Abstract
Hey1 is a member of the basic helix-loop-helix-Orange family of transcriptional repressors that mediate Notch signaling. Here we show that transcription from androgen-dependent target genes is inhibited by Hey1 and that expression of a constitutively active form of Notch is capable of repressing transactivation by the endogenous androgen receptor (AR). Our results indicate that Hey1 functions as a corepressor for AF1 in the AR, providing a mechanism for cross talk between Notch and androgen-signaling pathways. Hey1 colocalizes with AR in the epithelia of patients with benign prostatic hyperplasia, where it is found in both the cytoplasm and the nucleus. In marked contrast, we demonstrate that Hey1 is excluded from the nucleus in most human prostate cancers, raising the possibility that an abnormal Hey1 subcellular distribution may have a role in the aberrant hormonal responses observed in prostate cancer.
Androgens play critical roles in a wide range of developmental and physiological processes, particularly in male organs (18). Androgens regulate prostate epithelial cell growth, and survival and alterations in androgen-dependent signaling contribute to the development of prostate carcinoma, the most frequently diagnosed neoplasm and the second leading cause of cancer-related death in men in Western countries (15). The most common prostate cancer therapy is androgen elimination combined with antiandrogen treatment. However, most prostate tumors eventually become insensitive to this treatment and proliferate (9). The elucidation of mechanisms by which cancers become androgen independent is a crucial step towards developing successful therapies for prostate cancer.
The biological actions of androgens are mediated by the androgen receptor (AR), a member of the nuclear receptor (NR) superfamily of ligand-dependent transcription factors (21). NRs share a common domain structure, comprising an N-terminal activation domain (activation function 1 [AF1]), a central DNA-binding domain (DBD), and a C-terminal ligand-binding domain (LBD) that usually contains a second activation domain (AF2). Unlike many members of the NR superfamily, the AF1 domain contributes most of AR transactivation functions. Upon ligand binding, ARs adopt an active conformation, release chaperone heat shock proteins, and bind as homodimers to specific DNA sequences in the promoters of responsive genes, where they recruit cofactors that regulate the transcription of target genes (22). The ability of NRs to activate gene transcription depends on the recruitment of coactivator protein complexes with enzymatic activities that reorganize chromatin. Among them are members of the p160 family of coactivators, SRC1, TIF2/GRIP1, and RAC3/AIB1/ACTR/pCIP (19). The p160 coactivators interact directly with NRs via conserved LXXLL motifs (10, 28), and they act as platform proteins recruiting enzymes that catalyze posttranslational modifications in histones, including histone acetyltransferases (HATs) like CBP/p300 and pCAF and methyltransferases like CARM-1. The p160 proteins also contribute to the recruitment of ATP-dependent chromatin remodeling complexes (1). These chromatin modifications are reversible, and corepressor complexes with opposing enzymatic activities switch off gene transcription and maintain genes in a silenced state. AR, like other NRs, appears to recruit corepressors that target enzymatic activities such as histone deacetylases (HDACs) to promoters and thereby reorganize the chromatin structure to suppress transcription. Little is known regarding the mechanisms involved in AR-dependent gene repression, but recently a number of putative AR corepressors, including cyclin D1, HBO1, Pyk2, and PIASy, have been identified (18).
To investigate the function of the highly conserved basic helix-loop-helix (bHLH)-PAS domain found in the p160 coactivators, we performed a Saccharomyces cerevisiae two-hybrid screen using the bHLH-PAS domain in SRC1 as bait. Here we present evidence of a novel functional interaction between SRC1 and Hairy/Enhancer of split related with YRPW motif 1 (Hey1, also named Hesr1, HERP2, HRT1, and CHF2), a member of the vertebrate bHLH-Orange (bHLH-O) family of transcriptional repressors (6). Hey1 interacts directly with SRC1 and AR and specifically represses transcription from AR-dependent promoters. Hey1 is a downstream mediator of Notch-dependent signals, and our findings demonstrate that there is a cross talk between the Notch and AR-dependent pathways in target tissues.
MATERIALS AND METHODS
Two-hybrid screening.
Yeast two-hybrid screening, using SRC1 as bait and a mouse embryo (9.5 to 12.5 dpc) cDNA library, has been described previously (2).
Plasmids.
The complete open reading frames of full-length murine Hey1, human Hey1, human Hey2, and Hey1 deletion mutants (ΔY [containing amino acids 1 to 285], ΔY+O [amino acids 1 to 115], ΔY+O+H [amino acids 1 to 49], and ΔH [amino acids 116 to 299]) were amplified by PCR and subcloned into pSG5, pGEX-6P-1 (Amersham Pharmacia Biotech), or pSG-Gal (20).
The following plasmids have been described previously: pMT2-MOR, pSG5-SRC1e, pGL3-2ERE-PS2-LUC, GST-SRC1-(1-450), and pSG5-SRC1-(1-361) (1); pGL2-Lex-Gal-Luc and pSG5-Lex-VP16 (5); pSVAR (4); AR(1-653) (16); TAT-GRE-E1B-LUC and Probasin-LUC (29); pSG5-hGR and pSG5-hPR-B (17); and NICD (amino acids 1747 to 2531 of rat Notch1 subcloned into pEF1α-BOS) (24).
GST pull-down assays.
Expression vectors were transcribed and translated in vitro with [35S]methionine in reticulocyte lysate (Promega). Glutathione S-transferase (GST) fusion proteins were induced, purified, bound to Sepharose beads (Amersham), and incubated with translated proteins or whole-cell extracts as described previously (1) in NETN buffer (20 mM Tris-HCl [pH 8.0], 1 mM EDTA, 0.5% Nonidet P-40, 100 mM NaCl). After being washed extensively, the samples were separated on sodium dodecyl sulfate (SDS)-10% polyacrylamide gels. Gels were fixed and dried, and the 35S-labeled proteins were visualized by fluorography or blotted onto nitrocellulose and probed with antibodies.
Antibodies.
The antibodies used were rabbit anti-human Hey1 affinity-purified polyclonal antibody (Chemicon International), mouse anti-human monoclonal AR441 (DAKO), and rabbit polyclonal immunoglobulin G (IgG) anti-GAL4-DBD (Santa Cruz Biotechnology).
Immunoblotting.
Nitrocellulose membranes were blocked in TBS-T (20 mM Tris-HCl [pH 7.6], 137 mM NaCl, 0.1% Tween 20) containing 3% nonfat milk powder, washed with TBS-T, and incubated for 2 h with anti-Hey1, anti-AR, or anti-GAL4-DBD (1:1,000 dilution). After being washed, the membranes were incubated with goat anti-rabbit horseradish peroxidase or goat anti-mouse horseradish peroxidase (1:3,000; DAKO) and washed again with TBS-T. The bound immunoglobulins were visualized using the ECL detection system (Amersham Pharmacia Biotech).
Cell culture and transient-transfection experiments.
