Abstract
Iron-regulatory protein 2 (IRP2), a posttranscriptional regulator of iron metabolism, undergoes proteasomal degradation in iron-replete cells, while it is stabilized in iron deficiency or hypoxia. IRP2 also responds to nitric oxide (NO), as shown in various cell types exposed to pharmacological NO donors and in gamma interferon/lipopolysaccharide-stimulated macrophages. However, the diverse experimental systems have yielded conflicting results on whether NO activates or inhibits IRP2. We show here that a treatment of mouse B6 fibroblasts or human H1299 lung cancer cells with the NO-releasing drug S-nitroso-N-acetyl-penicillamine (SNAP) activates IRP2 expression. Moreover, the exposure of H1299 cells to SNAP leads to stabilization of hemagglutinin (HA)-tagged IRP2, with kinetics analogous to those elicited by the iron chelator desferrioxamine. Similar results were obtained with IRP2Δ73, a mutant lacking a conserved, IRP2-specific proline- and cysteine-rich domain. Importantly, SNAP fails to stabilize HA-tagged p53, suggesting that under the above experimental conditions, NO does not impair the capacity of the proteasome for protein degradation. Finally, by employing a coculture system of B6 and H1299 cells expressing NO synthase II or IRP2-HA cDNAs, respectively, we demonstrate that NO generated in B6 cells stabilizes IRP2-HA in target H1299 cells by passive diffusion. Thus, biologically synthesized NO promotes IRP2 stabilization without compromising the overall proteasomal activity. These results are consistent with the idea that NO may negatively affect the labile iron pool and thereby trigger responses to iron deficiency.
The iron-regulatory proteins IRP1 and IRP2 serve as intracellular iron sensors and control iron homeostasis at the posttranscriptional level. They interact with iron-responsive elements (IREs), phylogenetically conserved hairpin structures within the 5′- or 3′-untranslated region (UTR) of several mRNAs related to iron and energy metabolism (11, 21). IRP binding controls the translation or stability of IRE-containing transcripts, such as of transferrin receptor 1 (TfR1) and (H- and L-) ferritin, which encode crucial proteins for cellular iron acquisition and storage, respectively. In iron-starved cells, IRPs bind to multiple IREs within the 3′ UTR of TfR1 mRNA and to a single IRE in the 5′ UTR of ferritin mRNAs. The IRE-IRP interactions stabilize TfR1 mRNA against nucleolytic degradation and inhibit the translation of ferritin mRNAs in a coordinated fashion, resulting in increased iron uptake and reduced storage. Conversely, in iron-replete cells IRPs fail to bind to cognate mRNAs, eliciting opposite homeostatic responses.
The targeted disruption of IRP1 has yielded mice with a mild phenotype, restricted in the kidneys and in brown fat (20). On the other hand, IRP2 knockout mice misregulate iron metabolism in the intestine and the central nervous system and develop a neurodegenerative disorder (19). Thus, it appears that IRP2 has a central function in the control of systemic iron homeostasis. These findings have stimulated an interest in unravelling the regulatory mechanisms controlling IRP2 activity.
It is well established that iron promotes the proteasomal degradation of IRP2 (8), while IRP1 remains stable but assembles a cubane 4Fe-4S cluster that converts it to a cytosolic aconitase (9). The mechanism for IRP2 iron-dependent degradation is not well defined. Earlier work led to the conclusion that a proline- and cysteine-rich stretch of 73 amino acids (aa) encoded by a unique IRP2-specific exon is necessary and sufficient for IRP2 degradation by a mechanism involving oxidation of cysteines upon iron binding (13). More recent results have questioned the validity of the previous findings (1, 10, 27), and it has been proposed that the 73-aa domain plays a role in IRP2 degradation in response to heme (29). It should, however, be noted that an IRP2 deletion mutant lacking the entire 73-aa domain remains sensitive to iron, similar to wild-type IRP2 (10, 27). Interestingly, the iron-dependent degradation of IRP2 is, at least partially, blocked by inhibitors of 2-oxoglutarate-dependent oxygenases, suggesting a role of these enzymes in the underlying mechanism (10, 27).
Besides its apparent iron-sensing capacity, IRP2 also responds to nitric oxide (NO). However, conflicting data have been reported on whether NO activates or inhibits IRP2, which can to some extent be attributed to differences in the experimental approaches and the utilized sources of NO (23). Thus, the IRE-binding activity of IRP2 was induced in J774 macrophages treated with gamma interferon/lipopolysaccharide (IFN-γ/LPS) to stimulate the inducible NO synthase II (NOS II) (28) in B6.NOS cells stably transfected with an NOS II cDNA (22) and in Ltk− fibroblasts exposed to the NO donor S-nitroso-N-acetyl-penicillamine (SNAP) (24). However, in other studies employing IFN-γ/LPS-stimulated J774 (26) or RAW (2, 16) macrophages, IRP2 activity was diminished. Experiments with sodium nitroprusside (SNP), an iron-containing compound utilized for nitrosylation of protein thiols, have led to the conclusion that the redox status of NO is crucial for the IRP2 response (17). According to this view, NO+ promotes IRP2 proteasomal degradation following S-nitrosylation of C178, which lies within the 73-aa domain (18).
