Abstract
The process whereby the primitive vascular network develops into the mature vasculature, known as angiogenic vascular remodeling, is controlled by the Notch signaling pathway. Of the two mammalian Notch receptors expressed in vascular endothelium, Notch1 is broadly expressed in diverse cell types, whereas Notch4 is preferentially expressed in endothelial cells. As mechanisms that confer Notch4 expression were unknown, we investigated how NOTCH4 transcription is regulated in human endothelial cells and in transgenic mice. The NOTCH4 promoter and the 5′ portion of NOTCH4 assembled into an endothelial cell-specific histone modification pattern. Analysis of NOTCH4 primary transcripts in human umbilical vein endothelial cells by RNA fluorescence in situ hybridization revealed that 36% of the cells transcribed one or both NOTCH4 alleles. The NOTCH4 promoter was sufficient to confer endothelial cell-specific transcription in transfection assays, but intron 1 or upstream sequences were required for expression in the vasculature of transgenic mouse embryos. Cell-type-specific activator protein 1 (AP-1) complexes occupied NOTCH4 chromatin and conferred endothelial cell-specific transcription. Vascular angiogenic factors activated AP-1 and reprogrammed the endogenous NOTCH4 gene in HeLa cells from a repressed to a transcriptionally active state. These results reveal an AP-1-Notch4 pathway, which we propose to be crucial for transducing angiogenic signals and to be deregulated upon aberrant signal transduction in cancer.
The development and remodeling of blood vessels, which are termed vasculogenesis and angiogenesis, respectively, require integration of diverse cellular signals (79, 109). The Notch signaling pathway, in conjunction with secreted growth and differentiation factors, is critical for angiogenic vascular remodeling (29, 36), the process whereby the primitive vascular network is sculpted into mature vasculature. The binding of Notch ligands to transmembrane Notch receptors induces a proteolytic cascade that liberates the intracellular domain of Notch (NIC) from the plasma membrane (87). NIC translocates into the nucleus and forms a complex with the transcriptional repressor C promoter binding factor 1 (CBF1)/RBPJκ/suppressor of hairless/Lag-1 (CSL) (32, 37, 61, 84). Analogous to nuclear receptor-mediated coactivator-corepressor switches (107), NIC binding induces coactivator recruitment, precluding CSL interactions with corepressors (27, 33, 48, 100, 112, 113). Through its sequence-specific DNA binding function, CSL determines Notch target gene specificity. This canonical mechanism allows Notch to stringently control an ensemble of target genes and thus many developmental processes. NIC mutants defective in CSL binding retain certain activities, consistent with CSL-independent Notch signaling (69). However, such mutants can be tethered into CSL-containing complexes by the coregulator mastermind (38).
Loss-of-function and gain-of-function studies have provided evidence that two of the four mammalian Notch receptor subtypes expressed in vascular endothelium, Notch1 and Notch4, regulate vascular angiogenic remodeling (47, 96). Targeted deletion of the broadly expressed murine Notch1 gene yielded multiple developmental defects, including impaired vascular angiogenic remodeling (47, 91). Whereas targeted deletion of the Notch4 gene, which is expressed preferentially in endothelial cells (97), did not reveal overt phenotypes, the Notch1-Notch4 double knockout caused a more severe disruption of vascular angiogenic remodeling than the Notch1 single knockout (47). The genetic interaction of Notch4 with Notch1 provides strong evidence that physiological levels of Notch4 signaling control vascular development. Overexpression of NIC-1 and NIC-4 in transgenic mice and in cultured cells either inhibits (56, 60) or promotes (59, 92, 95) vascular morphogenesis and, in certain contexts, is oncogenic (3, 6, 10, 23, 76, 101).
Given the endothelial cell specificity of Notch4 expression (97), the Notch1-Notch4 genetic interaction (47), and the lack of nonvascular phenotypes in Notch4-null mice (47), Notch4 appears to have committed vascular functions. This attribute distinguishes Notch4 from other Notch subtypes, which are expressed more broadly (51, 102, 103). The vascular endothelium consists of functionally distinct endothelial cell subtypes (13), and Notch4 is not expressed in all endothelium. In situ hybridization analysis showed that Notch4 transcripts were highest in the vascular endothelium of embryonic day 9 (E9) and E13.5 mouse embryos and were detected in the pulmonary capillaries of adults (97). Another study reported that Notch4 transcripts were detected in arteries, but not veins, of the E13.5 embryo (99). Zebra fish notch3 is preferentially expressed in the dorsal aorta during embryogenesis and controls arterial-venous differentiation (52).
Besides endothelial cell-specific expression, Notch4 levels are dynamically regulated via cell signaling. Infection of human umbilical vein endothelial cells (HUVECs) with adenoviruses expressing vascular endothelial growth factor 121 (VEGF121) and fibroblast growth factor 2 (FGF-2) increased NOTCH4 mRNA levels (59). This effect was not specific for NOTCH4, as NOTCH1 mRNA was also induced. Treatment of rheumatoid arthritis synovial fibroblasts, but not normal synovial fibroblasts, with tumor necrosis factor induced NOTCH4 mRNA (1). Despite the selective expression and dynamic regulation of NOTCH4 in vascular endothelium, mechanisms underlying the endothelial cell specificity of NOTCH4 transcription have not been defined. Mechanisms controlling transcription of other Notch subtypes are also largely unknown.
