Abstract
The process of mRNA localization, often used for regulation of gene expression in polarized cells, requires recognition of cis-acting signals by components of the localization machinery. Many known RNA signals are active in the contexts of both the Drosophila ovary and the blastoderm embryo, suggesting a conserved recognition mechanism. We used variants of the bicoid mRNA localization signal to explore recognition requirements in the embryo. We found that bicoid stem-loop IV/V, which is sufficient for ovarian localization, was necessary but not sufficient for full embryonic localization. RNAs containing bicoid stem-loops III/IV/V did localize within the embryo, demonstrating a requirement for dimerization and other activities supplied by stem-loop III. Protein complexes that bound specifically to III/IV/V and fushi tarazu localization signals copurified through multiple fractionation steps, suggesting that they are related. Binding to these two signals was competitive but not equivalent. Thus, the binding complexes are not identical but appear to have some components in common. We have proposed a model for a conserved mechanism of localization signal recognition in multiple contexts.
Subcellular localization of mRNAs is a process widely used to concentrate accumulation of proteins at specific sites (3, 23, 26, 37, 45). In most cases, the mechanism of localization involves directed movement, and mRNAs appear to traffic along cytoskeletal elements. Molecular motors, including members of the dynein, kinesin, and myosin families, are required for localization of many mRNAs, and multiple motors may contribute to a single localization event (2, 4-7, 11, 20, 27, 36, 42, 49, 53). It is generally believed that the motors power translocation directly, moving along microtubules or microfilaments with mRNAs as cargo. The association of mRNAs with motors or other transport intermediates relies on signals contained within the localized mRNAs, most commonly found in 3′ untranslated regions (UTRs) (3, 45).
Developing Drosophila oocytes and embryos localize a large number of mRNAs, many of which display similar programs of movement (3). In the ovary, most localized mRNAs are efficiently transported into the oocyte from their sites of transcription in the associated nurse cells during early stages of oogenesis. In the syncytial blastoderm stage of embryogenesis, mRNAs encoding the pair rule segmentation proteins are localized apically, between the peripheral nuclei and the embryo cortex. Despite widely overlapping patterns of mRNA localization in each context, only one case of conservation between localization signals has been observed. Both the fs(1)K10 and orb mRNAs are concentrated in the oocyte early in oogenesis and accumulate transiently at the anterior of the oocyte during stage 7-8 (25, 40). An A- and U-rich sequence predicted to form a stem-loop structure has been identified as the fs(1)K10 localization signal (41), and a similar element is found within the less precisely mapped orb signal (25). None of the other mRNAs known to localize within the ovary appear to contain a similar signal. The general theme of localization signals with related functions but no obvious sequence similarities extends to the pair rule genes: signals for the hairy (h) and fushi tarazu (ftz) mRNAs have been mapped to their 3′ UTRs, but there are no obvious similarities (10, 13).
The absence of similarities among most localization signals suggests that each is recognized by dedicated localization factors. If so, it seems unlikely that a signal would retain activity in a foreign setting, where cognate recognition factors would presumably not be expressed. However, the demonstration that certain localized mRNAs from Drosophila germ line cells were localized when ectopically expressed in ovarian follicle cells provided the initial indication that this notion is incorrect (21). This phenomenon was investigated more closely by Bullock and Ish-Horowicz (9), who used an RNA microinjection assay to show that many localized mRNAs from ovaries can also be apically localized in embryos. Remarkably, the converse is also true, and pair rule mRNA signals that direct apical localization in embryos are competent to support the fs(1)K10-like program of early localization in ovaries. Furthermore, BicaudalD (BicD) and Egalitarian (Egl), localization factors that appear to mediate contacts between RNA signals and microtubule-based motors, are required for embryonic examples of mRNA localization and are implicated in mRNA localization events in the ovary as well (9, 14, 22, 33, 35, 48). Thus, there is at least partial conservation of both the signals and machinery used for mRNA localization. The apparent absence of similarities in the sequence of the signals active for both ovarian and embryonic localization is quite curious.
The bicoid (bcd) mRNA localization signal, which is normally active in the ovary and retains its activity in foreign settings, has been studied in detail and provides a good example to use in addressing questions about conservation of localization machinery. The early phase of bcd localization is directed by stem-loop IV/V, a subdomain of its 3′ UTR (30) (see Fig. 3). Subtle mutations that disrupt predicted secondary structural elements of IV/V block early localization, even when introduced into the complete bcd 3′ UTR (29). Thus, IV/V is both necessary and sufficient for the program of early ovarian localization similar to that supported by fs(1)K10 and heterologous embryonic localization signals. This apparent similarity between the activity of IV/V and that of embryonic signals is reinforced by the behavior of subtle mutations within IV/V that block activity in the ovary (29, 34) and also inhibit apical localization by the full-length bcd signal in the embryo (9; data presented here). If the recognition machinery is highly conserved between ovaries and embryos, as has been suggested, then the IV/V localization signal would be expected to support apical localization in the embryo.
FIG. 3.
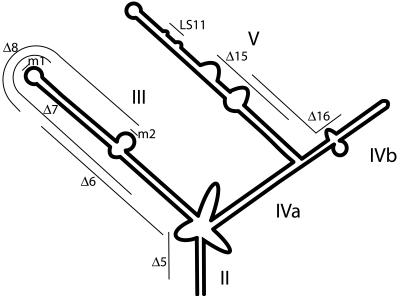
bcd 3′ UTR structure and mutations. Schematic representation of the structure of the bcd 3′ UTR showing stem-loops II to V. The regions removed by the Δ5, Δ6, Δ7, Δ8, Δ15, and Δ16 deletion mutations are shown, as is the site of the LS11 linker-scanning mutation. m1 and m2 are the sites of the substitution mutations that disrupt in vitro dimerization when they are individually present or restore in vitro dimerization when both are present.
We have tested that prediction and found that the IV/V domain of the bcd signal is necessary but not sufficient for robust apical localization within the embryo. Other regions of bcd RNA and the ability to dimerize are also required. On the basis of these results and the properties of isolated protein complexes that bind both ovarian and embryonic localization signals, we have suggested that a general strategy for recognition of localization signals is conserved, even though the recognition machinery does not appear to be highly conserved, and the signals themselves bear no obvious sequence similarity.
MATERIALS AND METHODS
Plasmids.
The plasmid used to synthesize bcd cRNA contained 22 nucleotides (nt) of the 5′ UTR and the full-length coding sequence and the 3′ UTR. Plasmids for transcription of bcd cDNA with subdomains of the bcd 3′ UTR were constructed by modification of a bcd cDNA with 22 nt of the 5′ UTR and the first 42 nt of the 3′ UTR. DNA fragments corresponding to 3′ UTR subdomains were inserted at the 3′ end of the truncated 3′ UTR and contained the following nucleotides: stem-loop III, 1890 to 2052; stem-loops IV/V, 2053 to 2327; stem-loops III/IV/V, 1890 to 2327 (coordinates of the sequence with GenBank accession no. nm_169157). The LS11 mutant of IV/V was from reference 34, and an LS11 mutant of III/IV/V was constructed by the same approach. The 3′ UTR deletion series Δ5 to Δ20 were derived from the corresponding plasmids in reference 31 and constructed by subcloning into a plasmid containing a bcd cDNA with 22 nt of its 5′ UTR. The series of plasmids used to test the importance of the bcd dimerization motif, p875(w,w), p875(m,w), p875(w,m), p875(m,m), p875(L,R), and p875(HIV) were generously provided by Christine Brunel and are described in reference 52. The plasmid encoding the bcd coding sequence is described in reference 1, and the green fluorescent protein (GFP) sequence is described in reference 43.
