused by Notch-1 inactivation (16). The reason for this discrepancy is not clear, but the latter results suggest that Deltex-1 may actually antagonize Notch signaling in common lymphocyte precursors to promote a B-cell fate. Deltex-1 may be important for later steps of B-cell differentiation as well, since it is highly expressed in mouse marginal-zone B cells (41) and human germinal-center B cells (13).
To investigate the physiological function of Deltex-1, we generated a mouse strain lacking the C-terminal half of Deltex-1, which contains the RING finger domain. To our surprise, Deltex-1Δ/Δ mice were viable and fertile and displayed normal lymphocyte differentiation and immune function. Possible compensatory mechanisms are discussed.
MATERIALS AND METHODS
Construction of targeting vector.
A mouse 129/SvJ genomic library (λFixII; Stratagene) was screened with a 1-kb probe located in the 3′ untranslated region of Deltex-1. A 15-kb phage encompassing exons 4 to 10 (Fig. 1) was selected. A 4.3-kb fragment upstream of exon 4 was amplified with Pfu-Turbo (Stratagene) and the primers GAGCCACGTGCTCCTGTTTG (forward) and GGCCTGGAACCCAACTATC (reverse). A 3.6-kb fragment downstream of the poly(A) signal was amplified with the primers CCAGGAGAATGAGGAAGACC (forward) and λFix5′ (reverse) (Fig. 1). Fragments were inserted by the use of restriction sites which were added by the primers into the SalI and XhoI cloning sites flanking the neomycin resistance gene (neoR) of a modified pLNTK vector (3).
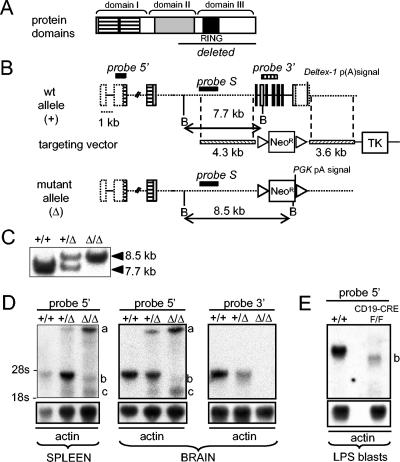
FIG. 1.
Targeted disruption of mouse Deltex-1 gene. (A) Schematic representation of Deltex-1 protein domains, with the deleted region including the RING finger domain underlined. (B) Schematic representation of wild-type and targeted Deltex-1 loci. Dotted boxes represent noncoding sequences within exons. Positions of BamHI sites (B) and of the probes used for Southern (S) and Northern (5′ and 3′ cDNA probes) blot analyses are indicated. PGK p(A) signal, polyadenylation signal of phosphoglycerate kinase gene; TK, herpes simplex virus thymidine kinase gene. Triangles flanking the neomycin resistance cassette (Neor) represent LoxP sites. (C) Southern blot analysis of BamHI-digested thymic DNAs from wild-type mice (+/+) and from mice that were heterozygous (+/Δ) and homozygous (Δ/Δ) for the targeted Deltex-1 gene. (D) Northern blot analysis of Deltex-1 expression in spleens and brains of wild-type, heterozygous, and homozygous mice by use of a cDNA probe outside (probe 5′) or inside (probe 3′) the deletion. “a,” “b,” and “c” mark the three truncated forms of the Deltex-1 transcript observed in mutant mice (see the text for details). Blots were normalized with an actin probe. (E) Northern blot analysis of B cells from Deltex-1Δ/Δ mice after excision of the neoRloxP gene (F/F). The analysis was performed on LPS-stimulated splenic B cells of Δ/Δ mice, with B-cell-restricted expression of the Cre recombinase (CD19-CRE).
Generation of gene-targeted mice.
E14.1 embryonic stem cells (ES) were transfected as described previously (47). G418- and ganciclovir-resistant clones were screened by PCRs (35 cycles with the Long Expand PCR system [Roche]) with the following external (E) and internal (I) primer sets: 5′(E) forward (ACAAGTTCCCAAGTCTTGCAGGAGC) with 5′(I) reverse (GCTGGACGTAAACTCCTCTTCAGAC) and 3′(I) forward (GTCTGAAGAGGAGTTTACGTCCAGC) with 3′(E) reverse (CTCACCCATGGGTTTACACTTAGCC). Homologous recombination was confirmed by Southern blot analyses of DNAs from ES clones and thymuses of gene-targeted mice (Fig. 1). Three recombinant clones (Deltex-1Δ/+) were obtained from 384 total clones and were injected into BALB/c or C57BL/6 blastocysts to generate chimeric mice for germ line transmission of the mutant allele. Deltex-1Δ/Δ mice of a mixed genetic background were analyzed after 6 weeks of age. The genotyping of mice was performed by PCR, with simultaneous amplification of wild-type (400 bp) and mutant (180 bp) alleles, by use of the primers CAGAGTGTTCTGCAGGAATCGATGC (forward), GGGATCCATAAGTGTGGACTCTATCGG (reverse), and GCTGGACGTAAACTCCTCTTCAGAC (reverse for Neor).
Analysis of Deltex-1 expression in gene-targeted mice.
Total RNAs were extracted from various tissues by the use of Trizol (Invitrogen). Lipopolysaccharide (LPS)-stimulated cells were obtained from T-cell-depleted splenic cells (EasySep; StemCell Technologies) after 3 days of stimulation in vitro with 25 μg of LPS/ml. Five micrograms of poly(A)+ mRNA isolated from the total RNA by the use of Micro Fast Track 2.0 (Invitrogen) was analyzed by Northern blotting. The blots were probed with 5′ (contained in exon 2) and 3′ (encompassing exons 5 to 8) probes (Fig. 1). The 5′ probe was amplified by reverse transcription-PCR (RT-PCR) with the primers CTAGGACAGACATTGCCTAC (forward) and GATGCATGGATTGTAGGTCGATG (reverse). The 3′ probe was a 500-bp digestion product of BamHI and EcoRI from an RT-PCR product cloned into pCR2.1-TOPO (Invitrogen) (the primers used were CTAGGACAGACATTGCCTAC [forward] and GCTGTGTCCCTGTCTTCTC [reverse]). The signals on the blots were normalized to those of β-actin.
Histology.
Organs were fixed in 4% paraformaldehyde for 48 h and then embedded in paraffin blocks. From these blocks, 5-μm-thick sections were cut and then stained with hematoxylin, eosin, and saffron.