Cell-type-specific transcription patterns can be achieved via utilization of a limited number of cell-type-specific transcription factors or through combinatorial interactions between multiple factors. Nearly all erythroid cell-specific genes are activated by the erythroid cell-specific factor GATA-1 (22, 93). While an endothelial cell-specific factor equivalent to GATA-1 that regulates most endothelial cell-specific genes has not been identified, GATA factors (41, 63, 65, 108), Ets factors (64), NF-κB (63), Egr1 (5), Vezf1 (105), HoxB5 (104), Sp1/Sp3 (11, 75), RTEF-1 (89), NFAT (111), and AP-1 (5, 41, 108) interact with and regulate promoters of endothelial cell-specific genes in transfection assays. No consensus has arisen regarding a single factor that might universally confer endothelial cell-specific transcription. Furthermore, no comprehensive studies of the native nucleoprotein structure (chromatin structure and bound trans-acting factors) of any endothelial cell-specific gene within its endogenous chromatin domain have been conducted.
The native nucleoprotein structures of endogenous loci reveal mechanistic insights that cannot be predicted from in vitro analysis (9, 39). Chromatin immunoprecipitation (ChIP) analysis has shown that the most abundant interactor in vitro is often not the actual endogenous interactor. Whereas c-myc occupies E-boxes in cells, the related factor USF (upstream stimulatory factor) is the predominant binder in vitro (8). GATA factors recognize simple (A/T)GATA(A/G) motifs distributed abundantly within chromosomal DNA (46, 62). However, ChIP analysis has revealed that the majority of such motifs are inaccessible to GATA factors in cells (28, 40, 73).
We used ChIP and functional assays to dissect the mechanism controlling NOTCH4 transcription in vascular endothelium. This analysis revealed an endothlelial cell-specific histone modification pattern localized in a highly restricted manner at the NOTCH4 promoter and the 5′ portion of NOTCH4. Analysis of trans-acting factors that confer NOTCH4 transcription in endothelial cells revealed an important role of cell-type-specific, signal-dependent AP-1 complexes. These results are discussed vis-à-vis a model of how signaling networks established by angiogenic factors target AP-1 to control NOTCH4 transcription and how disruption of the AP-1-Notch4 axis by aberrant signaling in cancer might facilitate tumor progression.
MATERIALS AND METHODS
Cell culture.
Primary HUVECs (Cascade Biologics) were maintained in Medium 200 (Cascade Biologics) containing 1% penicillin-streptomycin (Gibco/BRL) and Low Serum Growth Supplement (Cascade Biologics). HeLa cells, which were derived from an ovarian carcinoma and have epithelial properties, were maintained in Dulbecco's modified Eagle medium (DMEM) (Biofluids) containing 1% antibiotic-antimycotic (Gibco/BRL) and 10% fetal bovine serum (FBS).
Plasmids.
The bacterial artificial chromosome containing human NOTCH4 genomic DNA was generously provided by Monica Dors, Institute for Systems Biology. The pGL3basic and pGL3promoter (pGL3pro) reporter plasmids were obtained from Promega. Other luciferase reporter constructs for transient transfection assay were generated by the following methods. For pGL3 N4-pro, the human NOTCH4 promoter was amplified by PCR using the primer pair (5′ to 3′) TGACTCTCGAGACCAAGATTTCCCCAAAACC and CAGTCAAGCTTCAGGCAGGGACCCTC. The PCR product was digested with XhoI and HindIII and then inserted into the pGL3basic vector. For pGL3 N4-proIN1, intron-1 of NOTCH4 was amplified by PCR using the primer pair (5′ to 3′) AGTTAGGATCCTCAGTGGTCAGACCCAGAGG and TAGGTGTCGACATTGGCACAGGGTTCTGG. The PCR product was digested with SalI and BamHI and then inserted into pGL3 N4-pro construct. For pGL3 N4-CRpro, the NOTCH4 upstream conserved region was amplified by PCR using the primer pair (5′ to 3′) CTGATGCTAGCTACAGTGGCCTATTGCC and TCAGTCTCGAGCATGTTTAGGTGGGTCTC. The PCR product was digested with NheI and XhoI and then inserted into the pGL3 N4-pro construct. For pGL3 N4-UPpro, the NOTCH4 upstream region was amplified by PCR using primer pair (5′ to 3′) CTGATGCTAGCTACAGTGGCCTATTGCC and CAGTCAAGCTTCAGGCAGGGACCCTC. The PCR product was digested with NheI and HindIII and then inserted into the pGL3basic vector. A two-step PCR strategy was used to generate the AP-1 binding motif-mutated construct pGL3 N4-pro(mtAP-1) using primers (5′ to 3′) TGACTCTCGAGACCAAGATTTCCCCAAAACC, TTGTGGCTAGACGGAAACAGCTCAGACGTG, CACGTCTGAGCTGTTTCCGTCTAGCCACAA, and CAGTCAAGCTTCAGGCAGGGACCCTC. The PCR product was digested with XhoI and HindIII and then inserted into the pGL3basic vector.
The p4xAP-1 reporter and the AP-1 reporter plasmid pGL2AP-1, containing a collagenase promoter fragment (−73 to +67) with a single AP-1 binding motif in the luciferase reporter vector pGL2basic, were generously provided by Nancy Colburn, National Cancer Institute, National Institutes of Health. The expression vector encoding dominant-negative AP-1, A-Fos (71), was generously provided by Charles Vinson, National Cancer Institute, National Institutes of Health.
