Abstract
The p27Kip1 protein plays a critical role in the regulation of cell proliferation through the inhibition of cyclin-dependent kinase activity. Translation of p27Kip1 is directed by an internal ribosomal entry site (IRES) in the 5′ nontranslated region of p27Kip1 mRNA. Here, we report that polypyrimidine tract-binding protein (PTB) specifically enhances the IRES activity of p27Kip1 mRNA through an interaction with the IRES element. We found that addition of PTB to an in vitro translation system and overexpression of PTB in 293T cells augmented the IRES activity of p27Kip1 mRNA but that knockdown of PTB by introduction of PTB-specific small interfering RNAs (siRNAs) diminished the IRES activity of p27Kip1 mRNA. Moreover, the G1 phase in the cell cycle (which is maintained in part by p27Kip1) was shortened in cells depleted of PTB by siRNA knockdown. 12-O-Tetradecanoylphorbol-13-acetate (TPA)-induced differentiation in HL60 cells was used to examine PTB-induced modulation of p27Kip1 protein synthesis during differentiation. The IRES activity of p27Kip1 mRNA in HL60 cells was increased by TPA treatment (with a concomitant increase in PTB protein levels), but the levels of p27Kip1 mRNA remained unchanged. Together, these data suggest that PTB modulates cell cycle and differentiation, at least in part, by enhancing the IRES activity of p27Kip1 mRNA.
Cyclin-dependent kinases (CDKs), which are regulated by cyclin levels and specific inhibitory proteins (12, 49), play key roles in the regulation of cell proliferation (35). The p27Kip1 (hereinafter referred to as the p27) protein, a mammalian CDK inhibitor, plays a pivotal role in directing cells to arrest in G1 phase via an inhibition of the cyclin E/CDK2 complex that is both necessary and rate-limiting for S-phase entry (37, 45, 48). In normal cells, increased expression of p27 mediates transforming growth factor β-induced G1 arrest (42), contact inhibition (43), and growth in suspension (13). On the other hand, p27 is generally downregulated in cancer cells, and there is a strong inverse correlation between tumor progression and p27 levels (8, 29). The p27 protein is most abundant in G1 cells, decreases quickly as cells enter S phase, and remains at low levels throughout the remainder of the cell cycle (32). It is generally accepted that the level of p27 protein is regulated mainly by the rate of translational protein synthesis (1, 15) and/or by proteasome complex-mediated protein degradation (36, 38).
The p27 protein also plays an important role in the regulation of differentiation. In many cells and tissues, differentiation is accompanied by a profound increase in p27 protein content. Levels of p27 protein were shown to increase during the differentiation of HL60 promyelocytic leukemia cells (15), oligodendrocytes (11), neuroblastoma cells (26), and central glia-4 cells (50). In contrast, loss of p27 protein has been associated with poorly differentiated phenotypes in several human cell types (6, 10). Moreover, ectopic expression of p27 markedly enhanced the differentiation of HL60 cells (51), and augmented polyribosomal association with the p27 mRNA was observed in HL60 cells undergoing 12-O-tetradecanoylphorbol-13-acetate (TPA)-induced differentiation. The molecular basis of enhanced p27 translation during the differentiation of HL60 cells is not well understood, but it is known that the translation of p27 mRNA is directed by an internal ribosomal entry site (IRES) (33) and that the HuR RNA-binding protein represses the IRES activity of the p27 mRNA (28).
The IRES is a specialized RNA structure that recruits ribosomes to an mRNA in a cap-independent manner. Since the discovery of IRESs in the 5′ nontranslated regions (5′NTRs) of encephalomyocarditis virus (EMCV) (19) and poliovirus (40), many viral and cellular mRNAs have been shown to contain IRESs (http://www.rangueil.inserm.fr/iresdatabase). Studies of factors required for IRES-dependent translation have revealed that RNA-binding proteins such as polypyrimidine tract-binding protein (PTB), poly(rC)-binding protein, upstream of N-ras (unr), and the La autoantigen specifically enhance the translation of IRES-containing mRNAs.
PTB, also known as p57 and heterogeneous nuclear ribonucleoprotein I (hnRNP I), is an hnRNP family member that is known to shuttle between the nucleus and cytoplasm (30). PTB has four unconventional RNA recognition motifs (14) and exists as a homodimer in solution (41). PTB was originally identified as a protein binding to the polypyrimidine tracts of adenoviral major late and α-tropomyosin pre-mRNAs and was proposed to act as a splicing factor (30, 39). Independently, PTB was shown to interact specifically with the IRESs of several viral and cellular mRNAs, stimulating or inhibiting the activity of both viral and cellular IRES elements (18). Therefore, it is likely that PTB regulates alternative splicing in the nucleus and modulates translation via IRES elements in the cytoplasm.
Here we show that PTB binds to the IRES element of the p27 mRNA and augments its IRES-dependent translation, as shown through in vitro and in vivo assay systems based on overexpression and/or knockdown of PTB. We also observed that the population of cells at the G1 phase decreased as the level of PTB protein decreased following small interfering RNA (siRNA)-induced knockdown of PTB. Moreover, the population of cells at the G1 or G0 phase increased when the protein level of PTB increased during TPA-induced differentiation of HL60 cells. Together, these results suggest that PTB plays an important role in controlling the cell cycle and differentiation through the modulation of p27 IRES activity.
MATERIALS AND METHODS
Plasmid construction.
The 5′NTR of the p27 mRNA was obtained via PCR of a human Chang liver cDNA library using primers 5′-ACG CGT CGA CGC CAT ATT GGG CCA C-3′ and 5′-CCG GAA TTC ACT CGC ACG TTT GAC-3′. The resulting PCR product was digested with SalI and EcoRI and cloned into the SalI/EcoRI site of plasmid pSK(−) to yield pSK/p27. The sequence of the p27 5′NTR was confirmed by DNA sequencing and then subcloned into the intercistronic region of a pRF vector containing tandem copies of the Renilla and firefly luciferase (RLuc and FLuc, respectively) genes (23), yielding pR/p27/F. To map the PTB-binding site in the p27 5′NTR and to determine the presence of cis-acting IRES element in the p27 5′NTR, serial deletions of the p27 5′NTR were generated from pSK/p27 and pR/p27/F (the A in the initiation codon is referred to as nucleotide +1 hereinafter). To construct plasmids pSK/p27(−447 to −331), pSK/p27(−331 to −169), and pSK/p27(−169 to −1), the p27 fragment was digested with restriction enzymes SalI/SacII, SacII/BssHII, and BssHII/EcoRI, respectively, and then inserted into pSK(−) treated with the same restriction enzymes. To generate plasmids pR/p27(−331 to −1)/F, pR/p27(−169 to −1)/F, pR/27(−447 to −331)/F, and pR/p27(Δ−331 to −169)/F, plasmid pR/p27/F was digested with restriction enzymes SalI/SacII, SalI/BssHII, SacII/EcoRI, and SacII/BssHII, respectively, and then the DNA fragments were self-ligated. Plasmid pR/p27(−447 to −169)/F was digested with the restriction enzymes SalI and SacII, and then the DNA fragments were self-ligated to construct pR/p27(−331 to −169)/F. Plasmids pR/HCV/F and pR/EMCV/F were constructed as previously described (7), and plasmids pEGFP-PTB1, pEGFP-PTB4, pT7-7-PTB1, and pT7-7-PTB4 were generated as described by Back et al. (2). To construct pEGFP-NSAP1, plasmids pEGFP and pSK(−)/NSAP1 were treated with EcoRI-Klenow fragment-Asp718 and NheI-Klenow fragment-Asp718, respectively, and the resulting DNA fragments were ligated together. To construct plasmid pEGFP/HuR, the full-length HuR cDNA was amplified by PCR from a human Chang liver cDNA library using primers 5′-CCG GAA TTC CAT GTC TAA TGG TTA TGA AGA C-3′ and 5′-CGG GGT ACC TTA TTT GTG GGA CTT GTT G-3′. The resulting PCR product was digested with EcoRI/KpnI and inserted into the EcoRI/KpnI site of plasmid pSK(−). The nucleotide sequence of the entire HuR gene was confirmed by DNA sequencing. Plasmid pEGFP/HuR was constructed by digesting pSK/HuR with EcoRI/KpnI and inserting the resulting HuR gene fragment into the EcoRI/KpnI site of pEGFP.
To construct plasmids expressing siRNAs against PTB, we selected target sites on the PTB mRNA corresponding to nucleotides (nt) 730 to 748 and 802 to 820 downstream of the initiation codon. DNA duplexes corresponding to PTB (nt 730 to 748) were generated with the complementary oligonucleotides 5′-TCG ACG AAC ATC TAC AAC GCC TGC TTC AAG AGA GCA GGC GTT GTA GAT GTT CTT TTT TT-3′ and 5′-CTA GAA AAA AAG AAC ATC TAC AAC GCC TGC TCT CTT GAA GCA GGC GTT GTA GAT GTT CG-3′. DNA duplexes corresponding to PTB (nt 802 to 820) were generated with complementary oligonucleotides (5′-TCG ACC AAT GAC AAG AGC CGT GAC TTC AAG AGA GTC ACG GCT CTT GTC ATT GTT TTT TT-3′ and 5′-CTA GAA AAA AAC AAT GAC AAG AGC CGT GAC TCT CTT GAA GTC ACG GCT CTT GTC ATT GG-3′). The respective DNA pairs were annealed and ligated into plasmid pEBV-U6+p27, as described elsewhere (24).
UV cross-linking, immunoprecipitation of UV cross-linked proteins, and competition experiment.