Flow cytometry analyses.
Single-cell suspensions from spleens, Peyer's patches, thymuses, bone marrow, and peritoneal cavities were analyzed by use of a FACStar apparatus and CellQuest software (BD Biosciences) after staining with the following reagents: goat anti-mouse immunoglobulin M-fluorescein isothiocyanate (IgM-FITC) (Southern Biotechnology Associated); anti-CD16/CD32 (clone 2.4G2), anti-B220-phycoerythrin (anti-B220-PE) (clone RA3-6B2), anti-CD19-biotin (clone 1D3), anti-CD21-FITC (clone 7G6), anti-CD23-PE (clone B3B4), anti-CD5-FITC (53-7-3), anti-CD4-PE (clone RM 4-5), anti-CD8-FITC (clone 53-6.7), anti-CD25-FITC (7D4), and streptavidin-CyChrome (all from BD Biosciences); and peanut agglutinin (PNA)-FITC (Vector).
Immunizations and determinations of immunoglobulin titers.
Deltex-1Δ/Δ and littermate control mice were immunized by intraperitoneal injections of 100 μg of trinitrophenyl (TNP)-Ficoll or 100 μg of alum-precipitated TNP-keyhole limpet hemocyanin (TNP-KLH; Biosearch Technologies). TNP-KLH-immunized mice were given a booster 21 days later. Serum samples were collected before immunization and on day 7 after TNP-Ficoll immunization or on days 14 and 28 after TNP-KLH immunization. Plates were incubated overnight at 4°C with a TNP-bovine serum albumin capture antigen (Biosearch Technologies) (50 μg/ml) and then saturated with phosphate-buffered saline-1% bovine serum albumin (1 h at 37°C). Serial dilutions of serum samples were added to the wells for 2 h at room temperature, washed, and incubated with horseradish peroxidase-conjugated goat anti-mouse isotypes diluted 1/500 (Southern Biotechnology Associated). After revelation with the ABTS [2,2′-azinobis(3-ethyl benzo thiadine sulfonic acid)] substrate, the optical density at 405 nm was recorded. Serum from a pool of immunized wild-type mouse sera served as a standard control for the plates.
Basal immunoglobulin levels in sera were quantified by an enzyme-linked immunosorbent assay (ELISA) using the goat anti-mouse Ig heavy plus light chains and the SBA clonotyping system with HRP from Southern Biotechnology Associated.
For studies of somatic hypermutation, mice were immunized with phenyl-oxazolone, and splenic cells were treated as described previously (9).
Analysis of gene expression through semiquantitative RT-PCR.
Splenic marginal-zone B cells (CD19+ CD21hi CD23−/lo) and follicular B cells (CD19+ CD21int CD23hi) were sorted with a FACSVantage SE apparatus (BD Biosciences). Total RNAs were extracted from 2 × 105 cells by use of an RNeasy kit (QIAGEN), and cDNAs were synthesized by use of a ProSTAR First-Strand RT-PCR kit (Stratagene). RT-PCRs were performed with Advantage 2 polymerase (BD Biosciences) and the following sets of primers: Deltex-1 forward (GGTGGCCATGTACTCCATG) and Deltex-1 reverse (TTGGCCATGGCCTCAGAAAC), Deltex-2 forward (CAATGCTACCTGCCAGATAG) and Deltex-2 reverse (AAGAAGCTGACCTGAAGCTG), and Deltex-4 forward (TTGTTACCTTCCAGACAGCGAG) and Deltex-4 reverse (CCTTGACTACCCAGAACTGAAG). Semiquantitative PCRs were performed with serial dilutions of the templates. Reaction products were separated by electrophoresis, transferred onto Hybond N+ membranes (Amersham), and hybridized with internal 32P-labeled oligonucleotides. Quantitation was obtained with a Storm 840 phosphorimager (Molecular Dynamics).
RESULTS
Generation of a mouse strain lacking the Deltex-1 RING finger domain (Deltex-1Δ/Δ mice).
We engineered a gene-targeting vector that replaced exons 4 to 10 of Deltex-1 with a neomycin resistance cassette (Neor) (Fig. 1A). These exons code for the C-terminal half of Deltex-1 (G 321 to A 626), which contains one of the proline-rich sequences, the RING finger domain, and a motif that is highly conserved across all Deltex proteins (18). Exon 10 also contains the 3′ untranslated region of Deltex-1 with the polyadenylation signal.
Homologous recombination was confirmed by PCRs on both sides of the construct (data not shown) and by Southern blot analyses of DNAs from the three ES clones chosen for injection and from thymus DNAs of the resulting heterozygous (Deltex-1+/Δ) and homozygous (Deltex-1Δ/Δ) mice (Fig. 1C). RT-PCR and Northern blotting confirmed the expected deletion, while showing the appearance of three new RNA products in Deltex-1+/Δ and Deltex-1Δ/Δ mice (Fig. 1D and data not shown). The largest (a) and smallest (c) forms hybridized with a Neor probe and disappeared after removal of the Neor cassette from B cells by the mating of Deltex-1Δ/Δ mice with CD19-CRE mice (Fig. 1E and data not shown). Given its size, the largest form probably corresponded to a partially spliced pre-mRNA stabilized by the polyadenylation signal supplied by the Neor cassette. Sequencing of an RT-PCR product suggested that the smallest truncated form was created by splicing of the donor site of the third exon to a cryptic acceptor site in the Neor cassette, which created an in-frame stop codon seven amino acids downstream (data not shown). The intermediate truncated product (b) was still present after removal of the Neor cassette (Fig. 1E). Both truncated products, b and c, were at least three to five times less abundant than the wild-type Deltex-1 mRNA (Fig. 1D and E). However, due to the lack of an antibody against the Deltex-1 N terminus, we could not evaluate whether a truncated protein devoid of the RING finger domain was expressed. As a conservative estimate, we therefore qualified our mutant mice as having a deletion of the RING finger domain of Deltex-1 (Deltex-1Δ/Δ).
Gross phenotypic and histologic analyses of Deltex-1Δ/Δ mice.
