Abstract
Deltex is known as a Notch signal mediator, but its physiological action mechanism is poorly understood. Here we identified a new regulatory role of Deltex in T-cell activation. Deltex expression was constitutive in resting T cells and was reduced upon T-cell receptor (TCR)-stimulated activation. The biological role of Deltex is supported by the enhanced T-cell activation when Deltex1 was down-regulated by small interfering RNA. Overexpression of Deltex1 suppressed T-cell activation but not the proximal TCR activation events. The impaired activation of mitogen-activated protein kinase by Deltex could be partly attributed to a selective down-regulation of MEKK1 protein in T cells. We further found that Deltex promoted degradation of the C-terminal catalytic fragment of MEKK1 [MEKK1(C)]. Deltex1 interacted directly with MEKK1(C) and stimulated the ubiquitination of MEKK1(C) as shown by in vivo and in vitro ubiquitination analysis. Therefore, MEKK1(C), the dominant form of MEKK1 in T cells, is a target of Deltex E3 ubiquitin ligase. Our results reveal a novel mechanism as to how Deltex selectively suppresses T-cell activation through degradation of a key signaling molecule, MEKK1.
Deltex was first identified in Drosophila spp. for its interaction with Notch. Deltex contains an N-terminal basic region, for binding to the ankyrin repeats of the Notch intracellular region, a proline-rich motif, and a RING finger domain at its C terminus. Deltex is a positive modulator of Notch in Drosophila (30). The mammalian Deltex1 is the homologue most closely related to the Drosophila Deltex (31). Deltex1 has been shown to be a transcription target gene of Notch (7). Interestingly, overexpression of Deltex1 directs lymphoid development towards B-cell production concurrent with suppression of T-cell development (17, 53). Consistent with these observations, Deltex1 is overexpressed in marginal zone B cells (38) and in germinal center B cells (12). The precise action mechanism of Deltex remains elusive (28). Deltex binds to Grb2 and has been shown to inhibit E2A (31). Deltex inhibits Jun-mediated transcription, suggested to be at the stage of Ras-dependent Jun N-terminal protein kinase (JNK) activation (35). Deltex1, acting as a Notch downstream transcription regulator, has also been shown to interact with the transcription coactivator p300 and to inhibit transcription activation mediated by the neural specific transcription factor MASH1 (50). However, these reported effects cannot account for the biological events mediated by Deltex (17).
The ubiquitin-proteasome system controls intracellular protein degradation (13, 37). Ubiquitination starts from ATP-dependent and ubiquitin-activating enzyme (E1)-mediated ubiquitin conjugation to E1, followed by ubiquitin-conjugation enzyme (E2)-catalyzed transesterification of the E1-attached ubiquitin to E2. Ubiquitin-protein isopeptide ligase (E3), binding specifically to both E2 and the target protein, then promotes the transfer of ubiquitin to the target protein. From their structures, E3 enzymes have been classified into two distinct types: those possessing a HECT domain and those with a RING finger (18, 20). RING fingers are zinc-binding motifs with eight conserved cysteine and histidine residues. As RING finger proteins, Deltex family members were recently found to possess ubiquitin-protein isopeptide ligase (E3) activity (41). Through its own E3 activity, Deltex promotes self-ubiquitination (41); however, the in vivo substrate of DTX E3 ligase remains unknown.
Mitogen-activated protein kinase (MAPK) cascades mediate signal transduction from external stimulus to the nuclei (25, 36). MAP kinase kinase kinase 1 (MEKK1) is a 195-kDa protein with a C-terminal protein kinase domain and a large noncatalytic N terminus (26, 48). MEKK1 is processed by caspase-3-like protease to generate the 91-kDa C terminus kinase domain [MEKK1(C)] (43). Although MEKK1 is a primary activator for JNK through phosphorylation of stress-activated protein kinase/extracellular signal-regulated kinase (ERK) kinase (SEK, MKK4) (33, 43, 48, 51), MEKK1 can also regulate the ERK pathway (22, 49). Target gene disruption of MEKK1 has been shown to eliminate stress-induced JNK activation and reduce hyperosmolarity- and serum-stimulated ERK activation (47, 52). MEKK1 binds to a number of signaling proteins such as JNK, ERK2, MKK4, Raf-1, RhoA, TRAF2, and germinal center kinase (2, 3, 10, 22, 47, 49). Among these interactions, the simultaneous binding of MEKK1 to TRAF2 and germinal center kinase results in the activation of MEKK1 (2, 3). However, the in vivo significance of most MEKK1-dependent interactions remains unclear (10).
In the present study, we observed that Deltex is a physiological regulator of T-cell activation. Deltex inhibited T-cell activation not at the stage of T-cell receptor engagement but at the levels of MAPK activation. One of the defects identified in Deltex-overexpressing T cells was the diminished levels of MEKK1(C). We further demonstrated that Deltex is a ubiquitin E3 ligase for MEKK1(C). Our results reveal a novel function of Deltex through selective degradation of MEKK1.
MATERIALS AND METHODS
Reagents and antibodies.
12-O-Tetradecanoylphorbol-13-acetate and A23187 were purchased from Sigma (St. Louis, Mo.). MG132 was obtained from Calbiochem (San Diego, Calif.). The following antibodies were obtained from Santa Cruz Biotech (Santa Cruz, Calif.): ERK2 (C-14), JNK1 (C-17), c-Jun (N), p-c-Jun (KM-1), p38α (C-20), c-Fos (H-125), MEKK1 (C-22), MEKK2 (N-19), HPK1 (N-19), MKK1 (H-8), MKK3 (I-20), MKK4 (C-20), MKK6 (K-19), MKK7 (C-19), Ub (FL-76), Myc (9E10), His (H-3). Anti-Raf-1 (c-Raf-1, clone 52) was obtained from Transduction Lab. Anti-phospho-ERK (T202/Y204), anti-phospho (T180/Y182)-p38 MAPK, anti-phosphor-LAT (Y191), anti-IκBα, and anti-phospho-PKCθ (T538) were purchased from Cell Signaling (Beverly, Mass.). Goat anti-glutathione S-transferase (GST) was acquired from Amersham (Buckinghamshire, United Kingdom). Anti-β-tubulin was obtained from Upstate (Lake Placid, N.Y.). Rabbit polyclonal serum against GST-Deltex1 was obtained from IgMedica Biotech (Taipei, Taiwan). Interleukin-2 (IL-2)-chloramphenicol acetyltransferase (CAT) (34) was a gift from Ellen Rothenberg (California Institute of Technology, Pasadena), and CAT assays were conducted as previously described (14). Full-length MEKK1 cDNA (48) was a gift from Melanie H. Cobb (University of Texas, Dallas). MEKK1 active-form MEKK (3′)-5′EE-CMV [abbreviated as MEKK1(C) here] (51) was a gift from Dennis Templeton (Case Western Reserve University, Cleveland, Ohio). PCR primers for detecting Deltex1 are as follows: 5′ GTA AGG CTT CAA GGG GTC GCT and 3′ CTC AGC TTG ATG CGT GTA TAG CC. PCR primers for detecting MEKK1 are as follows: 5′ TGG CTG TGA AAC AGG TGA and 3′ AAG TTC TAA GCA GCG CAC.
Retroviral infection of T cells.
Mouse Deltex1 cDNA (24) was a gift from Hideyuki Okano (Keiko University, Tokyo, Japan). pGCIRES-YFP, a homologue of pGCIRES-GFP (5), was a gift from Gina Costa (Stanford University, Stanford, Calif.) and was obtained through Nan-Shih Liao (Academia Sinica, Taipei, Taiwan). Myc-tagged Deltex1 was cloned into pGC-IRES-YFP. Retroviruses were produced by transfecting Phoenix-Eco cells (gifts of Garry P. Nolan, Stanford University) with 10 μg of pGC-IRES-YFP or pGC-DTX1-IRES-YFP plasmids. Phoenix cell supernatants containing retrovirus were collected 48 h and 72 h after transfection. Viral titers were determined using NIH 3T3 cells, and virus stocks with titers greater than 1 × 106 were used for spin infection of DO11.10 T and splenic T cells (9). Splenic T cells were isolated by extensive panning against anti-mouse immunoglobulin and then activated by plate-bound anti-CD3 plus anti-CD28 in the presence of IL-2 (2 U/ml) for 2 days. The infection efficiency was 60% for DO11.10 and 20% for activated splenic T cells. Forty-eight hours after infection, yellow fluorescent protein (YFP)-expressing DO11.10 cells and splenic T cells were isolated by sorting on a FACSVantage SE sytstem (Becton Dickinson, Mountain View, Calif.).
Protein kinase assay.
Cell lysates were prepared 24 h after transfection, and 100 to 200 μg of lysate was precipitated with 1 μl of anti-JNK1 Ab101 (32) (gift of Tse-Hua Tan, Baylor Medical School) followed by 20 μl of protein A-Sepharose. The kinase activity of the immune complexes was determined by using GST-c-Jun (1-79) as substrates (4). The reaction mixtures were resolved on SDS-PAGE, followed by autoradiography, and quantitated by PhosphorImager (Molecular Dynamics).
siRNA.