All the experiments were performed as previously described (23) except that 32P-labeled RNA corresponding to the p27 IRES element and purified PTB were used as the interacting RNA and protein, respectively.
In vitro translation with RRL.
In vitro translation assays were carried out with rabbit reticulocyte lysates (RRL). To investigate the effect of PTB on p27 IRES-dependent translation, the dicistronic reporter mRNA Rp27F was incubated with RRL containing extra PTB1, PTB4, or La protein. The proteins were synthesized by in vitro translation as described by Kim et al. (23).
Cell culture and reporter assay.
293T and HeLa cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (Clontech). HL60 cells were maintained in RPMI medium (Invitrogen) supplemented with 10% fetal bovine serum. For the reporter assay, 293T cells transfected with dicistronic reporter plasmids were lysed in passive lysis buffer (Promega) at 48 h posttransfection, and then firefly and Renilla luciferase activities were determined using the dual-luciferase reporter assay system (Promega). The β-galactosidase activity was determined from the same lysate using the β-galactosidase enzyme assay system (Promega). To analyze the IRES activity of p27 mRNA in differentiating HL60 cells, the cells were treated with 30 nM TPA (Calbiochem) for various times (1, 3, 5, 9, 15, 24, and 36 h) and then transfected with the above-mentioned capped dicistronic reporter RNA, Rp27F. Three hours posttransfection, cell lysates were prepared and luciferase activities were measured.
Establishment of cell clones expressing PTB siRNAs.
Plasmids (1 μg each) expressing siRNAs [pEBV-U6+27, pEBV-U6+27/PTB (nt 730 to 748), and pEBV-U6+27/PTB (nt 802 to 820)] were transfected into HeLa cells by electroporation. From 48 h posttransfection, cells were maintained in Dulbecco's modified Eagle's medium containing hygromycin (300 μg/ml; Calbiochem). After 1 month of selection, hygromycin-resistant cell colonies were pooled together and cultivated for further analysis.
Western blot analysis.
Immunoblot analysis was performed with monoclonal anti-green fluorescent protein (GFP; Santa Cruz), anti-PTB (kindly provided by E. Wimmer), anti-HuR (Santa Cruz), anti-La (kindly provided by M. Bachmann), antiactin (ICN), and anti-cdc2 (Santa Cruz) as primary antibodies and horseradish peroxidase-conjugated anti-mouse immunoglobulin G as the secondary antibody. For detection of p27 protein, an anti-p27 polyclonal antibody (Santa Cruz) and horseradish peroxidase-conjugated anti-rabbit immunoglobulin G were used as the primary and secondary antibodies, respectively. The secondary antibodies were visualized by enhanced chemiluminescence (Amersham-Pharmacia Biotech).
Metabolic radiolabeling and immunoprecipitation.
HL60 cells were treated with 30 nM TPA for various times (6, 12, 24, 36, and 48 h). Cells were washed twice with PBS and then incubated in methionine- and cysteine-free RPMI medium for 60 min. Cells were further incubated for 60 min after supplementation of [35S]methionine ([35S]Met) and [35S]cysteine ([35S]Cys) (500 μCi/ml; NEN Life Science Products). Cells were washed, harvested, and lysed for immunoprecipitation. Cell lysates (2 × 105 cpm) were subjected to immunoprecipitation with a rabbit polyclonal antibody against p27 (Santa Cruz).
Fluorescence-activated cell sorter (FACS) analysis.
HeLa cells expressing siRNA against PTB (nt 730 to 748) or PTB (nt 802 to 820) were washed twice with phosphate-buffered saline and fixed in 100% cold ethanol. Fixed cells were stained with propidium iodide (40 μg/ml; Sigma) and treated with RNase A (50 μg/ml; Sigma) for 30 min at room temperature. About 2 × 104 cells were analyzed for DNA content on a FACScan (Becton Dickinson).
RESULTS
PTB interacts with the IRES of p27 mRNA.
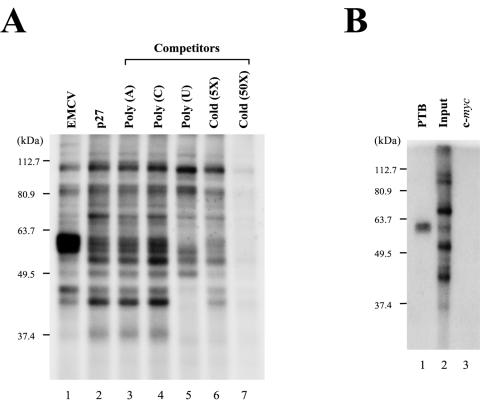
In an effort to better understand the molecular basis of the IRES-dependent translation of p27 mRNA, we sought to identify a cellular factor(s) interacting with the p27 IRES. For this purpose, a UV cross-linking experiment was performed with a cytoplasmic extract of HeLa S3 cells and a 32P-labeled RNA corresponding to the p27 IRES. Cellular proteins with apparent molecular masses of 37, 42, 52, 57, 60, 65, and 110 kDa were found to bind to the p27 IRES. The 37- and 42-kDa proteins were identified as HuR and hnRNP C1/C2, respectively, by Millard et al. (31). We focused our research on the 57- and 60-kDa proteins, as these appeared to share several characteristics of the PTB protein, which is known to enhance the translation of many IRES elements. First, the 57- and 60-kDa proteins showed the same mobilities as PTB isoforms that have been identified as the strongest bands exposed by the EMCV IRES element on a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel (Fig. 1A, compare lane 2 with lane 1) (4, 20). Second, the 57- and 60-kDa proteins showed high affinities to homopolymeric uracil [poly(W)]. In competition experiments using homopolymeric RNAs (lanes 3 to 5 in Fig. 1A), the 57- and 60-kDa protein bands disappeared in the presence of poly(U) RNA (lane 5 in Fig. 1A). The identities of the 57- and 60-kDa proteins were confirmed by immunoprecipitation of the UV cross-linked proteins by 32P-labeled p27 IRES RNA. The 57- and 60-kDa proteins were precipitated with a specific monoclonal antibody against PTB (DH17) (Fig. 1B, lane 1). In contrast, no band was detected when the negative control (monoclonal antibody against the c-myc protein) was used (Fig. 1B, lane 3). A direct interaction between PTB and p27 IRES RNA was further confirmed by a UV cross-linking experiment using purified PTB4 protein and 32P-labeled p27 IRES RNA (Fig. 2B, ii). Together, these data indicate that PTB proteins directly bind to the p27 IRES.
FIG. 1.
Identification of PTB as a cellular protein interacting with the human p27 IRES. (A) UV cross-linking experiments were performed with S10 extracts of HeLa S3 cells (20 μg each) and 32P-labeled RNAs (3 × 105 cpm each) corresponding to nt 260 to 833 of the EMCV IRES (lane 1) and to nt −447 to −1 of the p27 IRES (lanes 2 to 7). The first nucleotide in the EMCV genomic RNA is designated nt 1. For p27 mRNA, the adenine in the initiation codon is designated nt 1, and upstream sequences are denoted by the (−) symbol. Competition experiments were carried out with various competitor RNAs, including 50 ng of homopolymeric RNAs [poly(A), poly(C), and poly(U)] (lanes 3 to 5) and 5- and 50-fold molar excesses of unlabeled p27 IRES RNA (lanes 6 and 7). After cross-linking reactions, samples were treated with RNase cocktail and then analyzed by SDS-12% PAGE. (B) Immunoprecipitation of proteins labeled by UV cross-linking with the 32P-labeled p27 RNA probe. After RNase treatment, samples were precleared with protein G-agarose resin and then reacted with 2 μg of monoclonal anti-PTB antibody (DH17) (lane 1) or anti-c-myc antibody (lane 3). Resin-bound proteins were resolved by SDS-10% PAGE. Lane 2 shows the labeled proteins prior to immunoprecipitation.
FIG. 2.
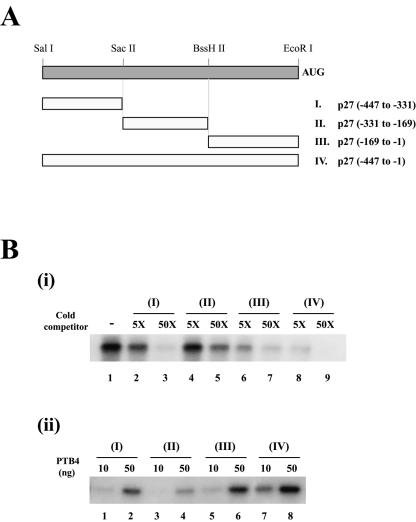
Determination of the PTB-binding site on the p27 IRES. (A) Schematic diagram of the p27 RNAs used in the various competition experiments. AUG denotes the initiator AUG codon. (B, i) Competition of RNAs for interaction with PTB. Competition experiments were carried out with purified PTB4 (100 ng), 32P-labeled RNA corresponding to probe IV of panel A (lane 1), and cold competitor RNAs (probe I [lanes 2 and 3], probe II [lanes 4 and 5], probe III [lanes 6 and 7], and probe IV [lanes 8 and 9]) in a 5-fold (lanes 2, 4, 6, and 8) or 50-fold (lanes 3, 5, 7, and 9) molar excess. (ii) Binding of PTB to various regions of the p27 5′NTR. UV cross-linking experiments were performed with 10 ng (lanes 1, 3, 5, and 7) or 50 ng (lanes 2, 4, 6, and 8) of purified PTB4 and 32P-labeled RNAs corresponding to probe I (lanes 1 and 2), probe II (lanes 3 and 4), probe III (lanes 5 and 6), and probe IV (lanes 7 and 8).