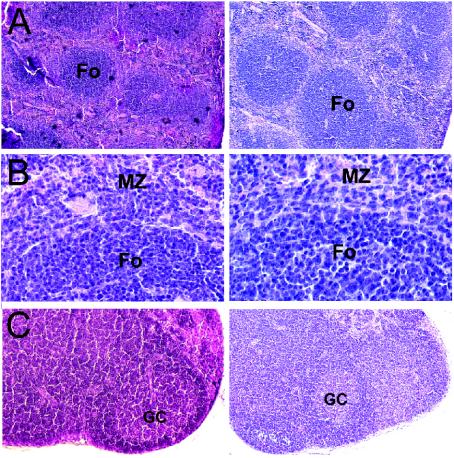
Homozygous Deltex-1Δ/Δ mice were viable and fertile and showed no apparent defects. Since Deltex-1 is expressed in a wide array of organs (18, 26, 30), we performed a detailed histological analysis of four adult Deltex-1Δ/Δ mice. Given the mixed genetic backgrounds of these mice, their wild-type littermates were used as a control. No abnormalities were detected in brains, spinal cords, eyes, livers, kidneys, urinary bladders, pancreases, salivary glands, lungs, testes, ovaries, uteruses, mammary glands, skin, aortas, and bone marrow (data not shown). Analyses of spleens and mesenteric lymph nodes did not reveal any differences between wild-type and Deltex-1Δ/Δ mice, since primary follicles as well as germinal centers were present in normal numbers (Fig. 2).
FIG. 2.
Deltex-1Δ/Δ mice have normal lymphoid organ structures. Sections from spleens (A and B) and mesenteric lymph nodes (C) of wild-type (left) and Deltex-1Δ/Δ (right) mice were stained with hematoxylin, eosin, and saffron and then photographed under a light microscope at a magnification of ×40. (B) Higher magnification of the images shown in panel A, showing primary B-cell follicles (Fo) and adjacent marginal zones (MZ). GC, germinal center.
Normal development of lymphocytes in Deltex-1Δ/Δ mice.
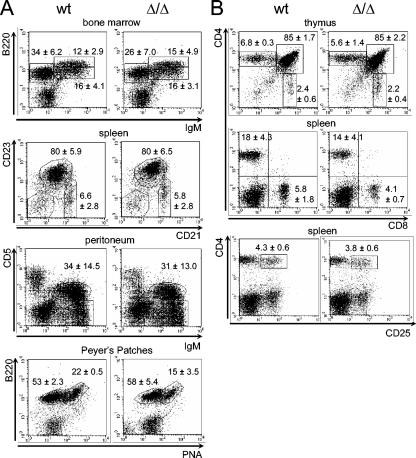
Deltex-1 overexpression in mouse hematopoietic stem cells inhibits T-cell development while inducing ectopic B-cell development in the thymus (16), which suggests that Deltex-1 antagonizes Notch-1 signaling to promote B-cell development. Thus, we first examined lymphocyte differentiation in the thymuses and bone marrow of Deltex-1Δ/Δ mice. The distribution of bone marrow B-cell subpopulations (pro-B/pre-B B220lo IgM−, immature B220lo IgM+, and mature B220hi IgM+ cells) in mutant mice was indistinguishable from that in wild-type mice (Fig. 3A). Similarly, as shown in Fig. 3B, the proportions of thymic T-cell subpopulations (CD4− CD8−, CD4+ CD8+, CD4+ CD8−, and CD4− CD8+ cells) were not altered in mutant mice, and the numbers of ectopic B cells in the thymus remained unchanged (data not shown).
FIG. 3.
Normal lymphoid development in Deltex-1Δ/Δ mice. (A) B-cell subpopulations in the bone marrow, spleen, peritoneal cavity, and Peyer's patches were detected by flow cytometry. (B) Thymic and splenic T-cell subpopulations were detected by flow cytometry. Mean values and standard deviations for at least five animals of each genotype are given.
We next checked the T-cell populations in the spleen because the Notch pathway is known to influence peripheral T-cell development and activation (1, 7, 29). In particular, Deltex-1 was recently shown to be constitutively expressed in human CD4+ CD25+ regulatory T cells and downregulated after activation of these cells with anti-CD3 (31). However, wild-type and mutant mice displayed similar numbers of CD4+ and CD8+ T cells in the spleen, and the splenic CD4+ CD25+ population was not affected in Deltex-1Δ/Δ mice (Fig. 3B).
Deltex-1 was also suggested to play a role in late B-cell differentiation (13, 41). Firstly, Deltex-1 is highly expressed in mouse marginal-zone B cells in a Notch-2 dependent fashion (41). We therefore examined splenic B-cell subsets. Three populations of B cells can be distinguished according to their relative expression of CD21 and CD23, i.e., transitional B-cell type 1 (CD19+ CD21− CD23−), marginal-zone B cells (CD19+ CD21hi CD23−/lo), and follicular B cells (comprising transitional B-cell type 2 and long-lived recirculating mature B cells [CD19+ CD21int CD23hi]) (23). In contrast with the selective loss of marginal-zone B cells observed for Notch-2−/− (41) or RBP-J−/− mice (45), all three populations were present in the spleens of Deltex-1Δ/Δ mice in normal percentages (Fig. 3A). In contrast to what was observed for Notch-2−/− mice (41), CD21 levels were not decreased on the surface of splenic B cells (data not shown). Since Deltex-1 has been reported to modulate the activity of E2A (16, 18, 34), a transcription factor which is essential for early and late B-cell development, we checked the E2A activity in vitro by using a reporter assay (16). Deltex-1Δ/Δ and wild-type splenic B cells stimulated with LPS had similar levels of E2A activity (data not shown). Secondly, we have reported that Deltex-1 is highly expressed in sheep and human germinal-center B cells as well as in their malignant counterparts (13). We thus investigated whether chronic germinal-center formation occurred normally in Peyer's patches of mutant mice by staining cells with PNA, which selectively binds centroblasts and centrocytes. The B220+ PNAhi germinal-center B-cell populations were comparable between wild-type and Deltex-1Δ/Δ mice. Thirdly, Notch-2 has also been proposed to play a role in peritoneal B1 cell development (49), potentially through an RBP-J-independent pathway (45). We did not observe any difference in the sizes of the B1 cell populations between wild-type and mutant mice (Fig. 3A).
Together, all of these data show that the Deltex-1 RING finger domain is dispensable for both early and late lymphocyte development in mice.
Deltex-1Δ/Δ mice have normal humoral immune responses.