COS-1, HeLa, C2C12, and MCF-7 cells were cultured in Dulbecco's modified Eagle's medium, and LNCaP cells were cultured in RPMI 1640 supplemented with 10% fetal bovine serum. Twenty-four hors before transfection, cells were plated in 96-well plates in phenol red-free medium with 5% dextran charcoal-stripped serum (the serum was incubated, twice, with activated charcoal and dextran for 30 min at 55°C). Cells were transfected using FuGENE 6 (Roche). The transfected DNA (measured in nanograms per well) included the pRL-CMV (Promega) control plasmid (1 ng), the reporters TAT-GRE-E1B-LUC (20 ng), Probasin-LUC (20 ng), pGL3-2ERE-PS2-LUC (10 ng), pGL2-Lex-Gal-Luc (20 ng), and the vectors pSVAR (5 ng), AR(1-653) (5 ng), pMT2-MOR (2.5 ng), pSG5-hPR-B (5 ng), pSG5-GR (5 ng), pSG5-SRC1e (10 ng), pSG5-Lex-VP16 (10 ng), pEF-BOS-NICD (10 ng, except where indicated otherwise), pSG5-Gal4-DBD or pSG5-Gal-Hey1 fusions (10 ng/well), and pSG5-Hey1 or pSG5-Hey2 (10 ng, except where indicated otherwise). Empty vectors were used to normalize DNA amounts. After incubation for 16 h, cells were washed and treated with hormones for 24 h (10 nM mibolerone for AR, 10 nM 17β-estradiol for estrogen receptor α [ERα], 10 nM R5020 for the progesterone receptor [PR], and 10 nM dexamethasone for the glucocorticoid receptor [GR]). Cell extracts were assayed for luciferase activity using a dual reporter assay as described previously (2). The results shown represent the averages of the results of at least two independent experiments assayed in quadruplicate plus standard deviations (SD).
Coimmunoprecipitation assay.
MCF-7 cells growing in 90-mm-diameter dishes in phenol red-free medium with 5% dextran charcoal-stripped serum were transfected with 5 μg of NICD plasmid or mock transfected using Lipofectamine 2000 (Invitrogen). After incubation for 5 h, cells were washed and treated with 10 nM mibolerone for 24 h. Cells were washed twice with ice-cold phosphate-buffered saline and immediately lysed by incubation for 20 min, at 4°C, in IP buffer (containing 50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% Nonidet P-40, 1 mM dithiothreitol, and complete protease inhibitors that were EDTA free [Roche]). After centrifugation at 14,000 × g for 20 min at 4°C, the supernatants were used for immunoprecipitation with nonimmune rabbit IgG or anti-Hey1 rabbit polyclonal antibody at 4°C for 90 min; immune complexes were then captured using protein A-Sepharose. Complexes were washed three times with IP buffer, and proteins were released by boiling the solution for 5 min in SDS loading buffer. The immunoprecipitated material was separated on SDS-10% polyacrylamide gels and blotted onto nitrocellulose. The membrane was probed using anti-AR antibody as described above.
siRNA.
hHey1 small interfering RNAs (siRNAs) were from Dharmacon. The target sequences were 5′-AAAAUGCUGCAUACGGCAGGA-3′ and 5′-AACAGUUUGUCUGAGCUGAGA-3′. MCF-7 cells were cotransfected in 24-well plates with both human Hey1 siRNAs (125 ng of each siRNA) or 250 ng of negative-control siRNA (QIAGEN), 100 ng of TAT-GRE-E1B-LUC and pSG5-Hey1 (10 ng) or pEF-BOS-NICD (0.5 or 1 ng) using Lipofectamine 2000 (Invitrogen) and antibiotic-free medium, as described previously (8). RNA was extracted using Trizol (GIBCO), and Hey1 expression was studied by real-time PCR. The primer sequences were 5′-GCTGGTACCCAGTGCTTTTGAG-3′ and 5′-TGCAGGATCTCGGCTTTTTCT-3′. For luciferase assay, after transfection, cells were washed and incubated in fresh medium for 16 h. Subsequently, cells were incubated for 24 h with vehicle or 10 nM mibolerone. Cell lysates were then assayed using a dual luciferase reporter system
Immunohistochemistry.
Prostate samples from 24 separate patients with prostate cancer or benign prostatic hyperplasia (BPH) were collected at surgery with the approval of St. Mary's National Health Service Trust local research ethics committee. Four-micrometer-thick sections of formalin-fixed, paraffin-embedded prostate tissue were dewaxed in xylene and rehydrated in decreasing concentrations of ethanol, and endogenous peroxidase activity was blocked using 2% hydrogen peroxide. Antigen retrieval was carried out by microwaving at 750 W in 0.01 M trisodium citrate, pH 6, three times for 5 min. Sections were blocked with goat serum (1:10 dilution) before incubation overnight at 4°C with the primary antibody: rabbit polyclonal anti-human AR441 (1:150 dilution) or rabbit anti-human Hey1 (1:1,000 dilution). After being washed, sections were incubated with biotinylated goat anti-rabbit IgG (1:200, 45 min; DAKO), followed by peroxidase conjugated with streptavidin (1:100, 30 min; DAKO). Sections were then washed, and enzyme activity was developed in 1 mg of 3,3′-diaminobenzidine tetrahydrochloride (DAKO) per ml and counterstained with hematoxylin (Vector Laboratories).
RESULTS
Hey1 interacts with SRC1 and AR.
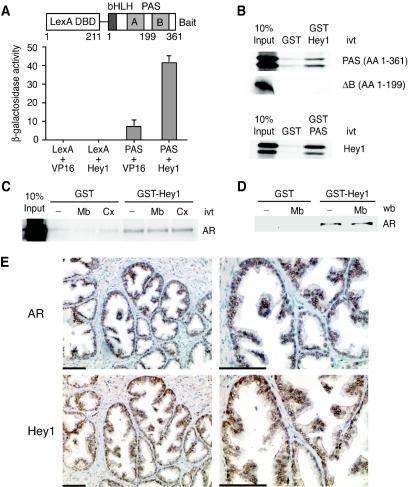
We have previously employed a yeast two-hybrid screen to identify proteins that interact with the bHLH-PAS domain of SRC1 (2). One of the clones encodes full-length Hey1, a member of the bHLH-O family of transcriptional repressors. The specificity of the interaction was confirmed by retransforming yeast with expression vectors for the fusion proteins shown in Fig. 1A. The increase in β-galactosidase indicates that the bHLH-PAS domain of SRC1 interacts with Hey1 in intact cells. We also confirmed that the bHLH-PAS domain was able to interact with Hey 1 in vitro by using GST pull-down assays (Fig. 1B). However, an in vitro-translated SRC1 deletion mutant comprising the bHLH domain and only half of the PAS domain (amino acids 1 to 199) (Fig. 1A) showed no interaction with Hey1 (Fig. 1B). This result indicates that the interaction requires an intact PAS domain and does not reflect only an interaction between the HLH domains present in both proteins. Expressed sequence tag databases indicate that Hey1 is highly expressed in prostate, an androgen-dependent tissue. Furthermore, the observation that AR interacts with amino-terminal enhancer of split (31), a member of the transducin-like enhancer of split family of transcriptional repressors, downstream effectors for several members of the bHLH-O family, prompted us to determine whether Hey1 was able to interact with AR. Using GST pull-down experiments, we detected a specific interaction between in vitro-translated full-length AR and GST-Hey1 (Fig. 1C) that was unaffected by the presence of AR agonists or antagonists. Similarly, endogenous AR, expressed in LNCaP prostate cells, bound to GST-Hey1 in a ligand-independent manner (Fig. 1D).
FIG. 1.