Murine deltex1 small interfering RNA (siRNA) sequence (D-010470-02; Dharmacon, Lafayette, Colo.) was subcloned into pSuper.gfp/neo (DNAengine, Seattle, Wash.), in which enhanced green fluorescent protein (EGFP) expression was driven by Phosphoglycerate kinase promoter. T cells were transfected with pSuper or pSuper-DTXsiRNA by electroporation. The EGFP-expressing fraction was sorted on a FACSVantage SE 48 h after transfection. A fraction of cells were collected for cell lysate preparation and RNA isolation, and the rest of the cells were stimulated with plate-bound CD3/CD28 antibodies and IL-2 quantitated 16 h after activation. For murine MEKK1, a pool of four specific siRNA duplexes (M-010471-00) were synthesized by Dharmacon. siRNA (500 nM) (containing four duplexes) was mixed with 2 × 106 T cells (DO11.10 or EL4) in 100 μl of Cell Line Nucleofector Solution V (Amaxa, Koeln, Germany), and electroporation was conducted on a Nucleofector. Forty-eight hours after transfection, the levels of MEKK1 and IL-2 production were similarly determined. The densitometry measurements were performed on a LAS-1000 luminescent image analyzer (Fuji Photo Film Co., Tokyo, Japan) with Image Gauge software (version 3.2; Fuji). Quant mode was used to select the reading area and to subtract background. The reading of the sample from the control T cells (mock transfected or pSuper vector transfected) was then set as 1 to get the preliminary n-fold of change for the siRNA-transfected T-cell samples. The final n-fold of change was obtained after normalization with β-tubulin contents.
GST pull-down assay.
His6-MEKK1(C) and His6-MEKK1(FL) were produced by subcloning MEKK1(C) (874 to 1493 amino acids) and MEKK1(FL) (1 to 1493 amino acids), respectively, into pRSET-a (Invitrogen) for N-terminal His6 flanking. Recombinant His6-MEKK1(C) and His6-MEKK1(FL) were then purified on Ni-nitrilotriacetic acid (NTA) agarose (QIAGEN). Cell lysates for the GST pull-down assay were prepared in GSH binding buffer (50 mM potassium phosphate buffer, pH 7.5, 150 mM KCl, 1 mM MgCl2, 10% glycerol, 1% Triton X-100, 1% aprotinin, 1 mM phenylmethylsulfonyl fluoride). Five micrograms of GST-fusion protein (GST-Deltex) was loaded onto GSH agarose (20 μl, 50% slurry) and incubated with 100 to 200 μg of cell lysates overnight at 4°C or with 5 μg of His6-MEKK1(C) or His6-MEKK1(FL) for 2 h at 4°C. GST agarose beads were then washed three times with binding buffer and analyzed on SDS-PAGE.
In vitro ubiquitination assay.
The in vitro ubiquitination reaction mixture contained bovine ubiquitin (1 μg), rabbit E1 (0.2 μg), UbcH5c (0.2 μg), GST-Deltex (0.2 μg), and His6-MEKK1(C) (0.5 μg) in 20 μl of reaction buffer (25 mM Tris, pH 7.5, 50 mM NaCl, 10 mM MgCl2, 2 mM ATP, 0.5 mM dithiothreitol). The reaction proceeded at 30°C for 30 min. Eighty microliters of reaction buffer and 20 μl of 50% slurry Ni-NTA agarose were then added and mixed at 4°C for 2 h to pull down His6-MEKK1(C). Ni-NTA agarose was then washed three times with ice-cold reaction buffer, resuspended in SDS sample buffer, and resolved on SDS-PAGE. The amount of ubiquitin associated was determined by blotting with ubiquitin antibody.
RESULTS
Physiological role of Deltex in T-cell activation.
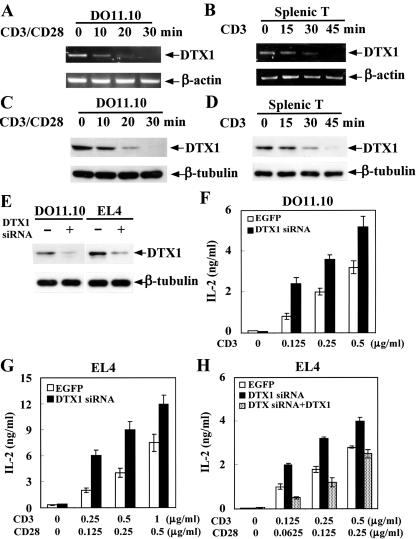
To delineate the potential function of Deltex, we first examined the expression of Deltex T lymphocytes. Deltex1 mRNA and protein were constitutively expressed in resting DO11.10 T hybridomas, EL4 T lymphomas, and normal splenic T cells (Fig. 1A to E). A notable trait of Deltex1 expression is its high sensitivity to T-cell receptor engagement. Anti-CD3 and anti-CD28 treatment resulted in immediate reduction of Deltex levels in DO11.10 T cells, as shown by disappearance of Deltex mRNA 30 min after TCR activation, corresponding to a largely diminished Deltex1 protein level 30 min after TCR ligation (Fig. 1A and C). A similar attenuation of Deltex1 expression was found in splenic T cells upon anti-CD3 stimulation (Fig. 1B and D). TCR-mediated decrease of Deltex suggests a negative role of Deltex in T-cell activation. To further confirm the physiological role of Deltex as a regulator for T-cell activation, we used siRNA to down-regulate the endogenous Deltex1 expression. Transfection of Deltex1-specific siRNA effectively reduced the levels of Deltex1 mRNA and protein in both DO11.10 and EL4 T cells (Fig. 1E; data not shown for mRNA). Down-regulation of Deltex expression resulted in enhanced TCR-stimulated IL-2 production in both DO11.10 and EL4 T cells (Fig. 1F and G). The specificity of Deltex siRNA was illustrated by the fact that expression of Deltex1 reversed the stimulatory effect of Deltex siRNA in EL4 cells (Fig. 1H). Therefore, Deltex is an antagonist of T-cell activation in vivo.
FIG. 1.
Deltex1 is a physiological negative regulator of T-cell activation. (A and B) Deltex1 mRNA expression was constitutive in resting T cells and was diminished by T-cell receptor ligation. DO11.10 T hybridomas (A) and purified splenic T cells (B) were activated by plate-bound anti-CD28 (2.5 μg/ml) and/or anti-CD3 (5 μg/ml), and total RNA was isolated at the indicated time points. Deltex1 (DTX1) mRNA was detected by reverse transcription-PCR. (C and D) Constitutive Deltex1 protein expression was eliminated by TCR stimulation. DO11.10 (C) and purified splenic T cells (D) were activated as in panels A and B. Deltex1 was detected using polyclonal antibody raised against GST-Deltex1. (E-G) Down-regulation of endogenous Deltex1 expression by specific siRNA resulted in enhanced T-cell activation. DO11.10 and EL4 cells were transfected with pSuper vector (EGFP, open column) or pSuper-Deltex1 siRNA (DTX1 siRNA, solid column) by electroporation. Transfected cells, marked by EGFP expression, were isolated 48 h after transfection by sorting on a FACSVantage SE. Total cell lysates were then prepared and Deltex1 protein expression was assessed by immunoblotting (E). A fraction of the sorted DO11.10 (F) and EL4 (G) cells were stimulated with CD3 (F) or CD3/CD28 (G), and culture supernatants were collected and the amounts of IL-2 produced were determined using the indicator cell line HT-2. (H) Deltex1 expression reversed the effect of Deltex siRNA in T cells. EL4 T cells were transfected with pSuper, pSuper-Deltex1 siRNA, or pSuper-Deltex1 siRNA plus pcDNA4-Deltex (DTX siRNA + DTX, dotted column). Transfected cells were sorted and IL-2 expression was determined as in panel G.
Deltex inhibits TCR-mediated IL-2 production in T cells.
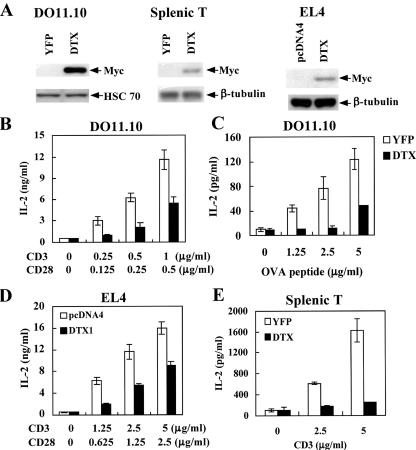
To examine how Deltex modulates the activation of T cells, Myc-tagged murine Deltex1 was introduced into DO11.10, EL4, and preactivated splenic T cells. The retroviral infected DO11.10 and splenic T cells, marked by YFP expression, were sorted on a flow cytometer. The expression of deltex1 was confirmed by Myc-specific antibody (Fig. 2A). T cells were stimulated either with immobilized CD3/CD28 antibodies or with antigenic peptide presented by A20 B cells. The effect of Deltex expression was assessed by difference in activation between YFP- and Deltex1-DO11.10 T cells. The level of IL-2 produced after CD3/CD28 engagement (Fig. 2B) or antigen stimulation (Fig. 2C) was significantly reduced in DO11.10 T cells transduced with Deltex1 compared with the control YFP cells. Similarly, Deltex expression suppressed T-cell activation in EL4 cells (Fig. 2D). Inhibition of T-cell activation by Deltex was not restricted to transformed T cells. Expression of Deltex1 in purified splenic T cells profoundly inhibited CD3-triggered IL-2 secretion (Fig. 2E).