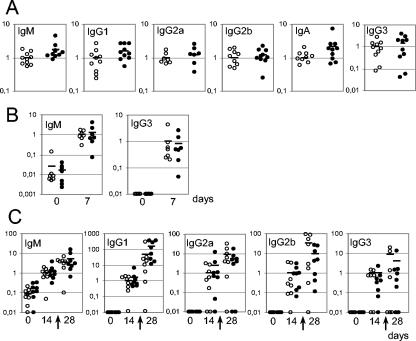
In order to look for a role of Deltex-1 in the terminal differentiation of B lymphocytes, we checked the ability of Deltex-1Δ/Δ mice to mount humoral immune responses. First, we found that Deltex-1Δ/Δ splenic B lymphocytes proliferated in response to LPS stimulation and were able to undergo class-switch recombination in vitro upon stimulation with LPS and interleukin-4 (data not shown). A comparison of the concentrations of different immunoglobulin classes in sera did not reveal any significant differences between Deltex-1Δ/Δ and wild-type mice (Fig. 4A). We then compared the humoral responses of mutant and wild-type mice in vivo by challenging them with a T-independent type 2 antigen, TNP-Ficoll, and a T-dependent antigen, TNP-KLH. The titers of IgM and IgG3 were similar for both groups of mice after immunization with TNP-Ficoll (Fig. 4B). Similarly, mutant and wild-type mice displayed comparable IgM, IgG1, IgG2a, IgG2b, and IgG3 anti-TNP antibody titers after primary and secondary immunizations with TNP-KLH (Fig. 4C).
FIG. 4.
Deltex-1Δ/Δ mice have normal basal immunoglobulin levels in serum and mount normal humoral responses. The plotted values represent concentrations in serum for each mouse relative to the average concentration of all wild-type mice. (A) Concentrations of indicated serum immunoglobulin isotypes of 10 Deltex-1Δ/Δ (filled circles) and 10 littermate (open circles) control mice as measured by ELISA. (B) T-independent type 2 response. Seven wild-type and seven Deltex-1Δ/Δ mice were immunized with TNP-Ficoll, and their sera were assayed for the presence of TNP-specific antibodies of the IgM and IgG3 isotypes by ELISA. (C) T-dependent response. Eight wild-type and eight Deltex-1Δ/Δ mice were immunized with TNP-KLH and boosted at day 21, as marked by arrows. Sera were quantified for the presence of TNP-specific antibodies of the IgM, IgG1, IgG2a, IgG2b, and IgG3 isotypes by ELISA.
Deltex-1 was found to be strongly expressed in hypermutating lymphocytes of sheep ileal Peyer's patches and human germinal-center centroblasts (13). We therefore investigated whether somatic hypermutation occurs normally in Deltex-1Δ/Δ mice. To this end, we immunized mice with the hapten phenyl-oxazolone, which elicits a well-characterized antibody response, and sequenced the rearranged VκOx1 gene segments of B220+ PNAhi splenic B cells. Deltex-1Δ/Δ mice showed a mutation rate that was equivalent to that of wild-type mice (data not shown).
Together, these data show that Deltex-1Δ/Δ mice mount normal T-dependent and T-independent type 2 humoral responses in vivo.
KIAA0937 is a fourth mammalian Deltex protein (Deltex-4).
The absence of an obvious phenotype for mice lacking the Deltex-1 RING finger domain prompted us to study the expression profiles of other Deltex family members that may have redundant functions. Three mouse Deltex genes were initially described (18). While the Deltex-2 protein sequence is quite similar to that of Deltex-1, Deltex-3 is far more divergent since it lacks domain I and does not bind Notch proteins in vitro (18). Thus, only Deltex-2 is likely to compensate for the absence of Deltex-1.
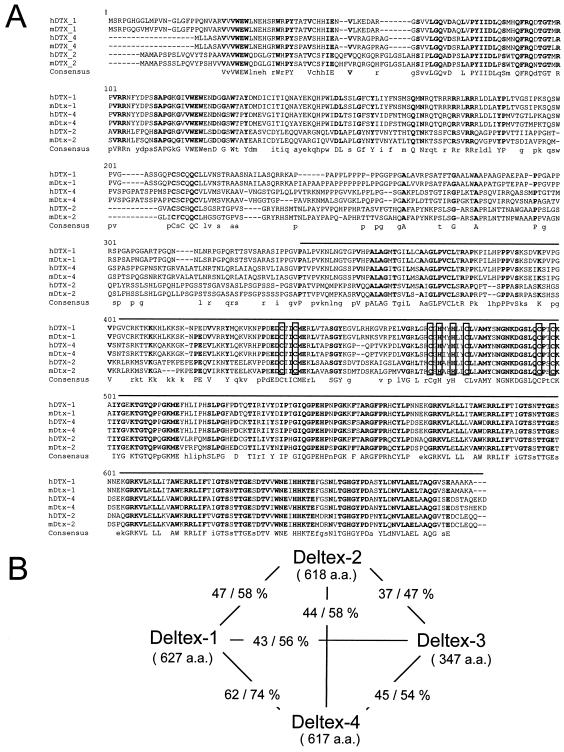
A chicken Deltex gene was cloned (cDTX2) and shown to be an orthologue of human KIAA0937 (10). KIAA0937 was therefore called Deltex-2. A sequence comparison actually demonstrated that KIAA0937 (and its mouse orthologue [NM_172442]) is a genuine fourth mammalian Deltex gene which encodes a protein with a sequence even closer than the Deltex-2 sequence to that of Deltex-1 (Fig. 5A and B). Therefore, we propose that this gene be named Deltex-4.
FIG. 5.
Deltex-4, a new member of the deltex gene family. (A) Comparison of human and mouse Deltex-1 (accession no. NP_004407 and BAB18939), Deltex-2 (NP_065943 and BAB18940), and Deltex-4 (XP_166213 and AAH58647) proteins, performed with the Multalin program (4). Conserved amino acids are indicated with bold uppercase letters in the consensus sequence line, with amino acids conserved between two Deltex members shown in lowercase. Open boxes represent the RING finger motif. The line above the sequence indicates the region that is deleted in Deltex-1Δ/Δ mice. (B) Two-by-two comparison of mouse Deltex-1 (BAB18939), Deltex-2 (BAB18940), Deltex-3 (BAB18942), and Deltex-4 (AAH58647) proteins, performed with the BLAST program. The number of amino acids and identity (left) and similarity (right) values are indicated for each pair of proteins.