The β-galactosidase reporter constructs for transient transgenic analysis were generated by the following methods. For pSVβ N4-pro, the NOTCH4 promoter was amplified by PCR using the primer pair (5′ to 3′) TGACTGAATTCAAGACCAAGATTTCCCCAAAACC and CTGACCTCGAGGCAGGGACCCTCAGAGCT. The PCR product was digested with EcoRI and XhoI and then inserted into the pSVβ vector (Clontech), which lacks a promoter (72). The pSVβ vector contains the full-length β-galactosidase gene with a poly(A) site in a 6.9-kb plasmid. For pSVβ N4-UPpro, the NOTCH4 upstream region was amplified by PCR using the primer pair (5′ to 3′) CTGATGCTAGCTACAGTGGCCTATTGCC and CTGACCTCGAGGCAGGGACCCTCAGAGCT. The PCR product was digested with NheI and XhoI and then inserted into the pSVβ vector. For pSVβ N4-proIN1, intron 1 of NOTCH4 was amplified by PCR using primer pair (5′ to 3′) AGTTAGTCGACTCAGTGGTCAGACCCAGAGG and TTGCTAAGCTTGGCACAGGGTTCTGGG. The PCR product was digested with SalI and HindIII and then inserted into pSVβ N4-pro. For pSVβ N4-pro(mAP-1)IN1, the AP-1 binding motif-mutated NOTCH4 promoter was amplified by PCR from pGL3 N4-pro(mtAP-1) using the primer pair (5′ to 3′) TGACTGAATTCAAGACCAAGATTTCCCCAAAACC and CTGACCTCGAGGCAGGGACCCTCAGAGCT. The PCR product was digested with EcoRI and XhoI and then substituted within the promoter region of pSVβ N4-proIN1. The Tie-2-LacZ construct (Tie-2-LacZ) containing the Tie-2 enhancer and promoter was generously provided by Tom Sato, University of Texas Southwestern Medical Center.
Antibodies.
The antibodies used in ChIP analysis were as follows. Rabbit anti-di-acetylated histone H3 (anti-di-acH3) (06-599), anti-tetra-acH4 (06-866), and anti-histone H3 methylation at lysine 4 (anti-H3-meK4) (07-030) antibodies were obtained from Upstate Biotechnology; rabbit anti-polymerase II (Pol II) (N-20) was obtained from Santa Cruz Biotechnology; rabbit immunoglobulin G (IgG) (Sigma) and preimmune serum (Covance) were used as controls. The AP-1 antibodies rabbit anti-c-Fos (K-25), anti-c-Fos (H-125), anti-FosB (102), anti-Fra-1 (R-20), anti-Fra-2 (L-15), anti-ATF2 (N-96), anti-c-Jun (D), anti-c-Jun (H-79), anti-c-Jun (N), anti-JunB (N-17), and anti-JunD (329) were obtained from Santa Cruz Biotechnology. Mouse antitubulin (Ab-1) antibody was obtained from Oncogene Research Products. The secondary antibodies goat-anti-rabbit IgG-horseradish peroxidase (HRP) and goat-anti-mouse IgG-HRP were obtained from Santa Cruz Biotechnology.
Quantitative real-time RT-PCR.
Total RNA was purified from cell cultures with Trizol (Invitrogen). cDNA was synthesized by annealing RNA (2 μg) with 50 ng of random hexamer and 200 ng of oligo(dT) primer (Promega) at 68°C for 10 min. After denaturation, the samples were incubated with Moloney murine leukemia virus reverse transcriptase (10 U/μl; Invitrogen) combined with 20 mM dithiothreitol, 1 mM deoxynucleoside triphosphates, and RNasin (2 U/μl; Promega) at 42°C for 1 h. The reaction mixture was heat inactivated at 95 to 100°C for 5 min and diluted to a final volume of 200 μl. The samples were analyzed by quantitative real-time reverse transcription-PCR (RT-PCR) (ABI Prism 7000) with primers designed with Primer Express 1.0 software (PE Applied Biosystems) to amplify regions of 50 to 150 bp. Quantitative real time RT-PCR mixtures (25 μl) contained 2 μl of cDNA, 12.5 μl of SYBR Green (Applied Biosystems), and the indicated primers. Product accumulation was monitored by the levels of SYBR Green fluorescence. Relative expression levels were determined from a standard curve of serial dilutions of cDNA samples. Analysis of product denaturation curves postamplification showed that primer pairs generated single products. Forward and reverse primers for real-time RT-PCR (5′-3′) were as follows: NOTCH4, GAGGACAGCATTGGTCTCAAGG and CAACTCCATCCTCATCAACTTCTG; NOTCH4-2, CAGCCCAAGCAGATATGTAAGGA and CGTCCAACCCACGTCACA; NOTCH1, CTGCATGCGGCTGTGTCT and CTCGGTTCCGGATCAGGAT; TSBP, AAAGTGCTAGACTTCTGGACTATGAGG and TGATATGCATGTCGGGATCCT; G18, CTTTACAGCACTATCCTCAGTCACCA and GCTCTGACCGCTGGGCT; HPRT, ATTGGTGGAGATGATCTCTCAACTTT and GCCAGTGTCAATTATATCTTCCACAA; eNOS, AATCAACGTGGCCGTGCT and ACGATGGTGACTTTGGCTAGCT; vWF, CCTCAAAGGCGGTGGTCAT and CCAATAGGGAACACTGTCACTCTG; and Flk-1, GAGGAGAAGTCCCTCAGTGATGTAG and CCTTATACAGATCTTCAGGAGCTTCC.
Quantitative ChIP analysis.