FIG. 2.
Deltex1 suppresses TCR-stimulated IL-2 production. (A) Overexpression of Deltex1 in T cells. DO11.10 and activated splenic T cells were transduced with pGC-DTX-IRES-YFP or pGC-IRES-YFP by retroviral infection. YFP-expressing T cells were sorted on a FACSVantage SE. EL4 T cells were transfected with pcDNA4 or pcDNA4-DTX by electroporation using Nucleofector (Amaxa). Deltex1-Myc expression was confirmed using anti-Myc antibody, with Hsc 70 or β-tubulin as the internal control. (B-D) Deltex1 inhibited TCR-stimulated IL-2 production in DO11.10 and EL4 T cells. DO11.10 (B and C) and EL4 T cells (D), expressing Deltex1 (DTX) (solid column) or YFP/pcDNA4 control (open column), were activated by the indicated concentrations of ovalbumin (323-339) peptide and A20 cells (C) or with anti-CD3 and -CD28 antibody immobilized at the concentration indicated (B and D), and the IL-2 production was quantitated 16 h later. (E) Deltex1 inhibited TCR-stimulated IL-2 production in normal T cells. Splenic T cells expressing Deltex1 or YFP were activated with anti-CD3 and the generated IL-2 was quantitated 16 h after activation.
Deltex suppresses TCR-mediated activation of JNK, ERK, and p38 MAPK but not of LAT and PKCθ.
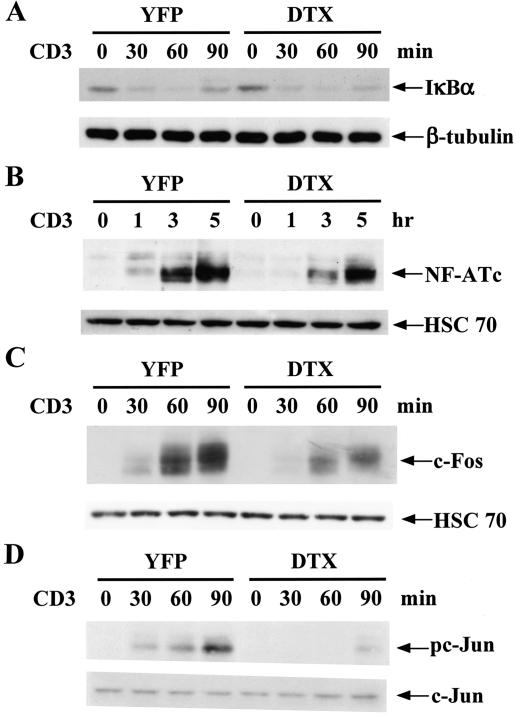
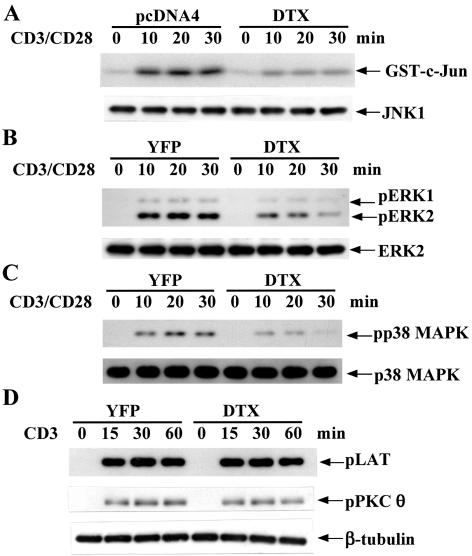
Since the expression of IL-2 in naive T cells is determined primarily by transcription activation of the IL-2 gene promoter (19), we next determined whether activation of the major transcription elements, NF-κB, NFAT, and AP-1 on the IL-2 gene promoter were affected by Deltex1 cotransfection. Deltex did not affect the activation of NF-κB (Fig. 3A), while it partially inhibited the induction of NFATc (Fig. 3B) and c-Fos (Fig. 3C) and effectively prevented c-Jun phosphorylation (Fig. 3D). Consistent with results on the nuclear appearance of the specific transcription factor, overexpression of Deltex in T cells repressed the activation of IL-2-CAT and AP-1-CAT (data not shown). JNK, ERK, and p38 MAPK contribute to c-Fos induction, c-Jun activation, and NFATc expression (23, 25, 46). We then determined the effect of Deltex on activation of JNK, ERK, and p38 MAPK. Consistent with a reduction in c-Jun Ser63 phosphorylation, CD3/CD28-triggered JNK activation, measured by phosphorylation of GST c-Jun (1-79), was largely attenuated in EL4 and DO11.10 T cells expressing Deltex1 (Fig. 4A; data not shown for DO11.10). A moderate decrease in ERK activation, assessed by phosphorylation of ERK, was also found in DO11.10 and EL4 T cells expressing Deltex1 compared with the YFP control (Fig. 4B; data not shown for EL4). An effective inhibition of p38 MAPK was observed in Deltex-expressing T cells (Fig. 4C). We thus studied whether defective MAPK activation could be attributed to an inhibition on proximal TCR activation by Deltex. TCR engagement is immediately followed by recruitment and tyrosine phosphorylation of the adaptor protein LAT; LAT is pivotal for activation of AP-1 and ERK (40). Figure 4D illustrates that anti-CD3 stimulation of T cells led to extensive LAT phosphorylation, identical in Deltex1-DO11.10 and YFP-DO11.10 cells, suggesting that Deltex1 does not affect early T-cell activation at the stage of LAT phosphorylation. We also determined the activation of PKCθ, another key signaling molecule in early T-cell activation (16). PKCθ has been shown to stimulate Ras-dependent transcription activation of AP-1 in T cells and to cooperate with calcineurin for JNK activation and IL-2 gene expression (1, 42). The level of PKCθ phosphorylation after TCR ligation was also similar for the Deltex-expressing T cells and the YFP control (Fig. 4D). Therefore, the stages of TCR activation that are regulated by Deltex1 should be downstream of the LAT and PKCθ activation.
FIG. 3.
Deltex suppresses TCR-stimulated activation of NFATc, c-Fos, and c-Jun. DO11.10 T cells expressing YFP vector or Deltex1 were stimulated with anti-CD3 (5 μg/ml), and nuclear extracts and cytosolic extracts were isolated at the indicated time. (A) IκB degradation was not affected by Deltex expression. The levels of IκB in cytoplasm were determined by immunoblotting with β-tubulin as an internal standard. (B) Deltex partially interfered with the induction of NFATc. The appearance of NFATc in the nucleus after TCR stimulation was assessed by immunoblotting. (C and D) Deltex inhibited c-Fos induction and c-Jun phosphorylation at serine 63. The levels of c-Fos (C) and c-Jun and the extent of c-Jun phosphorylation (D) in nuclear extracts were determined using anti-c-Fos, anti-c-Jun, and anti-phospho-c-Jun, respectively.
FIG. 4.
Deltex interferes with activation of JNK, ERK, and p38 MAPK but not of LAT and PKCθ. (A) Inhibition of CD3/CD28-simulated JNK activation by Deltex1. EL4 T cells, transfected with pcDNA4 or pcDNA4-Deltex, were stimulated with anti-CD3 (5 μg/ml) and anti-CD28 (2.5 μg/ml) and total cellular extracts were prepared at the indicated time points. JNK1 was then immunoprecipitated from cellular extracts (100 μg), and the kinase activity of the immune complexes was determined using GST-c-Jun (1-79) as substrate. The amount of JNK1 in 20% of the immune complexes was determined by immunoblotting. (B and C) Deltex interfered with p42/p44 ERK activation and p38 MAPK stimulation. ERK and p38 activation in DO11.10 T cells before and after CD3/CD28 (5 versus 2.5 μg/ml) stimulation was determined using anti-phosphorylated T202/Y204 ERK and anti-phospho-T180/Y182 p38 MAPK, respectively. The levels of ERK2 and p38α MAPK were used as an internal control. (D) Deltex did not affect TCR-mediated LAT and PKCθ activation. LAT and PKCθ activation in YFP- or Deltex1-DO11.10 T cells were determined using anti-phospho-LAT (Y191) and anti-phospho-PKCθ (T538). The protein level of β-tubulin was used as an internal control.
Deltex down-regulates protein expression, but not mRNA expression, of MEKK1(C).