Here, we utilize SNAP as a widely applied NO donor to assess the effects of NO on IRP2 expression and stability. In addition, we establish a coculture system of B6.NOS cells and H1299 cells expressing hemagglutinin (HA) epitope-tagged IRP2 (27) to evaluate the response of IRP2 to NO generated by a biologically relevant source in the absence of confounding pharmacological or cytokine-related side effects.
MATERIALS AND METHODS
Materials.
SNAP, NG-monomethyl-l-arginine (l-NMMA), and MG132 were obtained from Sigma (St. Louis, Mo.). Desferrioxamine (DFO) was from Novartis (Dorval, Canada), and l-arginine was from BioShop (Burlington, Ontario, Canada).
Cell culture.
Human H1299 lung cancer cells and murine B6 and B6.NOS fibroblasts were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum, 2 mM glutamine, 100 U of penicillin/ml, and 0.1 ng of streptomycin/ml. The B6.NOS cells express a murine NOS II cDNA from a simian virus 40-driven vector (22). H1299 clones expressing HA-tagged wide-type IRP2wt, mutant IRP2Δ73 (27), or wide-type p53 (3) (a gift from Xinbin Chen, University of Alabama, Birmingham), under the control of a tetracycline-inducible promoter, were maintained in DMEM containing 2 μg of tetracycline/ml, 2 μg of puromycin/ml, and 250 μg of G418/ml.
Western blotting.
Cells were washed twice in cold phosphate-buffered saline (PBS) and lysed in cytoplasmic lysis buffer (1% Triton X-100, 40 mM KCl, 25 mM Tris-Cl, pH 7.4). Cell debris was cleared by centrifugation, and protein concentration was measured with the Bradford reagent (Bio-Rad). For the analysis of p53-HA, extracts were prepared by direct lysis of cells in Laemmli sample buffer (50 μl of buffer/106 cells) and boiling for 5 min. Cell lysates (30 μg) were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 7.5% gels (or 11% gels for p53-HA), and proteins were transferred onto nitrocellulose filters. The blots were saturated with 10% nonfat milk in PBS containing 0.1% Tween 20 (PBS-T) and probed with polyclonal antibodies against IRP2 (27), HA (1:1,000 dilution; Santa Cruz), β-actin (1:1,000 dilution; Sigma), or monoclonal antibody against NOS II (1:500 dilution; BD Pharmingen). For the analysis of p53-HA, the blots were probed with the culture supernatant of 12CA5 hybridoma cells containing anti-HA monoclonal antibody. Following a wash with PBS-T, the blots were incubated with peroxidase-coupled goat anti-rabbit immunoglobulin G (1:5,000 dilution; Sigma) or rabbit anti-mouse immunoglobulin G (1:4,000 dilution; Sigma). Peroxidase-coupled antibodies were detected by the enhanced chemiluminescence method (Amersham). The blots were quantified by densitometric scanning.
Pulse-chase and immunoprecipitation.
Cells were metabolically labeled with (50 μCi/ml) trans-35S label, a mixture of 70:30 [35S]methionine-cysteine (ICN), and were chased in cold media. The cells were washed twice with PBS and lysed in a buffer containing 1% Triton X-100, 50 mM Tris-Cl (pH 7.4), and 300 mM NaCl. Cell debris was cleared by centrifugation, and cell lysates were subjected to quantitative immunoprecipitation with the HA antibody (Santa Cruz). Immunoprecipitated proteins were analyzed by SDS-PAGE and visualized by autoradiography. Radioactive bands were quantified by phosphorimaging.
Northern blotting.
The cells were washed twice in PBS and lysed with the Trizol reagent (Gibco BRL), and RNA was prepared according to the manufacturer's recommendation. Total cellular RNA (10 μg) was electrophoretically resolved on denaturing agarose gels, transferred onto nylon membranes, and hybridized to radiolabeled human IRP2 or rat GAPDH cDNA probes. Radioactive bands were visualized by autoradiography.
Nitrite assay.
Culture medium (0.25 ml) was mixed with 0.75 ml of Griess reagent (Merck, Darmstadt, Germany). After 10 min of incubation at room temperature, the absorbance was measured at 543 nm.
RESULTS
Nitric oxide increases the half-life of IRP2.
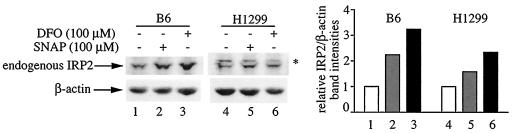
To evaluate the effects of NO on IRP2 expression, B6 and H1299 cells were exposed to 100 μM SNAP for 8 h. Under these conditions, SNAP is expected to release ∼1.4 μM NO/min (5). Because the half-life of SNAP in aqueous media at 37°C is 5 h (12), a fresh bolus of the drug was added after 4 h. This treatment resulted in 2.2- and 1.6-fold increases of IRP2 steady-state levels in B6 and H1299 cells, respectively (Fig. 1, lanes 2 and 5). The parallel administration of the iron chelator DFO (100 μM) activated IRP2 expression 3.2-fold in B6 cells and 2.3-fold in H1299 cells.