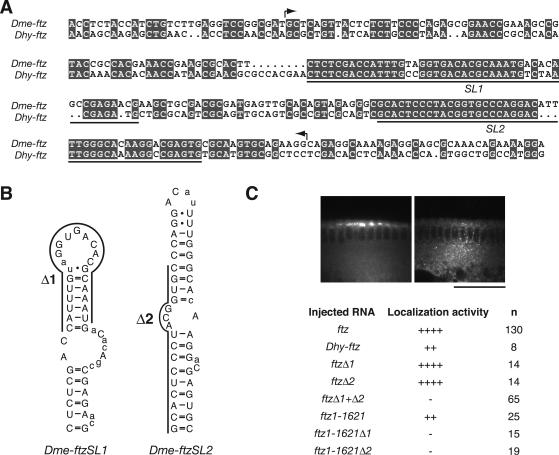
The D. melanogaster ftz RNAs for injection were prepared from a full-length cDNA or from a cDNA lacking the 3′ 121 nt. The Δ1 and Δ2 mutations (see Fig. 5B) were introduced into these template DNAs by Quikchange (Stratagene) mutagenesis. The Drosophila hydei ftz DNA template corresponds to a 538-bp HindIII/NcoI fragment of genomic sequence (GenBank accession number X79494) containing the predicted ftz localization element (FLE) and 333 bp of 5′ flanking sequences.
FIG. 5.
Two stem-loops are essential components of the FLE. (A) Alignment of partial DNA sequences from the ftz 3′ UTRs of D. melanogaster (Dme) and D. hydei (Dhy), which diverged from one another ∼60 million years ago. ftz sequences from both species supported localization upon injection into D. melanogaster embryos (see panel C). Arrows bracket the FLE (nt 1374 to 1579), which is the minimal region necessary and sufficient to mediate localization (9). Identical nucleotides are shaded; underlining indicates the positions of two highly conserved blocks, SL1 and SL2, that are thermodynamically predicted to form stem-loop structures (http://www.bioinfo.rpi.edu/applications/mfold/). (B) Predicted RNA secondary structures of D. melanogaster ftz SL1 and SL2. Positions conserved with respect to D. hydei ftz are in uppercase. Most of the nonconserved positions are found within predicted single-stranded regions, consistent with the importance of double-stranded regions for hairy pair rule transcript localization (10). Bases deleted in Δ1 and Δ2 are indicated. (C) Summary of injection of ftz transcript variants (D. melanogaster ftz, unless stated otherwise), showing the role of SL1 and SL2 in localization (n = number of embryos scored; see Table 1 for categorization of localization efficiency). In the context of the complete ftz mRNA, mutation of either SL1 or SL2 alone had little effect, while mutation of both blocked localization. In contrast, either mutation eliminated localization of the ftz1-1621 transcript, which lacks 121 nt from the 3′ end (numbered in accordance with reference 9; position 1621 corresponds to position 2671 in the ftz gene [GenBank accession no. X00854]). These 3′ sequences are not sufficient for localization (9) but can make contributions that are somewhat redundant with the FLE. Images show representative examples of ftz and ftz Δ1+Δ2 mRNA distribution 10 min after injection. Scale bar = 50 μm.
For RNA binding assays, the ftz RNA used encompassed most of the 3′ UTR (nt 2392 to 2835 of the ftz gene; accession no. X00854), either in the wild-type form, with the Δ1+Δ2 deletion mutations (Fig. 5B), or with nt 2478 to 2680 deleted in ftz ΔFLE.
Fly stocks.
w1118 Drosophila melanogaster was used as the wild type, and P[sryDB56]/+; sry δ14/Df(3R) X3F (38) was used in bcd RNA null experiments.
Fluorescent RNA synthesis.
Fluorescent RNAs were synthesized in accordance with the manufacturer's (Molecular Probes) instructions. Typically, the transcription reaction contained 500 ng of linearized plasmid DNA, T7 RNA polymerase, 0.5 mM ATP, 0.5 mM CTP, 0.5 mM GTP, 0.35 mM UTP, 2 mM 7mG(5′)ppp(5′)G cap analogue, and 0.13 mM Alexa 488-labeled UTP (Molecular Probes). RNAs were treated with DNase I, spun through a Sephadex G50 column, extracted with phenol chloroform, precipitated with ethanol in the presence of ammonium acetate, and resuspended in water.
Fluorescent RNA injections.
Dechorionated embryos were affixed to glass coverslips and injected with 500 ng of fluorescent RNA per μl under series 700 halocarbon oil (Halocarbon Products Corporation, River Edge, N.J.). RNAs were injected into the yolk-filled cytoplasm closest to the dorsal surface of syncytial blastoderm embryos and examined for localization 10 to 12 min after injection or monitored via time-lapse microscopy over a 10-min period with a Leica TCS SP2 inverted confocal microscope. The results of the injections were consistent between injections on different days and from RNA produced in different transcription reactions. In a single injection experiment, RNAs were characterized as having strong apical localization activity if the RNA was concentrated at the apical surface (this apical concentration may be less than the wild-type concentration in some cases), as weakly localized if particles were present but not concentrated in the apical cytoplasm, or as having no apical localization (see Table 1).
TABLE 1.
Localization activity of injected RNAsa
| Injected RNA | Nucleotides in constructa | Embryonic apical localization
|
Anterior oocyte localization
|
||
|---|---|---|---|---|---|
| Activity | n | Activity | n | ||
| bcd cDNA | 185-2493 | ++++ | 13 | ++ | 7 |
| Coding sequence + IV/V | 185-1717, 2053-2327 | ± | 34 | ++ | 11 |
| bcd cDNA in bcd RNA null | 185-2493 | ++ | 7 | ||
| bcd coding sequence + IV/V in bcd RNA null | 185-1717, 2053-2327 | ++ | 15 | ||
| bcd 3′ UTR | 1676-2493 | ++++ | 44 | ||
| − | 28 | ||||
| GFP coding sequence + LS11 IV/V | 185-1717, 2053-2327 | − | 23 | ||
| GFP coding sequence + III/IV/V | 185-1717, 1890-2327 | ++ | 36 | ||
| GFP coding sequence + LS11 III/IV/V | 185-1717, 1890-2327 | − | 33 | ||
| GFP coding sequence + III | 185-1717, 1890-2052 | − | 27 | ||
| Δ5 (II) | 185-1844, 1889-2493 | +++ | 21 | ||
| Δ6 (III) | 185-1890, 1939-2493 | ± | 42 | ||
| Δ7 (III) | 185-1906, 1958-2493 | + | 70 | ||
| Δ8 (III) | 185-1945, 1993-2493 | + | 18 | ||
| Δ9 (III) | 185-1987, 2038-2493 | + | 38 | ||
| Δ10 (IVa) | 185-2025, 2063-2493 | ++ | 13 | ||
| Δ11 (IVa/V) | 185-2054, 2096-2493 | ± | 4 | ||
| Δ12 (V) | 185-2085, 2128-2493 | − | 5 | ||
| Δ13 (V) | 185-2122, 2160-2493 | − | 10 | ||
| Δ14 (V) | 185-2132, 2181-2493 | − | 28 | ||
| Δ15 (V) | 185-2172, 2216-2493 | +++ | 23 | ||
| Δ16 (IVb/V) | 185-2189, 2242-2493 | − | 82 | ||
| Δ17 (IVb) | 185-2245, 2268-2493 | ± | 52 | ||
| Δ18 (IVa/IVb) | 185-2258, 2307-2493 | − | 9 | ||
| Δ19 (IVa/IVb) | 185-2284, 2323-2493 | − | 8 | ||
| Δ20 (II/IVa) | 185-2313, 2390-2493 | + | 7 | ||
| Dimerization mutant w,m | 1676-2493 | ± | 37 | ||
| Dimerization mutant m,w | 1676-2493 | ± | 31 | ||
| Double mutant m,m | 1676-2493 | + | 77 | ||
| bcd 3′ UTR + HIV dimerization motif | 1676-2493 | + | 57 | ||
| bcd 3′ UTR + group 1 intron dimerization motif | 1676-2493 | ± | 46 | ||
| FLE | 2077-2835 | + | 12 | ||
++++, 90 to 100% have strong localization; +++, 75 to 90% have strong localization; ++, 40 to 75% have strong localization; +, 10 to 40% have strong localization; ±, less than 10% show localization and many show weak localization; −, no localization.