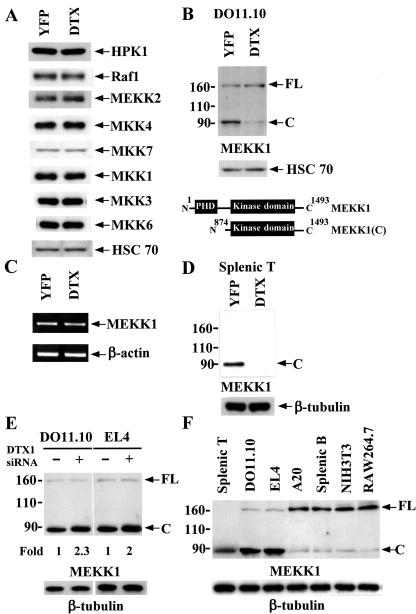
We next examined the signaling proteins immediately upstream of JNK, ERK, and p38 MAPK in Deltex1-expressing T cells, including Raf, MEKK1, MEKK2, HPK1, MKK1, MKK3, MKK4, MKK6, and MKK7. The protein levels of MEKK2, HPK1, Raf-1, MKK1, MKK3, MKK4, MKK6, and MKK7 were identical for YFP-DO11.10 and Deltex-DO11.10 cells (Fig. 5A). MEKK1 protein is present in two forms in T cells, as the full-length 196-kDa protein [MEKK1(FL)] and the 91-kDa C-terminal kinase domain-containing fragment [MEKK1(C)], with MEKK1(C) much more abundant than MEKK1(FL) in DO11.10 T cells (Fig. 5B). A significant reduction in MEKK1(C) was found in Deltex1-DO11.10 T cells compared to the control YFP T cells. In contrast, the protein levels of MEKK1(FL) were not altered by Deltex1 expression relative to the control (Fig. 5B). MEKK1 mRNA expression also was not affected in Deltex1-DO11.10 T cells (Fig. 5C), suggesting that the reduced protein levels of MEKK1(C) were not due to transcription inhibition by Deltex. A similar extent of MEKK1(C) down-regulation by Deltex1 was observed in EL4 T cells (data not shown). We also examined the effect of Deltex on normal T cells. In purified splenic T cells, almost all MEKK1 was processed into the catalytic active form [MEKK1(C)], with no MEKK1(FL) detected (Fig. 5D). Ectopic expression of Deltex1 led to the complete elimination of MEKK1(C) in splenic T cells. We further examined whether reduction of Deltex1 would enhance MEKK1(C) levels in resting T cells. Consistent with the prominent decrease of endogenous Deltex by the transfected Deltex1 siRNA (Fig. 1E), MEKK1(C) was increased in DO11.10 and EL4 cells expressing Deltex1 siRNA (Fig. 5E). Densitometry reading indicated a twofold increase of MEKK1(C) in T cells when endogenous Deltex was knocked down. Because the presence of MEKK1(C) as the dominant form of MEKK1 in T cells has not been previously documented, we also determined whether our experimental procedures may lead to procession of MEKK1(FL) to MEKK1(C) and overrepresentation of MEKK(C) in our T-cell preparations. Figure 5F illustrates that MEKK1(FL) was the major form in non-T cells including A20, splenic B cells, NIH 3T3, and RAW264.7, for which cell lysates were isolated simultaneously with the T-cell preparations in the same blot. The distinct MEKK1 expression pattern between T cells and non-T cells suggests that the predominant presence of MEKK1(C) in T cells is not an artifact of cell extract preparation. Since Deltex1 is down-regulated by TCR ligation (Fig. 1A to D), we also determined changes in MEKK1 levels after T cells were stimulated. Parallel to the reduction of Deltex1 following anti-CD3 treatment (Fig. 1), MEKK1 protein was proportionally increased in splenic T cells (Fig. 6A). TCR-initiated increase of MEKK1 expression was restricted to the protein level; MEKK1 mRNA remained unchanged after TCR engagement (Fig. 6B). Therefore, Deltex specifically down-regulates MEKK1(C) protein in T cells.
FIG. 5.
Deltex expression results in selective reduction of MEKK1(C) in T cells. (A) The levels of HPK1, MEKK2, Raf-1, MKK1, MKK3, MKK4, MKK6, and MKK7 were normal in DO11.10 cells expressing Deltex1. Total cell extracts from YFP- and Deltex1-DO11.10 T cells were prepared, and the levels of HPK1, MEKK2, Raf-1, MKK1, MKK3, MKK4, MKK6, and MKK7 were determined by their specific antibodies. (B) MEKK1(C), but not MEKK1(FL), was selectively down-regulated in Deltex1-expressing DO11.10 T cells. MEKK1 levels in YFP- and Deltex1-DO11.10 cells were determined by immunoblotting by using antibody against the C terminus of MEKK1 (C-22; Santa Cruz). Schematic representations of MEKK1(FL) and MEKK1(C) are shown in the lower panel. (C) Expression of MEKK1 mRNA was not affected by Deltex1. Total RNA of YFP- and Deltex1-DO11.10 T cells were isolated. The amounts of MEKK1 and β-actin mRNA were determined by reverse transcription-PCR. (D) MEKK1(C) was down-regulated in Deltex1-expressing normal T cells. Splenic T cells, transduced with Deltex1 or YFP only, were analyzed for their MEKK1 expression. (E) Down-regulation of endogenous Deltex1 increased MEKK1(C) levels. Deltex1 siRNA was expressed in DO11.10 and EL4 T cells as described in the legend for Fig. 1, and MEKK1 protein contents were determined by immunoblotting. The densitometry reading of MEKK1(C) levels in T cells transfected with pSuper vector was set as 1. (F) MEKK1(C) was the major form of MEKK1 in T cells but not in non-T cells. Cell lysates from the indicated cells were prepared in the same time and the contents of MEKK1 were assessed by immunoblotting.
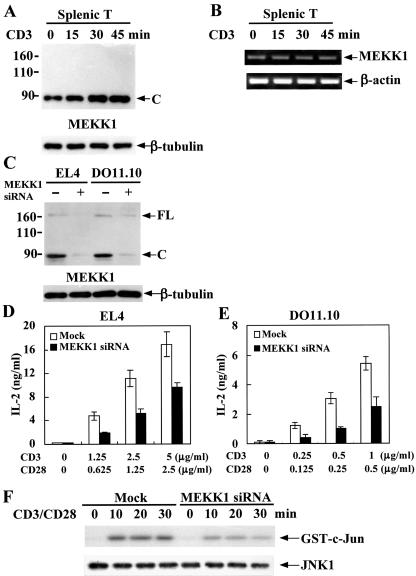
FIG. 6.
MEKK1 is increased during TCR stimulation while down-regulation of MEKK1 by siRNA inhibits T-cell activation. (A) TCR-mediated Deltex1 down-regulation was accompanied by an increase in MEKK1 protein levels. Splenic T cells were treated as described in the legend to Fig. 1D and MEKK1 contents were determined by immunoblotting. (B) TCR activation did not increase MEKK1 mRNA expression. Splenic T cells were stimulated as described in the legend to Fig. 1B and MEKK1 mRNA levels were assessed by reverse transcription-PCR. (C) Selective reduction of endogenous MEKK1 by siRNA. DO11.10 and EL4 cells were transfected with MEKK1-specific siRNA by electroporation with a Nucleofactor (Amaxa). The protein contents of MEKK1 were determined 48 h after transfection. (D and E) Decreased MEKK1 expression led to attenuated T-cell activation. EL4 (D) and DO11.10 cells (E), transfected with MEKK1-specific siRNA (solid column) or mock transfected (open column), were activated with plate-bound anti-CD3 and anti-CD28 48 h after transfection and IL-2 production was quantitated after another 16 h. (F) Down-regulation of MEKK1 impaired JNK activation. EL4 cells, transfected with MEKK1 siRNA or mock transfected, were stimulated through CD3/CD28, and JNK1 activity was determined as in Fig. 4A.
MEKK1 down-regulation by siRNA results in T-cell inactivation.
Even though Deltex specifically reduced MEKK1 expression, how T-cell activation would be affected could not be deduced because MEKK1 has not been demonstrated to be essential for T-cell activation. We thus studied whether MEKK1 down-regulation by siRNA would lead to an inhibition of IL-2 expression similar to that seen on Deltex-expressing T cells. MEKK1-specific siRNA was introduced into DO11.10 and EL4 cells by electroporation. There was a more than 80% reduction in the protein level of MEKK1 by siRNA transfection (Fig. 6C). This was accompanied by a significant decrease in CD3/CD28-stimulated IL-2 expression in both EL4 and DO11.10 cells (Fig. 6D and E). In addition, CD3/CD28-stimulated JNK activity was attenuated when MEKK1 was knocked down by siRNA (Fig. 6F). Therefore, MEKK1 is indispensable for JNK stimulation and T-cell activation. The Deltex-mediated T-cell inactivation could be attributed, at least in part, to the down-regulation of MEKK1.
Ubiquitin-mediated MEKK1(C) degradation in Deltex-expressing T cells.