FIG. 1.
NO activates the expression of endogenous IRP2. B6 or H1299 cells were either left untreated (lanes 1 and 4) or were exposed for 8 h to 100 μM SNAP (lanes 2 and 4) or DFO (lanes 3 and 6). A fresh bolus of SNAP was added after 4 h. Cell lysates were subjected to Western blotting with antibodies against IRP2 (top) and β-actin (bottom). The immunoreactive bands were quantified by densitometric scanning. The IRP2/β-actin ratios are plotted on the right. The asterisk denotes an apparently nonspecific band.
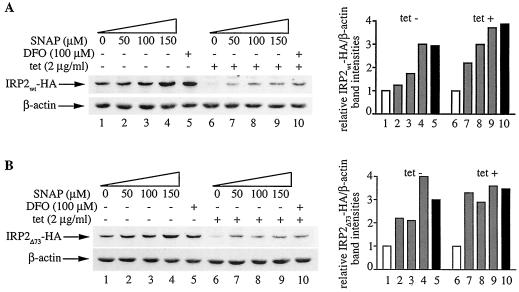
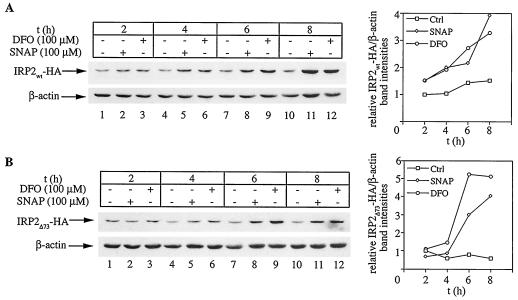
We hypothesized that NO may positively affect the half-life of IRP2, which is subjected to iron- and oxygen-dependent regulation (8, 10, 27). To examine this, we utilized H1299 cells expressing C-terminally HA-tagged wild-type or mutant IRP2 (lacking the 73-aa domain) under the control of a tetracycline-inducible promoter. We previously demonstrated that both IRP2wt-HA and IRP2Δ73-HA are active in IRE binding and are sensitive to iron-dependent degradation (27). Following removal of tetracycline for 24 h to allow the expression of IRP2wt-HA or IRP2Δ73-HA (tet-off system), the cells were exposed to increasing concentrations of SNAP for 8 h (Fig. 2). This resulted in a dose-dependent activation of IRP2wt-HA (Fig. 2A) and IRP2Δ73-HA (Fig. 2B), up to approximately threefold and fourfold, respectively (lanes 1 to 4). In quantitative terms, IRP2wt-HA and IRP2Δ73-HA were activated to almost the same degree by 150 μM SNAP and 100 μM DFO (lanes 4 and 5). Adding back tetracycline is expected to block the transcription of the mRNAs encoding the inducible proteins, thereby allowing a more direct assessment of the stability of the synthesized IRP2wt-HA and IRP2Δ73-HA pools. Under these conditions, SNAP stabilized both IRP2wt-HA and IRP2Δ73-HA in a dose-dependent manner by up to ∼3.5-fold, by analogy to DFO (lanes 6 to 10) and consistently with the data shown in lanes 1 to 5.
FIG. 2.
Dose-dependent increase in IRP2 expression by NO. H1299 cells were plated for 24 h in tetracycline (tet)-free media to express IRP2wt-HA (A) or IRP2Δ73-HA (B). Tetracycline (2 μg/ml) was then added back to half of the cells (lanes 6 to 10) to shut off the transcription of the IRP2-HA cDNAs. After 1 h, the cells were either left untreated (lanes 1 and 6) or exposed to increasing doses of SNAP (lanes 2 to 4 and 7 to 9) or to 100 μM DFO (lanes 5 and 10) for another 8 h. A fresh bolus of SNAP was added after 4 h. Cell lysates were subjected to Western blotting with HA (top) and β-actin antibodies (bottom). The immunoreactive bands were quantified by densitometric scanning. The IRP2wt-HA/β-actin and IRP2Δ73-HA/β-actin ratios are plotted on the right.
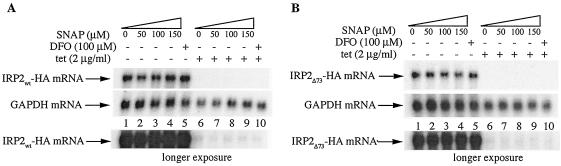
Northern analysis confirmed that the addition of tetracycline dramatically inhibits the expression of the mRNAs encoding IRP2wt-HA and IRP2Δ73-HA (Fig. 3A and B, respectively; compare lanes 1 to 5 to lanes 6 to 10). This experiment also revealed that the treatments with SNAP (lanes 1 to 4) or DFO (lane 5) do not affect the steady-state levels of IRP2wt-HA and IRP2Δ73-HA mRNAs. Moreover, a longer exposure of the blots shows no SNAP-dependent alterations in the residual amounts of IRP2wt-HA and IRP2Δ73-HA mRNAs (bottom panels). Taken together, the data in Fig. 2 and 3 suggest that NO activates IRP2 expression at the level of protein stability.
FIG. 3.