In order to map the site at which PTB binds the p27 IRES, we performed a UV cross-linking experiment in the presence of cold competitor RNAs corresponding to different portions of the p27 IRES (Fig. 2A). Purified PTB4 protein and 32P-labeled RNAs spanning nt −447 to +3 of the p27 mRNA were UV cross-linked in the presence of cold competitor RNAs. Addition of a 50-fold molar excess of RNAs I (corresponding to nt −447 to −331) and III (corresponding to nt −169 to −1) strongly inhibited the binding of PTB4 to the probe (Fig. 2B, i, lanes 3 and 7). RNA III showed the strongest competition for binding with PTB4 when a fivefold molar excess of competitor RNA was used (Fig. 2B, i, compare lane 6 with lanes 2 and 4). RNA II (corresponding to nt −331 to −169) weakly competed for interaction with PTB4 (Fig. 2B, i, lanes 4 and 5). Purified PTB1 also showed a similar pattern of competition (data not shown). Furthermore, we performed UV cross-linking experiments with purified PTB4 protein and 32P-labeled RNAs corresponding to different regions of the p27 IRES (Fig. 2A). PTB4 interacted strongly with RNAs III and IV, to a lesser extent with RNA I, and weakly with RNA II (Fig. 2B, ii). These data collectively indicate that PTB proteins strongly bind to a region proximal to the translation start codon of the p27 mRNA.
The region from nt −169 to −1 of the p27 IRES is required for strong IRES activity in the p27 mRNA.
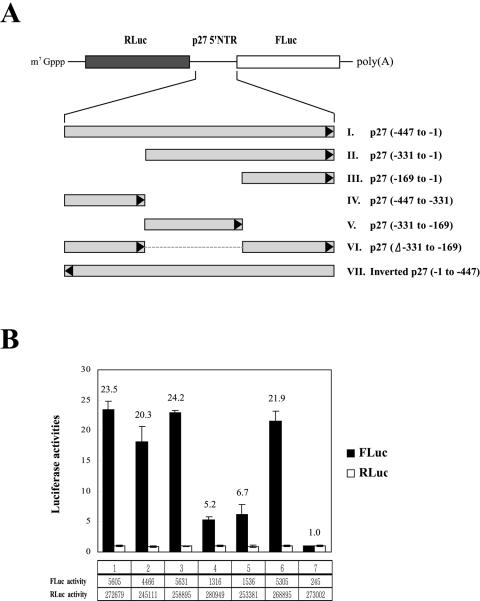
To determine the presence of cis-acting elements in the p27 IRES element, we investigated the effect of deletions in the 5′NTR of p27 mRNA on translation in mammalian cells, using dicistronic mRNAs containing different portions of the 5′NTR in the intercistronic region (Fig. 3A). In these constructs, the translation of RLuc from the first cistron is directed by ribosomal scanning, but translation of FLuc is directed by the modified IRES elements located in the intercistronic region. The 5′NTR of the p27 mRNA from nt −447 to −1, which was reported to contain a fully active IRES element, enhanced the translation of FLuc by 23.5-fold (Fig. 3B, lane 1) compared with that of a negative-control mRNA with an inverted sequence of the p27 mRNA from nt −447 to −1 (RNA VII in Fig. 3A; Fig. 3B, lane 7). All mRNAs containing nt −169 to −1 of the p27 5′NTR showed IRES activities similar to that of p27 mRNA (−447 to −1) (Fig. 3B, compare lane 1 with lanes 2, 3, and 6), indicating that nt −169 to −1 of the p27 5′NTR contains a functional IRES element. The region from nt −169 to −1 of p27 IRES has a high pyrimidine content and is particularly U rich. Curiously, RNA IV (spanning nt −447 to −331) and RNA V (spanning nt −331 to −169) also showed IRES activity, though to a lesser degree than the main p27 IRES (−169 to −1) (Fig. 3B, compare lanes 4 and 5 with lane 3). The physiological role of this possibly redundant IRES element remains to be elucidated, but the main p27 IRES element appears to span nt −169 to −1, which is also the region that strongly binds PTB (see above).
FIG. 3.
The region between nucleotides −169 to −1 is required for efficient IRES function of the p27 mRNA. (A) Schematic diagram of the dicistronic mRNAs used for monitoring the IRES activities of truncated p27 mRNAs. Various regions of the p27 5′NTR were inserted into the intercistronic regions of dicistronic mRNAs composed of the Renilla (RLuc) and firefly (FLuc) luciferase genes. Black arrowheads represent the orientation of the RNA sequence. RNA VII contains the inverted p27 RNA sequence (nt −1 to −447) at the intercistronic region. (B) IRES activities of various regions of the p27 5′NTR. 293T cells were transfected with plasmids expressing the dicistronic mRNAs shown in panel A and the control plasmid pCMV•SPORT-βgal (Invitrogen). Forty-eight hours posttransfection, cells were harvested and firefly and Renilla luciferase activities were measured. Firefly and Renilla luciferase activities were normalized with β-galactosidase activity to adjust the transfection efficiency. Black and white bars depict firefly and Renilla luciferase activities, respectively. The relative ratios of firefly luciferase to Renilla luciferase (FLuc/RLuc) are also depicted by numbers on the top of each bar. The ratio of the luciferase activities (FLuc/RLuc) in cells containing the dicistronic mRNA (RNA VII in panel A) with an inverted p27 IRES (−1 to −447) is set to 1. The activities of firefly and Renilla luciferases are depicted in boxes below the graph.
PTB enhances IRES-dependent translation of the p27 mRNA.
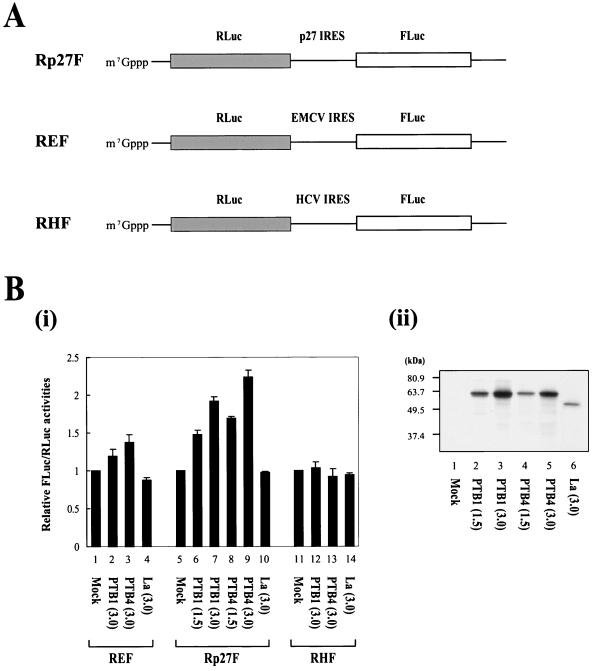
PTB has been shown to function as an IRES trans-acting factor (ITAF) for viral and cellular IRESs. Therefore, we investigated whether PTB augments the IRES-dependent translation of p27 mRNA in RRL, an in vitro translation system. We generated the effector proteins PTB1, PTB4, and La (as a control RNA-binding protein), through in vitro transcription by T7 RNA polymerase and in vitro translation of each mRNA in RRL. RRL containing the newly produced effector proteins were mixed with RRL containing dicistronic reporter mRNAs (Fig. 4A), and in vitro translation reactions were carried out as described by Kim et al. (23). Addition of PTB1 (Fig. 4B, i, lane 2) and PTB4 (Fig. 4B, i, lane 3) augmented translation directed by the EMCV IRES, the activity of which is known to be enhanced by PTB proteins (21). Similarly, addition of PTB1 (Fig. 4B, i, lanes 6 and 7) and PTB4 (Fig. 4B, i, lanes 8 and 9) proteins into the in vitro translation mixtures increased the IRES activity of the p27 mRNA in a dose-dependent manner (Fig. 4B, i, lanes 6 to 9). Interestingly, PTB4 has higher translational enhancing activity than PTB1 on the p27 IRES (compare lanes 8 and 9 with lanes 6 and 7 in Fig. 4B, i, respectively). This effect was obvious considering the translational efficiencies of PTB1 and PTB4 in RRL. The amount of PTB1 was larger than that of PTB4 (compare lanes 4 and 5 with lanes 2 and 3 in Fig. 4B, ii, respectively). The PTB-induced enhancement of the p27 IRES was greater in magnitude than the effect of PTB on the EMCV IRES. In contrast, addition of RRL containing newly synthesized La did not affect the IRES activities of the p27 and EMCV mRNAs (Fig. 4B, i, lanes 4 and 10). Note that the amount of La protein was underrepresented by the autoradiograph shown in Fig. 4B, ii, since the newly synthesized proteins were labeled by [35S]Met. La proteins and PTB isoforms contain 5 and 11 methionine residues, respectively. Translation directed by the hepatitis C virus (HCV) IRES was not affected by the PTBs or La proteins under these experimental conditions (Fig. 4B, i, lanes 11 to 14). Together, these data strongly suggest that PTB1 and PTB4 enhance IRES-dependent translation of the p27 mRNA.
FIG. 4.