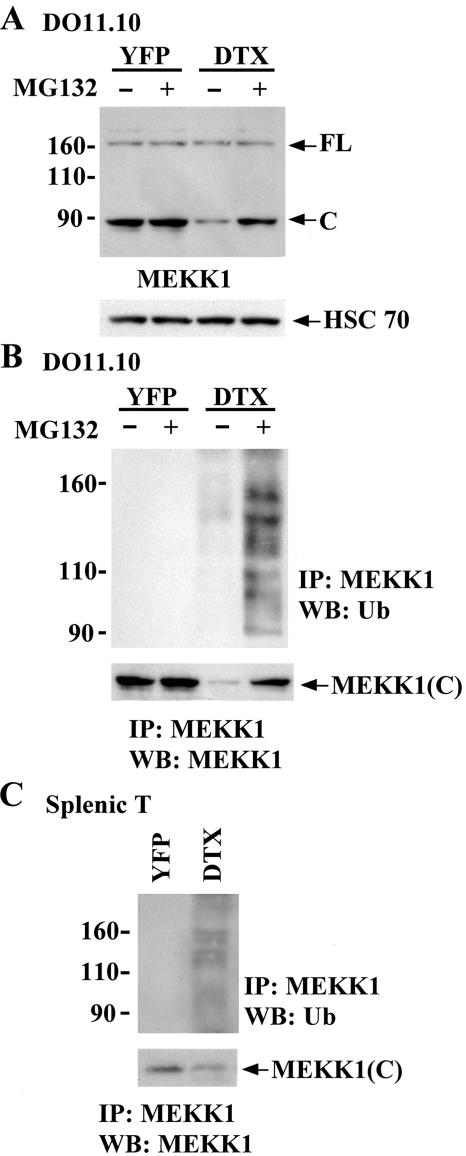
To examine whether the Deltex-directed MEKK1(C) reduction was due to proteasome-dependent degradation, T cells were treated with the proteasome inhibitor MG132. MG132 treatment had no effect on the level of MEKK1(FL) and MEKK1(C) in YFP control T cells (Fig. 7A). In contrast, MG132 restored MEKK1(C) expression in Deltex1-expressing T cells, indicating that Deltex1 triggered MEKK1(C) degradation in a proteasome-dependent manner (Fig. 7A). We next determined whether Deltex1 promoted the ubiquitination of MEKK1(C). MEKK1 was precipitated from T cell lysate and immunoblotted with antiubiquitin. MEKK1 was not ubiquitinated in control YFP T cells (Fig. 7B). However, ubiquitin modification of MEKK1(C), characterized by the presence of multiple MEKK1 protein bands, was evident in T cells expressing Deltex1. The level of MEKK1(C) ubiquitination was prominent when protein degradation was blocked by addition of MG132 to Deltex1-DO11.10 T cells (Fig. 7B). Similar results were found in splenic T cells, where Deltex1 expression resulted in extensive MEKK1 ubiquitination (Fig. 7C). Since MEKK1(C) is the only form of MEKK1 in splenic T cells (Fig. 5D), the high-molecular-weight ubiquitin conjugates (Fig. 7C) apparently came from MEKK1(C). These results suggest that Deltex1 targets MEKK1(C) for ubiquitin conjugation and concomitant proteasome-mediated degradation in T cells.
FIG. 7.
Down-regulation of MEKK1(C) by Deltex due to proteasome-mediated degradation in T cells. (A) Inhibition of proteasome-mediated degradation restored the protein expression of MEKK1(C) in Deltex1-DO11.10 T cells. YFP- and deltex1-DO11.10 cells were untreated or treated with MG132 (50 μM) for 6 h and the levels of MEKK1(FL) and MEKK1(C) were determined by immunoblotting. (B) Ubiquitination of MEKK1(C) in Deltex1-expressing DO11.10 T cells but not in YFP controls. Total cellular extracts (250 μg) from panel A were immunoprecipitated using anti-MEKK1 antibody and protein-A Sepharose; the immune precipitates were resolved on SDS-PAGE and blotted with anti-ubiquitin (Ub) and anti-MEKK1(C) antibodies. IP, immunoprecipitation; WB, Western blot. (C) Ubiquitination of MEKK1(C) in Deltex-expressing normal T cells. Ubiquitination of MEKK1 in splenic T cells expressing Deltex1 or YFP were analyzed in the presence of MG132 as described for panel B.
Deltex interacted with MEKK1(C) but poorly with the full-length MEKK1.
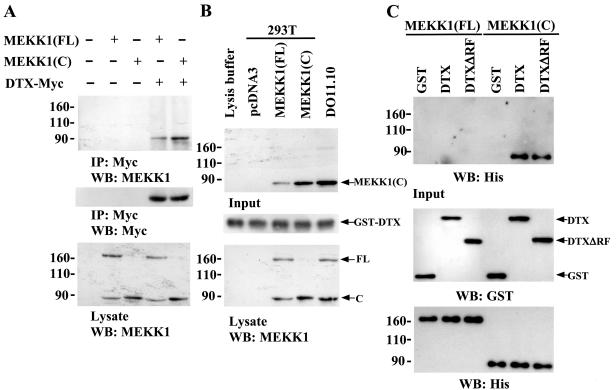
Inasmuch as Deltex1 promoted MEKK1 ubiquitination, we elucidated whether there is a direct association between Deltex1 and MEKK1. For overexpression purposes, 293T cells were used. MEKK1(FL) or MEKK1(C) was transfected into 293T cells in the presence or absence of Myc-tagged Deltex1. Overexpression of MEKK1(FL) resulted in the appearance of both full-length and the catalytic active MEKK1 (Fig. 8A). Immunoprecipitation with anti-Myc antibodies brought down Myc-tagged Deltex1 and MEKK1(C) but not the full-length MEKK1 (Fig. 8A), suggesting that Deltex1 interacts with MEKK1(C) but poorly with MEKK1(FL). We further used a GST pull-down assay to examine the protein interaction between Deltex and MEKK1(C). Incubation of bacterial expressed GST-Deltex1 with lysates from 293T cells overexpressing MEKK1(FL) or MEKK1(C) brought down MEKK1(C) (Fig. 8B). Endogenous MEKK1(C) from DO11.10 T cells was also pulled down by GST-Deltex1 (Fig. 8B). The direct binding of Deltex to MEKK1(C) was confirmed by the association of GST-Deltex with His-MEKK1(C) when only these two recombinant proteins were presented (Fig. 8C). Removal of the RING finger domain from Deltex did not affect its interaction with MEKK1(C). In contrast, no interaction between GST-Deltex1 and His-MEKK1(FL) was detected in the same assay (Fig. 8C). Taken together, Deltex1 interacted with the C-terminal fragment of MEKK1.
FIG. 8.
Interaction of Deltex with MEKK1(C). (A) Association of DTX with MEKK1(C) demonstrated by coprecipitation. 293T cells were transfected with pcDNA3 vector, MEKK1(FL), or MEKK1(C) in the presence or absence of Deltex-Myc and total cellular extracts were prepared 24 h after transfection. Extracts (200 μg) were immunoprecipitated using anti-Myc (9E10) antibody. 80% of the precipitate was separated on SDS-PAGE and the level of MEKK1 was determined by immunoblotting (upper panel). Twenty percent of the precipitate was similarly resolved and the contents of Deltex-Myc were detected by anti-Myc (middle panel). The expression of MEKK1(FL) and MEKK1(C) in transfected 293T cell was determined by immunoblotting with anti-MEKK1 (lower panel). (B) Association between Deltex and MEKK1(C) detected by GST pull-down assay. Five micrograms of GST-Deltex (GST-DTX) fusion proteins were first bound to glutathione agarose beads and incubated with 200-μg lysates of 293T cells transfected with the indicated plasmid or with 100-μg extracts of DO11.10 cells. After washing agarose beads, the amounts of MEKK1(C) pulled down were analyzed by immunoblotting with anti-MEKK1 (upper panel), and the quantity of GST-DTX on agarose was assessed by anti-GST (middle panel). The amounts of the transfected and endogenous MEKK1(FL) and MEKK1(C) present in the input cell lysates are shown in the bottom panel. (C) Direct Deltex-MEKK1(C) but not Deltex-MEKK1(FL) binding. Five micrograms of GST, GST-DTX, or GST-DTXΔRF were loaded onto glutathione agarose beads and incubated with 5 μg of His-MEKK1(FL) or His-MEKK1(C). The amounts of His-MEKK1(C) or His-MEKK1(FL) brought down by agarose were determined by immunoblotting with anti-His antibodies (upper panel). Input of GST, GST-DTX, and GST-DTXΔRF was determined by anti-GST antibodies (middle panel), while input of His-MEKK1(FL) and His-MEKK1(C) was assessed by anti-His antibodies (bottom panel).
Deltex induces MEKK1(C) ubiquitination in vivo.
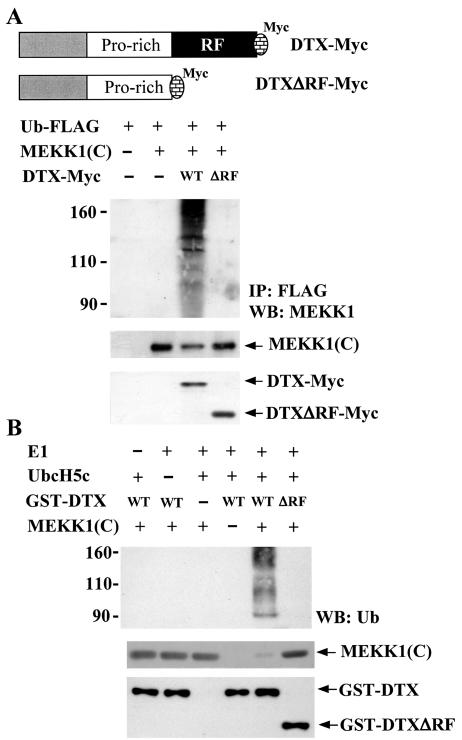
The C-terminal region of Deltex contains a RING finger, a domain shared by several ubiquitin E3 ligases. Human Deltex family members have been shown to function as an E3 ligase in vitro (41). The observed association between Deltex1 and MEKK1(C) (Fig. 8) and the down-regulation of MEKK1(C) by Deltex1 (Fig. 7) suggested that Deltex may act as an E3 ligase which binds MEKK1(C) and mediates the transfer of ubiquitin to MEKK1(C), leading to degradation of MEKK1(C) by the proteasome. To assess whether Deltex could target MEKK1(C) for ubiquitination, we transfected 293T cells with FLAG-tagged ubiquitin (FLAG-Ub) and MEKK1(C) in the presence or absence of Deltex-Myc. Ubiquitinated proteins were immunoprecipitated with FLAG-M2 affinity gel and the level of ubiquitinated MEKK1 was analyzed by using MEKK1 antibodies. Figure 9A demonstrates that MEKK1(C) was ubiquitinated only with the coexpression of Deltex. To determine if the RING finger domain of Deltex is required for the MEKK1(C) ubiquitination, in vivo ubiquitination analysis was also conducted on a Deltex1 mutant with its RING finger deleted (DTXΔRF-Myc). Deletion of RING finger domain did not affect the binding of Deltex to MEKK1(C) (Fig. 8C) but abolished the capacity of Deltex1 to trigger MEKK1(C) ubiquitination (Fig. 9A). These results demonstrated that DTX acts as an E3 ligase for the ubiquitination of the C-terminal of MEKK1 in vivo.