NO does not affect the expression of IRP2 mRNA. Cells were treated in the same manner as those shown in Fig. 2, and the expression of the mRNAs encoding IRP2wt-HA (A) or IRP2Δ73-HA (B) was analyzed by Northern blotting. A longer exposure of the blots is shown at the bottom. Hybridization with a GAPDH probe served as control.
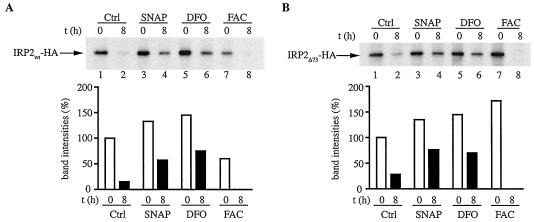
To directly examine whether NO inhibits the decay of IRP2, a pulse-chase experiment was performed. The cells were pulsed for 2 h with [35S]methionine/cysteine and harvested or chased in cold media for 8 h. The levels of radiolabeled IRP2wt-HA or IRP2Δ73-HA were assessed by quantitative immunoprecipitation. A treatment of the cells with 100 μM SNAP during the pulse-chase stabilized both IRP2wt-HA (Fig. 4A) and IRP2Δ73-HA (Fig. 4B) by approximately four- and threefold, respectively (compare lanes 1 and 2 to lanes 3 and 4). A similar effect was observed with 100 μM DFO (lanes 5 and 6), while, as expected, a treatment with 30 μg of ferric ammonium citrate (FAC)/ml elicited opposite responses (lanes 7 and 8). We conclude that NO stabilizes IRP2 by a mechanism that does not involve its 73-aa domain. Interestingly, the presence of SNAP during the pulse appears to stimulate the de novo synthesis of both IRP2wt-HA and IRP2Δ73-HA by 30 to 40% (compare lanes 1 and 3), indicative of a small translational contribution to the NO-mediated activation of IRP2. The same holds true for DFO, in agreement with previous observations (27). However, considering that the mRNA constructs encoding the HA-tagged IRP2 proteins do not retain the authentic IRP2 sequences in their untranslated regions, the physiological relevance of this finding is questionable.
FIG. 4.
Stabilization of IRP2 by NO. H1299 cells engineered to express IRP2wt-HA (A) or IRP2Δ73-HA (B) were grown for 3 days in media lacking tetracycline. The cells were metabolically labeled for 2 h with trans-35S label (50 μCi/ml) in the absence or presence of 100 μM of SNAP, 100 μM DFO, or 30 μg of ferric ammonium citrate (FAC)/ml. Subsequently, the cells were either harvested or chased for 8 h in cold media in the absence or presence of the indicated drugs, and cytoplasmic lysates (500 μg) were subjected to quantitative immunoprecipitation with 1 μg of polyclonal anti-HA antibody (Santa Cruz). Immunoprecipitated proteins were analyzed by SDS-PAGE on a 7.5% gel and were visualized by autoradiography. The radioactive bands were quantified by phosphorimaging, and the intensities are shown in the bar graphs. Ctrl, control; t, time.
Having established that a treatment with SNAP promotes IRP2 stabilization, we analyzed the time course of IRP2 accumulation in response to 100 μM SNAP or DFO. Both the NO-releasing SNAP and the iron chelator DFO activate the expression of IRP2wt-HA (Fig. 5A) as well as IRP2Δ73-HA (Fig. 5B) within 4 to 8 h, with strikingly similar kinetics. Thus, nitric oxide inhibits the degradation of IRP2, resulting in its stabilization and accumulation.
FIG. 5.
Kinetics of activation in IRP2 expression by NO and iron chelation. H1299 cells were plated for 24 h in tetracycline-free media to express IRP2wt-HA (A) or IRP2Δ73-HA (B). Subsequently, the cells were either left untreated (lanes 1, 4, 7, and 10) or were exposed for the indicated time intervals to 100 μM SNAP (lanes 2, 5, 8, and 11) or DFO (lanes 3, 6, 9, and 12). A fresh bolus of SNAP was added after 4 h (lanes 8 and 11). Cell lysates were analyzed by Western blotting with HA (top) and β-actin antibodies (bottom). The immunoreactive bands were quantified by densitometric scanning. The IRP2wt-HA/β-actin and IRP2Δ73-HA/β-actin ratios are plotted against the time (t) on the right. Ctrl, control.
Nitric oxide does not inhibit the proteasomal degradation machinery.