Ovaries were dissected from well-fed 3-day-old females into series 95 halocarbon oil on glass coverslips. Individual egg chambers were teased apart with forceps and transferred to the stage of a confocal microscope, where nurse cells were injected with RNA. The degree of anterior RNA localization was determined after 30 min of observation. Injections into P[sryDB56]/+; sry δ14/Df(3R)X3F were performed with egg chambers removed from females raised at 18°C (containing little or no bcd mRNA [38]) that were injected at room temperature. No difference was detected between the results from these injections and those from injections into females with the same genotype raised at room temperature (data not shown).
RNA binding assays.
Preparation of probe and competitor RNAs and mobility shift assays were performed as described previously (1), except for the reaction conditions when the FLE was used as a probe: 35 mM HEPES (pH 7.4)-1 mM MgCl2-70 μg of heparin per ml-60 μg of yeast tRNA per ml-1.2 mM spermidine-150 mM KCl-1% glycerol. ftz-derived RNAs were folded by heating to 50°C in the same buffer and slowly cooling to room temperature.
Purification of embryonic recognition complexes.
Complexes were purified from unstaged embryo extract under the same conditions and by the same procedure previously used for purification of ovarian complexes (1) up through the second density gradient sedimentation: ammonium sulfate fractionation, DEAE anion exchange, Sephacryl S-400 (Pharmacia) gel filtration, and 5 to 35% Nycodenz (Sigma) density gradients.
RESULTS
bcd IV/V RNA is not sufficient for efficient apical embryonic localization.
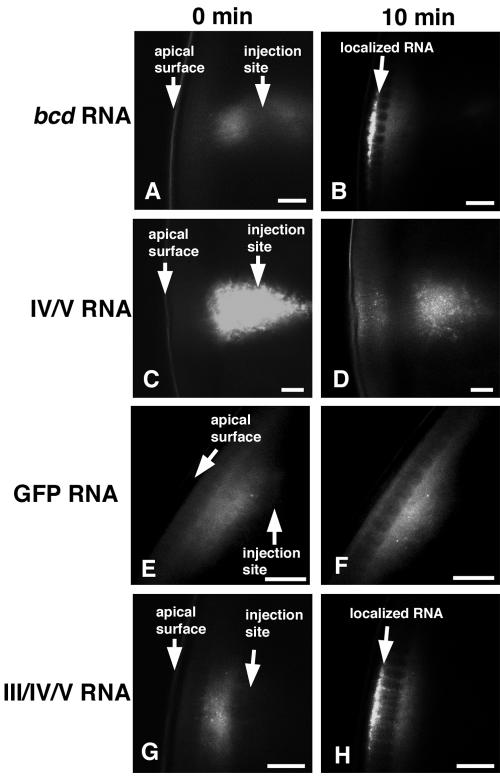
Previous demonstration of apical localization in the embryo by the bcd localization signal relied on an assay in which fluorescent RNAs were injected into a subcortical zone of live syncytial blastoderm embryos and tracked by time-lapse microscopy or viewed after fixation (9, 10, 24, 53). We also found that labeled full-length bcd transcripts injected into embryos rapidly formed particles that moved unidirectionally toward the apical surface, becoming apically enriched after 2 min and displaying a heavy apical concentration within 10 min (10, 53) (Fig. 1 and Table 1). Control RNAs lacking a localization signal, such as the GFP coding region, could also form particles, but these particles moved only randomly in the cytoplasm. While they sometimes approached the embryo periphery, GFP RNA particles never became concentrated between the apical side of the nuclei and the cortex. Thus, the injection assay robustly differentiates between localized and unlocalized RNAs.
FIG. 1.
Apical embryonic localization of bcd RNA requires stem-loop III. Representative confocal images of fluorescent RNA injected into live syncytial blastoderm embryos. At the left are images obtained immediately after injection, and at the right are images obtained 10 min later. bcd transcripts (A and B) localized strongly to the apical surface 10 min after injection. Constructs containing stem-loops IV/V of bcd RNA localized weakly to the apical surface (C and D). GFP RNA formed particles, but they never became concentrated apically (E and F). Constructs containing stem-loops III/IV/V of the bcd 3′ UTR were concentrated apically after injection (G and H). The injected RNA was often not visible at the injection site, presumably because of refraction of light by yolk, but the RNAs were always visible after they migrated into the yolk-free apical cytoplasm. Scale bars are 20 μm.
To test the localizing activity of the IV/V localization signal by the injection assay, we generated bcd transcripts in which the 3′ UTR was replaced with the IV/V region. Unlike the full-length bcd RNA, the bcd IV/V RNA did not direct strong apical localization. After injection, it appeared in particles, but only a small portion of these moved toward the apical surface. By 10 min postinjection, this limited localization appeared as weak subapical fluorescence clearly different from the strong apical localization of the full-length bcd RNA (Fig. 1 and Table 1). The bcd IV/V RNA was also distinct from control RNAs with no localization activity, which never displayed even a weak concentration at the apical surface. Varying the concentration of RNA injected did not alter the weak localization of the bcd IV/V RNA (data not shown). The residual low activity of this subdomain of the bcd localization signal appeared to be similar to that of certain mutant forms of the hairy signal, which are considerably impaired but not completely inactivated (10).
Stem-loops IV/V are sufficient for localization in the oocyte.