FIG. 9.
Ubiquitination of MEKK1(C) by Deltex but not DeltexΔRF in vivo and in vitro. (A) 293T cells were transfected with Ub-FLAG, MEKK1(C), DTX-Myc, or DTXΔRF-Myc as indicated. Schematic representations of DTX-Myc and DTXΔRF-Myc are shown. Forty-eight hours after transfection, 293T cells were treated with MG132 (25 μM) for 5 h and total cellular extracts were prepared. Ubiquitinated proteins were immunoprecipitated from 200 μg of total cellular extracts by FLAG-M2 affinity gel. The precipitates were resolved on SDS-PAGE and the presence of MEKK1(C) [representing ubiquitinated MEKK1(C)] determined using anti-MEKK1 (upper panel). Protein levels of MEKK1(C), DTX, or DTXΔRF in the cell extracts were detected using anti-MEKK1 or anti-Myc (lower panels). (B) In vitro ubiquitination assays were performed in reaction mixtures containing bovine ubiquitin, E1, E2 (UbcH5c), His-MEKK1(C), GST-DTX, or GST-DTXΔRF fusion protein as indicated. Reactions were incubated at 30°C for 30 min. His-MEKK1(C) was then captured by Ni-NTA agarose, separated on a SDS-8% PAGE, and blotted with ubiquitin antibody (upper panel). Ubiquitination of MEKK1(C) led to the appearance of multiple forms of higher-molecular-weight MEKK1(C). The input of GST-DTX (including GST-DTXΔRF) and His-MEKK1(C) fusion proteins in the reaction mixture is shown in the bottom panel.
Deltex promotes MEKK1(C) ubiquitination in vitro.
We further conducted the in vitro ubiquitination analysis to confirm the role of DTX as a ubiquitin E3 ligase for MEKK1(C). In mixtures containing only ubiquitin, E1, E2 (UbcH5c), and recombinant MEKK1(C), no ubiquitination of MEKK1(C) was detected (Fig. 9B). The addition of recombinant GST-Deltex to the same reaction mixture stimulated the ubiquitination of MEKK1(C). In contrast, no ubiquitination of MEKK1(C) was detected if the Deltex1 mutant with the RING finger deleted (GST-DTXΔRF) was added to the same reaction, suggesting that the RING finger domain is required for Deltex to induce MEKK1(C) ubiquitination.
DISCUSSION
Despite the fact that many physiological functions are assigned to Deltex, the action mechanisms of Deltex remain largely elusive. In the present study, we demonstrated a new function of Deltex in T cells and a novel mechanism of Deltex in regulating T-cell signaling. Deltex expression was constitutive in resting T cells (Fig. 1A to E) and was subjected to a profound down-regulation by TCR ligation. The levels of Deltex1 mRNA and protein were immediately decreased following TCR stimulation and were nearly undetectable in T cells 45 min after anti-CD3 stimulation. TCR-mediated Deltex1 reduction could be due to an inhibitory effect on Deltex1 mRNA expression (Fig. 1A and B). The exact activation signals that contributed to Deltex down-regulation are being elucidated. Since T cells were activated by plate-bound antibody, in the absence of contact with antigen-presenting cells (Fig. 1), the regulation of Deltex expression by TCR stimulation was apparently independent of Notch signals.
We also used Deltex-specific siRNA to demonstrate the physiological role of Deltex in regulating T-cell activation (Fig. 1E to G). Even if Deltex1 levels were attenuated by TCR stimulation (Fig. 1A to D), a reduced Deltex1 expression prior to T-cell activation further enhanced TCR-stimulated IL-2 production, suggesting a pivotal role of Deltex in T-cell activation events. Several signaling pathways affected by Deltex, including activation of AP-1 and NFATc and stimulation of JNK, p38 MAPK, and ERK, were identified by Deltex1 overexpression (Fig. 3 and 4). Most of these activation defects are known to be involved in T-cell activation. For example, previous studies have established a critical role of p38 MAPK in T-cell activation (15, 29, 39, 46), that interference of p38 MAPK activation severely impairs T-cell activation. For NFAT and AP-1, their coordinated association on the IL-2 gene promoter dictates the expression of the IL-2 gene (19). Interestingly, with the profound inhibition of quite a few downstream signals, the proximal TCR activation events were mostly unaffected by Deltex1. The activation of LAT and PKCθ was normal in the presence of Deltex overexpression (Fig. 4D). Instead, one of the defects was mapped to selective degradation of MEKK1 (Fig. 5B). This was best illustrated by the exact inverse correlation between the expression of Deltex1 and the levels of MEKK1 in T cells. In the absence of any Deltex1 overexpression, TCR stimulation down-regulated Deltex1, with a simultaneous up-regulation of MEKK1 (Fig. 1A to D and Fig. 6A). In addition, reduced Deltex1 expression by siRNA in T cells (Fig. 1E) led to a significant increase of MEKK1(C) protein (Fig. 5E).
Interference of JNK activation by Deltex has been previously suggested, based on the suppression of Ras-dependent c-Jun activation by Deltex (35). The present study provides the first direct proof that JNK activation is profoundly attenuated by Deltex (Fig. 4A). We also found that ERK activation was partially inhibited by Deltex1 (Fig. 4B). It has been shown that MEKK1 gene knockout results in severe reduction in JNK activity and mild inhibition of ERK activity (52), a phenotype similar to our Deltex1-expressing T cells. Despite the fact that the link between MEKK1 and p38 MAPK is less direct (52), p38 MAPK has been shown to be activated by MEKK1 in many different cells (6, 8, 11, 44, 46). Therefore, defects in the activation of JNK, p38 MAPK, and ERK in Deltex1-expressing T cells could be attributed, at least in part, to the down-regulation of MEKK1(C). It has to be noted that we do not exclude the possibility that Deltex expression leads to defects in additional signal molecules and contributes to the aberrant MAPK activation observed here.
Our results further demonstrated, likely for the first time, that MEKK1 is essential for TCR signal transduction. There are more than 12 different MAP kinase kinases in mammalian cells (25, 36), suggesting a possible signaling redundancy among several MAP kinase kinases, which makes defining the specific involvement of a MAP kinase kinase downstream of the T-cell receptor difficult. Activation of MEKK1 by TCR ligation has been previously demonstrated (1, 21). Whether MEKK1 is essential for T-cell activation, however, remains unclear. Our observations that down-regulation of MEKK1 by Deltex resulted in inactivation of JNK and p38 MAPK and a decrease in IL-2 generation (Fig. 2, 4, and 5), together with the finding that T-cell activation was suppressed when MEKK1 was specifically reduced by siRNA (Fig. 6), clearly support an indispensable role of MEKK1 in T-cell activation.
A critical observation from the present study is the identification of MEKK1 as a substrate of Deltex E3 ligase. The C terminus of Deltex is a RING finger, a domain present in many E3 ligases and functioning to bind E2 (18, 20). Deltex1 has since been shown to exhibit E3 ligase activity based on its ability to promote self-ubiquitination (41), despite the fact that the in vivo targets of Deltex1 E3 activity remain unknown. Our results illustrate the in vivo E3 ligase activity of Deltex1 and identify MEKK1(C) as the first known physiological target of Deltex E3 ligase. Deltex interacted directly with MEKK1(C), as shown by coimmunoprecipitation and GST pull-down analysis (Fig. 8). Deltex1 bound substrate MEKK1(C) through a region distinct from its RING finger (Fig. 8C), consistent with the function of the RING finger for E2 association. Deltex1 promoted MEKK1(C) ubiquitination when coexpressed with MEKK1(C) in cells (Fig. 9A) or coincubated with recombinant MEKK1(C) in the presence of E1 and E2 (Fig. 9B). Deletion of the RING finger, the E2-binding domain, abolished the ability of Deltex1 to stimulate MEKK1(C) ubiquitination (Fig. 9B) despite the fact that Deltex1 still interacted with MEKK(C) (Fig. 8C). Deltex1 fulfills all the criteria for an E3 ligase specific for MEKK1(C).