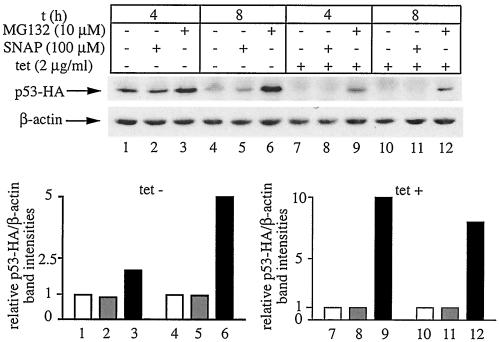
The NO-mediated stabilization of IRP2 could be a result of a global impairment of the proteasomal degradation machinery, or an IRP2-specific phenomenon. In light of reports that NO inhibits the proteasomal pathway in cell extracts (7), we sought to address the former scenario. We thus employed H1299 cells expressing a C-terminally HA-tagged version of p53 from a tetracycline-dependent promoter (3) to examine the effects of NO on p53-HA stability. It is well established that p53 is a short-lived protein (with a half-life of ∼5 to 20 min) that undergoes degradation via the ubiquitin-proteasome system (6). The cells were grown for 24 h in the absence of tetracycline to induce the expression of p53-HA (tet-off system). Subsequent treatment with 100 μM SNAP for 4 or 8 h failed to increase p53-HA steady-state levels (Fig. 6, lanes 1 and 2 as well as 4 and 5). Likewise, a similar treatment following readdition of tetracycline to shut off the transcription of p53-HA mRNA failed to stabilize the synthesized p53-HA pool (lanes 7 and 8 as well as 10 and 11). As expected, the proteasomal inhibitor MG132 profoundly inhibited the degradation of p53-HA (lanes 3, 6, 9, and 12). Thus, NO does not globally inhibit the proteasomal degradation machinery, at least under the above experimental conditions. Considering that an almost identical experimental setting was employed to study the effects of NO on IRP2-HA turnover, we conclude that NO stabilizes IRP2 in an apparently specific manner.
FIG. 6.
Proteasomal degradation of p53 is not impaired by NO. H1299 cells were plated for 24 h in tetracycline (tet)-free media to express p53-HA. Tetracycline (2 μg/ml) was then added back to half of the cells (lanes 7 to 12) to shut off the transcription of p53-HA cDNA. The cells were either left untreated (lanes 1, 4, 7, and 10), treated with 100 μM SNAP for 4 h (lanes 2 and 8) or 8 h (lanes 5 and 11), or treated with 10 μM MG132 for 4 h (lanes 3 and 9) and 8 h (lanes 6 and 12), respectively. When necessary, a fresh bolus of SNAP was added after 4 h. Cell lysates were subjected to Western blotting with HA (top) and β-actin antibodies (bottom). The immunoreactive bands were quantified by densitometric scanning. The p53-HA/β-actin ratios are plotted at the bottom. t, time.
IRP2 stabilization by physiologically synthesized NO.
The experiments with SNAP, an NO donor with established pharmacological properties (5, 12), clearly suggest that NO antagonizes the degradation of IRP2. These results are in agreement with earlier findings showing that NO stimulates the IRE-binding activity of IRP2 (22, 24). Nevertheless, on the basis of recent experiments with SNP, it has been proposed that the oxidized nitrosonium cation NO+ promotes the proteasomal degradation of IRP2 (17). Because it is not clear whether SNAP or SNP recapitulate physiological conditions, we employed a coculture system to elucidate the response of IRP2 to biologically synthesized NO. Previously characterized B6.NOS cells (22, 24), which express an NOS II cDNA (Fig. 7, lanes 6 to 10), or control parent B6 cells were cocultured with H1299 cells expressing IRP2wt-HA at a ratio of 3:1. We expected that NO synthesized in B6.NOS cells would diffuse and modulate the expression of IRP2wt-HA in target H1299 cells. To directly assess IRP2wt-HA stability, the cells were initially grown in tetracycline-free media. Subsequently, tetracycline was added back for 1 h, and either the cells were harvested or incubation under different experimental conditions was continued for another 8 h.
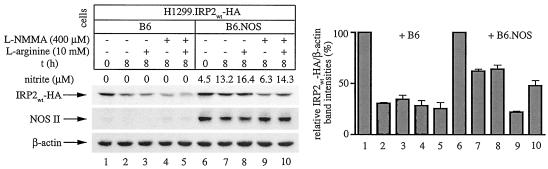
FIG. 7.
Stabilization of IRP2 by physiologically generated NO via intercellular signaling. H1299 cells (106) expressing IRP2wt-HA (indicated as H1299.IRP2wt-HA) were mixed with either 3 × 106 control B6 cells (lanes 1 to 5) or 3 × 106 B6.NOS cells (lanes 6 to 10) and evenly plated onto 100-mm-diameter dishes. The cells were cultivated in tetracycline (tet)-free media to activate the expression of IRP2wt-HA. After 24 h, 5 mM sodium butyrate was added and incubation was continued for another 12 h to augment the expression of NOS II (22). Subsequently, tetracycline (2 μg/ml) was added back to turn off the synthesis of IRP2wt-HA, and, after an 1 h incubation, the cells were either harvested (lanes 1 and 6) or further incubated for 8 h under following conditions: no additives (lanes 2 and 7), with 10 mM l-arginine (lanes 3 and 8), or with 400 μM l-NMMA in the absence (lanes 4 and 9) or presence of 10 mM l-arginine (lanes 5 and 10) to modulate NO production. Nitrite levels in the culture supernatant were measured with Griess reagent. The levels of IRP2wt-HA (expressed in H1299.IRP2wt-HA cells), NOS II (expressed in B6.NOS cells), and control β-actin (expressed in H1299.IRP2wt-HA, B6.NOS, and control B6 cells) were analyzed by Western blotting with HA (top), NOS II (middle), and β-actin antibodies (bottom). The immunoreactive bands were quantified by densitometric scanning. The IRP2wt-HA/β-actin ratios from three independent experiments (± standard deviations) are plotted at the bottom. t, time.