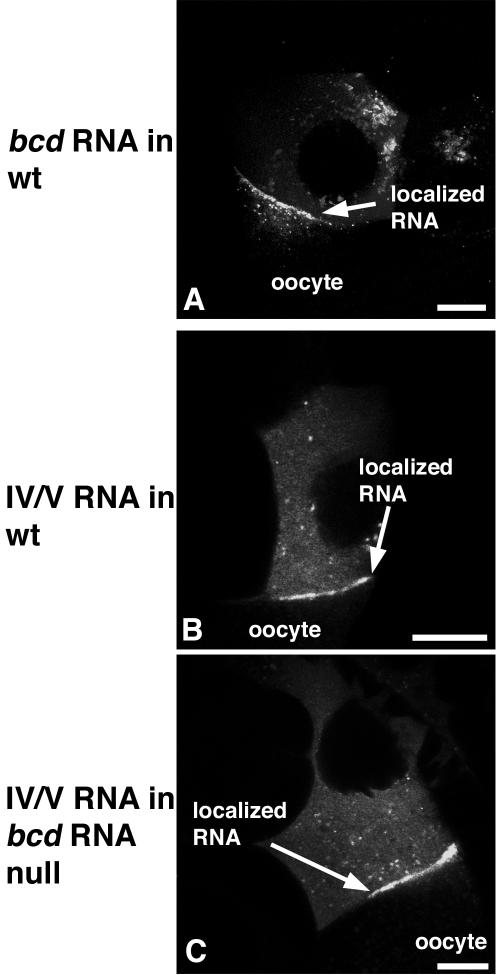
The weak localization activity of the IV/V signal in the embryo could signify differences in the machinery used for the ovarian and embryonic localization programs. Alternatively, the strong activity of the IV/V signal—observed when it was expressed in transgenic ovaries—might be impaired when the RNA is introduced by injection. To address this possibility, we compared the full-length bcd RNA to the bcd IV/V RNA after injection into stage 9 nurse cells and found that the two RNAs had similar localization activities: each formed particles in the cytoplasm that moved rapidly, clustered at and passed through ring canals, and localized to the anterior of the oocyte. Anterior concentration of both RNAs was observed within 10 min and was very strong after 30 min. Although the full-length bcd RNA localized somewhat more consistently (5 of 7 injections) than the bcd IV/V RNA (6 of 11 injections) (Fig. 2) (negatively scored injections included those in which the injection procedure disrupts the nurse cell), the extreme difference between the activities of the two signals observed in the embryo injection assay cannot be attributed to an inherently weak localization activity of the IV/V RNA or to significantly reduced activity in injection assays.
FIG. 2.
Stem-loops IV/V are sufficient for anterior oocyte localization in the presence or absence of endogenous bcd RNA. Fluorescent RNA constructs were injected into the cytoplasm of nurse cells and imaged 30 min after injection. bcd RNA formed particles in the nurse cell cytoplasm, which passed through ring canals and become localized to the anterior margin of the oocyte (A). Mutant bcd RNA in which the 3′ UTR was limited to just stem-loops IV/V localized efficiently to the anterior margin in both wild-type (wt) egg chambers (B) and egg chambers that lacked endogenous bcd RNA, P[sryDB56]/+;sry δ14/Df(3R) X3F (C). Scale bars are 20 μm.
Another potential explanation of the weak activity of the IV/V signal for apical localization in embryos is that the IV/V RNA mediates association with the bcd RNA, such that its apparent localization activity in the ovary reflects association with endogenous bcd transcripts. Indeed, exogenously expressed osk 3′ UTR RNA is dependent upon the presence of endogenous osk RNA for its localization (18). Furthermore, interactions between two or more bcd mRNA molecules are possible since the bcd 3′ UTR contains a dimerization domain (51, 52) and there is evidence of dimerization after injection into early-stage embryos (16). Although the IV/V RNA lacks this dimerization domain, it might nevertheless assemble into particles that contain endogenous bcd transcripts and thus be colocalized. The very weak localization activity of IV/V in blastoderm embryos would then simply reflect the absence of endogenous bcd mRNA at the site of injection (maternally contributed bcd mRNA is largely gone in the late-stage blastoderm embryos used for the injection assay, and any bcd mRNA remaining is concentrated at the anterior pole, distant from the site of injection). To address this possibility, the nurse cell injection assays were repeated with recipient ovaries from flies in which bcd mRNA transcription is largely eliminated (38). There was no significant difference in these ovaries between the strength of localization for the full-length bcd and bcd IV/V RNAs, which occurred in the majority of injections (5 of 7 for full-length bcd RNA and 10 of 15 for bcd IV/V RNA) (Fig. 2). We conclude that endogenous bcd RNA does not assist the localization of IV/V RNA in the ovary and that IV/V has only very weak intrinsic localization activity in the embryo.
Stem-loops III and IV/V direct apical localization.
Although the IV/V RNA has very limited ability to direct apical embryonic localization, it does appear to be required for the activity of the full-length bcd RNA. A point mutation that largely eliminates IV/V activity in the ovary also impairs apical localization (9, 30), and multiple deletions within the IV/V region also block apical localization of the full-length bcd RNA (see below). Thus, another region of the 3′ UTR must work together with IV/V to direct apical localization. The organization of the bcd 3′ UTR includes four highly structured domains (stem II and stem-loops III, IV, and V; Fig. 3), as well as one relatively unstructured domain that is dispensable for localization (8, 28, 32). To identify regions that are important for apical localization, we used the injection assay to test deletion mutants removing segments of the 3′ UTR in roughly 50-nt increments (31).
Stem II is not essential for apical localization, as the Δ5 mutant, which lacks most of one strand, retained substantial activity. A mutation that removes the complementary strand of stem II (Δ20) showed inhibited localization, but this mutant also lacks a substantial portion of stem-loop IV. Mutations within stem-loop III, which include Δ6 through Δ9, consistently displayed reduced localization activity. The most severe defects were observed for deletions in IV/V, with the single exception of Δ15, which retained strong localization activity and thus defines a nonessential portion of stem-loop V. Any deletion mutant removing part of the remainder of stem-loop V, or the distal part of stem-loop IV, strongly impaired or abolished apical localization (Table 1).
The deletion mutants indicate that the IV/V region is of primary importance for localization. Because mutations in other regions do not produce as severe defects, there may be some redundancy in the other required elements. Nevertheless, on the basis of the severity of the defects of the deletion mutants, stem-loop III is the best candidate subdomain to constitute, together with IV/V, a functional apical localization signal. To test this prediction, we made a bcd RNA with a III/IV/V 3′ UTR. This bcd III/IV/V RNA had readily detectable apical localization activity, although it was not as active as the full-length bcd RNA (Fig. 1 and Table 1). In contrast, stem-loop III alone had no localization activity, as bcd III transcripts were not localized (Table 1). We conclude that most of the bcd 3′ UTR is required for full activity in apical localization but that III/IV/V is a minimal signal required to provide substantial activity.
A dimerization motif is required for full apical bcd RNA localization.
The role of stem-loop III in apical localization could be to provide binding sites for localization factors other than those that recognize IV/V. Alternatively, the contribution of stem-loop III may be its ability to dimerize. Localization elements with weak or undetectable activity in isolation can be dramatically strengthened when expressed as multimers (10, 17, 31), and therefore dimerization of IV/V might increase its activity. Dimerization of stem-loop III involves base pairing between complementary sequences in the terminal loop and a bulge on the 3′ strand of the stem. When either sequence is altered, as in mutant w,m or m,w constructs (m and w indicate mutant and wild type, respectively), dimerization is largely eliminated. However, in the m,m double mutant with compensatory changes to restore complementarity, dimerization is largely restored (16, 51, 52).
We used these mutants to assess the contribution of dimerization to apical localization. When introduced into the full-length bcd 3′ UTR, both the w,m and m,w configurations strongly inhibited apical localization. The m,m configuration restored a low level of localization, but this mutant remained substantially less active than the wild type (Table 1). These results suggest that dimerization contributes to the activity of the bcd signal but that the mutations in the dimerization motif have additional consequences that affect localization.
To determine if stem-loop III can be replaced with other dimerization motifs, we tested bcd RNAs in which stem-loop III was replaced with sequences that promote dimerization of human immunodeficiency virus (HIV) RNA (44) or group I introns (12, 19). Injected RNAs containing the HIV dimerization motif had a low level of localization activity, similar to that of the m,m mutant and much less than that of wild-type bcd RNA. The bcd RNA with the group I intron motif had even less activity (Table 1). Collectively, the data indicate that dimerization is important for apical bcd localization but that the stem-loop III dimerization motif provides additional required features.
Identification of an embryonic III/IV/V binding activity.