Hey1 interacts with SRC1 and AR. (A) The L40a yeast strain expressing either LexA-DBD (LexA) or LexA-DBD fused to the SRC1 bHLH-PAS domain (PAS) was transformed with either an empty pASV3 plasmid (VP16) or pASV3 expressing Hey1 fused to the VP16 activation domain (Hey1). β-Galactosidase activity in each yeast extract was measured in duplicate. Data represent the means + SD of results with two independent transformants. The schematic representation of the LexA chimera used as bait in the yeast two-hybrid screening is shown above. (B) Hey1 interacts in vitro with the SRC1 bHLH-PAS domain. GST fusion proteins coupled with Sepharose beads were incubated with in vitro-translated (ivt) [35S]methionine-labeled Hey1 or SRC1 fragments as indicated. After being washed extensively, samples were boiled and separated by SDS-10% polyacrylamide gel electrophoresis. Gels were fixed and dried, and the labeled proteins were detected by fluorography. AA, amino acids. (C) Hey1 interacts in vitro with AR. GST alone or GST-Hey1 were incubated with 35S-labeled AR, in the presence of vehicle (−), 100 nM mibolerone (Mb), or 10 μM Casodex (Cx). (D) Whole-cell extracts from LNCaP cells were incubated with GST alone or GST-Hey1 bound to glutathione-Sepharose in the presence of vehicle (−) or 100 nM Mb. The associated AR was detected by immunoblotting using anti-AR antibody. wb, Western blot. (E) AR and Hey1 colocalize in prostate epithelial cells. Adjacent sections of human BPH samples were stained with anti-AR antibodies (top panels) or anti-Hey1 antibodies (bottom panels). Two different magnifications are shown (scale bar = 100 μm). The brown color reflects positive staining for AR or Hey1, and negative nuclei are blue.
Next we investigated whether Hey1 and AR are expressed in the same regions in the prostate. We analyzed sections of human BPH by immunostaining and detected both proteins in luminal epithelial cells (Fig. 1E). Taken together our data raise the possibility that Hey1 may be involved in the regulation of AR activity.
Hey1 represses AR-dependent gene expression.
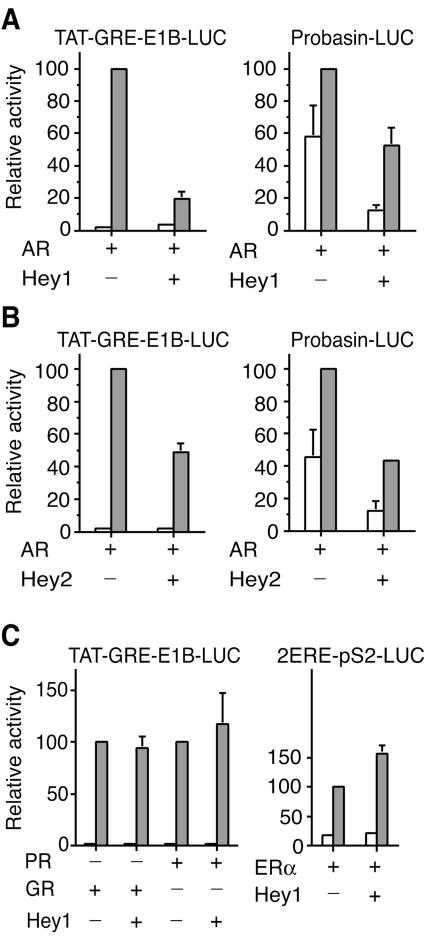
In view of the interaction between Hey1 and AR, we tested the effect of Hey1 expression on the ability of AR to stimulate transcription from luciferase reporter genes in transfected cells. The expression of Hey1 reduced ligand-dependent activation from two androgen-responsive promoters, Probasin-LUC and TAT-GRE-E1B-LUC (Fig. 2A). Basal transcription from the Probasin-LUC reporter was also reduced in the presence of Hey1, but only in the presence of AR (data not shown), indicating that the repression was specific for the AR and not a general effect. Another member of the Hey family, Hey2, was also able to repress AR-dependent transcriptional activity from the two androgen-responsive promoters tested, indicating that the high homology at sequence level reflects similar functional characteristics (Fig. 2B).
FIG. 2.
Hey1 represses AR-mediated transactivation in vivo. (A) COS-1 cells were cotransfected with 20 ng of TAT-GRE-E1B-LUC or Probasin-LUC and expression vectors for AR (5 ng) and Hey1 (10 ng). (B) COS-1 cells were cotransfected with 20 ng of TAT-GRE-E1B-LUC or Probasin-LUC and expression vectors for AR (5 ng) and Hey2 (10 ng). (C) COS-1 cells were cotransfected with 20 ng of TAT-GRE-E1B-LUC and expression vectors for Hey1 (10 ng) and PR (5 ng) or GR (5 ng), or with 10 ng of 2ERE-pS2-LUC and expression vectors for ERα (2.5 ng) and Hey1 (10 ng). After transfection, cells were washed and incubated for 24 h in the presence of vehicle (white bars) or a 10 nM concentration of hormones (gray bars; mibolerone for AR, 17β-estradiol for ERα, R5020 for PR, and dexamethasone for GR). Subsequently, cell lysates were assayed using a dual luciferase reporter system. Normalized values are expressed relative to the activity of AR, PR, GR, or ER alone in the presence of their respective ligands. The results shown represent the averages of results of at least two independent experiments assayed in quadruplicate + SD.
To investigate whether Hey1 and Hey2 were able to function as corepressors for other steroid receptors, we examined transcription from the TAT-GRE-E1B-LUC reporter in the presence of the progesterone receptor (PR) or the glucocorticoid receptor (GR). Both these receptors stimulated the reporter gene in the presence of their cognate ligands, but neither Hey1 (Fig. 2C, left panel) nor Hey2 (data not shown) were inhibitory. Similarly, we found that Hey1 did not repress the transcriptional activity of estrogen receptor α (ERα) (Fig. 2C, right panel). Although these receptors were insensitive to the inhibitory effects of Hey1, we were able to detect a very weak interaction between them in GST pull-down assays (data not shown), which is presumably insufficient to mediate repression. Therefore, we conclude that the AR is the only steroid receptor sensitive to the inhibitory effects of Hey1.
Hey1 represses AR activation function-1.
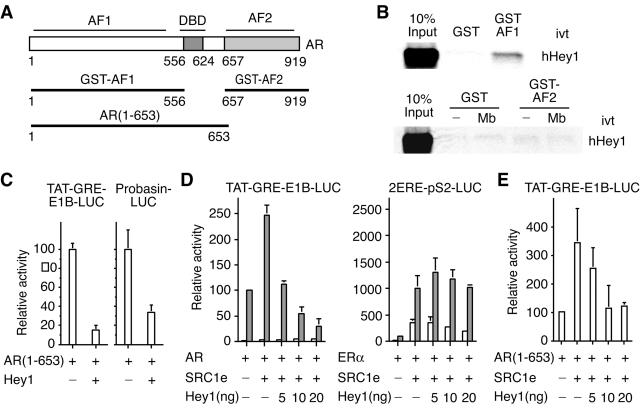
AR contains two activation domains, AF1, which seems to be responsible for most of AR transcriptional activity (reference 3 and references therein) and AF2 in the LBD. To investigate the interaction between Hey1 and these domains, we used GST pull-down experiments (constructs are shown in Fig. 3A) and found that GST-AF1, but not GST-AF2, bound in vitro-translated full-length Hey1 (Fig. 3B).
FIG. 3.