Effects of PTB1 and PTB4 on translation directed by the p27 IRES element. (A) Schematic diagram of dicistronic mRNAs used in monitoring IRES activities in vitro. Dicistronic mRNAs Rp27F, REF, and RHF contained the p27 IRES, EMCV IRES, and HCV IRES, respectively. The dicistronic mRNAs were synthesized by T7 RNA polymerase in the presence of 7methyl-GpppG for addition of cap structures at the 5′ ends. (B, i) Translation reactions were carried out as described in Materials and Methods. Various amounts of PTB1, PTB4, and La (a control RNA-binding protein) were generated by in vitro translation of effector mRNAs encoding PTB1, PTB4, and La, respectively. Dicistronic mRNAs were preincubated with RRL containing the effector proteins, and then translation reactions were carried out by addition of fresh RRL and amino acids. After translation, firefly and Renilla luciferase activities were measured, and the relative IRES activities were calculated. The value obtained by the addition of effector mRNA-free RRL was set to 1 for each mRNA (bars 1, 5, and 11). The numbers in parentheses indicate the volume (in microliters) of effector-containing RRL added. (ii) The amounts of effector proteins newly synthesized in RRL were monitored by labeling newly synthesized proteins with [35S]Met. The labeled proteins were analyzed by SDS-12% PAGE.
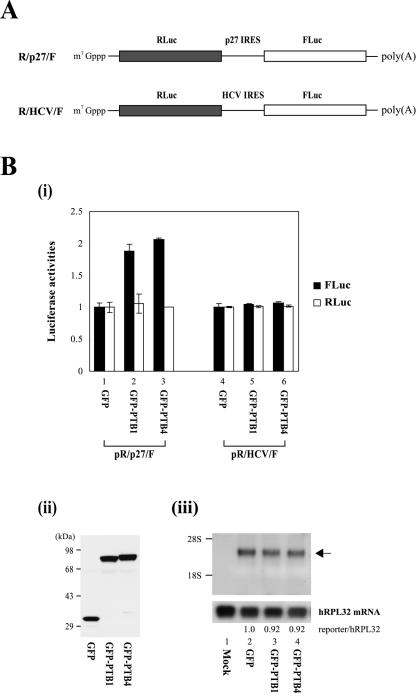
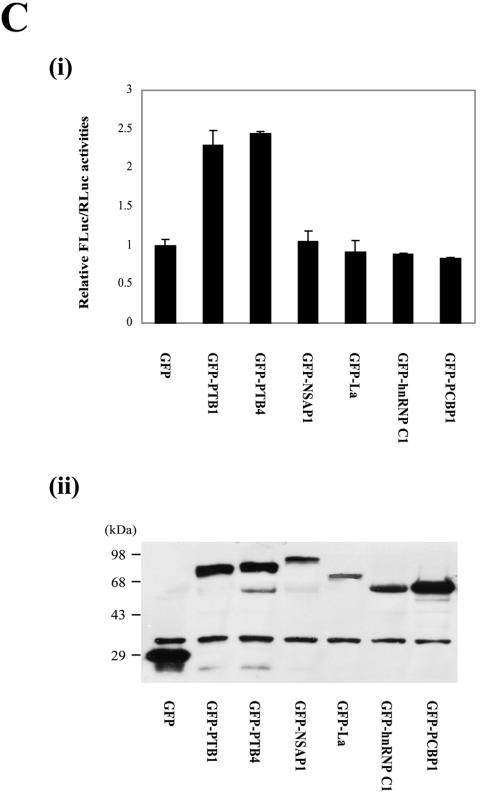
We also investigated the effect of PTB on p27 IRES-dependent translation in mammalian cells through cotransfection of reporter plasmids producing the dicistronic mRNAs R/p27/F and R/HCV/F (Fig. 5A) and effector plasmids producing PTB1 or PTB4 fused with GFP. Overexpression of GFP-PTB1 or GFP-PTB4 in 293T cells enhanced the IRES activity of the p27 mRNA by about twofold (Fig. 5B, i, lanes 2 and 3), whereas the IRES activity of the HCV mRNA was not affected under the same conditions (Fig. 5B, i, lanes 5 and 6). To exclude the possibility that the increase of firefly luciferase activity by PTB proteins can be attributed to an alteration of integrity and/or stability of reporter mRNA, we monitored the dicistronic reporter mRNA R/p27/F by Northern blot analysis with a 32P-labeled probe corresponding to the firefly luciferase gene. Overexpression of GFP-PTB1 and GFP-PTB4 had no significant effect on the amount or integrity of the dicistronic mRNA. (Fig. 5B, iii, compare lanes 3 and 4 with lane 2). No mRNA of monocistronic mRNA size, which might be produced by a putative cryptic promoter or cryptic splicing site, was detected. Similar amounts of GFP, GFP-PTB1, and GFP-PTB4 proteins were expressed in the transfected cells, as detected by Western blotting with a monoclonal anti-GFP antibody (Fig. 5B, ii). Consistent with the results of an in vitro experiment (Fig. 4B, i, compare lanes 6 and 7 with lanes 8 and 9), PTB4 showed a slightly higher translational enhancement of the p27 IRES than did PTB1 (Fig. 5B, i, compare lane 2 with lane 3). Next, the effects of other well-known ITAFs (NSAP1, La, hnRNP C1, and PCBP1) on the p27 IRES function were investigated through cotransfection of dicistronic reporter plasmid pR/p27/F and effector plasmids producing GFP-fused ITAFs (Fig. 5C). Only PTB1 and PTB4 showed significant effects on p27 IRES activity (Fig. 5C, i), even though the various effector proteins were expressed at acceptable levels in the transfected cells (Fig. 5C, ii). Taken together, these results indicate that PTB proteins specifically enhance IRES-dependent translation of the p27 mRNA in vitro and in vivo.
FIG. 5.
PTB1 and PTB4 enhance the IRES activity of p27 mRNA in vivo. (A) Schematic diagram of dicistronic mRNAs used for in vivo monitoring of the effect of PTB on IRES activities. The dicistronic mRNA containing the HCV IRES was used as a control. (B, i) 293T cells were cotransfected with a reporter plasmid (pR/p27/F or pR/HCV/F), an effector plasmid expressing GFP, GFP-PTB1, or GFP-PTB4, and the control plasmid pCMV•SPORT-βgal. Forty-eight hours posttransfection, cells were harvested and luciferase activities were measured. Firefly and Renilla luciferase activities were normalized with β-galactosidase activity to adjust transfection efficiency. Black and white bars depict firefly and Renilla luciferase activities, respectively. Luciferase activities in cells expressing GFP is set to 1 (lanes 1 and 4). (ii) Lysates were Western blotted with a monoclonal anti-GFP antibody. (iii) Northern blot analysis of reporter dicistronic mRNA R/p27/F produced in the transfected cells. Three micrograms of poly(A)+ RNAs purified from transfected cells were subjected to Northern blotting with a 32P-labeled probe corresponding to the firefly luciferase gene. The positions of 28S and 18S rRNAs are indicated by 28S and 18S, respectively. The hRPL32 blot was used as an internal control of poly(A)+ mRNAs. The arrow indicates the position of the reporter mRNA. Radioactivities of the bands corresponding to dicistronic reporter mRNA (R/p27/F) were normalized to those corresponding to hRPL32 mRNAs. Numbers under the panel reporter/hRPL32 indicate the relative radioactivities of dicistronic reporter mRNA (R/p27/F) bands normalized with hPRL32 bands. The value of mRNAs from the cells expressing GFP was set to 1 (lane 2). (C) 293T cells were cotransfected with reporter plasmid pR/p27/F and effector plasmids expressing GFP (lane GFP) and ITAFs fused with GFP. The identities of the various ITAFs are indicated below the graph (i) and the gel (ii). Forty-eight hours posttransfection, the relative luciferase activities were determined. The ratio of firefly luciferase activity to Renilla luciferase activity in cells transfected with effector GFP was set to 1 (i). (ii) Lysates were Western blotted with a polyclonal anti-GFP antibody.
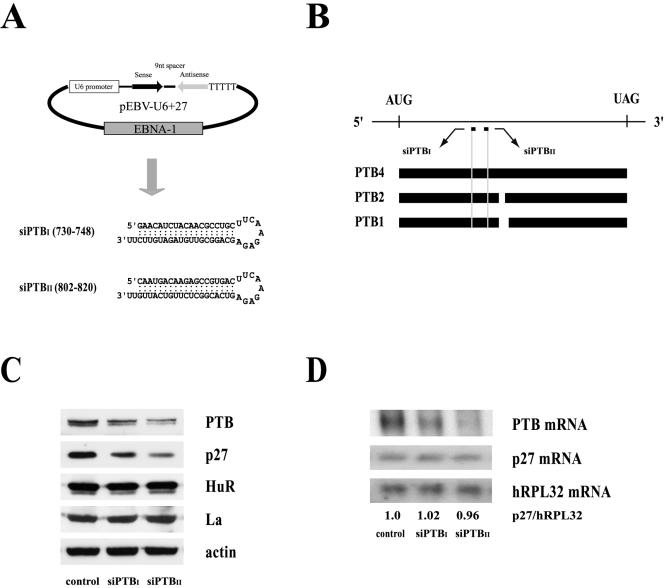
Knockdown of PTB by siRNAs inhibits IRES-dependent translation of the p27 mRNA.
We further analyzed the effect of PTBs on p27 IRES activity by reducing cellular PTB protein levels with PTB-specific siRNAs. We established HeLa cell clones constitutively expressing two siRNAs, siPTBI and siPTBII, which corresponded to nt 730 to 748 and 802 to 820 of PTB, respectively. These regions are present in PTB isoforms PTB1, PTB2, and PTB4 (Fig. 6A and B). To exclude observational bias resulting from the use of a single clone, we pooled hygromycin-resistant cell colonies and used the combined pools for our analyses. Cells expressing siPTBI and siPTBII showed dramatically reduced PTB levels compared with those of cell clones containing the negative-control plasmid (Fig. 6C, panel PTB). Interestingly, siPTBII showed a stronger knockdown effect than did siPTBI, as shown by levels of PTB protein (Fig. 6C, panel PTB; compare lanes control, siPTBI, and siPTBII) and mRNA (Fig. 6D, panel PTB mRNA; compare lanes control, siPTBI, and siPTBII). Consistent with the translational activation effect of PTB on the p27 IRES, p27 protein levels decreased dose dependently as the PTB levels decreased due to siRNA knockdown (Fig. 6C, panel p27; compare lanes control, siPTBI, and siPTBII). In contrast, the levels of other proteins, such as two well-known ITAFs (HuR and La) and the negative control (actin), were not affected by the presence of the PTB-specific siRNA.