HUVECs or HeLa cells were incubated in a culture plate with medium containing 0.4% (for analysis of histone modifications) or 1% (for analysis of AP-1 occupancy) formaldehyde for 10 min at room temperature. The cross-linking reaction was terminated by incubation with 0.125 M glycine for 5 min. Cells were harvested by scraping, collected by centrifugation at 400 × g for 8 min, and washed in phosphate-buffered saline (PBS). Real-time PCR-based quantitative ChIP analysis was preformed as previously described (35). Nuclei were isolated by a 10-min incubation in cell lysis buffer (10 mM Tris, 10 mM NaCl, 0.2% NP-40 [pH 8.0]) on ice, followed by centrifugation at 500 × g for 5 min. Nuclei were lysed in nucleus lysis buffer (50 mM Tris, 10 mM EDTA, 1% sodium dodecyl sulfate [SDS] [pH 8.0]) for 10 min on ice. The lysate was diluted with IP dilution buffer (20 mM Tris, 150 mM NaCl, 2 mM EDTA, 0.01% SDS, 1% Triton X-100 [pH 8.0]) and sonicated with eight 30-s pulses at 50 to 60% of maximum power with a HeatWave Systems W185F sonicator (Ultrasonics, Inc., Plainview, N.Y.) equipped with a microtip. Sonicated chromatin fragments were an average size of ∼300 to 400 bp. Soluble chromatin was precleared by the addition of 50 μl of preimmune serum, followed by 100 μl of protein A-Sepharose. Precleared chromatin (180 μl) was removed (input) and used in the subsequent PCR analysis. The remainder of the chromatin was aliquoted and incubated with the indicated antibodies in a final volume of 900 μl for 3 h at 4°C. Immune complexes were collected by incubation with 30 μl of protein A-Sepharose for 2 h at 4°C. Protein A-Sepharose pellets were washed twice with 500-μl aliquots of IP wash buffer 1 (20 mM Tris, 50 mM NaCl, 2 mM EDTA, 0.1% SDS, 1% Triton X-100 [pH 8.0]), once with IP wash buffer 2 (10 mM Tris, 0.25 mM LiCl, 1 mM EDTA, 1% NP-40, 1% desoxycholate [pH 8.0]), and twice with TE (10 mM Tris, 1 mM EDTA [pH 8.0]). Immune complexes were eluted twice with 150 μl of IP elution buffer (0.1 mM NaHCO3, 1% SDS). RNase A (0.03 mg/ml) and NaCl (0.3 m/liter) were added, and cross-links were reversed by incubation for 4 to 5 h at 65°C. DNA was then digested with proteinase K (0.24 mg/ml) for at least 2 h at 45°C and purified by two extractions with phenol-chloroform, followed by ethanol precipitation. Purified DNA was resuspended in 30 μl of water. Aliquots (1 μl) were analyzed by quantitative real-time PCR with the indicated primer pairs. The amounts of products were determined relative to a standard curve generated from a titration of input chromatin. Measurements were made under conditions in which signals were in the linear range, and analysis of denaturation curves postamplification showed that primer pairs generated single products (data not shown). Forward and reverse primers for real-time PCR (5′ to 3′) ChIP analysis were as follows: N4-1, CCTCTCTGTCTCCGTTGCAAT and GGCACTAGGCTGGAGCATGA; N4-2, ACCATTATAAATGATGCTGGCTCAC and AGTACCAGTGTATCACATTTGGAAGC; N4-3, GTGTTTGTTACAGACAATTCAGACTGC and TGAACATAGTCTACCCTAAATTTTGCA; N4-pro1, CAGCCACCTTGCAATTCTCA and CAGCCCTGCTGTTTGTTGATC; N4-pro2, GGACATTGTGTGACTCAGGAAACA and CCTCGGCCTGCTGCAA; N4-in1, CTGTTGTCTTGCTTCCGAGAGAT and TTATTCTCTGGCCTCCCAAGTC; N4-ex3, TGGCTTCACTGGCGAGAGAT and GGCCCCTTTTGGAACAGAA; N4-in3, GGGTCCTCCAGACTTTTGCAT and ATGGCTCCCTCCACTCAGAAT; N4-in8, CAGACTCCTCAGGCAAGAAAAGA and TGGGATCAACCTCTGGACCTT; N4-ex11, GAAGGGCCACGCTGTCAA and CAACGGGACATGGGTCACTC; N4-ex18, GGATTCCAAGGCAGCCTGT and CCTGGACTCACATGGGTTCAC; N4-in20, TCTCTGTTGCCCCCTATGCT and CCCTTCTGGGATTCCAACTGA; N4-ex30, CCTGCGATAATGCGAGGAA and AATCACAGGGCCAGTCATCC; cyclophilin pro, GTCTATAGGCCAGATGCACTGTCA and CCAATCGGGTCTGCGACTT; β-globin pro, AGTGCCAGAAGAGCCAAGGA and CAGGGTGAGGTCTAAGTGATGACA; IL-8 pro, GGGATGGGCCATCAGTTG and CCTCATCTTTTCATTATGTCAGAGGAA; and N4 pro(AP-1), CCCCCATTACTAGGGTGTCCA and TGCTGCAAGCCTCACGTC.
Transient transfection assay.
HUVECs or HeLa cells were plated 1 day before transfection and were ∼60 to 70% confluent at the time of transfection. The indicated amounts of plasmid DNA were added to 100 μl of Opti-MEM (Gibco/BRL), incubated with Lipofectin reagent (6 μl/1 μg of DNA; Invitrogen) for 15 min at room temperature, and then added to the cells. The cells were incubated with the transfection mixture for 5 h before the addition of normal medium. The cells were harvested 48 h posttransfection. Cell lysates were assayed for luciferase activity by a luciferase assay system (Promega). The luciferase activity was normalized by the protein content of the lysates, as determined by a Bradford assay (Bio-Rad) with γ-globulin as a standard.
Nuclear extract preparation.