Another important finding here is that Deltex is a ubiquitin ligase for MEKK1(C) but much less so for MEKK1(FL). In contrast to the effective interaction between MEKK1(C) and Deltex1, the binding of MEKK1(FL) to Deltex was minimum (Fig. 8). The ability of a substrate to bind to a specific E3 is a prerequisite for ubiquitin transfer mediated by the given E3. The differential effect of Deltex1 on the degradation of MEKK1(C) and MEKK1(FL), therefore, could be attributed to the distinct binding affinity for Deltex1 between MEKK1(C) and MEKK1(FL). At present, the cause for the poor interaction between MEKK1(FL) and Deltex is not understood. In addition to the C-terminal kinase domain, the full-length MEKK1 contains a long N-terminal domain (48) with proline-rich domains for binding to SH3, sequences for binding to 14-3-3, and pleckstrin homology domains (25, 36). A possibility is that interaction of MEKK1(FL) with other signaling molecules in vivo prevents the binding of Deltex to MEKK1(FL). Alternatively, the long N-terminal region of MEKK1(FL) may shield the Deltex-binding area of MEKK1(C) from Deltex interaction. A study is ongoing to delineate these two possibilities.
It is interesting that recent findings show that the plant homeodomain of MEKK1 is an E3 ubiquitin ligase and mediates the degradation of ERK (27, 45). Together with results from the present study, there are two parallel ubiquitin conjugation pathways centered on MEKK1; MEKK1(C) is degraded through ubiquitination by Deltex E3, while MEKK1(FL) acts as an E3 for other signaling proteins. A potential physiological significance of the resistance of MEKK1(FL) to Deltex1-mediated ubiquitination could be an assurance of the operation of the MEKK1(FL) E3 pathway by preventing unnecessary degradation of MEKK1(FL).
Both full-length MEKK1 and MEKK1(C) activate JNK; the processing of full-length MEKK1 to MEKK1(C) is not essential for activation of JNK (43). Blockage of caspase-mediated MEKK1 cleavage by specific inhibitors does not interfere with the downstream JNK and ERK activation, and neither does mutation of the caspase-3 recognition sites (DEVE→A860 and DTVD→A874) on MEKK1 affect JNK activation (43). Therefore, MEKK1(FL) may convey sufficient JNK activation in the absence of MEKK1(C) generation. MEKK1(C), however, is the dominant form of MEKK1 in T cells (Fig. 5F). Deltex-mediated degradation of MEKK1(C) eliminates most of MEKK1 in T cells (Fig. 5B and D), leading to defective T-cell activation.
In summary, we identified a novel pathway regulating MEKK1 stability and T-lymphocyte activation by the Notch signal mediator Deltex. As an E3 ligase, Deltex1 binds specifically to its target MEKK1(C) and mediates the transfer of ubiquitin from the RING finger-associated E2 to MEKK1(C). MEKK1(C) is a key to T-cell activation (Fig. 6) and its degradation by proteasome results in defective T-cell activation. The ubiquitin E3 ligase activity of Deltex is clearly dissociated from the transcription activity reported for Deltex (50) and may contribute to physiological activities previously thought to be Deltex independent. We are currently working to identify other substrates targeted by Deltex E3 ligase, which will help unveil additional functional roles of Deltex.
Acknowledgments
We thank Hideyuki Okano, Melanie Cobb, Dennis Templeton, Gina Costa, Garry Nolan, Ellen Rothenberg, Tse-Hua Tan, and Nan-Shih Liao for providing key materials used in this study and Ken Deen for editorial correction of the manuscript.
This work was supported by grant 92-2320-B001-006 from the National Science Council, Taiwan, grant NHRI-EX93-9217BI from the National Health Research Institute, Taiwan, and a grant from Academia Sinica.
REFERENCES
- 1.Avraham, A., S. Jung, Y. Samuels, R. Seger, and Y. Ben-Neriah. 1998. Co-stimulation-dependent activation of a JNK-kinase in T lymphocytes. Eur. J. Immunol. 28:2320-2330. [DOI] [PubMed] [Google Scholar]
- 2.Baud, V., Z. G. Liu, B. Bennett, N. Suzuki, Y. Xia, and M. Karin. 1999. Signaling by proinflammatory cytokines: oligomerization of TRAF2 and TRAF6 is sufficient for JNK and IKK activation and target gene induction via an amino-terminal effector domain. Genes Dev. 13:1297-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chadee, D. N., T. Yuasa, and J. M. Kyiakis. 2002. Direct activation of mitogen-activated protein kinase kinase kinase MEKK1 by the Ste20p homologue GCK and the adapter protein TRAF2. Mol. Cell. Biol. 22:737-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, Y., and M. Z. Lai. 2001. JNK-induced apoptosis is independent of Fas ligand in Jurkat T cells. J. Biol. Chem. 276:8350-8357. [DOI] [PubMed] [Google Scholar]
- 5.Costa, G. L., J. M. Benson, C. M. Seroogy, P. Achacoso, C. J. Fathman, and G. P. Nolan. 2000. Targeting rare populations of murine antigen-specific T lymphocytes by retroviral transduction for potential application in gene therapy for autoimmune disease. J. Immunol. 164:3581-3590. [DOI] [PubMed] [Google Scholar]
- 6.Cuenda, A., and D. S. Dorow. 1998. Differential activation of stress-activated protein kinase kinases SKK4/MKK7 and SKK1/MKK4 by the mixed-lineage kinase-2 and mitogen-activated protein kinase kinase (MKK) kinase-1. Biochem. J. 333:11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deftos, M. L., Y. W. He, E. W. Ojala, and M. J. Bevan. 1998. Correlating notch signaling with thymocyte maturation. Immunity 9:777-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Efimova, T., P. LaCelle, J. F. Welter, and R. L. Eckert. 1998. Regulation of human involucrin promoter activity by a protein kinase C, Ras, MEKK1, MEK3, p38/RK, AP1 signal transduction pathway. J. Biol. Chem. 273:24387-24395. [DOI] [PubMed] [Google Scholar]
- 9.Fang, L. W., T. S. Tai, W. N. Yu, F. Liao, and M. Z. Lai. 2004. Phosphatidylinositide 3-kinase priming couples c-FLIP to T cell activation. J. Biol. Chem. 279:13-18. [DOI] [PubMed] [Google Scholar]
- 10.Gallagher, E. D., S. Gutowski, P. C. Sternweis, and M. H. Cobb. 2004. RhoA binds to the amino terminus of MEKK1 and regulates its kinase activity. J. Biol. Chem. 279:1872-1877. [DOI] [PubMed] [Google Scholar]
- 11.Guan, Z., S. Y. Buckman, A. P. Pentland, D. J. Templeton, and A. R. Morrison. 1998. Induction of cyclooxygenase-2 by the activated MEKK1 → SEK1/MKK4 → p38 mitogen-activated protein kinase pathway. J. Biol. Chem. 273:12901-12908. [DOI] [PubMed] [Google Scholar]
- 12.Gupta-Rossi, N., S. Storck, P. J. Griebel, C. A. Reynaud, J. C. Weill, and A. Dahan. 2003. Specific over-expression of DTX and a new Kelch-like protein in human germinal center B cells. Mol. Immunol. 39:791-799. [DOI] [PubMed] [Google Scholar]
- 13.Hershko, A., A. Ciechanover, and A. Varshavsky. 2000. The ubiquitin system. Nat. Med. 6:1073-1081. [DOI] [PubMed] [Google Scholar]
- 14.Ho, H. Y., H. H. Lee, and M. Z. Lai. 1997. Overexpression of mitogen-activated protein kinase kinase kinase reverses cAMP inhibition of NF-κB in T cells. Eur. J. Immunol. 27:222-226. [DOI] [PubMed] [Google Scholar]
- 15.Hsu, S. C., M. Gavrilin, M. H. Tsai, J. Han, and M. Z. Lai. 1999. p38 mitogen activated protein kinase is involved in Fas ligand expression. J. Biol. Chem. 274:25769-25776. [DOI] [PubMed] [Google Scholar]
- 16.Isakov, N., and A. Altman. 2002. Protein kinase Cθ in T cell activation. Annu. Rev. Immunol. 20:761-794. [DOI] [PubMed] [Google Scholar]
- 17.Izon, D. J., J. C. Aster, Y. He, A. Weng, F. G. Karnell, V. Patriub, L. Xu, S. Bakkour, C. Rodriguez, D. Allman, and W. S. Pear. 2002. DTX1 redirects lymphoid progenitors to the B cell lineage by antagonizing Notch1. Immunity 16:231-243. [DOI] [PubMed] [Google Scholar]
- 18.Jackson, P. K., A. G. Eldridge, E. Freed, L. Furstenthal, J. Y. Hsu, B. K. Kaiser, and J. D. Reimann. 2000. The lore of the RINGs: substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol. 10:429-439. [DOI] [PubMed] [Google Scholar]
- 19.Jain, J., C. Loh, and A. Rao. 1995. Transcriptional regulation of the IL-2 gene. Curr. Opin. Immunol. 7:333-342. [DOI] [PubMed] [Google Scholar]