Characterization of Hey1 as an AR corepressor. (A-C) AF1 activity is repressed via direct interaction with Hey1. (A) Schematic representation of full-length AR and deletion mutants. (B) Binding of GST fusion proteins of AR deletion mutants to 35S-labeled Hey1. Where appropriate, assays were performed in the presence of vehicle (−) or 100 nM mibolerone (Mb). Bound proteins were visualized as described in the legend for Fig. 1B. ivt, in vitro translated. (C) COS-1 cells were cotransfected with 20 ng of TAT-GRE-E1B-LUC or Probasin-LUC and expression vectors for Hey1 (10 ng) and AR(1-653) (5 ng). (D) Hey1 overcomes SRC1 coactivation of AR. COS-1 cells were cotransfected with 20 ng of TAT-GRE-E1B-LUC and expression vectors for AR (5 ng) and SRC1e (10 ng) or with 10 ng of 2ERE-pS2-LUC and expression vectors for ERα (2.5 ng) and SRC1e (10 ng). In each case, increasing amounts of the Hey1 expression vector (5, 10, or 20 ng) were added. (E) Hey1 overcomes SRC1 coactivation of AR AF1. COS-1 cells were cotransfected with 20 ng of TAT-GRE-E1B-LUC, expression vectors for AR(1-653) (5 ng) and SRC1e (10 ng), and increasing amounts of the Hey1 expression vector (5, 10, or 20 ng). After transfection, cells were washed and incubated for 24 h in the presence of vehicle (white bars) or a 10 nM concentration of hormones (gray bars; 10 nM mibolerone for AR and 17β-estradiol for ERα). Subsequently, cell lysates were assayed using a dual luciferase reporter system. Data are presented as described for Fig. 2. The results shown represent the averages of results of two independent experiments assayed in quadruplicate + SD.
Deletion of the LBD results in a constitutively active receptor that activates ARE-containing reporters in a ligand-independent manner with the same potency as the full-length AR (27). We investigated whether this AF1-dependent transcriptional activity is sensitive to Hey1 repression. When we cotransfected AR(1-653) together with TAT-GRE-E1B-LUC or Probasin-LUC, there was a marked increase in luciferase activity that was dramatically reduced when Hey1 was coexpressed (Fig. 3C). These results suggest that the ability of Hey1 to inhibit AR activity is mediated by a functional interaction with AF1.
Hey1 overcomes SRC1 coactivation of AR.
Given the interaction between SRC1 and Hey1, we investigated whether there was a functional link in transiently transfected COS-1 cells. SRC1 promotes the ability of AR to stimulate transcription from the TAT-GRE-E1B-LUC reporter gene (Fig. 3D), but this was abolished in a dose-dependent manner when Hey1 was expressed. Interestingly, Hey1 did not affect the ability of SRC1 to potentiate transcriptional activation by ERα (Fig. 3D), suggesting that Hey1 does not merely sequester SRC1 to prevent activation. Our results indicate that the ability of SRC1 to function as an AR coactivator depends on the relative concentration of Hey1 by a mechanism that does not affect other steroid receptors. According to our previous results, Hey1 seems to target AF1 in the AR; therefore, we investigated whether Hey1 was able to overcome SRC1 coactivation of the deletion mutant AR(1-653), which lacks AF2. Hey1 inhibited SRC1 coactivation of the AR deletion mutant in a dose-dependent manner (Fig. 3E), again suggesting a role for Hey1 in the modulation of AR activity through the AF1 domain.
Characterization of Hey1 as a transcriptional repressor.
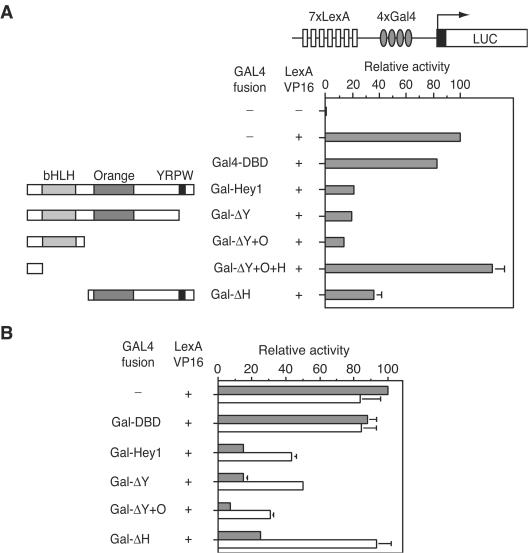
Hey1, Hey2, and HeyL form the HEY subfamily of transcriptional repressors (12). To characterize the repressive activity of Hey1, we investigated its ability to repress in trans the activity of a strong transcription factor in a trans-repression assay. Seven LexA and five Gal4 binding elements were fused in tandem upstream of a luciferase reporter (Fig. 4A). Then, the VP16 transcription factor fused to LexA-DBD was cotransfected with Gal4-DBD alone as a control or with Gal4-DBD fused to full-length Hey1 or deletion mutants. The strong luciferase activity induced by LexA-VP16 was dramatically reduced in the presence of a Gal4-Hey1 fusion, whereas expression of Gal4-DBD alone affected the luciferase activity only marginally (Fig. 4A). In order to determine the contribution of individual regions of the protein to the repression observed in our assay, we fused Hey1 deletion mutants to GAL4-DBD. Western blotting control experiments showed that all deletion mutants were expressed at similar levels (data not shown). The deletion of the YRPW motif (Gal-ΔY) or both YRPW and the orange domain (Gal-ΔY+O) did not significantly reduce the repression of LexA-VP16 activity (Fig. 4A). However, further C-terminal deletion of the HLH domain (Gal-ΔY+O+H) completely abolished the repression (Fig. 4A). Interestingly, the N-terminal deletion mutant lacking the HLH domain (Gal-ΔH) repressed the luciferase activity observed by 70% (Fig. 4A). These results indicate that Hey1 contains at least two distinct repressive domains, one in the N-terminal region and the second in the C-terminal half of the protein.
FIG. 4.
Characterization of Hey1 as a transcriptional repressor. (A) Mapping of autonomous repression domains in Hey1. HeLa cells were cotransfected with 20 ng of pGL2-Lex-Gal-Luc and expression vectors for LexA-VP16 (10 ng) and Gal4-DBD fusion proteins (10 ng) as indicated. (B) Effects of HDAC inhibitors in Hey1-mediated repression. COS-1 cells were cotransfected as indicated above before being incubated for 24 h in the presence of a vehicle (gray bars) or 500 nM TSA (white bars). Subsequently, cell lysates were assayed using a dual luciferase reporter system. Normalized values are expressed relative to the activity observed in the presence of LexA-Vp16 alone. The results shown represent the averages of results of two independent experiments assayed in quadruplicate + SD.
To investigate whether HDACs play a role in Hey1-mediated repression, we performed the trans-repression experiments with full-length Hey1 or deletion mutants in the presence of the HDAC inhibitor Tricostatin A (TSA). The repression activity of Gal-ΔH was completely reversed by TSA treatment, whereas that of full-length Gal-Hey1, Gal-ΔY, and Gal-ΔO was only partially reversed (30 to 40%) (Fig. 4B). These observations suggest that multiple repression mechanisms contribute to the repression by full-length Hey1. We conclude that the C-terminal repression domain in Hey1, which is TSA sensitive, is HDAC dependent but that the repression by the HLH domain may involve both HDAC-dependent and HDAC-independent mechanisms.
Contribution of Hey1 domains to the repression of AR.