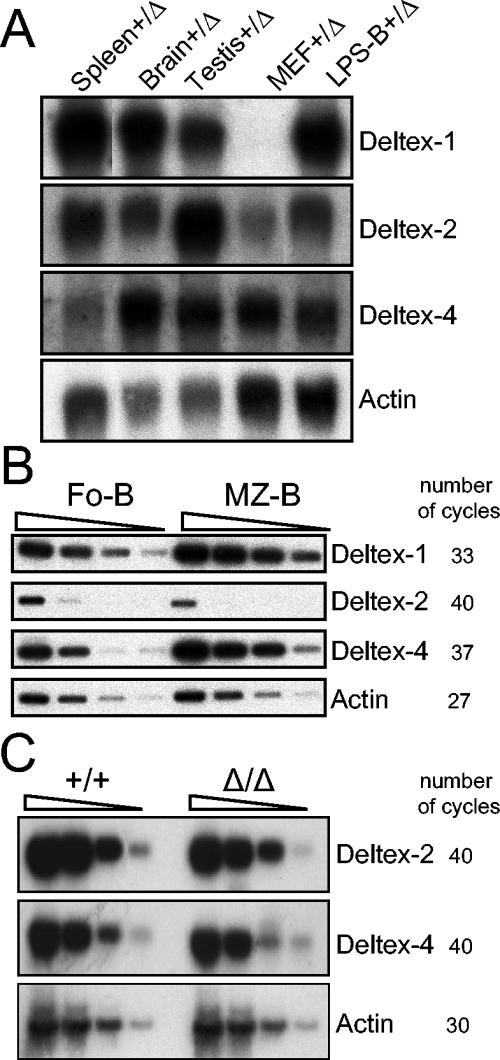
We first determined the relative levels of expression of Deltex-1, Deltex-2, and Deltex-4 in different adult organs by Northern blotting. All three Deltex genes had very different, though partially overlapping, expression patterns (Fig. 6A). A search of expressed sequence tag (EST) sequences revealed that human Deltex-4 (XM_166213) is expressed in many fetal, adult, and cancerous tissues, particularly in the brain (35 ESTs), heart (17 ESTs), colon (11 ESTs), stomach (8 ESTs), and lungs (7 ESTs). Mouse ESTs for Deltex-4 can be found in the brain (38 ESTs), eyes (9 ESTs), and thymus (4 ESTs), as well as other organs. We then determined the relative levels of Deltex-1, Deltex-2, and Deltex-4 in marginal-zone B cells and follicular B cells by semiquantitative RT-PCR. As previously described, Deltex-1 was expressed far more (ninefold) in marginal-zone B cells (Fig. 6B). Interestingly, Deltex-4 was hardly detectable in the spleen by Northern blotting. However, this gene showed a similar expression bias (ninefold) in favor of marginal-zone B cells (Fig. 6B). In contrast, Deltex-2 was hardly detectable in any splenic B-cell subset (Fig. 6B), suggesting that T cells are major contributors to its splenic expression level. We next investigated whether Deltex-1 inactivation induces the upregulation of a Deltex-2 or Deltex-4 transcript that could compensate for its absence. Deltex-2 and Deltex-4 transcript levels remained unchanged in the brains, testes, and spleens of Deltex-1Δ/Δ mice and were comparable to those in wild-type mice (Fig. 6C and data not shown).
FIG. 6.
Expression profiles of Deltex-1, Deltex-2, and Deltex-4 genes. (A) Northern blot analysis of mouse Deltex-1, Deltex-2, and Deltex-4 expression. Spleens, brains, testes, mouse embryonic fibroblasts (MEF), and LPS-stimulated B cells (LPS-B) were prepared from Deltex-1+/Δ mice. The exposure times were 7 days (Deltex-1), 5 days (Deltex-2), and 6 days (Deltex-4). Normalization was performed by use of an actin probe. (B) Semiquantitative RT-PCR analysis of Deltex-1, Deltex-2, and Deltex-4 expression in follicular B cells (FoB; CD19+ CD21int CD23hi) and marginal-zone B cells (MZ-B; C19+ CD21hi CD23−/lo). Threefold serial dilutions of reverse transcription products were analyzed. The number of cycles used for each reaction is indicated. (C) Semiquantitative RT-PCR analysis of Deltex-2 and Deltex-4 expression in splenic cells from Deltex-1+/+ and Deltex-1Δ/Δ mice. Tenfold serial dilutions of reverse transcription products were analyzed. The number of cycles used for each reaction is indicated.
DISCUSSION
During the course of cDNA subtraction, we identified Deltex-1, a modulator of the Notch signaling pathway, as being highly expressed in human and sheep germinal-center B cells (13). Meanwhile, Deltex-1 was shown to be highly expressed in mouse marginal-zone B cells (41), and overexpression studies suggested that Deltex-1 promotes commitment to the B-cell lineage (16). Moreover, its expression profile indicated a potential role for Deltex-1 in T-cell differentiation (6, 31, 34) and several other developmental processes (5, 15, 18, 26, 30, 43).
For this study, we generated a mouse strain with a defect in the Deltex-1 gene. We chose to delete the C-terminal RING finger domain responsible for the ubiquitin ligase activity of Deltex-1 (44). Since residual expression of the mRNA coding for the N-terminal half of the protein was observed in the mutant mice thus obtained, we refer to these strains as having a deletion of the RING finger domain (Deltex-1Δ/Δ). Our results demonstrated that this domain is dispensable for mouse development and normal immune system functions. Firstly, Deltex-1Δ/Δ mice have normal lymphoid development in the thymus and bone marrow, and peripheral B- and T-cell subpopulations are present in expected proportions. In particular, neither marginal-zone B cells nor germinal-center B cells were affected by this mutation. Secondly, Deltex-1Δ/Δ mice mount efficient T-independent type 2 and T-dependent humoral immune responses, which suggests that the Deltex-1 RING finger domain is also dispensable for terminal B-cell differentiation and function. Considering the potential role of Deltex-1 in lymphopoiesis and embryonic development, these results are quite surprising. Three hypotheses can account for this lack of an overt phenotype.