- 20.Joazeiro, C. A., and A. M. Weissman. 2000. RING finger proteins: mediators of ubiquitin ligase activity. Cell 102:549-552. [DOI] [PubMed] [Google Scholar]
- 21.Kaga, S., S. Ragg, K. A. Rogers, and A. Ochi. 1998. Activation of p21-CDC42/Rac-activated kinases by CD28 signaling: p21-activated kinase (PAK) and MEK kinase 1 (MEKK1) may mediate the interplay between CD3 and CD28 signals. J. Immunol. 160:4182-4189. [PubMed] [Google Scholar]
- 22.Karandikar, M., S. Xu, and M. H. Cobb. 2000. MEKK1 binds raf-1 and the ERK2 cascade components. J. Biol. Chem. 275:40120-40127. [DOI] [PubMed] [Google Scholar]
- 23.Karin, M., Z. Liu, and E. Zandi. 1997. AP-1 function and regulation. Curr. Opin. Cell Biol. 9:240-246. [DOI] [PubMed] [Google Scholar]
- 24.Kishi, N., Z. Tang, Y. Maeda, A. Hirai, R. Mo, M. Ito, S. Suzuki, K. Nakao, T. Kinoshita, T. Kadesch, C. Hui, S. Artavanis-Tsakonas, H. Okano, and K. Matsuno. 2001. Murine homologs of DTX define a novel gene family involved in vertebrate Notch signaling and neurogenesis. Int. J. Dev. Neurosci. 19:21-35. [DOI] [PubMed] [Google Scholar]
- 25.Kyriakis, J. M., and J. Avruch. 2001. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol. Rev. 81:807-869. [DOI] [PubMed] [Google Scholar]
- 26.Lange-Carter, C. A., C. M. Pleiman, A. M. Gardner, K. J. Blumer, and G. L. Johnson. 1993. A divergence in the MAP kinase regulatory network defined by MEK kinase and Raf. Science 260:315-319. [DOI] [PubMed] [Google Scholar]
- 27.Lu, Z., S. Xu, C. Joazeiro, M. H. Cobb, and T. Hunter. 2002. The PHD domain of MEKK1 acts as an E3 ubiquitin ligase and mediates ubiquitination and degradation of ERK1/2. Mol. Cell 9:945-956. [DOI] [PubMed] [Google Scholar]
- 28.Maillard, I., S. H. Adler, and W. S. Pear. 2003. Notch and the immune system. Immunity 19:781-791. [DOI] [PubMed] [Google Scholar]
- 29.Matsuda, S., T. Moriguchi, S. Koyasu, and E. Nishida. 1998. T lymphocyte activation signal for interleukin-2 production involve activation of MKK6-p38 and MKK7-SAPK-JNK signaling pathways sensitive to cyclosporin A. J. Biol. Chem. 273:12378-12382. [DOI] [PubMed] [Google Scholar]
- 30.Matsuno, K., R. J. Diederich, M. J. Go, C. M. Blaumueller, and S. Artavanis-Tsakonas. 1995. DTX acts as a positive regulator of Notch signaling through interactions with the Notch ankyrin repeats. Development 121:2633-2644. [DOI] [PubMed] [Google Scholar]
- 31.Matsuno, K., D. Eastman, T. Mitsiades, A. M. Quinn, M. L. Carcanciu, P. Ordentlich, T. Kadesch, and S. Artavanis-Tsakonas. 1998. Human DTX is a conserved regulator of Notch signalling. Nat. Genet. 19:74-78. [DOI] [PubMed] [Google Scholar]
- 32.Meyer, C. F., X. Wang, C. Chang, D. Templeton, and T. H. Tan. 1996. Interaction between c-Rel and the mitogen-activated protein kinase kinase kinase 1 signaling cascade in mediating κB enhancer activation. J. Biol. Chem. 271:8971-8976. [DOI] [PubMed] [Google Scholar]
- 33.Minden, A., A. Lin, M. McMahon, C. Lange-Carter, B. Derijard, R. J. Davis, G. L. Johnson, and M. Karin. 1994. Differential activation of ERK and JNK mitogen-activated protein kinases by Raf-1 and MEKK. Science 266:1719-1723. [DOI] [PubMed] [Google Scholar]
- 34.Novak, T. J., P. M. White, and E. V. Rothenberg. 1990. Regulatory anatomy of the murine interleukin-2 gene. Nucleic Acids Res. 18:4523-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ordentlich, P., A. Lin, C. P. Shen, C. Blaumueller, K. Matsuno, S. Artavanis-Tsaonas, and T. Kadesch. 1998. Notch inhibition of E47 supports the existence of a novel signaling pathway. Mol. Cell. Biol. 18:2230-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pearson, G., F. Robinson, T. B. Gibson, B. E. Xu, M. Karandikar, K. Berman, and M. H. Cobb. 2001. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr. Rev. 22:153-183. [DOI] [PubMed] [Google Scholar]
- 37.Pickart, C. M. 2004. Back to the future with ubiquitin. Cell 116:181-190. [DOI] [PubMed] [Google Scholar]
- 38.Saito, T., S. Chiba, M. Ichikawa, A. Kunisato, T. Asai, K. Shimizu, T. Tamaguchi, G. Yamamoto, S. Seo, K. Kumano, E. Nakagami-Yamaguchi, Y. Hamada, S. Aizawa, and H. Hirai. 2003. Notch2 is preferentially expressed in mature B cells and indispensable for marginal zone B lineage development. Immunity 18:675-685. [DOI] [PubMed] [Google Scholar]
- 39.Salojin, K. V., J. Zhang, and T. L. Delovitch. 1999. TCR and CD28 are coupled via ZAP-70 to the activation of the Vav/Rac-1-/PAK-1/p38 MAPK signaling pathway. J. Immunol. 163:844-853. [PubMed] [Google Scholar]
- 40.Samelson, L. E. 2002. Signal transduction mediated by the T cell antigen receptor: the role of adaptor protein. Annu. Rev. Immunol. 20:371-394. [DOI] [PubMed] [Google Scholar]
- 41.Takeyama, K., R. C. Aguiar, L. Gu, C. He, G. J. Freeman, J. L. Kutok, J. C. Aster, and M. A. Shipp. 2003. The BAL-binding protein BBAP and related DTX family members exhibit ubiquitin-protein isopeptide ligase activity. J. Biol. Chem. 278:21930-21937. [DOI] [PubMed] [Google Scholar]
- 42.Werlen, G., E. Jacinto, Y. Xia, and M. Karin. 1998. Calcineurin preferentially synergizes with PKC-theta to activate JNK and IL-2 promoter in T lymphocytes. EMBO J. 17:3101-3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Widmann, C., P. Gerwins, N. L. Johnson, M. B. Jarpe, and G. L. Johnson. 1998. MEK kinase 1, a substrate for DEVD-directed caspases, is involved in genotoxin-induced apoptosis. Mol. Cell. Biol. 18:2416-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winzen, R., M. Kracht, B. Ritter, A. Wilhelm, C. Y. Chen, A. B. Shyu, M. Muller, M. Gaestel, K. Resch, and H. Holtmann. 1999. The p38 MAP kinase pathway signals for cytokine-induced mRNA stabilization via MAP kinase-activated protein kinase 2 and an AU-rich region-targeted mechanism. EMBO J. 18:4969-4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Witowsky, J. A., and G. L. Johnson. 2003. Ubiquitination of MEKK1 inhibits its phosphorylation of MKK1 and MKK4 and activation of the ERK1/2 and JNK pathways. J. Biol. Chem. 278:1403-1406. [DOI] [PubMed] [Google Scholar]
- 46.Wu, C. C., S. C. Hsu, H. M. Shih, and M. Z. Lai. 2003. NFATc is a target of p38 mitogen activated protein kinase in T cells. Mol. Cell. Biol. 23:6442-6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xia, Y., Z. Wu, B. Su, B. Murray, and M. Karin. 1998. JNKK1 organizes a MAP kinase module through specific and sequential interactions with upstream and downstream components mediated by its amino-terminal extension. Genes Dev. 12:3369-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu, S., D. J. Robbins, L. B. Christerson, J. M. English, C. A. Vanderbilt, and M. H. Cobb. 1996. Cloning of rat MEK kinase 1 cDNA reveals an endogenous membrane-associated 195-kDa protein with a large regulatory domain. Proc. Natl. Acad. Sci. USA 93:5291-5295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu, S., and M. H. Cobb. 1997. MEKK1 binds directly to the c-Jun N-terminal kinases stress-activated protein kinases. J. Biol. Chem. 272:32056-32060. [DOI] [PubMed] [Google Scholar]
- 50.Yamamoto, N., S. Yamamoto, F. Inagaki, M. Kawaichi, A. Fukamizu, N. Kishi, K. Matsuno, K. Nakamura, G. Weinmaster, H. Okano, and M. Nakafuku. 2001. Role of DTX-1 as a transcriptional regulator downstream of the Notch receptor. J. Biol. Chem. 276:45031-45040. [DOI] [PubMed] [Google Scholar]
- 51.Yan, M., T. Dal, J. C. Deak, J. M. Kyriakis, L. I. Zon, J. R. Woodgett, and D. J. Templeton. 1994. Activation of stress-activated protein kinase by MEKK1 phosphorylation of its activator SEK1. Nature 372:798-800. [DOI] [PubMed] [Google Scholar]
- 52.Yujiri, T., S. Sather, G. R. Fanger, and G. L. Johnson. 1998. Role of MEKK1 in cell survival and activation of JNK and ERK pathways defined by targeted gene disruption. Science 282:1911-1914. [DOI] [PubMed] [Google Scholar]
- 53.Yun, T. J., and M. J. Bevan. 2003. Notch-regulated ankyrin-repeat protein inhibits Notch1 signaling: multiple Notch1 signaling pathways involved in T cell development. J. Immunol. 170:5834-5841. [DOI] [PubMed] [Google Scholar]