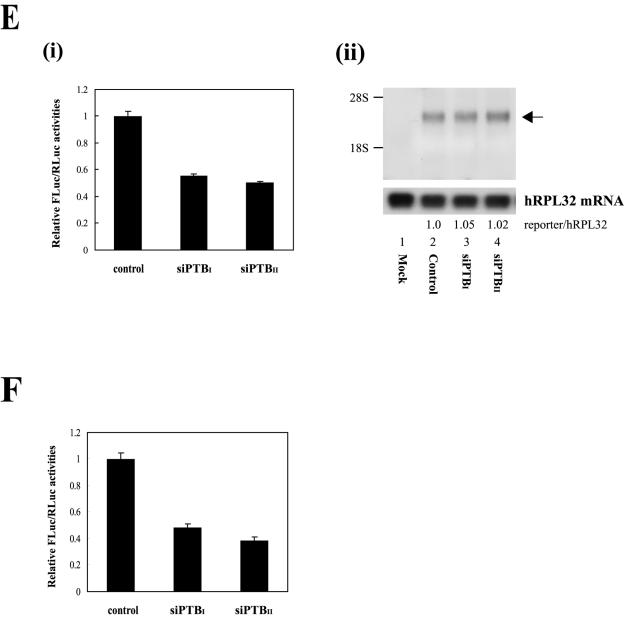
FIG. 6.
Effects of PTB-specific siRNA on p27 IRES activity. (A) Schematic diagram of the pEBV-U6+27 siRNA-expressing plasmid (upper panel) and the predicted secondary structures of siRNAs generated from pEBV-U6+27/PTBI (nt 730 to 748) and -U6+27/PTBII (nt 802 to 820) (lower panel). (B) The positions of the siRNA target sites (nt 730 to 748 and 802 to 820) on the PTB mRNAs (PTB1, PTB2, and PTB4) are depicted by the short bars below the mRNA. AUG and UAG represent the initiation and termination codons of the PTB mRNA, respectively. The open reading frames of PTB4, PTB2, and PTB1 are depicted by solid and broken bars. (C) Western blot analysis of HeLa cells expressing the two siRNAs, siPTBI and siPTBII. Stably transformed HeLa cells were generated by transfection of the control vector and vectors encoding siPTBI and siPTBII. Cells were harvested, and protein levels were analyzed by immunoblotting with anti-PTB, -p27, -HuR, -La, and -actin antibodies. (D) Northern blot analysis of HeLa cells expressing siRNAs against PTB. Thirty micrograms of total RNA was resolved on a denaturing gel and immobilized onto a nylon membrane. The filter was consecutively hybridized with 32P-labeled probes specific for the p27, PTB, and hRPL32 mRNAs. Radioactivities of the bands corresponding to p27 mRNAs were normalized to those corresponding to hRPL32 mRNAs. Numbers under the panel p27/hRPL32 indicate the relative radioactivities of p27 bands normalized with hPRL32 bands. The value of mRNAs from the cells containing control vector was set to 1. (E, i) The effects of siRNAs on p27 IRES activity monitored by transfection of DNA expressing a dicistronic mRNA R/p27/F (as shown in Fig. 5A). Cells with or without siRNAs were transfected with plasmid pR/p27/F, and the IRES activity was measured by firefly luciferase activity 48 h posttransfection. The DNA transfection efficiency was normalized in terms of Renilla luciferase activity directed by the cap-dependent translation, and the ratios of firefly luciferase to Renilla luciferase activity were calculated. The value of siRNA-lacking control cells was set to 1. (ii) Northern blot analysis of dicistronic mRNAs produced from HeLa cells expressing siRNAs against PTB. Three micrograms of poly(A)+ RNAs prepared from transfected cells was subjected to Northern blotting with a 32P-labeled probe corresponding to the firefly luciferase gene. The positions of 28S and 18S ribosomal RNAs are indicated by 28S and 18S, respectively. The hRPL32 blot was used as an internal control of poly(A)+ mRNAs. The arrow indicates the position of the reporter mRNA. Relative radioactivities of bands corresponding to dicistronic reporter mRNA (R/p27/F) and hRPL32 mRNAs were calculated as described in the legend to Fig. 5B, iii. The value of mRNA from cells containing vector was set to 1 (lane 2). (F) The effects of siRNAs on p27 IRES activity monitored by transfection of the Rp27F dicistronic RNA (shown in Fig. 4A). Relative IRES activities were measured and calculated as described for panel Ei, except that cells were harvested 3 h posttransfection.
The effects of the PTB-specific siRNAs on the PTB and p27 mRNAs were monitored by Northern blot analysis (Fig. 6D). As described previously, the levels of PTB mRNA were drastically reduced by expression of the PTB-specific siRNAs (Fig. 6D, panel PTB mRNA). In contrast, the levels of p27 mRNA and human ribosomal protein large-subunit 32 (hRPL32) mRNA (negative control) were not affected by the siRNAs (Fig. 6D, panels p27 mRNA and hRPL32 mRNA). The effects of siRNA-induced PTB protein knockdown on p27 IRES activity were further monitored via transfection of plasmid pR/p27/F (encoding a dicistronic mRNA) (Fig. 3A and 6E, i) or an in vitro-transcribed dicistronic mRNA (Fig. 4A and 6F). The RNA transfection method was used to rule out the possibility of a monocistronic mRNA being produced from a putative cryptic promoter or cryptic splicing acceptor in the 5′NTR of the p27 mRNA. Both the DNA (Fig. 6E, i) and RNA (Fig. 6F) transfection experiments revealed that the reduction of PTB protein levels by siRNAs decreased the IRES activity of the p27 5′NTR. Interestingly, cells expressing siPTBII, which had a stronger siRNA activity than cells expressing siPTBI, showed weaker IRES activities toward the p27 5′NTR (Fig. 6E, i, and F, compare the relative FLuc and RLuc activities in cells expressing siPTBI with those in cells expressing siPTBII). The amount and the integrity of the dicistronic reporter mRNA in the cells containing siRNAs were monitored by Northern blot analysis with a 32P-labeled probe corresponding to the firefly luciferase gene. PTB knockdown affected neither the amount nor the integrity of reporter mRNAs (Fig. 6E, ii, compare lanes 3 and 4 with lane 2). Together, these data indicate that PTB regulates the expression of endogenous p27 at the translational level via an IRES element in the p27 5′NTR.
TPA-induced enhancement of p27 in HL60 cells correlates with cellular PTB levels and the activity of the p27 IRES.
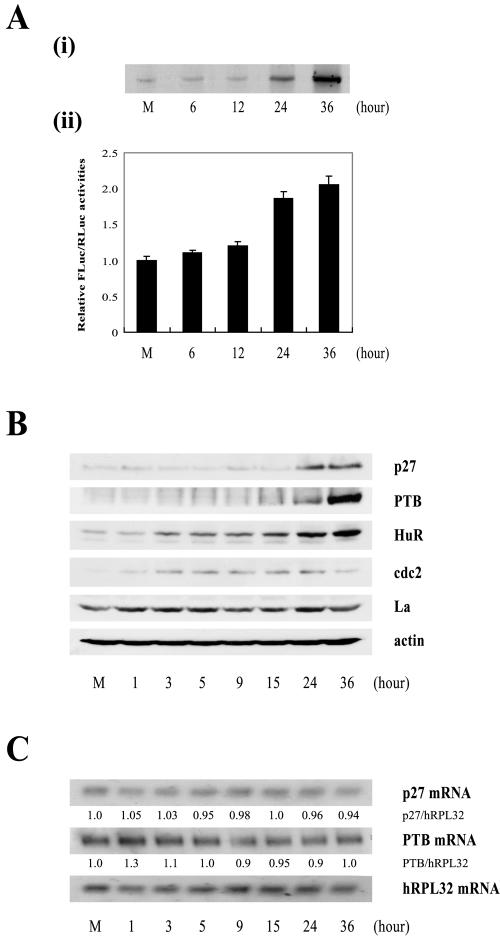
HL60 cells have been used to investigate the molecular mechanism of cellular differentiation. In the presence of TPA, HL60 cells cease proliferating and differentiate into monocytes. TPA treatment of HL60 cells has also been associated with increases in p27 protein levels due to increased levels of polysomally associated p27, suggesting that TPA treatment might increase the translation of p27 mRNA (32). To confirm the translational regulation of p27 mRNA during the differentiation of HL60 cells, we examined the newly synthesized p27 protein by metabolic radiolabeling of HL60 cells for 60 min at various times after TPA treatment (Fig. 7A, i). Newly synthesized p27 protein was observed by immunoprecipitation of 35S-labeled proteins with an antibody against p27. The numbers indicate the commencement time of metabolic labeling after TPA treatment. The newly synthesized p27 protein dramatically increased at 24 h after TPA treatment and reached maximum at 36 h after TPA treatment. Furthermore, we investigated the IRES activity of the p27 mRNA by transfecting a dicistronic mRNA Rp27F (Fig. 4A) into HL60 cells at various times after TPA treatment (Fig. 7A, ii). The efficiency of translation through the p27 IRES was compared to that occurring through scanning, as measured by the relative activities of FLuc to RLuc in cells 3 h posttransfection with the reporter RNA Rp27F (Fig. 7A, ii). The results revealed that the IRES activity of the p27 mRNA gradually increased following TPA treatment (Fig. 7A, ii, compare bars 6, 12, 24, and 36 with bar M), with an unusually drastic increase of p27 IRES activity seen 24 h after TPA treatment. We also measured the levels of endogenous p27 protein in the TPA-treated HL60 cells by Western blotting. Consistent with the IRES activity of the p27 mRNA, p27 protein levels were drastically increased 24 h after TPA treatment (Fig. 7B, panel p27; compare lane 24 with lanes M, 1, 3, 5, 9, and 15). Curiously, however, the level of p27 protein 36 h after TPA treatment was not increased compared with that 24 h after TPA treatment. In other words, the p27 protein level after 36 h of TPA treatment (Fig. 7B, panel p27) underrepresents the rate of protein synthesis shown by the immunoprecipitation of newly synthesized p27 protein (Fig. 7A, i) and by p27 IRES activity (Fig. 7A, ii). The discrepancy may be attributed to the increased degradation rate of p27 protein after 36 h of TPA treatment through an unknown mechanism. After TPA treatment, the levels of p27 mRNA remained unchanged, as analyzed by Northern blot analysis (Fig. 7C, panel p27 mRNA). Together, these data strongly suggest that translation through the p27 IRES increases greatly 24 h after TPA treatment of HL60 cells.