One explanation may be that the Deltex-1 RING finger domain is dispensable for Deltex-1 functions in vivo. Prior experiments supported this hypothesis, since Deltex domain I was able to rescue the deltex phenotype of Drosophila mutants on its own (25). However, the nature of the mutation has not been determined for these mutants, which are likely hypomorphic and may be specifically impaired in some function of Deltex that relies on the sole domain I. Similarly, the overexpression of Deltex-1 domain I in cell lines has been shown to antagonize the transcriptional activity of the NICD/RBP-J complex (16). However, several articles have reported that Deltex proteins lacking a RING finger domain behave as a weak form of Deltex (25, 27) or even as a dominant-negative form of Deltex in vitro (53) and in vivo (5, 8, 15). Lastly, it should be noted that experiments investigating the effects of Deltex-1 on NICD overexpression may not be fully relevant and may result in nonphysiological effects, such as competition with other factors for binding to Notch. Indeed, Deltex is likely to act downstream of full-length Notch and upstream of an activated NICD (27). Since Deltex-1 is a bona fide ubiquitin ligase (44), one possibility is that Deltex-1 ubiquitinates a membrane-anchored form of Notch through its RING finger to control its stability and/or subcellular localization (42). Moreover, it should be noted that the level of the truncated Deltex-1 transcript that we observed in Deltex-1Δ/Δ mice was quite low compared to that of the normal transcript in wild-type mice. Preliminary data showed that excision of the Neor cassette in B lymphocytes by mating Deltex-1Δ/Δ mice with CD19-CRE mice results in a stronger decrease in the expression of the truncated mRNAs, with still no alteration in early and late B-cell development (our unpublished results). We therefore think that the lack of a phenotype observed for Deltex-1Δ/Δ mice is unlikely to originate from incomplete inactivation of the gene.
A second hypothesis is that gene redundancy compensates for the absence of Deltex-1. Deltex-1 does indeed belong to a multigenic family (18). During the course of this study, we identified a fourth Deltex gene, Deltex-4, which is the closest homolog of Deltex-1, with 62% identity at the protein level. Deltex-2 also has a high level of homology with Deltex-1 (47% identity), whereas Deltex-3 is more divergent and lacks the putative Notch-binding domain. Since the expression profile of Deltex-2 overlaps only partially with that of Deltex-1, this gene appears unlikely to compensate for the loss of Deltex-1 function, at least in the immune system. Interestingly, Deltex-4 showed 5- to 10-fold higher expression in splenic marginal-zone B cells than in follicular B cells, similar to Deltex-1. However, its absolute expression level was quite low, as it was barely detectable in total spleens by Northern blotting. Moreover, its expression was not upregulated in Deltex-1Δ/Δ animals. It is therefore unclear for the moment whether Deltex-4 and Deltex-1 have redundant functions in lymphoid tissues.
Another possibility is that the Deltex-1 deficiency is compensated for by an unrelated protein that displays functional convergence, i.e., by other ubiquitin ligases that substitute for the Deltex-1 function. Such a situation might indeed exist between Neuralized and Mind Bomb, two unrelated ubiquitin ligases that are both involved in the endocytosis of a Notch ligand (discussed in reference 22).
If an imperfect compensation process takes place, one might envision the possibility of uncovering more subtle phenotypes associated with specific differentiation processes, possibly by mating Deltex-1Δ/Δ mice with strains that are heterozygous for a mutation in another component of the Notch pathway.
Acknowledgments
We thank Annie De Smet for excellent technical assistance with ES cell handling and cytometry analysis, Rachid Zoubairi for mouse breeding and handling, Patricia Wattier for preparation of histological sections, Corinne Garcia for performing cell sorting, and the Service d’Experimentation animale et de transgénèse for the generation of mutant mice. We thank Simon Fillatreau for his advice on immunizations and ELISA tests, Barbara Bertocci for advice on cytometry analysis, and Michel Cogné for help and advice on the induction of isotype switching in vitro. We thank Warren Pear for providing the E2A reporter and Meinrad Busslinger for providing CD19-CRE mice. We also thank Auriel Dahan for his continuous support throughout this work.
This work was supported by grants from the Ministère de la Recherche (ACI Biologie du Développement et Physiologie Intégrative) and the Fondation Princesse Grace. S.S. was supported by grants from the Ministère de l'Education Nationale de la Recherche et de la Technologie and the Association pour la Recherche contre le Cancer.
REFERENCES
- 1.Amsen, D., J. M. Blander, G. R. Lee, K. Tanigaki, T. Honjo, and R. A. Flavell. 2004. Instruction of distinct CD4 T helper cell fates by different notch ligands on antigen-presenting cells. Cell 117:515-526. [DOI] [PubMed] [Google Scholar]
- 2.Artavanis-Tsakonas, S., M. D. Rand, and R. J. Lake. 1999. Notch signaling: cell fate control and signal integration in development. Science 284:770-776. [DOI] [PubMed] [Google Scholar]
- 3.Bertocci, B., A. De Smet, E. Flatter, A. Dahan, J. C. Bories, C. Landreau, J. C. Weill, and C. A. Reynaud. 2002. DNA polymerases mu and lambda are dispensable for Ig gene hypermutation. J. Immunol. 168:3702-3706. [DOI] [PubMed] [Google Scholar]