Nuclear extracts were prepared as described previously (49). HUVECs or HeLa cells were harvested by scraping and collected by centrifugation at 400 × g for 8 min. Cells were washed once with ice-cold PBS and resuspended in 1.5 volumes of nucleus lysis buffer (10 mM Tris-HCl [pH 7.5], 10 mM NaCl, 3 mM MgCl2, and 0.2% NP-40) on ice for 3 min. Nuclei were collected by centrifugation for 5 min at 400 × g. Nuclei were washed by gentle resuspension in 1.5 volumes of nucleus wash buffer (10 mM Tris-HCl [pH 7.5], 10 mM NaCl, and 3 mM MgCl2) and then collected by centrifugation for 4 min at 400 × g. Nuclei were immediately resuspended in an equal volume of low-KCl extract buffer (20 mM HEPES [pH 7.5], 20 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA, and 25% glycerol), and 1.33 volumes of the same buffer containing 1.2 M KCl was added dropwise. Nuclei were extracted for 45 min at 4°C with constant mixing. The suspension was then centrifuged for 30 min at 150,000 × g. Aliquots of the supernatant were frozen on dry ice and stored at −80°C. The protein concentration was measured by the Bradford assay with γ-globulin as a standard. Dithiothreitol (5 mM), phenylmethylsulfonyl fluoride (0.5 mM), and leupeptin (20 μg/ml) were included in all buffers.
Electrophoretic mobility shift assay.
Electrophoretic mobility shift assays (EMSAs) were conducted as described previously (50). Aliquots of HUVEC or HeLa nuclear extracts (3 μg) were incubated in 50 mM HEPES (pH 7.8), 300 mM KCl, 50% glycerol, 5 mM MgC12, 5 mM dithiothreitol, and 1 μg of poly(dI-dC) with or without 4 nmol of unlabeled double-stranded oligonucleotide or 4 μl of antibody in a final volume of 16 μl for 15 min at 4°C. End-labeled, double-stranded oligonucleotide (40 fmol) was added, and reaction mixtures (20 μl) were incubated for 20 min at 25°C. Samples were resolved on 6.5% nondenaturing polyacrylamide gels in 0.75× Tris-acetate-EDTA running buffer (30 mM Tris-acetate, 0.75 mM EDTA [pH 8.0]) at 180 V for 3 h at 4°C. Gels were preelectrophoresed for at least 10 min at 4°C. DNA binding activity was detected by analyzing dried gels with a PhosphorImager (Molecular Dynamics).
Western blotting.
To detect the expression levels of AP-1 subunits, nuclear extracts were prepared from HUVECs or HeLa cells under the indicated conditions. Total protein (20 μg) of nuclear extract was resolved by SDS-polyacrylamide gel electrophoresis on a 10% acrylamide gel. The proteins were transferred to an Immobilon P membrane (Millipore) and detected by immunoblotting with the indicated antibody. Proteins were visualized with ECL-Plus (Amersham).
RNA fluorescence in situ hybridization (FISH) analysis.
RNA FISH was performed as described previously (28a). Endogenous NOTCH4 transcription was detected with an antisense, digoxigenin-labeled, single-stranded DNA probe visualized with fluorescein isothiocyanate-labeled antibodies. The 1,420-bp probe fragment, spanning NOTCH4 introns 15 to 17, was generated by PCR with the primer pair CAGACAGGTGAGCAGGGCCCAAAGA and ATTCCTGGGTGGAGACTGGTCTGGG.
Analysis of lacZ fusion constructs in transgenic mice.
DNA constructs for F0 transgenic analysis were linearized, purified with the QIAEX II gel purification kit (QIAGEN), and microinjected into fertilized mouse oocytes. For whole-mount analysis, 5-bromo-4-chloro-3-indolyl β-galactoside (X-Gal) staining was performed with E10.5 embryos as described previously (72). Embryos were fixed with 1% formaldehyde, 0.2% glutaraldehyde, and 0.06% Igepal CA-630 (Sigma) in PBS for 40 min at 4°C. Samples were washed twice with PBS and then incubated for 4 h at 37°C in 2 mM MgCl2, 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, and X-Gal (1 mg/ml) in PBS. After X-Gal staining, embryos were washed twice with PBS and refixed with 10% formaldehyde overnight at 4°C. For tissue sections, the refixed samples were dehydrated with 40 and 80% methanol in PBS and then in 100% methanol. Samples were embedded into paraffin and were dried for 3 days at 4°C before being sectioned. The sectioned samples (each, 4 μm) were counterstained with Kernechtrot staining solution. Genomic DNA was purified from the embryos, and integration of the transgenes was verified by PCR. The following primers were used for the analysis: β-gal-5′ (5′-ACCGACTACACAAATCAGCG-3′) and β-gal-3′ (5′-CAACCACCGCACGATAGAGA-3′).
RESULTS AND DISCUSSION
Endothelial cell-specific histone modification pattern of the endogenous NOTCH4 locus.
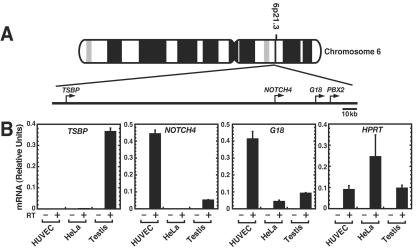
To investigate mechanisms underlying the endothelial cell specificity of NOTCH4 transcription, we compared the expression of NOTCH4 and genes flanking NOTCH4 on chromosome 6 (Fig. 1A) in HUVECs and nonendothelial HeLa cells. NOTCH4 transcript levels were high in HUVECs, undetectable in HeLa cells, and low in testis (Fig. 1B). The low-level expression likely originates from the endothelium of the vascularized testis. The TSBP gene, a testis-specific gene of unknown function (106), resides approximately 100 kb to the 5′ side of NOTCH4. TSBP transcripts were not detected in HUVECs or HeLa cells (Fig. 1B). Immediately downstream of NOTCH4 resides G18, which is also of unknown function (42). G18 transcript levels were high in HUVECs and lower in HeLa cells and in testis (Fig. 1B). As expected, the broadly expressed HPRT gene was expressed in HUVECs, HeLa cells, and testis.