A large multiprotein complex from ovaries binds specifically to IV/V RNA and has been strongly implicated in its localization (1). Some of the IV/V binding complex components are not present in the embryo, so the exact same complex cannot contribute to embryonic localization directed by III/IV/V. Nevertheless, subtle mutations inhibit both IV/V activity in the ovary and the activity of the full-length signal in the embryo (9, 29, 30), and the results presented above clearly imply that IV/V has a major role in apical localization in the embryo. Consequently, there may be a related binding complex in the embryo. To search for such a complex that binds III/IV/V RNA, we used the gel mobility shift assay that allowed detection of the ovarian IV/V RNA binding complex.
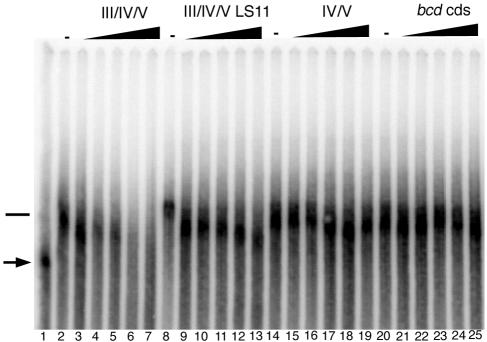
Factors present in Drosophila embryo extracts bound to radiolabeled III/IV/V RNA and retarded its mobility through a native polyacrylamide gel (data not shown). This binding activity was extensively purified by fractionation of embryo extracts, by the purification scheme originally used for the ovarian complex (see Materials and Methods). To assess the specificity of binding in highly purified fractions, competition binding experiments were performed by adding increasing amounts of unlabeled competitor RNA prior to the addition of radiolabeled III/IV/V RNA probe (Fig. 4). In such competition binding assays, unlabeled III/IV/V RNA competed effectively for binding with the III/IV/V probe (Fig. 4, lanes 2 to 7). In contrast, a III/IV/V RNA bearing the LS11 linker scanning mutation, which inhibits activity of both IV/V in ovarian localization (34) and III/IV/V in embryo apical localization (Table 1), was an ineffective competitor (lanes 8 to 13). IV/V RNA alone was also a poor competitor (lanes 14 to 19), as was a fragment from the bcd coding region (lanes 20 to 25).
FIG. 4.
Gel mobility shift assays detecting a complex that binds specifically to localized bcd RNAs. The migration of radiolabeled bcd III/IV/V RNA in a native gel (lane 1, probe indicated by an arrow) was retarded by a complex present in the purified embryonic fraction (lanes 2, 8, 14, and 20); the position of the RNA associated with the complex is indicated by a horizontal bar. To determine the specificity of binding, aliquots of extracts were incubated with 3.5, 7, 11, 16, or 22 pmol of unlabeled III/IV/V (lane 3 to 7), III/IV/V LS11 (lanes 9 to 13), IV/V (lanes 15 to 19), or bcd coding sequence (cds) (lanes 21 to 25) RNA before addition of 1.2 pmol of labeled III/IV/V RNA.
These results show that the binding activity has specificity for III/IV/V RNA. Moreover, the sensitivity of binding to the LS11 mutation strongly suggests that the activity contributes to localization. The inability of IV/V to compete for binding is consistent with its very limited apical localization activity and appears to place restrictions on the role of IV/V when it acts in the context of III/IV/V. Specifically, it seems unlikely that III and IV/V act as independent binding domains. If they did, the IV/V competitor would be expected to at least alter the mobility of the bound complex.
Some of the components of the ovarian IV/V binding complex have been identified by Western blot analysis and include Modulo (Mod), Swallow (Sww), poly(A) binding protein, Smooth, and Nod. Mod is the most abundant of these (1). Preliminary mass spectrographic analysis of the embryonic III/IV/V binding activity has failed to detect any of these proteins. Thus, the two complexes appear to be substantially different.
ftz and III/IV/V binding activities cofractionate and overlap.
Dynein, BicD, and Egl are all implicated in apical localization of both pair rule and bcd mRNAs, indicating that bcd relies on at least some of the same localization machinery used by the pair rule mRNAs (9, 53). None of these factors is thought to bind directly to localization signals, and the possibility that the different transcripts also rely on shared recognition factors is open. The pair rule mRNA localization signals are relatively large (greater than 100 nt), suggesting that they may be recognized by multiple factors, possibly by a large multiprotein complex similar to the ovarian complex involved in IV/V recognition. However, there are no obvious sequence similarities between the characterized pair rule localization elements and the bcd signals.
To address the issue of signal recognition and determine whether related or distinct factors bind to the different RNAs, we focused on the ftz signal, which is similar in size to III/IV/V and thus ideal for competitive binding assays. This signal directs anterior localization in oocytes, both when expressed as a transgene (9) and when injected (Table 1). A comparison of the D. hydei and D. melanogaster ftz RNAs, both of which localize in D. melanogaster embryos (Fig. 5C), revealed two conserved regions within a minimal localization element (the FLE) (9) that are predicted to form stem-loop structures (Fig. 5A and B). Deletions that disrupt a single stem-loop had no effect on localization within the context of the full-length transcript, but disruption of both stem-loops (in ftz Δ1+Δ2) eliminated localization activity (Fig. 5C). Thus, the ftz Δ1+Δ2 RNA is likely to be defective in binding to some or all of the factors involved in the localization of ftz mRNA.
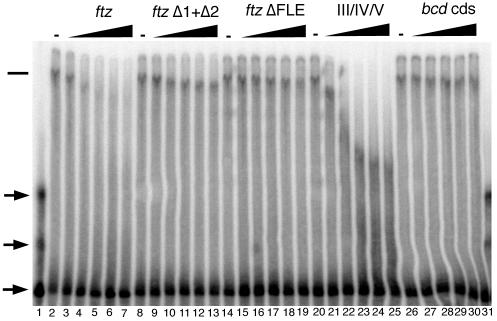
We examined whether the most highly purified fractions of specific III/IV/V binding activity also had FLE binding activity. Initial experiments with FLE RNA prepared by the method used for III/IV/V RNAs did not reveal binding (data not shown). However, the bcd RNA is much more structured than predicted for the FLE RNA, which may not fold correctly under the same conditions. Consequently, various folding regimens were tested. When folded in the presence of 1 mM magnesium ion, FLE RNA was bound by factors in the purified embryo fractions, and this binding was effectively competed by addition of unlabeled FLE competitor RNA (Fig. 6, lanes 1 to 7). The binding activity is specific and is sensitive to mutations that affect localization activity. The ftz Δ1+Δ2 RNA was a very poor competitor for binding (lanes 8 to 13), as was an ftz RNA with the entire localization signal deleted, ftz ΔFLE (lanes 14 to 19). Similarly, an unrelated RNA from the bcd coding region also failed to compete for binding (lanes 26 to 31). The specificity of the binding activity and its sensitivity to mutations that inhibit localization suggest that it acts in recognizing ftz RNA for apical localization.
FIG. 6.
Binding of embryonic proteins to the ftz localization signal is partially competed by bcd III/IV/V RNA. Electrophoretic migration of radiolabeled FLE RNA in a native gel. In the absence of added protein (lane 1), the RNA appears in one predominant band, as well as several less abundant bands (arrows); the multiple bands probably represent different conformers. The FLE RNA was bound by factors present in an embryonic extract (lanes 2, 8, 14, 20, and 26); the position of the RNA associated with the complex is indicated by the horizontal bar. To determine the specificity of binding, aliquots of extracts were incubated with 3.5, 7, 11, 16, or 22 pmol of unlabeled ftz (lanes 3 to 7), ftz Δ1+Δ2 (lanes 9 to 13), ftz ΔFLE (lanes 15 to 19), III/IV/V (lanes 21 to 25), or bcd coding sequence (cds) (lanes 27 to 31) RNA before addition of 1.2 fmol of labeled ftz RNA.