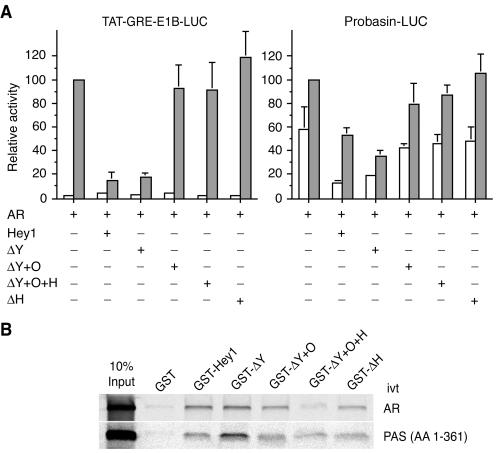
We then investigated which repressive domains in Hey1 were required for the transcriptional repression of AR using Hey1 deletion mutants corresponding to the same regions fused to GAL4-DBD in the trans-repression assay. The C-terminal deletion mutant lacking the YRPW motif repressed AR activity to the same degree as full-length Hey1 in both reporters tested (Fig. 5A). Surprisingly, none of the other deletion mutants, even those with intrinsic repressive properties, significantly inhibited AR-dependent reporter activity (Fig. 5A). A similar level of expression was observed for all deletion mutants in control Western blotting experiments (data not shown). To check whether the lack of repression by Hey1 deletion mutants simply reflected the loss of the domains required for the interaction with AR and/or SRC1, we fused the same Hey1 regions to GST and performed in vitro GST pull-down experiments. Only the mutant GST-ΔY+O+H failed to interact with in vitro-translated AR, and all the mutants interacted to some extent with the in vitro-translated SRC1-PAS domain (Fig. 5B). Thus, the mutants Gal-ΔY+O and Gal-ΔH, which contain intrinsic repressive domains and interact in vitro with AR and SRC1, do not inhibit AR-dependent transcriptional activity. These observations suggest that Hey1 requires the concerted action of both the N-terminal bHLH and the C-terminal repression domains in order to act as a corepressor for the AR.
FIG. 5.
Contribution of Hey1 domains to the repression of AR-dependent transcriptional activity. (A) COS-1 cells were cotransfected with 20 ng of TAT-GRE-E1B-LUC or Probasin-LUC reporters and expression vectors for AR (5 ng) and Hey1 (10 ng) deletion mutants. After transfection, cells were washed and incubated for 24 h in the presence of vehicle (white bars) or 10 nM mibolerone (gray bars). Subsequently, cell lysates were assayed using a dual luciferase reporter system. Data are presented as described for Fig. 2. The results shown represent the averages of results of two independent experiments assayed in quadruplicate + SD. (B) In vitro interaction of Hey1 deletion mutants with AR and SRC1. GST fusion proteins of full-length Hey1 or deletion mutants were incubated with 35S-labeled AR or the SRC1 bHLH-PAS domain. Bound proteins were visualized as described in the legend for Fig. 1B. ivt, in vitro translated; AA, amino acids.
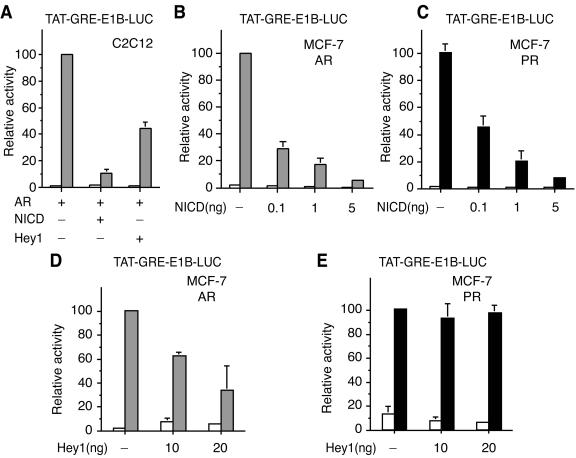
Activation of Notch pathways represses AR-dependent gene expression.
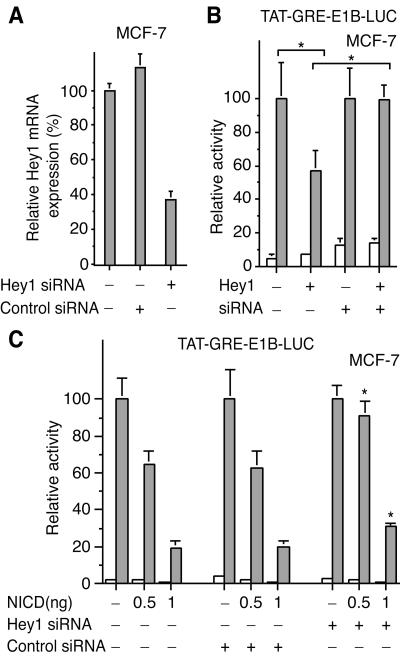
Hey1 is known to be a target gene for Notch signaling in C2C12 cells (13). Given the reported increase in expression and nuclear accumulation of Hey1 upon Notch activation, we investigated whether expression of the Notch intracellular domain (NICD), which is constitutively active, would inhibit the ability of AR to stimulate transcription. We found that expression of NICD dramatically inhibited AR-dependent transcriptional activity. This repression was even more potent than that caused by Hey1 (Fig. 6A), perhaps reflecting the induction of other repressors, like Hes1, known to synergistically cooperate with Hey1 to repress gene transcription (14). Since our C2C12 cell line does not express functional AR, we screened cell lines reported to be AR positive, including LNCaP, ZR-75-1, and MCF-7 cells, for expression of both AR and Hey1, as judged by real-time PCR (data not shown) and found that only the human breast cancer MCF-7 cell line fulfilled these criteria. The ability of endogenous AR to stimulate transcription from the TAT-GRE-E1B-LUC reporter gene in these cells was markedly reduced in the presence of increasing amounts of NICD (Fig. 6B) and again the repression was greater than that observed transfecting Hey1 (Fig. 6D). NICD also inhibited endogenous PR-dependent transcription in MCF-7 cells (Fig. 6C). However, Hey1 did not inhibit PR in this cell line (Fig. 6E), in agreement with the results observed with COS-1 cells (Fig. 2C), suggesting that the repressive effect of Hey1 is selective for the AR. NICD is not a general repressor for all steroid receptors because in parallel experiments we observed that expression of NICD did not affect ERα-dependent transcription (data not shown). We confirmed that endogenous levels of Hey1 mRNA increased when we overexpressed constitutively active NICD in MCF-7 cells (data not shown). According to our previous results, we predicted that the repressive effect of NICD on AR-dependent transcription was mediated, at least in part, by Hey1. To test this possibility, we used siRNAs that reduced Hey1 mRNA by approximately 60% (Fig. 7A). We found that the Hey1 siRNA was able to reverse the inhibitory effects of Hey1 (Fig. 7B) and partially reversed the inhibition induced by Notch activation (Fig. 7C). It seems likely that this partial reversal reflects the reduction in endogenous Hey1 levels but may be incomplete because of the presence of other Hey1 family members or the residual levels of Hey1 expression. Our results suggest that there is a functional interaction between the endogenous Hey1 and AR in MCF-7 cells that modulates transcription from androgen target genes.
FIG. 6.