FIG. 1.
Distinct cell-type-specific transcription patterns of NOTCH4 and adjacent genes. (A) Human NOTCH4 locus on chromosome 6. The nearby genes, testis-specific basic protein gene (TSBP), G18, and the pre-B-cell leukemia transcription factor-2 gene (PBX2) are also shown. (B) Quantitative real-time RT-PCR analysis of NOTCH4, TSBP, and G18 mRNA expression in HUVECs, HeLa cells, and testis. HPRT mRNA transcripts were measured as a control. Relative expression levels were normalized by total RNA (mean ± standard error of the mean [SEM]; three independent experiments).
Given the distinct expression patterns of NOTCH4 and its flanking genes, we reasoned that NOTCH4 might assemble a histone modification pattern that segregates regulatory elements associated with these genes. The rationale for this assumption is illustrated by analysis of the chicken β-globin locus. A 15-kb hypoacetylated region lies between the chicken β-globin locus and an upstream folate receptor gene, which has a distinct expression pattern (77). A chromatin insulator, characterized by a sharp peak of histone acetylation, resides at the junction of the 5′ end of the β-globin locus and the 3′-end of the hypoacetylated region (17, 58, 77). Additional examples of broad histone modification patterns that demarcate transcriptional regulatory regions include the high enrichments in histone acetylation and H3-meK4 present at upstream locus control regions of the β-globin (25, 45, 85) and growth hormone (21, 31) loci.
Quantitative ChIP analysis was used to determine if endogenous NOTCH4 in HUVECs assembles a broad pattern of acH3, acH4, and H3-meK4 that is distinct from that of HeLa cells. Histone acetylation increases factor access to chromatin (53, 98) and counteracts higher-order chromatin folding (94), which masks cis elements. Only a threefold increase in acetylation is required to unfold the higher-order structure of a reconstituted chromatin template (94). H3-meK4 has a distribution similar to that of acH3 and acH4 (45, 58, 70), although establishment and maintenance of these modifications can be differentially regulated (45). These studies utilized chromatin fragments averaging 300 to 400 bp, and real-time PCR was used to quantitate the relative levels of immunoprecipated DNA fragments under conditions of linearity (data not shown).
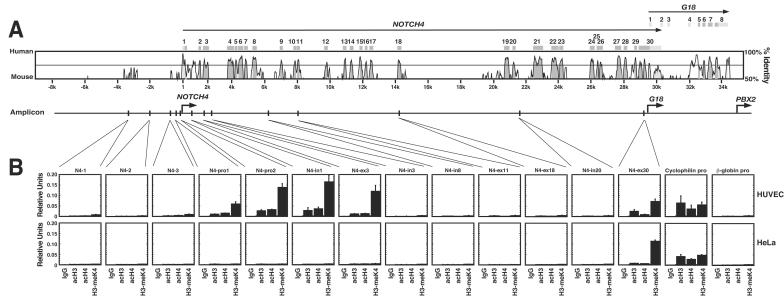
The chromosomal region upstream of the NOTCH4 promoter extending to TSBP is saturated with repetitive DNA elements and has only one major region of sequence conservation approximately 4 kb upstream of the NOTCH4 promoter (Fig. 2A). Little or no enrichment in acH3, acH4, and H3-meK4 was detected at this region (N4-1) or at two nonconserved regions at the 3′ side of the conserved region (N4-2 and -3) in either HUVECs or HeLa cells (Fig. 2B). By contrast, amplicons at the promoter (N4-pro1 and N4-pro2), intron 1 (N4-in1), and exon 3 (N4-ex3) revealed enrichments of acH3, acH4, and H3-meK4 in HUVECs but not in HeLa cells. Analysis of amplicons at intron 3, intron 8, exon 11, exon 18, and intron 20 (N4-in3, N4-in8, N4-ex11, N4-ex18, and N4-in20, respectively) revealed little or no enrichment of these histone modifications in HUVECs or HeLa cells. Thus, acH3, acH4, and H3-meK4 were not enriched throughout the open reading frame of NOTCH4. Recently, a genomics analysis indicated that multiple human genes lack high enrichments of these modifications downstream of the transcription start site (57). AcH3, acH4, and H3-meK4 were enriched at exon 30 in both HUVECs and HeLa cells. As exon 30 and the G18 promoter overlap, the enrichments at N4-ex30 likely reflect the transcriptionally active state of the G18 promoter. Histone modifications at the broadly active cyclophilin promoter and the erythroid cell-specific β-globin promoter were analyzed as positive and negative controls, respectively. The enrichments of acH3, acH4, and H3-meK4 at the cyclophilin promoter were comparable to enrichments at the NOTCH4 promoter and at the 5′ portion of the NOTCH4 open reading frame in HUVECs. Unlike the absolute endothelial cell-specific histone modification pattern of NOTCH4, the enrichments of histone modifications at the cyclophilin promoter were comparable in HUVECs and HeLa cells. No enrichments were detected at the β-globin promoter, consistent with the erythroid cell-specific histone modification pattern and expression of β-globin (25).
FIG. 2.
Endothelial cell-specific histone modification pattern of the endogenous NOTCH4 locus. (A) VISTA plot comparison of the human and murine NOTCH4 sequences. The amplicons analyzed by ChIP analysis are indicated at the bottom. (B) ChIP analysis of acH3, acH4, and H3-meK4 at the NOTCH4 locus in HUVECs and HeLa cells. Histone modifications were analyzed at the broadly expressed cyclophilin and erythroid cell-specific β-globin promoters as controls (mean ± SEM; at least three independent experiments). pro, promoter; in, intron; ex, exon.
NOTCH4 histone modification pattern delineates functional sequences.