The presence of an ftz localization signal binding activity in the same extensively purified fractions that contain III/IV/V-binding activity implies that the complexes have components in common or that they have physical and chemical properties in common that determine their fractionation behavior. To distinguish between these options, we examined whether the III/IV/V and FLE RNAs could compete with one another for binding. Both are effective competitors, although neither simply shifted the probe RNA to its initial position (Fig. 6, lanes 20 to 25, and data not shown). The simplest interpretation of these results is that the FLE binding activity is a multiprotein complex and that the III/IV/V RNA can compete for binding to a subset of the components. Thus, the novel position of the FLE probe RNA following competition either reflected a partial binding complex (lacking a subset of proteins stripped off by binding to III/IV/V) or a hybrid entity containing the intact ftz binding complex, the FLE RNA, and the III/IV/V RNA. In either case, the results demonstrate that the FLE and III/IV/V binding complexes are not identical but suggest that some components are common to both.
DISCUSSION
Recent evidence has pointed to the conservation of machinery and signals among different programs of mRNA localization in the Drosophila ovary and embryo. Transport to the oocyte and apical localization in the embryo both rely on microtubules (24, 39, 50, 53), and there are apparently shared requirements for two proteins, BicD and Egl, thought to be part of a complex linking localized mRNAs to cytoskeletal elements (9, 33, 35, 48). The evidence for a conserved system to recognize localization signals rests on the ability of localized mRNAs from one setting—ovary or blastoderm embryo—to function when introduced into the other by injection or ectopic expression. Moreover, for the better-characterized signals, including those from the fs(1)K10 and bcd mRNAs, cis-acting mutations that inhibit localization in the ovary also interfere with apical localization in blastoderm embryos (9). These features suggest that recognition systems in the different cell types may be related or identical. Our results argue very strongly against the notion of identical recognition systems (as did the presence of ovary-specific proteins in the IV/V RNA binding complex), since the IV/V signal has extremely weak activity for apical localization in the embryo despite its robust activity in the ovary. Nevertheless, the III/IV/V bcd signal is recognized and this could indicate that the recognition machinery is at least partially conserved. Alternatively, the apical localization of III/IV/V RNA in the embryo and the binding of III/IV/V RNA to the complex that recognizes the FLE could be fortuitous and an artifact.
The mutational analysis of the bcd localization signal argues that the recognition events in different contexts are related. Both instances of localization are sensitive to particular point and linker-scanning mutations in the IV/V region. Larger deletions within the IV/V region abolish apical embryonic localization. A subset of these, including Δ17 to Δ19, have been tested in the context of lacZ reporter transcripts, and all eliminate anterior localization in the oocyte (K. Kerr and P. M. Macdonald, unpublished results). Thus, it appears that similar bcd sequences are required in both the ovary and the embryo, suggesting that the recognition machinery is related. However, we cannot exclude the possibility that the mutants alter the structure of the signal, rather than affecting individual protein binding sites. If so, then the argument for shared recognition factors in the ovary and embryo is less compelling.
A general model for signal recognition.
Because the bcd mRNA is not expressed in the embryo at the time when apical localization occurs, its ability to be localized there requires that a recognition system normally used to bind apically localized mRNAs is co-opted or that a cryptic recognition system is present. Our demonstration that the bcd III/IV/V RNA can compete for binding of a protein complex specific for localization-competent FLE RNA very strongly supports the first model. The precise nature of the competition, however (conversion of the FLE RNA probe-protein complex to a complex with novel mobility in the gel), argues that the interactions of the different localization signals with the recognition complexes are not equivalent, since the III/IV/V competitor does not displace all factors bound to FLE RNA.
Why should the bcd localization signal, with no obvious sequence similarities to the ftz signal, direct an ftz-like program of localization and compete with the ftz signal for binding to a presumptive localization complex? One explanation follows from the model proposed for recognition of the bcd IV/V RNA (1). In the ovary, a large complex of proteins binds IV/V RNA with high affinity and specificity, and this binding activity appears to represent the sum of multiple low-affinity and low-specificity interactions by individual proteins. We propose that this general combinatorial recognition strategy is conserved, even if the complex components differ substantially in different cell types. The binding specificity of such a complex would be generated by the spatial juxtaposition of associated proteins with low specificities, or of proteins with high specificity for short (and thus low-complexity) sequences.
Conserved elements in functionally interchangeable binding substrates for localization recognition complexes could be difficult to identify, for three reasons. First, different signals may have different subsets of protein binding sites, reducing the likelihood of extensive sequence or structural identities. Second, conserved low-complexity binding sites could only be readily discerned if present in multiple copies, a situation that would only be expected if the recognition complexes had multiple identical subunits. Third, because the individual binding sites appear to be presented in the context of an ordered tertiary structure, they would not necessarily appear in a fixed order in the linear RNA sequence. Thus, we suspect that the bcd and ftz localization signals, as well as many others, do indeed have strong structural or sequence similarities, but they cannot be easily recognized by visual examination.
Contribution of stem-loop III to the function of the bcd localization signal.
How, specifically, does stem-loop III contribute to localization of bcd RNAs in the embryo? One known function of stem-loop III is to promote dimerization. Dimerization of the full-length bcd 3′ UTR is a two (or more)-step process (51, 52) initiated by base pairing between complementary nucleotides in the terminal loop of stem-loop III (LIIIb) and in a bulged loop on the 3′ strand of stem III (LIIIa). Notably, these predicted base pairs are conserved in bcd genes from all of the Drosophila species examined (28). In a second step, for which the required sequence elements in the 3′ UTR are not well defined but do not include most of IV/V, dimers are stabilized by an essentially irreversible reaction, presumably a conformational change.
Results from three different assays address the importance of dimerization for bcd RNA localization. The first assay, using transgenes expressed in the ovary, involved several mutants lacking parts of stem-loop III (31). For some transgenes, the only assay of activity was rescue of the bcd mutant phenotype: although all rescued, this need not imply that localization was completely normal, as small amounts of bcd localization can support wild-type embryonic development. The localization of one of the mutants, Δ7, was tested more directly by visualization of the distribution of an attached lacZ reporter RNA. This RNA spreads laterally from the anterior of the oocyte but remains enriched in the anterior of embryos. Thus, the mutant has at least a modest localization defect, although this could be due to inability to dimerize, loss of binding sites for recognition factors, or both.
The injection assays described here also addressed the role of dimerization. The III/IV/V RNA is the minimal segment from the bcd 3′ UTR that is sufficient for strong apical localization in the embryo, but it is not as active as the full-length 3′ UTR. Stem-loop III is expected to promote the initial step in dimerization of III/IV/V while lacking sequences required for the subsequent step (51). Thus, there is a correlation between dimerization and apical localization. Further evidence that dimerization is important is provided by the point mutations that disrupt the LIIIa-LIIIb base-pairing interaction and produce severely inhibited localization activity. However, the failure of compensatory changes that restore LIIIa-LIIIb pairing to fully restore apical localization indicates that stem-loop III contributes more to apical localization than dimerization activity alone. This conclusion is reinforced by the very limited activity of RNAs in which stem-loop III is replaced with other dimerization motifs.