An active form of Notch (NICD) represses AR in vivo. (A) C2C12 cells were cotransfected with 20 ng of TAT-GRE-E1B-LUC and expression vectors for AR (5 ng), NICD (10 ng), and Hey1 (10 ng). (B and C) MCF-7 cells were cotransfected with 20 ng of TAT-GRE-E1B-LUC and expression vectors for NICD (0.1, 1, or 5 ng). (D and E) MCF-7 cells were cotransfected with 20 ng of TAT-GRE-E1B-LUC and expression vectors for Hey1 (10 or 20 ng). After transfection, cells were washed and incubated in fresh medium for 16 h. Subsequently, cells were incubated for 24 h with vehicle (white bars), 10 nM mibolerone (gray bars), or 10 nM R5020 (black bars). Cell lysates were then assayed using a dual luciferase reporter system. Data are presented as described for Fig. 2. The results shown represent the averages of results from at least two independent experiments assayed in quadruplicate + SD.
FIG. 7.
Hey1 siRNA partially reverses the AR repression induced by the active form of Notch (NICD). (A) Hey1 siRNA decreases Hey1 expression. MCF-7 cells were mock transfected or transfected with 250 ng of a negative control (QIAGEN) or Hey1 siRNAs. Hey1 mRNA was quantified by real-time PCR. (B) MCF-7 cells were cotransfected with 100 ng of TAT-GRE-E1B-LUC, 10 ng of the Hey1 expression vector, and 250 ng of Hey1 siRNAs. *, by Student's t test, P was <0.05 (n = 4). (C) MCF-7 cells were cotransfected with 100 ng of TAT-GRE-E1B-LUC, expression vectors for NICD (0.5 or 1 ng), and 250 ng of a negative control (QIAGEN) or Hey1 siRNAs. *, by Student's t test, P was <0.005 compared with values for controls (n = 6). After transfection, cells were washed and incubated in fresh medium for 16 h. Subsequently, cells were incubated for 24 h with vehicle (white bars) or 10 nM mibolerone (gray bars). Cell lysates were then assayed using a dual luciferase reporter system. Data are presented as described for Fig. 2. The results shown represent the averages of results from at least two independent experiments assayed in quadruplicate + SD.
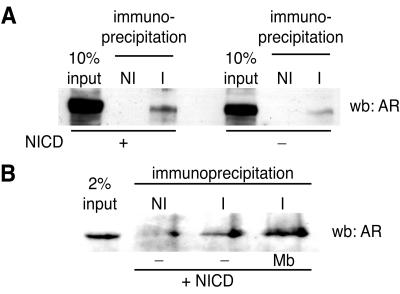
The characterization of a cell line coexpressing endogenous Hey1 and AR provided us with an opportunity to analyze whether the physical interactions between Hey1 and AR observed in vitro could also occur between the endogenous proteins in intact cells. Upon immunoprecipitation with antibodies against Hey1 and Western blotting using anti-AR antibodies, we detected a specific interaction between endogenous AR and Hey1 in MCF-7 whole-cell extracts (Fig. 8A). NICD expression had no significant effect on the interaction. Next we investigated whether this interaction was regulated by the presence of AR ligands. Consistent with the results presented in Fig. 1, we found that endogenous Hey1 and AR were able to interact in the absence of the AR agonist (Fig. 8B), although there was a slight increase in the presence of the agonist (Fig. 8B). The physical association between endogenous AR and Hey1 supports our proposal that this interaction plays a role in the regulation of AR transcriptional activity.
FIG. 8.
Coimmunoprecipitation of endogenous Hey1 and AR. (A) MCF-7 cells were mock transfected or transfected with 5 μg of NICD. After transfection, cells were washed and incubated for 24 h. Whole-cell extracts were then immunoprecipitated with antibodies against Hey1 (I) or nonimmune IgG (NI). The immunoprecipitated material was subjected to Western blotting analysis (wb) with anti-AR monoclonal IgG. (B) Effects of AR agonists in the interaction between Hey1 and AR. MCF-7 cells were transfected with 5 μg of NICD. After transfection, cells were washed and incubated for 24 h in the presence of 10 nM mibolerone (Mb) or vehicle (−) before an immunoprecipitation was performed as described above.
Prostate adenocarcinoma is associated with the nuclear exclusion of Hey1 protein.
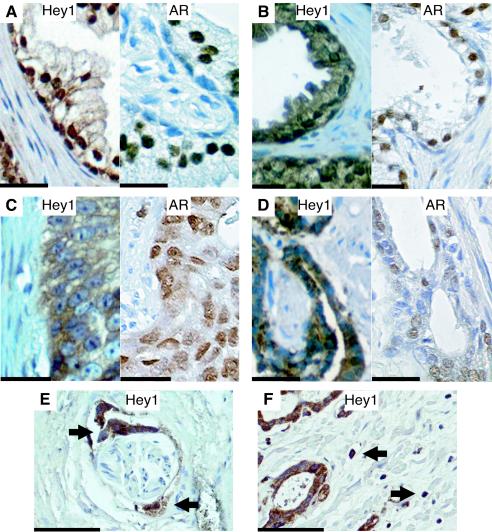
Amplification of chromosome band 8q occurs in a large fraction of PCs and correlates with the aggressiveness of tumors (7). Since Hey1 maps to this region, we examined its expression in a series of such tumors by immunocytochemistry and compared it with a series of genes from BPH samples (Fig. 9 and data summarized in Table 1), obtained from distinct patients. The expression of AR was also examined in the same samples. Hey1 expression was detected in both nuclear and cytoplasmic compartments of epithelial cells in 13 out 14 BPH samples (Table 1 and Fig. 9A and B, left panels). The relative proportions varied, but the majority of BPH samples showed strong nuclear Hey1 expression and nuclear exclusion was observed in only one sample. In contrast, we found that Hey1 was restricted to the cytoplasmic compartment of epithelial cells in 8 out 10 cancer samples (Table 1 and examples in Fig. 9C and D, left panels). Parallel immunostaining with anti-AR-specific antibody showed that the AR was localized preferentially in epithelial cell nuclei in both BPH and prostate cancer samples (Fig. 9A to D, right panels). The subcellular distribution of Hey1, therefore, does not seem to influence AR distribution. Hey1 nuclear exclusion was also observed in perineural invasion (Fig. 9E) and single-cell invasion of stroma (Fig. 9F) by malignant cells in cancer samples.
FIG. 9.
Nuclear exclusion of Hey1 staining in human prostate cancer tissue. Sections of human prostate tissue from patients with benign prostatic hyperplasia (A and B) or malignant adenocarcinomas of Gleason grades 5 (C) and 3 (D) were stained with anti-Hey1 antibody (left panels), and sections from the same samples were stained in parallel with anti-AR antibody (right panels). (E and F) Hey1 staining of sections from malignant epithelial cells infiltrating a nerve sheath (E) and the stromal compartment (F), indicated with arrows. Scale bars, 20 μm (A to D) or 100 μm (E and F). The brown color reflects positive staining for Hey1 or AR; negative nuclei are blue.
TABLE 1.