FIG. 7.
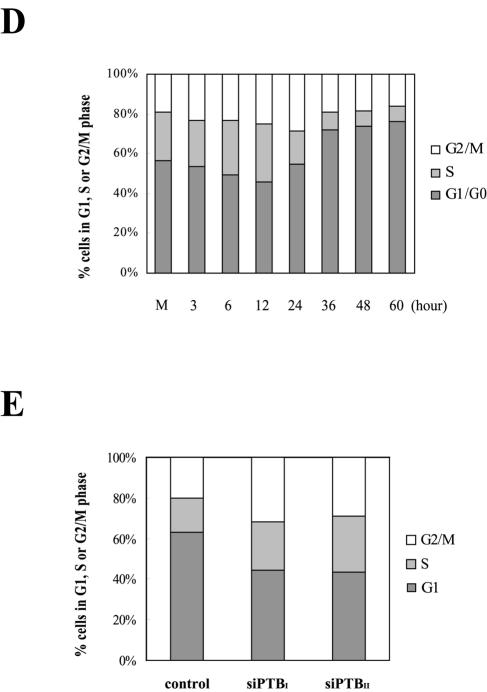
IRES activity of the p27 mRNA during differentiation of HL60 cells. (A, i) The newly synthesized p27 proteins were monitored by metabolic labeling of proteins with [35S]Met and [35S]Cys followed by immunoprecipitation with an antibody against p27. The immunoprecipitated samples were resolved by SDS-15% PAGE and then subjected to autoradiogram. (ii) Differentiation of HL60 cells was induced by TPA treatment (30 nM). The Rp27F dicistronic reporter RNA was transfected into HL60 cells 6, 12, 24, and 36 h after TPA was used to induce differentiation, and luciferase activities were measured 3 h posttransfection. The relative luciferase activity of untreated cells was set to 1. (B) Protein levels in HL60 cells during differentiation. HL60 cells were harvested 1, 3, 5, 9, 15, 24, and 36 h after TPA treatment (30 nM) and then subjected to Western blot analysis with anti-p27, -PTB, -HuR, -cdc2, -La, and -actin antibodies. (C) Northern blot analysis of p27, PTB, and hRPL32 mRNAs during differentiation of HL60 cells. Cells were treated with TPA (30 nM) for 1, 3, 5, 9, 15, 24, and 36 h, and total RNAs were isolated. The RNAs (30 μg each) were resolved on a denaturing agarose gel, transferred to a nylon membrane, and hybridized with 32P-labeled probes for p27, PTB, and hRPL32. Relative radioactivities of bands corresponding to p27 and PTB mRNAs were calculated as described in the legend to Fig. 6D. The relative radioactivities of undifferentiated cells were set to 1. (D) Changes in cell phase population proportions during differentiation of HL60 cells. HL60 cells were stained with propidium iodide before (lane M) and after treatment with TPA (30 nM) for the indicated times (6, 12, 24, 36, 48, and 60 h) and then subjected to FACS analysis. The relative proportions of cells in the different phases (G2/M, S, and G1/G0) are depicted in the graphs. (E) Effects of PTB-specific siRNAs on cell proliferation. The proportion of cell phases in control and siRNA-expressing cells was analyzed and depicted as described for panel D.
To examine the molecular basis of p27 mRNA translational activation, the protein levels of PTB were next examined by Western blotting. PTB protein levels (Fig. 7B, panel PTB) correlate well with the levels of newly synthesized p27 protein (Fig. 7A, i), indicating that TPA-induced increases in p27 protein levels occur, at least in part, through PTB-induced activation of the p27 IRES element. Curiously, the mRNA levels of both PTB and hRPL32 (control) were unchanged after TPA treatment of HL60 cells, as shown by the amounts of p27 and PTB mRNAs relative to that of hRPL32 mRNA (Fig. 7C, panels p27/hRPL32 and PTB/hRPL32), indicating that the PTB levels are also posttranscriptionally regulated. A study of the regulation of PTB gene expression is in progress. We also measured the levels of HuR proteins in HL60 cells after TPA treatment, since HuR was shown to repress the IRES activity of the p27 mRNA (28). To our surprise, HuR levels increased gradually following TPA treatment, reaching maximal levels 24 and 36 h after TPA treatment, when the IRES activity of p27 mRNA was also at its highest level (Fig. 7B, panel HuR). This indicates that HuR is not a major factor regulating p27 IRES activity during the TPA-induced differentiation of HL60 cells. Consistent with a previous report describing the behavior of cdc2, a crucial regulator of G2/M progression, during rapid proliferation of HL60 cells before maturation (5), we found that cdc2 fluctuated following TPA treatment (Fig. 7B, panel cdc2). In contrast, the levels of La and actin (controls) were unchanged following TPA treatment.
Finally, the physiological role of PTB in cell cycle regulation (possibly occurring through modulation of p27 expression) was monitored by cell cycle FACS analysis during TPA-induced differentiation of HL60 cells (Fig. 7D) and siRNA-induced knockdown of PTB (Fig. 7E). It has been demonstrated that p27 plays a key role in maintaining the G1 phase of the cell cycle (48). Therefore, higher levels of p27 generated through PTB-induced activation of the p27 IRES would likely result in larger populations of cells in the G1 and/or G0 phase. As shown in Fig. 7D, the G1 cell population started to increase 24 h after TPA treatment, concurrent with the putatively PTB-induced dramatic increase in p27 IRES activity (Fig. 7A and B). When cells were further incubated with TPA, the population at G1 and/or G0 reached as high as 80%. A reciprocal phenomenon was observed when the PTB level was reduced by PTB-specific siRNAs. Over 60% of control HeLa cells were in G1 phase of the cell cycle, whereas only about 40% of cells containing PTB-specific siRNAs were in the G1 phase of the cell cycle (Fig. 7E, columns siPTBI and siPTBII). This is most likely due to a blockage of PTB expression, which leads to decreased IRES activity of the p27 mRNA and consequently reduced p27 protein levels (Fig. 6C and E). Taken together, these data suggest that PTB plays an important role in regulating cell cycle progression by modulating the IRES activity of the p27 mRNA.
DISCUSSION
The p27 protein, a key regulator of cell cycle progression, is regulated both transcriptionally and posttranscriptionally (17, 44). Two posttranscriptional steps, translation and protein degradation, are mainly responsible for the oscillation of p27 levels during cell cycle and the induction of p27 in growth-arrested cells (1, 15, 32, 36, 38). According to recent reports, increased protein synthesis (rather than proteolytic degradation) plays a major role in determining p27 levels during TPA-induced differentiation of HL60 cells, HeLa cell growth arrest at G1 phase, and contact inhibition of human diploid fibroblasts (15, 28, 32).
It has been shown that an IRES element in the 5′NTR of p27 mRNA directs translation of the mRNA (28, 33). Therefore, it is likely that the IRES activity of the p27 mRNA varies in response to intracellular and extracellular changes that modulate the availability and/or activity of ITAFs. In this respect, IRES-binding proteins, such as HuR, HuD, and hnRNP C1/C2, which interact with the IRES element of the p27 mRNA (28, 31), are possible candidates for regulation of the p27 IRES. In fact, HuR and the related HuD protein were shown to repress IRES-dependent translation of the p27 mRNA through an interaction with a U-rich element in the IRES element (28). However, no previous work has identified an ITAF capable of stimulating the IRES element of the p27 mRNA.