In the coculture of H1299 with parent B6 cells, the addition of tetracycline resulted in a decrease of the IRP2wt-HA signal by ∼30% over 8 h (Fig. 7, lanes 1 to 5). By contrast, in the coculture of H1299 with B6.NOS cells, the addition of tetracycline was associated with a twofold stabilization of IRP2wt-HA (lanes 6 to 8). Importantly, this effect was eliminated by 400 μM l-NMMA, an NOS substrate analog (lane 9), and it was restored in the presence of an excess of 10 mM l-arginine, the substrate of NOS (lane 10). The capacity of B6.NOS cells to generate and release NO, as well as the effectiveness of the drug treatments in the modulation of NOS activity, are illustrated by the measurement of nitrite concentration in the culture supernatant. In conclusion, this experiment demonstrates that NO synthesized from a physiologically relevant source promotes the stabilization of IRP2.
DISCUSSION
Over the last 10 years, work from several laboratories has shown that NO has the potential to regulate IRP2. However, the actual response of IRP2 to NO remains thus far a subject of controversy. In different experimental settings, the generation of NO has been associated with either activation (22, 24, 28) or inactivation (16, 26) of IRP2. In other reports, the exposure of cells to an NO-releasing drug did not affect IRP2 expression (1, 15, 25). Conceivably, confounding variables related to diverse NO sources could account for these apparent discrepancies. Thus, the cautious choice of an appropriate and well-defined experimental system can be crucial to understanding the nature and molecular basis of the IRP2 response to NO.
By utilizing SNAP, a pharmacological NO donor with defined properties (5, 12), we show here that NO increases IRP2 steady-state levels (Fig. 1), mainly by stabilizing the protein against degradation (Fig. 2 to 5). These results not only corroborate earlier findings (22, 24) but also offer a mechanistic explanation for the previously reported activation of the IRE-binding activity of IRP2 as a result of SNAP-mediated NO release (24). Importantly, the kinetics of IRP2-HA stabilization by SNAP, which range between 4 and 8 h (Fig. 5), are in perfect agreement with the time course of endogenous IRP2 activation by SNAP (24). The data shown in Fig. 2 to 5 also suggest that the 73-aa domain of IRP2 is not necessary for its stabilization by NO.
The relatively slow induction of IRP2 (and IRP1) requires a constant supply of NO to cultured cells (24). Based on this notion and considering that the half-life of SNAP is 5 h (12), we thought that it is essential to replenish the cell cultures with fresh solutions of the drug for prolonged treatments. It is thus possible that the lack of IRP2 response following an exposure of 293 cells to single SNAP boluses for 16 h (1) could be related to a failure to sustain a critical NO threshold due to SNAP decay. In other reports, SNAP efficiently activated IRP1 but failed to activate IRP2 (15, 25). It is, however, apparent from the data presented in references 15 and 25 that DFO also failed to appreciably activate IRP2. Hence, under these experimental conditions, IRP2 may have already been stable prior to treatment.
The stimulation of RAW 264.7 or J774 macrophages with IFN-γ/LPS activates the inducible NOS II, which in turn generates NO. While it is generally acknowledged that this treatment results in IRP1 activation (2, 16, 26, 28), the response of IRP2 appears to be more complex. Contradictory data, reporting either IRP2 activation (28) or inactivation (26), have been obtained with J774 cells. On the other hand, experiments with RAW 264.7 seem to agree that the IFN-γ/LPS treatment inactivates IRP2 (16, 17), even though it has been disputed whether this effect is NO dependent (2). These discrepancies may be related to pleiotropic effects of cytokines, possibly combined with the growth conditions of the cells.
In a more defined experimental system, the data with SNAP (Fig. 1 to 5) offer compelling evidence that NO increases the half-life of IRP2 and promotes its stabilization. The employment of NO-releasing drugs certainly offers many advantages to dissect the mechanism of the IRP2 response to NO. Nevertheless, apparently conflicting data have also been obtained with pharmacological NO donors. Thus, the iron-containing compound SNP, which is frequently utilized as an S-nitrosylating agent, promotes IRP2 degradation by the proteasome, and it has been hypothesized that this response may mimic NO generation in immunologically stimulated macrophages (17). To better characterize the interplay between NO and IRP2 under physiological conditions, we employed a coculture system of B6.NOS cells generating NO by a transfected NOS II cDNA (22) and target H1299 cells expressing epitope-tagged IRP2wt-HA, which serve as a faithful model for the study of IRP2 regulation (27). This system recapitulates physiological NO synthesis, diffusion, and intercellular signaling in the absence of cytokine stimulation. Importantly, a similar approach previously revealed that NO regulates the expression of IRE-containing indicator mRNAs in target cells via iron-regulatory proteins (24). The data shown in Fig. 7 demonstrate that NO synthesized enzymatically, under physiologically relevant conditions in effector cells, stabilizes IRP2 in target cells. This result confirms the conclusions reached with the NO donor SNAP and substantiates the pharmacological approach in a physiological context.