In a different type of RNA injection assay (15, 16) dimerization appeared to be the only role of stem-loop III. When the bcd 3′ UTR RNA was injected into the anterior of early precellularization embryos, it recruited Staufen, a protein required for anchoring of endogenous bcd mRNA (46, 47). The resulting particles were transported along astral microtubules during mitosis (a novel program of localization not displayed by endogenous bcd mRNA). When base pairing between LIIIa and LIIIb was disrupted, Stau-containing particles still formed but astral transport was eliminated, reminiscent of the effect on apical localization. However, unlike the case of apical localization, astral transport was restored completely by compensatory mutation of LIIIa and LIIIb to restore base pairing. Moreover, heterologous dimerization domains can substitute for stem-loop III to promote astral transport (52), even though they do not for apical localization. It is also noteworthy that apical localization can be directed by the bcd 3′ UTR either in isolation or in the context of the entire bcd mRNA with the coding region, while only the isolated bcd 3′ UTR is active for astral transport. The basis for these differences is not clear.
Which assay provides a faithful picture of the role of dimerization in the normal program of bcd mRNA localization? Specifically, is the contribution of stem-loop III only in dimerization, or does stem-loop III have an additional role in recognition, as suggested by the apical localization assay? The failure of compensatory mutations or heterologous dimerization domains to fully restore apical localization of III/IV/V clearly indicates an additional role for stem-loop III. It seems unlikely that stem-loop III would fortuitously mediate a recognition interaction in apical localization that is completely unrelated to what occurs in the ovary. We suggest that both dimerization and the recognition of stem-loop III by binding factors are events that contribute to bcd localization and that the degree to which either is required in a particular heterologous assay will reflect the degree of conservation of the heterologous recognition system.
Acknowledgments
This work was supported by NIH Public Health Service grant GM42612 and by the Beit Memorial Trust.
We thank D. Ish-Horowicz for discussions, F. Schnorrer and A Vincent for fly stocks, and C. Brunel for plasmids. Thanks to members of the Macdonald lab for helpful discussion and comments.
REFERENCES
- 1.Arn, E. A., B. J. Cha, W. E. Theurkauf, and P. M. Macdonald. 2003. Recognition of a bicoid mRNA localization signal by a protein complex containing Swallow, Nod, and RNA binding proteins. Dev. Cell. 4:41-51. [DOI] [PubMed] [Google Scholar]
- 2.Aronov, S., G. Aranda, L. Behar, and I. Ginzburg. 2002. Visualization of translated tau protein in the axons of neuronal P19 cells and characterization of tau RNP granules. J. Cell Sci. 115:3817-3827. [DOI] [PubMed] [Google Scholar]
- 3.Bashirullah, A., R. L. Cooperstock, and H. D. Lipshitz. 1998. RNA localization in development. Annu. Rev. Biochem. 67:335-394. [DOI] [PubMed] [Google Scholar]
- 4.Bertrand, E., P. Chartrand, M. Schaefer, S. M. Shenoy, R. H. Singer, and R. M. Long. 1998. Localization of ASH1 mRNA particles in living yeast. Mol. Cell 2:437-445. [DOI] [PubMed] [Google Scholar]
- 5.Betley, J. N., B. Heinrich, I. Vernos, C. Sardet, F. Prodon, and J. O. Deshler. 2004. Kinesin II mediates Vg1 mRNA transport in Xenopus oocytes. Curr. Biol. 14:219-224. [DOI] [PubMed] [Google Scholar]
- 6.Bobola, N., R. P. Jansen, T. H. Shin, and K. Nasmyth. 1996. Asymmetric accumulation of Ash1p in postanaphase nuclei depends on a myosin and restricts yeast mating-type switching to mother cells. Cell 84:699-709. [DOI] [PubMed] [Google Scholar]
- 7.Brendza, R. P., L. R. Serbus, J. B. Duffy, and W. M. Saxton. 2000. A function for kinesin I in the posterior transport of oskar mRNA and Staufen protein. Science 289:2120-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brunel, C., and C. Ehresmann. 2004. Secondary structure of the 3′ UTR of bicoid mRNA. Biochimie 86:91-104. [DOI] [PubMed] [Google Scholar]
- 9.Bullock, S. L., and D. Ish-Horowicz. 2001. Conserved signals and machinery for RNA transport in Drosophila oogenesis and embryogenesis. Nature 414:611-616. [DOI] [PubMed] [Google Scholar]
- 10.Bullock, S. L., D. Zicha, and D. Ish-Horowicz. 2003. The Drosophila hairy RNA localization signal modulates the kinetics of cytoplasmic mRNA transport. EMBO J. 22:2484-2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carson, J. H., K. Worboys, K. Ainger, and E. Barbarese. 1997. Translocation of myelin basic protein mRNA in oligodendrocytes requires microtubules and kinesin. Cell Motil. Cytoskelet. 38:318-328. [DOI] [PubMed] [Google Scholar]
- 12.Costa, M., and F. Michel. 1997. Rules for RNA recognition of GNRA tetraloops deduced by in vitro selection: comparison with in vivo evolution. EMBO J. 16:3289-3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis, I., and D. Ish-Horowicz. 1991. Apical localization of pair-rule transcripts requires 3′ sequences and limits protein diffusion in the Drosophila bastoderm embryo. Cell 67:927-940. [DOI] [PubMed] [Google Scholar]
- 14.Ephrussi, A., L. K. Dickinson, and R. Lehmann. 1991. oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell 66:37-50. [DOI] [PubMed] [Google Scholar]
- 15.Ferrandon, D., L. Elphick, C. Nüsslein-Volhard, and D. St Johnston. 1994. Staufen protein associates with the 3′UTR of bicoid mRNA to form particles that move in a microtubule-dependent manner. Cell 79:1221-1232. [DOI] [PubMed] [Google Scholar]
- 16.Ferrandon, D., I. Koch, E. Westhof, and C. Nüsslein-Volhard. 1997. RNA-RNA interaction is required for the formation of specific bicoid mRNA 3′ UTR-STAUFEN ribonucleoprotein particles. EMBO J. 16:1751-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gautreau, D., C. A. Cote, and K. L. Mowry. 1997. Two copies of a subelement from the Vg1 RNA localization sequence are sufficient to direct vegetal localization in Xenopus oocytes. Development 124:5013-5020. [DOI] [PubMed] [Google Scholar]
- 18.Hachet, O., and A. Ephrussi. 2004. Splicing of oskar RNA in the nucleus is coupled to its cytoplasmic localization. Nature 428:959-963. [DOI] [PubMed] [Google Scholar]
- 19.Jaeger, L., E. Westhof, and N. B. Leontis. 2001. TectoRNA: modular assembly units for the construction of RNA nano-objects. Nucleic Acids Res. 29:455-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Januschke, J., L. Gervais, S. Dass, J. A. Kaltschmidt, H. Lopez-Schier, D. St Johnston, A. H. Brand, S. Roth, and A. Guichet. 2002. Polar transport in the Drosophila oocyte requires dynein and kinesin I cooperation. Curr. Biol. 12:1971-1981. [DOI] [PubMed] [Google Scholar]