Here, we show that PTB enhances p27 IRES activity through an RNA-protein interaction. Several lines of evidence indicate that PTB augments p27 IRES function. First, PTB specifically interacts with the p27 IRES, as shown by UV cross-linking of HeLa cell extracts with 32P-labeled p27 IRES RNA followed by immunoprecipitation with an anti-PTB antibody (Fig. 1B), and further supported by UV cross-linking of purified PTB proteins with 32P-labeled p27 IRES RNAs in the presence or absence of various cold competitor RNAs (Fig. 2B). Second, addition of PTB to an in vitro translation reaction mixture increased the activity of a reporter under the control of the p27 IRES element. The effect of PTB on the p27 IRES was higher than that on the EMCV IRES, a positive-control IRES known to use PTB as an ITAF (Fig. 4B). Third, overexpression of PTB increased the activity of a reporter under the control of the p27 IRES element in an artificial dicistronic mRNA in vivo (Fig. 5B). Fourth, siRNA-mediated knockdown of PTB levels inhibited the IRES activity of p27, as shown by transfection of DNA (Fig. 6E, i) and RNA (Fig. 6F) reporters. And fifth, the level of p27 protein (which reflect the IRES activity of endogenous p27 mRNA) correlated well with the level of PTB protein during TPA-induced differentiation of HL60 cells (Fig. 7A and B).
It is not clear how PTB enhances translation of the p27 mRNA, but we can speculate several modes of action. First, a putative PTB-ribosome interaction may assist in binding or guiding ribosomes to the IRES. Alternatively, PTB may conformationally change the IRES into a structure that is better able to interact with the translational machinery, including the 40S ribosomal subunit. In support of this possibility, a previous study suggested that the RNA chaperone activity of PTB may favor rearrangement of the IRES structure to one that permits translation initiation (34). Second, it is possible that PTB recruits a canonical translation factor(s) through a putative protein-protein interaction. Third, PTB may activate IRES-dependent translation through interactions with other ITAFs. In this respect, it is noteworthy that many ITAFs are known to interact with each other (22).
Cells undergoing differentiation exhibit extensive changes in their patterns of gene expression and a profound reduction of global protein synthesis, as indicated by the release of most mRNAs from polysomes early during TPA-induced differentiation of HL60 cells (27). However, protein expression is activated in selective genes, as indicated by retention or even migration of a subset of transcripts onto polysomes during differentiation (27). The association of the p27 mRNA with polysomes is enhanced by TPA treatment of HL60 cells (32). Unexpectedly, the protein levels of HuR, which was shown to repress the activity of the p27 IRES (28), gradually increased following TPA treatment of HL60 cells, beginning at 3 h posttreatment and reaching maximal levels at 36 h posttreatment, when the p27 IRES activity was also maximal (Fig. 7A, ii, and B). This result strongly suggests that the increased IRES activity of the p27 mRNA is not associated with decreases in HuR protein levels during TPA-induced differentiation of HL60 cells. Instead, the activation of the p27 IRES function during differentiation is more likely due to the dramatic increase in PTB protein levels noted 24 h after TPA treatment (Fig. 7B, panel PTB). Interestingly PTB expression also seems to be regulated at the posttranscriptional level, since the level of PTB mRNA remained consistent during TPA-induced differentiation of HL60 cells, as indicated by Northern blot analysis (Fig. 7C, panel PTB mRNA). However, the molecular basis of posttranscriptional regulation of the PTB gene remains to be elucidated.
A few lines of evidence suggest that PTB functions in differentiation and cell cycle control. For instance, loss of Drosophila PTB specifically affects spermatid differentiation of Drosophila melanogaster (46), and PTB-like protein, a rat homolog of PTB, was shown to modulate the differentiation of PC12 cells into neuronal cells (16). However, this is the first report suggesting that PTB plays a role in cell cycle arrest during cellular differentiation. In this respect, it is noteworthy that the knockdown of PTB by specific siRNAs reduced the proportion of cells in G1 phase relative to those in G2/M and S phases (Fig. 7E). Conversely, TPA-induced differentiation of HL60 cells, which causes induction of PTB protein, increased the proportion of cells in G1/G0 phase relative to those in the G2/M and S phases (Fig. 7D). This result strongly suggests that the arrest of cell proliferation during TPA-induced differentiation of HL60 cells is most likely due at least in part to enhancement of p27 IRES activity via induction of PTB levels. However, the function of PTB should be further investigated to confirm its precise role during differentiation.
Some IRESs and ITAFs have been reported to regulate the translation of cellular mRNAs that play critical roles in cell cycle progression and differentiation. For example, the platelet-derived growth factor B (PDGF2/c-sis) mRNA contains an IRES element that shows increased activity during megakaryocytic differentiation of K562 cells (3). Sella et al. demonstrated that differentiation-dependent activation of the PDGF2/c-sis IRES correlated with binding of hnRNP C (47), and Koloteva-Levine et al. showed that Apc5, a component of anaphase-promoting complex/cyclosome (APC/C), inhibited the IRES activity of the PDGF2/c-sis mRNA in a differentiation-dependent manner (25). The IRES activity of the c-myc proto-oncogene, a key regulator of cell proliferation and apoptosis, is regulated during differentiation of a transgenic mouse (9), and HnRNP C was shown to enhance the activity of the c-myc IRES in a cell cycle phase-dependent manner (23). Here, we report that PTB enhances p27 IRES activity. Our data and analysis suggest that IRESs and ITAFs play important roles in fine-tuning the levels of pivotal regulatory proteins during cell proliferation and differentiation. Therefore, this work and future investigations into IRESs and ITAFs will increase our understanding of the molecular basis of cell differentiation and may contribute to the development of novel therapeutic strategies against cancer.
Acknowledgments
We thank E. Wimmer (State University of New York at Stony Brook) and M. Bachmann (Johannes-Gutenberg Universität, Mainz, Germany) for providing the anti-PTB and anti-La monoclonal antibodies, respectively. We are grateful to Tae-Don Kim for technical advices and helpful discussions.
The present study was supported in part by grants from the NRL (M10204000018-03J0000-01410), the MMRG (M10106000056-03B4500-01010) of MOST, the KHIDI (02-PJ2-PG1-CH16-0002), the KRF (KRF-2003-005-C00011), and POSCO.
REFERENCES
- 1.Agrawal, D., P. Hauser, F. McPherson, F. Dong, A. Garcia, and W. J. Pledger. 1996. Repression of p27kip1 synthesis by platelet-derived growth factor in BALB/c 3T3 cells. Mol. Cell. Biol. 16:4327-4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Back, S. H., Y. K. Kim, W. J. Kim, S. Cho, H. R. Oh, J. E. Kim, and S. K. Jang. 2002. Translation of polioviral mRNA is inhibited by cleavage of polypyrimidine tract-binding proteins executed by polioviral 3C(pro). J. Virol. 76:2529-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernstein, J., O. Sella, S. Y. Le, and O. Elroy-Stein. 1997. PDGF2/c-sis mRNA leader contains a differentiation-linked internal ribosomal entry site (D-IRES). J. Biol. Chem. 272:9356-9362. [DOI] [PubMed] [Google Scholar]
- 4.Borovjagin, A. V., A. G. Evstafieva, T. Ugarova, and I. N. Shatsky. 1990. A factor that specifically binds to the 5′-untranslated region of encephalomyocarditis virus RNA. FEBS Lett. 261:237-240. [DOI] [PubMed] [Google Scholar]
- 5.Brown, G., M. A. Choudhry, J. Durham, M. T. Drayson, and R. H. Michell. 1999. Monocytically differentiating HL60 cells proliferate rapidly before they mature. Exp. Cell Res. 253:511-518. [DOI] [PubMed] [Google Scholar]
- 6.Casaccia-Bonnefil, P., R. Tikoo, H. Kiyokawa, V. Friedrich, Jr., M. V. Chao, and A. Koff. 1997. Oligodendrocyte precursor differentiation is perturbed in the absence of the cyclin-dependent kinase inhibitor p27Kip1. Genes Dev. 11:2335-2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi, K., J. H. Kim, X. Li, K. Y. Paek, S. H. Ha, S. H. Ryu, E. Wimmer, and S. K. Jang. 2004. Identification of cellular proteins enhancing activities of internal ribosomal entry sites by competition with oligodeoxynucleotides. Nucleic Acids Res. 32:1308-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clurman, B. E., and P. Porter. 1998. New insights into the tumor suppression function of P27(kip1). Proc. Natl. Acad. Sci. USA 95:15158-15160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Creancier, L., P. Mercier, A. C. Prats, and D. Morello. 2001. c-myc internal ribosome entry site activity is developmentally controlled and subjected to a strong translational repression in adult transgenic mice. Mol. Cell. Biol. 21:1833-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drissi, H., D. Hushka, F. Aslam, Q. Nguyen, E. Buffone, A. Koff, A. van Wijnen, J. B. Lian, J. L. Stein, and G. S. Stein. 1999. The cell cycle regulator p27kip1 contributes to growth and differentiation of osteoblasts. Cancer Res. 59:3705-3711. [PubMed] [Google Scholar]