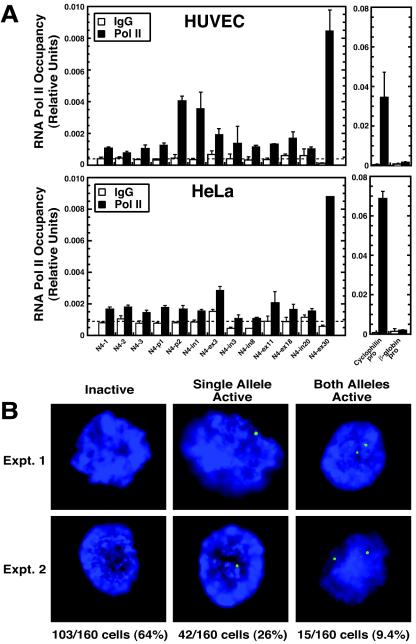
Based on the nearly undetectable acH3 and acH4 at the NOTCH4 locus in HeLa cells and the role of acetylation in increasing chromatin accessibility, it is unlikely that RNA Pol II would be able to access the NOTCH4 promoter in these cells, although there are reports of basal transcription complex and factor recognition of heterochromatin (12, 86). Quantitative ChIP was used to test whether Pol II occupies the locus in a cell-type-specific manner. The greatest enrichments in Pol II occupancy were detected at the promoter amplicon (N4-pro2) closest to the transcription start site and the exon-30 amplicon (N4-ex30), which overlaps the G18 promoter (Fig. 3A). Little or no enrichment was detected upstream of N4-pro2 or throughout the open reading frame. Little or no enrichment was detected in HeLa cells, with the exception of the N4-ex30 amplicon, in which the level of Pol II occupancy was comparable to the same region in HUVECs (Fig. 3A). Thus, Pol II occupancy of the NOTCH4 locus was specific to that of HUVECs and was greatest at the NOTCH4 and G18 promoters, in which the enrichments of acH3, acH4, and H3-meK4 were also high (Fig. 2B). Pol II occupancy was almost 10-fold higher at the broadly expressed cyclophilin promoter versus the NOTCH4 promoter, and no enrichments were detected at the β-globin promoter.
FIG. 3.
RNA Pol II selectively occupies the NOTCH4 locus in endothelial cells, but only a small percentage of the NOTCH4 alleles are transcriptionally active. (A) ChIP analysis of Pol II occupancy at the NOTCH4 locus in HUVECs (top) and HeLa cells (bottom). Pol II binding was analyzed at the broadly expressed cyclophilin and erythroid cell-specific β-globin promoters as controls (mean ± SEM; at least three independent experiments). N4-p1 and N4-p2, N4-pro1 and N4-pro2. (B) In situ RNA FISH analysis of endogenous NOTCH4 primary transcript levels in HUVECs. NOTCH4 transcription was detected by RNA FISH with an antisense, digoxigenin-labeled, single-stranded DNA probe and visualized with fluorescein isothiocyanate-labeled antibodies. Representative pictures (two independent experiments) of a transcriptionally inactive cell and cells expressing either one allele or both alleles of NOTCH4 are shown. The numbers and percentages of transcriptionally inactive cells and cells expressing either one or two alleles of NOTCH4 are shown at the bottom.
Although Pol II occupancy was clearly detected at the NOTCH4 promoter, the enrichment levels were relatively low, which might reflect a low transcription rate or a small percentage of cells expressing NOTCH4. To distinguish between these mechanisms, RNA FISH was used to measure primary NOTCH4 transcripts in single HUVECs. Transcription signals were detected at 23% (72 of 320) of NOTCH4 alleles (Fig. 3B), and 36% of the cells in the population transcribed one (42 of 160 cells) or both (15 of 160 cells) alleles. Thus, NOTCH4 is only transcribed in a small fraction of the HUVECs at any given time. These results are consistent with a mechanism in which NOTCH4 undergoes transcriptional oscillations, which has been shown to be common for mammalian genes in other contexts (45a, 72a). RNA FISH analysis of primary transcripts from the broadly expressed Rps18 gene approximately 30 Mb from NOTCH4, which encodes a riboprotein, revealed that 49% of Rps18 alleles were active in HUVECs (data not shown). Double-labeling analysis of both NOTCH4 and Rps18 expression revealed that 76% of HUVECs had active NOTCH4 and/or Rps18 alleles (data not shown), indicating that the RNA FISH assay was capable of detecting expression in at least 76% of the cells in the population.
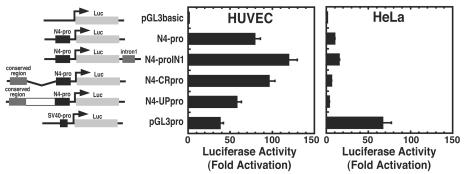
Strong enrichments of acH3, acH4, and H3-meK4 distal to a promoter are often hallmarks of transcriptional elements such as enhancers, locus control regions, and insulators (9). The lack of such epigenetic marks at the conserved region upstream of the NOTCH4 promoter (Fig. 2) suggested that the transcriptional determinants of NOTCH4 reside within the promoter and potentially at sites in the 5′ portion of the NOTCH4 open reading frame. To assess whether the promoter and/or other regions contain determinants of endothelial cell specificity, transient transfections were conducted with NOTCH4 promoter-luciferase reporter constructs in HUVECs and HeLa cells. The NOTCH4 promoter reporter (N4-pro) was 80-fold more active than the pGL3basic reporter in HUVECs, whereas it was only 8-fold more active than pGL3basic in HeLa cells (Fig. 4). Moreover, the NOTCH4 promoter was twice as active as the simian virus 40 (SV40) promoter (pGL3pro) in HUVECs, whereas it was ∼4-fold less active than the SV40 promoter in HeLa cells. Since intron 1 is highly conserved and resides within the restricted region of enriched histone modifications, we tested whether intron 1 regulates promoter activity. A ∼800-bp fragment of intron 1 cloned downstream of luciferase (N4-proIN1) induced a small increase in reporter activity in HUVECs and HeLa cells.