What is the molecular basis for the NO-mediated stabilization of IRP2? A global impairment of the proteasome pathway by NO could offer a reasonable mechanistic explanation. However, this scenario is unlikely, as NO fails to stabilize p53 (Fig. 6), which undergoes degradation by the proteasome (6), by analogy to the iron-dependent degradation of IRP2. Considering that the experimental conditions to examine the effects of NO on IRP2-HA and p53-HA were indistinguishable, we conclude that NO stabilizes IRP2 in an apparently specific manner.
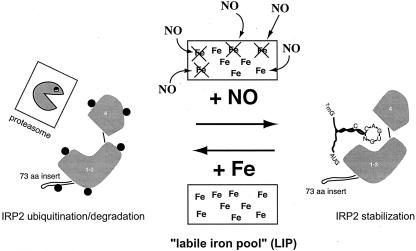
Taken together, the results presented here are consistent with those of an earlier proposed model (23, 24, 28). According to this earlier model, NO may control the levels of the labile iron pool (LIP), an ill-defined fraction of chelatable iron with an important regulatory function (14). An exposure of cells to NO may mobilize intracellular iron to form iron-nitrosyl complexes (4) and thereby decrease the size of the LIP. This would in turn activate homeostatic responses to iron starvation, including the stabilization of IRP2 (Fig. 8) and the removal of the iron-sulfur cluster from IRP1 (23, 24, 28). The striking similarities in the kinetics of IRP2 stabilization by SNAP and DFO provide indirect support to this model, which awaits further experimental validation.
FIG. 8.
A model for IRP2 stabilization via NO-mediated decrease of the labile iron pool.
Acknowledgments
We thank Xinbin Chen (Birmingham, Ala.) for the H1299 cells expressing p53-HA.
J.W. holds a fellowship from the Canadian Institutes of Health Research (CIHR). K.P. is a scholar of CIHR and a researcher of the Canada Foundation for Innovation.
REFERENCES
- 1.Bourdon, E., D. K. Kang, M. C. Ghosh, S. K. Drake, J. Wey, R. L. Levine, and T. A. Rouault. 2003. The role of endogenous heme synthesis and degradation domain cysteines in cellular iron-dependent degradation of IRP2. Blood Cells Mol. Dis. 31:247-255. [DOI] [PubMed] [Google Scholar]
- 2.Bouton, C., L. Oliveira, and J.-C. Drapier. 1998. Converse modulation of IRP-1 and IRP-2 by immunological stimuli in murine RAW 264.7 macrophages. J. Biol. Chem. 273:9403-9408. [DOI] [PubMed] [Google Scholar]
- 3.Chen, X., L. J. Ko, L. Jayaraman, and C. Prives. 1996. p53 levels, functional domains, and DNA damage determine the extent of the apoptotic response of tumor cells. Genes Dev. 10:2438-2451. [DOI] [PubMed] [Google Scholar]
- 4.Drapier, J. C., C. Pellat, and Y. Henry. 1991. Generation of EPR-detectable nitrosyl-iron complexes in tumor target cells cocultured with activated macrophages. J. Biol. Chem. 266:10162-10167. [PubMed] [Google Scholar]
- 5.Feelisch, M. 1991. The biochemical pathways of nitric oxide formation from nitrovasodilators: appropriate choice of exogenous NO donors and aspects of preparation and handling of aqueous NO solutions. J. Cardiovasc. Pharmacol. 17(Suppl. 3):S25—S33. [Google Scholar]
- 6.Giaccia, A. J., and M. B. Kastan. 1998. The complexity of p53 modulation: emerging patterns from divergent signals. Genes Dev. 12:2973-2983. [DOI] [PubMed] [Google Scholar]
- 7.Glockzin, S., A. von Knethen, M. Scheffner, and B. Brune. 1999. Activation of the cell death program by nitric oxide involves inhibition of the proteasome. J. Biol. Chem. 274:19581-19586. [DOI] [PubMed] [Google Scholar]
- 8.Guo, B., J. D. Phillips, Y. Yu, and E. A. Leibold. 1995. Iron regulates the intracellular degradation of iron regulatory protein 2 by the proteasome. J. Biol. Chem. 270:21645-21651. [DOI] [PubMed] [Google Scholar]
- 9.Haile, D. J., T. A. Rouault, C. K. Tang, J. Chin, J. B. Harford, and R. D. Klausner. 1992. Reciprocal control of RNA-binding and aconitase activity in the regulation of the iron-responsive element binding protein: role of the iron-sulfur cluster. Proc. Natl. Acad. Sci. USA 89:7536-7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanson, E. S., M. L. Rawlins, and E. A. Leibold. 2003. Oxygen and iron regulation of iron regulatory protein 2. J. Biol. Chem. 278:40337-40342. [DOI] [PubMed] [Google Scholar]
- 11.Hentze, M. W., M. U. Muckenthaler, and N. C. Andrews. 2004. Balancing acts; molecular control of mammalian iron metabolism. Cell 117:285-297. [DOI] [PubMed] [Google Scholar]