- 11.Durand, B., F. B. Gao, and M. Raff. 1997. Accumulation of the cyclin-dependent kinase inhibitor p27/Kip1 and the timing of oligodendrocyte differentiation. EMBO J. 16:306-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elledge, S. J., and J. W. Harper. 1994. Cdk inhibitors: on the threshold of checkpoints and development. Curr. Opin. Cell Biol. 6:847-852. [DOI] [PubMed] [Google Scholar]
- 13.Fang, F., G. Orend, N. Watanabe, T. Hunter, and E. Ruoslahti. 1996. Dependence of cyclin E-CDK2 kinase activity on cell anchorage. Science 271:499-502. [DOI] [PubMed] [Google Scholar]
- 14.Ghetti, A., S. Pinol-Roma, W. M. Michael, C. Morandi, and G. Dreyfuss. 1992. hnRNP I, the polypyrimidine tract-binding protein: distinct nuclear localization and association with hnRNAs. Nucleic Acids Res. 20:3671-3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hengst, L., and S. I. Reed. 1996. Translational control of p27Kip1 accumulation during the cell cycle. Science 271:1861-1864. [DOI] [PubMed] [Google Scholar]
- 16.Ichikawa, M., T. Kikuchi, H. Tateiwa, N. Gotoh, K. Ohta, J. Arai, and N. Yoshimura. 2002. Role of PTB-like protein, a neuronal RNA-binding protein, during the differentiation of PC12 cells. J. Biochem. (Tokyo) 131:861-868. [DOI] [PubMed] [Google Scholar]
- 17.Inoue, T., J. Kamiyama, and T. Sakai. 1999. Sp1 and NF-Y synergistically mediate the effect of vitamin D(3) in the p27(Kip1) gene promoter that lacks vitamin D response elements. J. Biol. Chem. 274:32309-32317. [DOI] [PubMed] [Google Scholar]
- 18.Jang, S. K., and E. Wimmer. 2001. The role of RNA-binding proteins in IRES-dependent translation, p. 1-33. In K. Sandberg and S. E. Mulroney (ed.), RNA binding proteins: new concepts in gene expression. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 19.Jang, S. K., H. G. Krausslich, M. J. Nicklin, G. M. Duke, A. C. Palmenberg, and E. Wimmer. 1988. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 62:2636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jang, S. K., and E. Wimmer. 1990. Cap-independent translation of encephalomyocarditis virus RNA: structural elements of the internal ribosomal entry site and involvement of a cellular 57-kD RNA-binding protein. Genes Dev. 4:1560-1572. [DOI] [PubMed] [Google Scholar]
- 21.Kaminski, A., S. L. Hunt, J. G. Patton, and R. J. Jackson. 1995. Direct evidence that polypyrimidine tract binding protein (PTB) is essential for internal initiation of translation of encephalomyocarditis virus RNA. RNA 1:924-938. [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, J. H., B. Hahm, Y. K. Kim, M. Choi, and S. K. Jang. 2000. Protein-protein interaction among hnRNPs shuttling between nucleus and cytoplasm. J. Mol. Biol. 298:395-405. [DOI] [PubMed] [Google Scholar]
- 23.Kim, J. H., K. Y. Paek, K. Choi, T. D. Kim, B. Hahm, K. T. Kim, and S. K. Jang. 2003. Heterogeneous nuclear ribonucleoprotein C modulates translation of c-myc mRNA in a cell cycle phase-dependent manner. Mol. Cell. Biol. 23:708-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, J. H., K. Y. Paek, S. H. Ha, S. Cho, K. Choi, C. S. Kim, S. H. Ryu, and S. K. Jang. 2004. A cellular RNA-binding protein enhances internal ribosomal entry site-dependent translation through an interaction downstream of the hepatitis C virus polyprotein initiation codon. Mol. Cell. Biol. 24:7878-7890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koloteva-Levine, N., D. Pinchasi, I. Pereman, A. Zur, M. Brandeis, and O. Elroy-Stein. 2004. The Apc5 subunit of the anaphase-promoting complex/cyclosome interacts with poly(A) binding protein and represses internal ribosome entry site-mediated translation. Mol. Cell. Biol. 24:3577-3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kranenburg, O., V. Scharnhorst, A. J. Van der Eb, and A. Zantema. 1995. Inhibition of cyclin-dependent kinase activity triggers neuronal differentiation of mouse neuroblastoma cells. J. Cell Biol. 131:227-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krichevsky, A. M., E. Metzer, and H. Rosen. 1999. Translational control of specific genes during differentiation of HL-60 cells. J. Biol. Chem. 274:14295-14305. [DOI] [PubMed] [Google Scholar]
- 28.Kullmann, M., U. Gopfert, B. Siewe, and L. Hengst. 2002. ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5′UTR. Genes Dev. 16:3087-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lloyd, R. V., L. A. Erickson, L. Jin, E. Kulig, X. Qian, J. C. Cheville, and B. W. Scheithauer. 1999. p27kip1: a multifunctional cyclin-dependent kinase inhibitor with prognostic significance in human cancers. Am. J. Pathol. 154:313-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michael, W. M., H. Siomi, M. Choi, S. Pinol-Roma, S. Nakielny, Q. Liu, and G. Dreyfuss. 1995. Signal sequences that target nuclear import and nuclear export of pre-mRNA-binding proteins. Cold Spring Harbor Symp. Quant. Biol. 60:663-668. [DOI] [PubMed] [Google Scholar]
- 31.Millard, S. S., A. Vidal, M. Markus, and A. Koff. 2000. A U-rich element in the 5′ untranslated region is necessary for the translation of p27 mRNA. Mol. Cell. Biol. 20:5947-5959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Millard, S. S., J. S. Yan, H. Nguyen, M. Pagano, H. Kiyokawa, and A. Koff. 1997. Enhanced ribosomal association of p27(Kip1) mRNA is a mechanism contributing to accumulation during growth arrest. J. Biol. Chem. 272:7093-7098. [DOI] [PubMed] [Google Scholar]
- 33.Miskimins, W. K., G. Wang, M. Hawkinson, and R. Miskimins. 2001. Control of cyclin-dependent kinase inhibitor p27 expression by Cap-independent translation. Mol. Cell. Biol. 21:4960-4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitchell, S. A., K. A. Spriggs, M. J. Coldwell, R. J. Jackson, and A. E. Willis. 2003. The Apaf-1 internal ribosome entry segment attains the correct structural conformation for function via interactions with PTB and unr. Mol. Cell 11:757-771. [DOI] [PubMed] [Google Scholar]
- 35.Morgan, D. O. 1997. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu. Rev. Cell Dev. Biol. 13:261-291. [DOI] [PubMed] [Google Scholar]
- 36.Nguyen, H., D. M. Gitig, and A. Koff. 1999. Cell-free degradation of p27kip1, a G1 cyclin-dependent kinase inhibitor, is dependent on CDK2 activity and the proteasome. Mol. Cell. Biol. 19:1190-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohtsubo, M., and J. M. Roberts. 1993. Cyclin-dependent regulation of G1 in mammalian fibroblasts. Science 259:1908-1912. [DOI] [PubMed] [Google Scholar]
- 38.Pagano, M., S. W. Tam, A. M. Theodoras, P. Beer-Romero, G. Del Sal, V. Chau, P. R. Yew, G. F. Draetta, and M. Rolfe. 1995. Role of the ubiquitin-proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science 269:682-685. [DOI] [PubMed] [Google Scholar]
- 39.Patton, J. G., S. A. Mayer, P. Tempst, and B. Nadal-Ginard. 1991. Characterization and molecular cloning of polypyrimidine tract-binding protein: a component of a complex necessary for pre-mRNA splicing. Genes Dev. 5:1237-1251. [DOI] [PubMed] [Google Scholar]
- 40.Pelletier, J., G. Kaplan, V. R. Racaniello, and N. Sonenberg. 1988. Cap-independent translation of poliovirus mRNA is conferred by sequence elements within the 5′ noncoding region. Mol. Cell. Biol. 8:1103-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perez, I., J. G. McAfee, and J. G. Patton. 1997. Multiple RRMs contribute to RNA binding specificity and affinity for polypyrimidine tract binding protein. Biochemistry 36:11881-11890. [DOI] [PubMed] [Google Scholar]
- 42.Polyak, K., J. Y. Kato, M. J. Solomon, C. J. Sherr, J. Massague, J. M. Roberts, and A. Koff. 1994. p27Kip1, a cyclin-Cdk inhibitor, links transforming growth factor-beta and contact inhibition to cell cycle arrest. Genes Dev. 8:9-22. [DOI] [PubMed] [Google Scholar]
- 43.Polyak, K., M. H. Lee, H. Erdjument-Bromage, A. Koff, J. M. Roberts, P. Tempst, and J. Massague. 1994. Cloning of p27Kip1, a cyclin-dependent kinase inhibitor and a potential mediator of extracellular antimitogenic signals. Cell 78:59-66. [DOI] [PubMed] [Google Scholar]
- 44.Qian, X., L. Jin, E. Kulig, and R. V. Lloyd. 1998. DNA methylation regulates p27kip1 expression in rodent pituitary cell lines. Am. J. Pathol. 153:1475-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Resnitzky, D., M. Gossen, H. Bujard, and S. I. Reed. 1994. Acceleration of the G1/S phase transition by expression of cyclins D1 and E with an inducible system. Mol. Cell. Biol. 14:1669-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robida, M. D., and R. Singh. 2003. Drosophila polypyrimidine-tract binding protein (PTB) functions specifically in the male germline. EMBO J. 22:2924-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sella, O., G. Gerlitz, S. Y. Le, and O. Elroy-Stein. 1999. Differentiation-induced internal translation of c-sis mRNA: analysis of the cis elements and their differentiation-linked binding to the hnRNP C protein. Mol. Cell. Biol. 19:5429-5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sherr, C. J., and J. M. Roberts. 1999. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 13:1501-1512. [DOI] [PubMed] [Google Scholar]
- 49.Sherr, C. J., and J. M. Roberts. 1995. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 9:1149-1163. [DOI] [PubMed] [Google Scholar]
- 50.Tikoo, R., P. Casaccia-Bonnefil, M. V. Chao, and A. Koff. 1997. Changes in cyclin-dependent kinase 2 and p27kip1 accompany glial cell differentiation of central glia-4 cells. J. Biol. Chem. 272:442-447. [DOI] [PubMed] [Google Scholar]
- 51.Zhou, P., Y. Yao, J. W. Soh, and I. B. Weinstein. 1999. Overexpression of p21Cip1 or p27Kip1 in the promyelocytic leukemia cell line HL60 accelerates its lineage-specific differentiation. Anticancer Res. 19:4935-4945. [PubMed] [Google Scholar]