Hey1 subcellular localization in human prostate luminal epithelial cells
| Sample | Diagnosis (Gleason grades) | Hey1 intensity ina:
|
|
|---|---|---|---|
| Nucleus | Cytoplasm | ||
| 1 | Cancer (2 + 3) | − | +++ |
| 2 | Cancer (3 + 3) | − | +++ |
| 3 | Cancer (3 + 4) | ++ | ++ |
| 4 | Cancer (3 + 4) | − | ++ |
| 5 | Cancer (4 + 5) | − | +++ |
| 6 | Cancer (4 + 5) | ++ | ++ |
| 7 | Cancer (4 + 5) | − | +++ |
| 8 | Cancer (5 + 5) | − | + |
| 9 | Cancer (5 + 5) | − | +++ |
| 10 | Cancer (5 + 5) | − | ++ |
| 11 | BPH | ++ | ++ |
| 12 | BPH | ++ | + |
| 13 | BPH | ++ | + |
| 14 | BPH | ++ | + |
| 15 | BPH | ++ | + |
| 16 | BPH | ++ | + |
| 17 | BPH | ++ | ++ |
| 18 | BPH | ++ | + |
| 19 | BPH | ++ | + |
| 20 | BPH | ++ | ++ |
| 21 | BPH | + | ++ |
| 22 | BPH | + | ++ |
| 23 | BPH | + | + |
| 24 | BPH | − | ++ |
−, not present; +, slight presence; ++, moderate presence; +++, strong presence.
DISCUSSION
Transcriptional regulation by AR is achieved through interaction with cofactors, proteins that are recruited to the AR and convey the enzymatic activities required for the chromatin reorganization around the androgen-responsive promoters. In addition, these cofactors contribute to the regulation of the assembly and activity of the RNA polymerase II transcription machinery. Therefore, the identification and characterization of these transcriptional cofactors is a crucial step towards the elucidation of the mechanisms that control AR-dependent gene expression. Here we demonstrate that Hey1 functionally interacts with the AR, acting as a specific corepressor. Our data show that Hey1 and AR are expressed in luminal epithelial prostate cells. Androgens play a central role in the control of proliferation, differentiation, and survival of this subpopulation of prostate cells, and alterations in the normal transmission of androgen-dependent signals initiate the majority of prostate adenocarcinomas. Our expression analysis uncovered a striking difference in the subcellular localizations of Hey1 in patients with prostate cancer and BPH. Most cancer samples show a total exclusion of Hey1 from the nucleus, whereas nuclear exclusion was detected in only 1 out of 14 BPH patient samples.
One of the fundamental challenges for researchers studying prostate cancer is the understanding of the pathways that lead to the transition to androgen independence in recurrent prostate cancer. Most advanced prostate tumors express AR, suggesting that AR-dependent signals are indispensable for cell survival. However, these tumors escape hormonal regulation and become refractory to antiandrogen therapy. Different models have been proposed to explain the development of androgen-independent prostate cancer, including mutations in AR, changes in the levels of AR and/or its coactivators, and ligand-independent activation of AR through cross talk with other signaling pathways (for a review, see reference 9). Prostate cancers often contain chromosomal abnormalities, and indeed a region of chromosome 8 that includes Hey1 is amplified in the majority of individuals with androgen-independent prostate cancer. At first sight, the overexpression of an AR corepressor seems contradictory with most current models proposed to explain androgen-independent prostate cancer, which postulate an abnormal activation of AR rather than its repression. However, the amplification of other chromosome 8 genes may be the critical event during tumorigenesis, and any increase in Hey1 levels may be a bystander effect. Importantly, the repressive effects of Hey1 in prostate cancer may be evaded by excluding it from the nucleus. In addition, if Hey1 is required for the repression of AR in the presence of antiandrogens, the cytoplasmic localization of Hey1 may, at least in part, explain why cancer cells are resistant to antiandrogen treatment.
Unlike most NRs, AF1 is the main domain responsible for AR transactivation properties, and many corepressors and coactivators interact preferentially with this region of the receptor. We have demonstrated that Hey1 is recruited to the AR by means of the AF1, which might explain why agonists or antagonists failed to modulate the in vitro interaction between Hey1 and AR. In vivo, the interaction between Hey1 and AR might be regulated by subcellular localization and the presence of chaperone proteins that prevent the AR translocating to the nucleus and interacting with cofactors. Also, AF1 shows high sequence diversity between different NRs, perhaps explaining why Hey1 failed to repress other steroid receptors.
The observed interaction between a coactivator and a corepressor might seem paradoxical, but it is tempting to speculate that it may be part of mechanisms that promote the exchange between coactivator and corepressor complexes when transcription must be switched off. The translocation of Hey1 into the nucleus may provide prostate cells with a mechanism to suppress the transmission of endocrine signals.
Hey1 contains at least two domains with cell-independent, autonomous repressive activity: the N-terminal bHLH region and the C-terminal half of the protein. Our experiments with HDAC inhibitors reveal that the bHLH region acts through HDAC-dependent and HDAC-independent mechanisms. The second repressive domain seems to be completely HDAC dependent. Interestingly, both isolated domains failed to repress transcription from androgen-responsive promoters, despite the fact that they contain autonomous repressive domains and can still interact in vitro with AR and SRC1. This observation suggests that Hey1 repression of AR needs the concerted action of both repressive domains, indicating a complex regulatory mechanism.
The main difference observed in Hey1 expression between prostate cancer and BPH is the exclusion of Hey1 protein from the nucleus in malignant cells. Hey1 expression is regulated by Notch signaling. Upon activation of Notch signaling, Hey1 expression increases and accumulates in the nucleus (11, 13). Notch pathways regulate cell fate determination mediated by local cell-cell contact and may play a role in tumorigenesis (25). Notch1 is expressed in normal prostate epithelial cells and in prostate cancer cells, and its expression is regulated during prostate development (26). Overexpression of constitutively active Notch1 inhibits the proliferation of various prostate cells (26), and we observed a dramatic inhibition of AR activity upon activation of Notch signaling. Transgenic mice revealed that Notch1-expressing cells may define the progenitor cells in prostate epithelium (30), and gene profiling data showed that the expression of Notch1 and its ligand jagged1 is regulated by AR in the prostate (23). Notch1 and jagged1 are downregulated upon testosterone treatment, and sel-1L, a negative regulator of Notch, is upregulated (23). These observations, together with our results, provide a direct link between an endocrine pathway and Notch signaling, suggesting two-way negative feedback between AR and Notch pathways. The characterization of these mechanisms and the abnormalities in Notch signaling leading to Hey1 exclusion from the nucleus may be critical to the understanding of the development of androgen-independent prostate cancer.
Acknowledgments
We thank Frank Claessens, Gerry Weinmaster, Guido Jenster, and Albert Brinkmann for gifts of plasmids; Anup Patel for supplying prostate samples; Alpna Lad for histological sectioning; and Paul Duffy from AstraZeneca for Casodex. We are grateful to all members of the Molecular Endocrinology Laboratory and the Prostate Cancer Research Group, Ana Aranda, and Jose M. Cosgaya for helpful discussion and critical reading of the manuscript.
This work was funded by the Wellcome Trust (grant 061930), the Prostate Cancer Charity, and the Association for International Cancer Research.