- 12.Ignarro, L. J., H. Lippton, J. C. Edwards, W. H. Baricos, A. L. Hyman, P. J. Kadowitz, and C. A. Gruetter. 1981. Mechanism of vascular smooth muscle relaxation by organic nitrates, nitrites, nitroprusside and nitric oxide: evidence for the involvement of S-nitrosothiols as active intermediates. J. Pharmacol. Exp. Ther. 218:739-749. [PubMed] [Google Scholar]
- 13.Iwai, K., R. D. Klausner, and T. A. Rouault. 1995. Requirements for iron-regulated degradation of the RNA binding protein, iron regulatory protein 2. EMBO J. 14:5350-5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kakhlon, O., and Z. I. Cabantchik. 2002. The labile iron pool: characterization, measurement, and participation in cellular processes. Free Radic. Biol. Med. 33:1037-1046. [DOI] [PubMed] [Google Scholar]
- 15.Kim, S., and P. Ponka. 1999. Control of transferrin receptor expression via nitric oxide-mediated modulation of iron-regulatory protein 2. J. Biol. Chem. 274:33035-33042. [DOI] [PubMed] [Google Scholar]
- 16.Kim, S., and P. Ponka. 2000. Effects of interferon-gamma and lipopolysaccharide on macrophage iron metabolism are mediated by nitric oxide-induced degradation of iron regulatory protein 2. J. Biol. Chem. 275:6220-6226. [DOI] [PubMed] [Google Scholar]
- 17.Kim, S., and P. Ponka. 2002. Nitrogen monoxide-mediated control of ferritin synthesis: implications for macrophage iron homeostasis. Proc. Natl. Acad. Sci. USA 99:12214-12219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, S., S. S. Wing, and P. Ponka. 2004. S-nitrosylation of IRP2 regulates its stability via the ubiquitin-proteasome pathway. Mol. Cell. Biol. 24:330-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LaVaute, T., S. Smith, S. Cooperman, K. Iwai, W. Land, E. Meyron-Holtz, S. K. Drake, G. Miller, M. Abu-Asab, M. Tsokos, R. Switzer III, A. Grinberg, P. Love, N. Tresser, and T. A. Rouault. 2001. Targeted deletion of the gene encoding iron regulatory protein-2 causes misregulation of iron metabolism and neurodegenerative disease in mice. Nat. Genet. 27:209-214. [DOI] [PubMed] [Google Scholar]
- 20.Meyron-Holtz, E. G., M. C. Ghosh, K. Iwai, T. LaVaute, X. Brazzolotto, U. V. Berger, W. Land, H. Ollivierre-Wilson, A. Grinberg, P. Love, and T. A. Rouault. 2004. Genetic ablations of iron regulatory proteins 1 and 2 reveal why iron regulatory protein 2 dominates iron homeostasis. EMBO J. 23:386-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pantopoulos, K. 2004. Iron metabolism and the IRE/IRP regulatory system: an update. Ann. N. Y. Acad. Sci. 1012:1-13. [DOI] [PubMed] [Google Scholar]
- 22.Pantopoulos, K., and M. W. Hentze. 1995. Nitric oxide signaling to iron-regulatory protein (IRP): direct control of ferritin mRNA translation and transferrin receptor mRNA stability in transfected fibroblasts. Proc. Natl. Acad. Sci. USA 92:1267-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pantopoulos, K., and M. W. Hentze. 2000. Nitric oxide, oxygen radicals and iron metabolism, p. 293-313. In L. Ignarro (ed.), Nitric oxide. Academic Press, San Diego, Calif.
- 24.Pantopoulos, K., G. Weiss, and M. W. Hentze. 1996. Nitric oxide and oxidative stress (H2O2) control mammalian iron metabolism by different pathways. Mol. Cell. Biol. 16:3781-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Phillips, J. D., D. V. Kinikini, Y. Yu, B. Guo, and E. A. Leibold. 1996. Differential regulation of IRP-1 and IRP-2 by nitric oxide in rat hepatoma cells. Blood 87:2983-2992. [PubMed] [Google Scholar]
- 26.Recalcati, S., D. Taramelli, D. Conte, and G. Cairo. 1998. Nitric oxide-mediated induction of ferritin synthesis in J774 macrophages by inflammatory cytokines: role of selective iron regulatory protein-2 downregulation. Blood 91:1059-1066. [PubMed] [Google Scholar]
- 27.Wang, J., G. Chen, M. Muckenthaler, B. Galy, M. W. Hentze, and K. Pantopoulos. 2004. Iron-mediated degradation of IRP2: an unexpected pathway involving a 2-oxoglutarate-dependent oxygenase activity. Mol. Cell. Biol. 24:954-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weiss, G., B. Goossen, W. Doppler, D. Fuchs, K. Pantopoulos, G. Werner-Felmayer, H. Wachter, and M. W. Hentze. 1993. Translational regulation via iron-responsive elements by the nitric oxide/NO-synthase pathway. EMBO J. 12:3651-3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamanaka, K., H. Ishikawa, Y. Megumi, F. Tokunaga, M. Kanie, T. A. Rouault, I. Morishima, N. Minato, K. Ishimori, and K. Iwai. 2003. Identification of the ubiquitin-protein ligase that recognizes oxidized IRP2. Nat. Cell Biol. 5:336-340. [DOI] [PubMed] [Google Scholar]