- 4.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cui, X. Y., Q. D. Hu, M. Tekaya, Y. Shimoda, B. T. Ang, D. Y. Nie, L. Sun, W. P. Hu, M. Karsak, T. Duka, Y. Takeda, L. Y. Ou, G. S. Dawe, F. G. Yu, S. Ahmed, L. H. Jin, M. Schachner, K. Watanabe, Y. Arsenijevic, and Z. C. Xiao. 2004. NB-3/Notch1 pathway via Deltex1 promotes neural progenitor cell differentiation into oligodendrocytes. J. Biol. Chem. 279:25858-25865. [DOI] [PubMed] [Google Scholar]
- 6.Deftos, M. L., Y. W. He, E. W. Ojala, and M. J. Bevan. 1998. Correlating notch signaling with thymocyte maturation. Immunity 9:777-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eagar, T. N., Q. Tang, M. Wolfe, Y. He, W. S. Pear, and J. A. Bluestone. 2004. Notch 1 signaling regulates peripheral T cell activation. Immunity 20:407-415. [DOI] [PubMed] [Google Scholar]
- 8.Endo, Y., N. Osumi, and Y. Wakamatsu. 2003. Deltex/Dtx mediates NOTCH signaling in regulation of Bmp4 expression in cranial neural crest formation during avian development. Dev. Growth Differ. 45:241-248. [DOI] [PubMed] [Google Scholar]
- 9.Frey, S., B. Bertocci, F. Delbos, L. Quint, J. C. Weill, and C. A. Reynaud. 1998. Mismatch repair deficiency interferes with the accumulation of mutations in chronically stimulated B cells and not with the hypermutation process. Immunity 9:127-134. [DOI] [PubMed] [Google Scholar]
- 10.Frolova, E., and D. Beebe. 2000. The expression pattern of a novel Deltex homologue during chicken embryogenesis. Mech. Dev. 92:285-289. [DOI] [PubMed] [Google Scholar]
- 11.Gorman, M. J., and J. R. Girton. 1992. A genetic analysis of deltex and its interaction with the Notch locus in Drosophila melanogaster. Genetics 131:99-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gupta-Rossi, N., O. Le Bail, H. Gonen, C. Brou, F. Logeat, E. Six, A. Ciechanover, and A. Israel. 2001. Functional interaction between SEL-10, an F-box protein, and the nuclear form of activated Notch1 receptor. J. Biol. Chem. 276:34371-34378. [DOI] [PubMed] [Google Scholar]
- 13.Gupta-Rossi, N., S. Storck, P. J. Griebel, C. A. Reynaud, J. C. Weill, and A. Dahan. 2003. Specific over-expression of deltex and a new Kelch-like protein in human germinal center B cells. Mol. Immunol. 39:791-799. [DOI] [PubMed] [Google Scholar]
- 14.Harman, B. C., E. J. Jenkinson, and G. Anderson. 2003. Entry into the thymic microenvironment triggers Notch activation in the earliest migrant T cell progenitors. J. Immunol. 170:1299-1303. [DOI] [PubMed] [Google Scholar]
- 15.Hu, Q. D., B. T. Ang, M. Karsak, W. P. Hu, X. Y. Cui, T. Duka, Y. Takeda, W. Chia, N. Sankar, Y. K. Ng, E. A. Ling, T. Maciag, D. Small, R. Trifonova, R. Kopan, H. Okano, M. Nakafuku, S. Chiba, H. Hirai, J. C. Aster, M. Schachner, C. J. Pallen, K. Watanabe, and Z. C. Xiao. 2003. F3/contactin acts as a functional ligand for Notch during oligodendrocyte maturation. Cell 115:163-175. [DOI] [PubMed] [Google Scholar]
- 16.Izon, D. J., J. C. Aster, Y. He, A. Weng, F. G. Karnell, V. Patriub, L. Xu, S. Bakkour, C. Rodriguez, D. Allman, and W. S. Pear. 2002. Deltex1 redirects lymphoid progenitors to the B cell lineage by antagonizing Notch1. Immunity 16:231-243. [DOI] [PubMed] [Google Scholar]
- 17.Jehn, B. M., I. Dittert, S. Beyer, K. von der Mark, and W. Bielke. 2002. c-Cbl binding and ubiquitin-dependent lysosomal degradation of membrane-associated Notch1. J. Biol. Chem. 277:8033-8040. [DOI] [PubMed] [Google Scholar]
- 18.Kishi, N., Z. Tang, Y. Maeda, A. Hirai, R. Mo, M. Ito, S. Suzuki, K. Nakao, T. Kinoshita, T. Kadesch, C. Hui, S. Artavanis-Tsakonas, H. Okano, and K. Matsuno. 2001. Murine homologs of deltex define a novel gene family involved in vertebrate Notch signaling and neurogenesis. Int. J. Dev. Neurosci. 19:21-35. [DOI] [PubMed] [Google Scholar]
- 19.Kuroda, K., H. Han, S. Tani, K. Tanigaki, T. Tun, T. Furukawa, Y. Taniguchi, H. Kurooka, Y. Hamada, S. Toyokuni, and T. Honjo. 2003. Regulation of marginal zone B cell development by MINT, a suppressor of Notch/RBP-J signaling pathway. Immunity 18:301-312. [DOI] [PubMed] [Google Scholar]
- 20.Lai, E. C. 2004. Notch signaling: control of cell communication and cell fate. Development 131:965-973. [DOI] [PubMed] [Google Scholar]
- 21.Lai, E. C. 2002. Protein degradation: four E3s for the notch pathway. Curr. Biol. 12:R74-R78. [DOI] [PubMed] [Google Scholar]
- 22.Le Borgne, R., and F. Schweisguth. 2003. Notch signaling: endocytosis makes delta signal better. Curr. Biol. 13:R273-R275. [DOI] [PubMed] [Google Scholar]
- 23.Loder, F., B. Mutschler, R. J. Ray, C. J. Paige, P. Sideras, R. Torres, M. C. Lamers, and R. Carsetti. 1999. B cell development in the spleen takes place in discrete steps and is determined by the quality of B cell receptor-derived signals. J. Exp. Med. 190:75-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinez Arias, A., V. Zecchini, and K. Brennan. 2002. CSL-independent Notch signalling: a checkpoint in cell fate decisions during development? Curr. Opin. Genet. Dev. 12:524-533. [DOI] [PubMed] [Google Scholar]
- 25.Matsuno, K., R. J. Diederich, M. J. Go, C. M. Blaumueller, and S. Artavanis-Tsakonas. 1995. Deltex acts as a positive regulator of Notch signaling through interactions with the Notch ankyrin repeats. Development 121:2633-2644. [DOI] [PubMed] [Google Scholar]
- 26.Matsuno, K., D. Eastman, T. Mitsiades, A. M. Quinn, M. L. Carcanciu, P. Ordentlich, T. Kadesch, and S. Artavanis-Tsakonas. 1998. Human deltex is a conserved regulator of Notch signalling. Nat. Genet. 19:74-78. [DOI] [PubMed] [Google Scholar]
- 27.Matsuno, K., M. Ito, K. Hori, F. Miyashita, S. Suzuki, N. Kishi, S. Artavanis-Tsakonas, and H. Okano. 2002. Involvement of a proline-rich motif and RING-H2 finger of Deltex in the regulation of Notch signaling. Development 129:1049-1059. [DOI] [PubMed] [Google Scholar]
- 28.McGill, M. A., and C. J. McGlade. 2003. Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. J. Biol. Chem. 278:23196-23203. [DOI] [PubMed] [Google Scholar]