REFERENCES
- 1.Belandia, B., R. L. Orford, H. C. Hurst, and M. G. Parker. 2002. Targeting of SWI/SNF chromatin remodelling complexes to estrogen-responsive genes. EMBO J. 21:4094-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belandia, B., and M. G. Parker. 2000. Functional interaction between the p160 coactivator proteins and the transcriptional enhancer factor family of transcription factors. J. Biol. Chem. 275:30801-30805. [DOI] [PubMed] [Google Scholar]
- 3.Bevan, C. L., S. Hoare, F. Claessens, D. M. Heery, and M. G. Parker. 1999. The AF1 and AF2 domains of the androgen receptor interact with distinct regions of SRC1. Mol. Cell. Biol. 19:8383-8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brinkmann, A. O., P. W. Faber, H. C. van Rooij, G. G. Kuiper, C. Ris, P. Klaassen, J. A. van der Korput, M. M. Voorhorst, J. H. van Laar, E. Mulder, et al. 1989. The human androgen receptor: domain structure, genomic organization and regulation of expression. J. Steroid Biochem. 34:307-310. [DOI] [PubMed] [Google Scholar]
- 5.Christian, M., J. M. Tullet, and M. G. Parker. 2004. Characterization of four autonomous repression domains in the corepressor receptor interacting protein 140. J. Biol. Chem. 279:15645-15651. [DOI] [PubMed] [Google Scholar]
- 6.Davis, R. L., and D. L. Turner. 2001. Vertebrate hairy and Enhancer of split related proteins: transcriptional repressors regulating cellular differentiation and embryonic patterning. Oncogene 20:8342-8357. [DOI] [PubMed] [Google Scholar]
- 7.DeMarzo, A. M., W. G. Nelson, W. B. Isaacs, and J. I. Epstein. 2003. Pathological and molecular aspects of prostate cancer. Lancet 361:955-964. [DOI] [PubMed] [Google Scholar]
- 8.Elbashir, S. M., J. Harborth, K. Weber, and T. Tuschl. 2002. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods 26:199-213. [DOI] [PubMed] [Google Scholar]
- 9.Feldman, B. J., and D. Feldman. 2001. The development of androgen-independent prostate cancer. Nat. Rev. Cancer 1:34-45. [DOI] [PubMed] [Google Scholar]
- 10.Heery, D. M., E. Kalkhoven, S. Hoare, and M. G. Parker. 1997. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387:733-736. [DOI] [PubMed] [Google Scholar]
- 11.Iso, T., G. Chung, Y. Hamamori, and L. Kedes. 2002. HERP1 is a cell type-specific primary target of Notch. J. Biol. Chem. 277:6598-6607. [DOI] [PubMed] [Google Scholar]
- 12.Iso, T., L. Kedes, and Y. Hamamori. 2003. HES and HERP families: multiple effectors of the Notch signaling pathway. J. Cell Physiol. 194:237-255. [DOI] [PubMed] [Google Scholar]
- 13.Iso, T., V. Sartorelli, G. Chung, T. Shichinohe, L. Kedes, and Y. Hamamori. 2001. HERP, a new primary target of Notch regulated by ligand binding. Mol. Cell. Biol. 21:6071-6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iso, T., V. Sartorelli, C. Poizat, S. Iezzi, H. Y. Wu, G. Chung, L. Kedes, and Y. Hamamori. 2001. HERP, a novel heterodimer partner of HES/E(spl) in Notch signaling. Mol. Cell. Biol. 21:6080-6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jemal, A., T. Murray, A. Samuels, A. Ghafoor, E. Ward, and M. J. Thun. 2003. Cancer statistics, 2003. CA Cancer J. Clin. 53:5-26. [DOI] [PubMed] [Google Scholar]
- 16.Jenster, G., H. A. van der Korput, C. van Vroonhoven, T. H. van der Kwast, J. Trapman, and A. O. Brinkmann. 1991. Domains of the human androgen receptor involved in steroid binding, transcriptional activation, and subcellular localization. Mol. Endocrinol. 5:1396-1404. [DOI] [PubMed] [Google Scholar]
- 17.Kalkhoven, E., L. Kwakkenbos-Isbrucker, S. W. de Laat, P. T. van der Saag, and B. van der Burg. 1994. Synthetic progestins induce proliferation of breast tumor cell lines via the progesterone or estrogen receptor. Mol. Cell. Endocrinol. 102:45-52. [DOI] [PubMed] [Google Scholar]
- 18.Lee, H. J., and C. Chang. 2003. Recent advances in androgen receptor action. Cell. Mol. Life Sci. 60:1613-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leo, C., and J. D. Chen. 2000. The SRC family of nuclear receptor coactivators. Gene 245:1-11. [DOI] [PubMed] [Google Scholar]
- 20.Mak, H. Y., S. Hoare, P. M. Henttu, and M. G. Parker. 1999. Molecular determinants of the estrogen receptor-coactivator interface. Mol. Cell. Biol. 19:3895-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mangelsdorf, D. J., C. Thummel, M. Beato, P. Herrlich, G. Schutz, K. Umesono, B. Blumberg, P. Kastner, M. Mark, P. Chambon, J. Wong, P. Tempst, and Y. Zhang. 1995. The nuclear receptor superfamily: the second decade. Cell 83:835-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKenna, N. J., R. B. Lanz, and B. W. O'Malley. 1999. Nuclear receptor coregulators: cellular and molecular biology. Endocr. Rev. 20:321-344. [DOI] [PubMed] [Google Scholar]
- 23.Nantermet, P. V., J. Xu, Y. Yu, P. Hodor, D. Holder, S. Adamski, M. A. Gentile, D. B. Kimmel, S. I. Harada, D. Gerhold, L. P. Freedman, and W. J. Ray. 2003. Identification of genetic pathways activated by the androgen receptor during the induction of proliferation in the ventral prostate gland. J. Biol. Chem. 23:23. [DOI] [PubMed] [Google Scholar]
- 24.Nofziger, D., A. Miyamoto, K. M. Lyons, and G. Weinmaster. 1999. Notch signaling imposes two distinct blocks in the differentiation of C2C12 myoblasts. Development 126:1689-1702. [DOI] [PubMed] [Google Scholar]
- 25.Radtke, F., and K. Raj. 2003. The role of Notch in tumorigenesis: oncogene or tumour suppressor? Nat. Rev. Cancer 3:756-767. [DOI] [PubMed] [Google Scholar]
- 26.Shou, J., S. Ross, H. Koeppen, F. J. de Sauvage, and W. Q. Gao. 2001. Dynamics of notch expression during murine prostate development and tumorigenesis. Cancer Res. 61:7291-7297. [PubMed] [Google Scholar]
- 27.Simental, J. A., M. Sar, M. V. Lane, F. S. French, and E. M. Wilson. 1991. Transcriptional activation and nuclear targeting signals of the human androgen receptor. J. Biol. Chem. 266:510-518. [PubMed] [Google Scholar]
- 28.Torchia, J., D. W. Rose, J. Inostroza, Y. Kamei, S. Westin, C. K. Glass, and M. G. Rosenfeld. 1997. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature 387:677-684. [DOI] [PubMed] [Google Scholar]
- 29.Verrijdt, G., E. Schoenmakers, A. Haelens, B. Peeters, G. Verhoeven, W. Rombauts, and F. Claessens. 2000. Change of specificity mutations in androgen-selective enhancers. Evidence for a role of differential DNA binding by the androgen receptor. J. Biol. Chem. 275:12298-12305. [DOI] [PubMed] [Google Scholar]
- 30.Wang, X. D., J. Shou, P. Wong, D. M. French, and W. Q. Gao. 2004. Notch1-expressing cells are indispensable for prostatic branching morphogenesis during development and regrowth following castration and androgen replacement. J. Biol. Chem. 279:24733-24744. [DOI] [PubMed] [Google Scholar]
- 31.Yu, X., P. Li, R. G. Roeder, and Z. Wang. 2001. Inhibition of androgen receptor-mediated transcription by amino-terminal enhancer of split. Mol. Cell. Biol. 21:4614-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]