- 21.Karlin-McGinness, M., T. L. Serano, and R. S. Cohen. 1996. Comparative analysis of the kinetics and dynamics of K10, bicoid, and oskar mRNA localization in the Drosophila oocyte. Dev. Genet. 19:238-248. [DOI] [PubMed] [Google Scholar]
- 22.Kim-Ha, J., J. L. Smith, and P. M. Macdonald. 1991. oskar mRNA is localized to the posterior pole of the Drosophila oocyte. Cell 66:23-35. [DOI] [PubMed] [Google Scholar]
- 23.Kloc, M., N. R. Zearfoss, and L. D. Etkin. 2002. Mechanisms of subcellular mRNA localization. Cell 108:533-544. [DOI] [PubMed] [Google Scholar]
- 24.Lall, S., H. Francis-Lang, A. Flament, A. Norvell, T. Schupbach, and D. Ish-Horowicz. 1999. Squid hnRNP protein promotes apical cytoplasmic transport and localization of Drosophila pair-rule transcripts. Cell 98:171-180. [DOI] [PubMed] [Google Scholar]
- 25.Lantz, V., and P. Schedl. 1994. Multiple cis-acting targeting sequences are required for orb mRNA localization during Drosophila oogenesis. Mol. Cell. Biol. 14:2235-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lipshitz, H. D., and C. A. Smibert. 2000. Mechanisms of RNA localization and translational regulation. Curr. Opin. Genet. Dev. 10:476-488. [DOI] [PubMed] [Google Scholar]
- 27.Long, R. M., R. H. Singer, X. Meng, I. Gonzalez, K. Nasmyth, and R.-P. Jansen. 1997. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science 277:383-387. [DOI] [PubMed] [Google Scholar]
- 28.Macdonald, P. M. 1990. bicoid mRNA localization signal: phylogenetic conservation of function and RNA secondary structure. Development 110:161-171. [DOI] [PubMed] [Google Scholar]
- 29.Macdonald, P. M., and K. Kerr. 1998. Mutational analysis of an RNA recognition element that mediates localization of bicoid mRNA. Mol. Cell. Biol. 18:3788-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Macdonald, P. M., and K. Kerr. 1997. Redundant RNA recognition events in bicoid mRNA localization. RNA 3:1413-1420. [PMC free article] [PubMed] [Google Scholar]
- 31.Macdonald, P. M., K. Kerr, J. L. Smith, and A. Leask. 1993. RNA regulatory element BLE1 directs the early steps of bicoid mRNA localization. Development 118:1233-1243. [DOI] [PubMed] [Google Scholar]
- 32.Macdonald, P. M., and G. Struhl. 1988. cis-acting sequences responsible for anterior localization of bicoid mRNA in Drosophila embryos. Nature 336:595-598. [DOI] [PubMed] [Google Scholar]
- 33.Mach, J. M., and R. Lehmann. 1997. An Egalitarian-BicaudalD complex is essential for oocyte specification and axis determination in Drosophila. Genes Dev. 11:423-435. [DOI] [PubMed] [Google Scholar]
- 34.Mancebo, R., X. Zhou, W. Shillinglaw, W. Henzel, and P. M. Macdonald. 2001. BSF binds specifically to the bicoid mRNA 3′ untranslated region and contributes to stabilization of bicoid mRNA. Mol. Cell. Biol. 21:3462-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Navarro, C., H. Puthalakath, J. M. Adams, A. Strasser, and R. Lehmann. 2004. Egalitarian binds dynein light chain to establish oocyte polarity and maintain oocyte fate. Nat. Cell Biol. 6:427-435. [DOI] [PubMed] [Google Scholar]
- 36.Oleynikov, Y., and R. H. Singer. 2003. Real-time visualization of ZBP1 association with beta-actin mRNA during transcription and localization. Curr. Biol. 13:199-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palacios, I. M., and D. St. Johnston. 2001. Getting the message across: the intracellular localization of mRNAs in higher eukaryotes. Annu. Rev. Cell Dev. Biol. 17:569-614. [DOI] [PubMed] [Google Scholar]
- 38.Payre, F., M. Crozatier, and A. Vincent. 1994. Direct control of transcription of the Drosophila morphogen bicoid by the serendipity delta zinc finger protein, as revealed by in vivo analysis of a finger swap. Genes Dev. 8:2718-2728. [DOI] [PubMed] [Google Scholar]
- 39.Pokrywka, N. J., and E. C. Stephenson. 1991. Microtubules mediate the localization of bicoid mRNA during Drosophila oogenesis. Development 113:55-66. [DOI] [PubMed] [Google Scholar]
- 40.Serano, T. L., and R. S. Cohen. 1995. Gratuitous mRNA localization in the Drosophila oocyte. Development 121:3013-3021. [DOI] [PubMed] [Google Scholar]
- 41.Serano, T. L., and R. S. Cohen. 1995. A small predicted stem-loop structure mediates oocyte localization of Drosophila K10 mRNA. Development 121:3809-3818. [DOI] [PubMed] [Google Scholar]
- 42.Severt, W. L., T. U. Biber, X. Wu, N. B. Hecht, R. J. DeLorenzo, and E. R. Jakoi. 1999. The suppression of testis-brain RNA binding protein and kinesin heavy chain disrupts mRNA sorting in dendrites. J. Cell Sci. 112:3691-3702. [DOI] [PubMed] [Google Scholar]
- 43.Siemering, K. R., R. Golbik, R. Sever, and J. Haseloff. 1996. Mutations that suppress the thermosensitivity of green fluorescent protein. Curr. Biol. 6:1653-1663. [DOI] [PubMed] [Google Scholar]
- 44.Skripkin, E., J. C. Paillart, R. Marquet, B. Ehresmann, and C. Ehresmann. 1994. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc. Natl. Acad. Sci. USA 91:4945-4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.St Johnston, D. 1995. The intracellular localization of messenger RNAs. Cell 81:161-170. [DOI] [PubMed] [Google Scholar]
- 46.St Johnston, D., D. Beuchle, and C. Nüsslein-Volhard. 1991. staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell 66:51-63. [DOI] [PubMed] [Google Scholar]
- 47.St Johnston, D., W. Driever, T. Berleth, S. Richstein, and C. Nüsslein-Volhard. 1989. Multiple steps in the localization of bicoid RNA to the anterior pole of the Drosophila oocyte. Development 107(Suppl.):13-19. [DOI] [PubMed] [Google Scholar]
- 48.Swan, A., and B. Suter. 1996. Role of Bicaudal-D in patterning the Drosophila egg chamber in mid-oogenesis. Development 122:3577-3586. [DOI] [PubMed] [Google Scholar]
- 49.Takizawa, P. A., A. Sil, J. R. Swedlow, I. Herskowitz, and R. D. Vale. 1997. Actin-dependent localization of an RNA encoding a cell-fate determinant in yeast. Nature 389:90-93. [DOI] [PubMed] [Google Scholar]
- 50.Theurkauf, W. E., B. M. Alberts, Y. N. Jan, and T. A. Jongens. 1993. A central role for microtubules in the differentiation of Drosophila oocytes. Development 118:1169-1180. [DOI] [PubMed] [Google Scholar]
- 51.Wagner, C., C. Ehresmann, B. Ehresmann, and C. Brunel. 2004. Mechanism of dimerization of bicoid mRNA: initiation and stabilization. J. Biol. Chem. 279:4560-4569. [DOI] [PubMed] [Google Scholar]
- 52.Wagner, C., I. Palacios, L. Jaeger, D. St Johnston, B. Ehresmann, C. Ehresmann, and C. Brunel. 2001. Dimerization of the 3′UTR of bicoid mRNA involves a two-step mechanism. J. Mol. Biol. 313:511-524. [DOI] [PubMed] [Google Scholar]
- 53.Wilkie, G. S., and I. Davis. 2001. Drosophila wingless and pair-rule transcripts localize apically by dynein-mediated transport of RNA particles. Cell 105:209-219. [DOI] [PubMed] [Google Scholar]