- 29.McKenzie, G. J., L. L. Young, E. Briend, J. R. Lamb, M. J. Dallman, and B. R. Champion. 2003. Notch signalling in the regulation of peripheral T-cell function. Semin. Cell Dev. Biol. 14:127-134. [DOI] [PubMed] [Google Scholar]
- 30.Mitsiadis, T. A., O. Gayet, N. Zhang, and P. Carroll. 2001. Expression of Deltex1 during mouse embryogenesis: comparison with Notch1, 2 and 3 expression. Mech. Dev. 109:399-403. [DOI] [PubMed] [Google Scholar]
- 31.Ng, W. F., P. J. Duggan, F. Ponchel, G. Matarese, G. Lombardi, A. D. Edwards, J. D. Isaacs, and R. I. Lechler. 2001. Human CD4(+) CD25(+) cells: a naturally occurring population of regulatory T cells. Blood 98:2736-2744. [DOI] [PubMed] [Google Scholar]
- 32.Nie, L., M. Xu, A. Vladimirova, and X. H. Sun. 2003. Notch-induced E2A ubiquitination and degradation are controlled by MAP kinase activities. EMBO J. 22:5780-5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oberg, C., J. Li, A. Pauley, E. Wolf, M. Gurney, and U. Lendahl. 2001. The Notch intracellular domain is ubiquitinated and negatively regulated by the mammalian Sel-10 homolog. J. Biol. Chem. 276:35847-35853. [DOI] [PubMed] [Google Scholar]
- 34.Ordentlich, P., A. Lin, C. P. Shen, C. Blaumueller, K. Matsuno, S. Artavanis-Tsakonas, and T. Kadesch. 1998. Notch inhibition of E47 supports the existence of a novel signaling pathway. Mol. Cell. Biol. 18:2230-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pampeno, C. L., and D. Meruelo. 1996. A novel cDNA transcript expressed in fractionated X-irradiation-induced murine thymomas. Cell Growth Differ. 7:1113-1123. [PubMed] [Google Scholar]
- 36.Pui, J. C., D. Allman, L. Xu, S. DeRocco, F. G. Karnell, S. Bakkour, J. Y. Lee, T. Kadesch, R. R. Hardy, J. C. Aster, and W. S. Pear. 1999. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity 11:299-308. [DOI] [PubMed] [Google Scholar]
- 37.Radtke, F., A. Wilson, S. J. Mancini, and H. R. MacDonald. 2004. Notch regulation of lymphocyte development and function. Nat. Immunol. 5:247-253. [DOI] [PubMed] [Google Scholar]
- 38.Radtke, F., A. Wilson, G. Stark, M. Bauer, J. van Meerwijk, H. R. MacDonald, and M. Aguet. 1999. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity 10:547-558. [DOI] [PubMed] [Google Scholar]
- 39.Ramain, P., K. Khechumian, L. Seugnet, N. Arbogast, C. Ackermann, and P. Heitzler. 2001. Novel Notch alleles reveal a Deltex-dependent pathway repressing neural fate. Curr. Biol. 11:1729-1738. [DOI] [PubMed] [Google Scholar]
- 40.Robey, E., D. Chang, A. Itano, D. Cado, H. Alexander, D. Lans, G. Weinmaster, and P. Salmon. 1996. An activated form of Notch influences the choice between CD4 and CD8 T cell lineages. Cell 87:483-492. [DOI] [PubMed] [Google Scholar]
- 41.Saito, T., S. Chiba, M. Ichikawa, A. Kunisato, T. Asai, K. Shimizu, T. Yamaguchi, G. Yamamoto, S. Seo, K. Kumano, E. Nakagami-Yamaguchi, Y. Hamada, S. Aizawa, and H. Hirai. 2003. Notch2 is preferentially expressed in mature B cells and indispensable for marginal zone B lineage development. Immunity 18:675-685. [DOI] [PubMed] [Google Scholar]
- 42.Schweisguth, F. 2004. Notch signaling activity. Curr. Biol. 14:R129-R138. [PubMed] [Google Scholar]
- 43.Sestan, N., S. Artavanis-Tsakonas, and P. Rakic. 1999. Contact-dependent inhibition of cortical neurite growth mediated by notch signaling. Science 286:741-746. [DOI] [PubMed] [Google Scholar]
- 44.Takeyama, K., R. C. Aguiar, L. Gu, C. He, G. J. Freeman, J. L. Kutok, J. C. Aster, and M. A. Shipp. 2003. The BAL-binding protein BBAP and related Deltex family members exhibit ubiquitin-protein isopeptide ligase activity. J. Biol. Chem. 278:21930-21937. [DOI] [PubMed] [Google Scholar]
- 45.Tanigaki, K., H. Han, N. Yamamoto, K. Tashiro, M. Ikegawa, K. Kuroda, A. Suzuki, T. Nakano, and T. Honjo. 2002. Notch-RBP-J signaling is involved in cell fate determination of marginal zone B cells. Nat. Immunol. 3:443-450. [DOI] [PubMed] [Google Scholar]
- 46.Tanigaki, K., M. Tsuji, N. Yamamoto, H. Han, J. Tsukada, H. Inoue, M. Kubo, and T. Honjo. 2004. Regulation of alphabeta/gammadelta T cell lineage commitment and peripheral T cell responses by Notch/RBP-J signaling. Immunity 20:611-622. [DOI] [PubMed] [Google Scholar]
- 47.Torres, R., and R. Kühn. 1997. Laboratory protocols for conditional gene targeting. Oxford University Press, Oxford, United Kingdom.
- 48.Wilson, A., H. R. MacDonald, and F. Radtke. 2001. Notch 1-deficient common lymphoid precursors adopt a B cell fate in the thymus. J. Exp. Med. 194:1003-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Witt, C. M., W. J. Won, V. Hurez, and C. A. Klug. 2003. Notch2 haploinsufficiency results in diminished B1 B cells and a severe reduction in marginal zone B cells. J. Immunol. 171:2783-2788. [DOI] [PubMed] [Google Scholar]
- 50.Wolfer, A., A. Wilson, M. Nemir, H. R. MacDonald, and F. Radtke. 2002. Inactivation of Notch1 impairs VDJbeta rearrangement and allows pre-TCR-independent survival of early alpha beta lineage thymocytes. Immunity 16:869-879. [DOI] [PubMed] [Google Scholar]
- 51.Wu, G., S. Lyapina, I. Das, J. Li, M. Gurney, A. Pauley, I. Chui, R. J. Deshaies, and J. Kitajewski. 2001. SEL-10 is an inhibitor of notch signaling that targets notch for ubiquitin-mediated protein degradation. Mol. Cell. Biol. 21:7403-7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu, T., and S. Artavanis-Tsakonas. 1990. Deltex, a locus interacting with the neurogenic genes, Notch, Delta and mastermind in Drosophila melanogaster. Genetics 126:665-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamamoto, N., S. Yamamoto, F. Inagaki, M. Kawaichi, A. Fukamizu, N. Kishi, K. Matsuno, K. Nakamura, G. Weinmaster, H. Okano, and M. Nakafuku. 2001. Role of Deltex-1 as a transcriptional regulator downstream of the Notch receptor. J. Biol. Chem. 276:45031-45040. [DOI] [PubMed] [Google Scholar]