FIG. 4.
Endothelial cell-specific NOTCH4 promoter activity. HUVECs and HeLa cells were transiently transfected with the following luciferase reporter constructs: pGL3basic that lacks a promoter, pGL3basic containing the NOTCH4 promoter (N4-pro), pGL3basic containing the NOTCH4 promoter and intron 1 (N4-proIN1), pGL3basic containing the NOTCH4 promoter and an upstream conserved region (N4-CRpro), pGL3basic containing the entire upstream region including the conserved region and the NOTCH4 promoter (N4-UPpro), and pGL3basic containing the SV40 promoter (pGL3pro). Luciferase activity was normalized by the protein content of the lysates (mean ± SEM; five independent experiments). Relative luciferase activities are shown as fold activation, with pGL3basic activity designated as 1.0.
Although the region ∼4 kb upstream of the promoter was not enriched in acH3, acH4, or H3-meK4, the high conservation of this region suggested that it might be functionally important. Additional constructs containing the NOTCH4 promoter with ∼4 kb of upstream sequence (N4-UPpro) or the NOTCH4 promoter with a ∼1-kb fragment spanning the kb −4 conserved region (N4-CRpro) were tested. These constructs had activities that differed only slightly from the NOTCH4 promoter alone in HUVECs and HeLa cells (Fig. 4). Thus, a ∼650-bp promoter fragment was sufficient to confer cell-type-specific transcriptional activity in HUVECs and HeLa cells in a transient transfection assay. Furthermore, intron 1 and the upstream conserved region lack enhancer activity, as defined by the ability to strongly activate a reporter gene in a transient transfection assay.
Cell-type-specific AP-1 complexes occupy the NOTCH4 promoter and confer high-level transcription in endothelial cells.
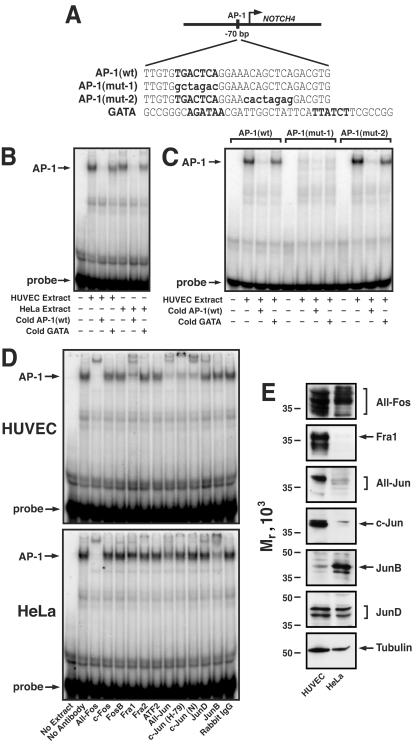
Analysis of conserved sequence motifs within the NOTCH4 promoter revealed several prospective cis elements, including an AP-1 motif. EMSA was performed with overlapping oligonucleotides spanning ∼300 bp of NOTCH4 promoter sequence to test whether these sequences mediate assembly of unique protein-DNA complexes with HUVEC versus HeLa nuclear extracts. Whereas no complexes were uniquely formed with HUVEC extracts (data not shown), the HUVEC complex that assembled on the AP-1(wt) oligonucleotide (Fig. 5A) had a slightly faster mobility in the gel than the corresponding HeLa complex (Fig. 5B). The complexes were abrogated by inclusion of a 100-fold excess of unlabeled AP-1(wt) probe, whereas an unrelated oligonucleotide containing two GATA motifs (GATA) had no effect.
FIG. 5.
Assembly of AP-1 complexes on a conserved AP-1 motif of the NOTCH4 promoter in vitro. (A) Sequences of oligonucleotides used in the EMSA: AP-1(wt), an oligonucleotide consisting of a NOTCH4 promoter sequence with a conserved AP-1 motif; AP-1(mut-1), an oligonucleotide with a mutated AP-1 motif; AP-1(mut-2), an oligonucleotide with a sequence downstream of the AP-1 motif mutated; GATA, an oligonucleotide with two GATA binding motifs. The conserved AP-1 motif and the GATA motifs are indicated in uppercase boldface type. The specific bases mutated are indicated in lowercase boldface type. The GATA oligonucleotide was used as a nonspecific competitor. (B) EMSA analysis of nucleoprotein complex assembly on the AP-1(wt) oligonucleotide with nuclear extracts from HUVECs or HeLa cells. Nuclear extracts (5 μg) were preincubated with or without a 100-fold excess of unlabeled AP-1(wt) or GATA oligonucleotides and then incubated with 32P-labeled AP-1(wt) oligonucleotide (40 fmol). (C) AP-1 motif-specific mutation abolishes its complex formation with HUVEC extract. Nuclear extract (5 μg) from HUVECs was preincubated with or without 100-fold excess of unlabeled AP-1(mut-1) or AP-1(mut-2) and then incubated with 32P-labeled oligonucleotide (40 fmol). (D) Unique AP-1 complexes assemble on the NOTCH4 promoter in HUVECs versus HeLa cells. Nuclear extract (5 μg) from HUVECs (top) or HeLa cells (bottom) was preincubated with the indicated antibodies (reading from left to right, lanes 3 to 14) and then incubated with 32P-labeled AP-1(wt) oligonucleotide (40 fmol). Lane 1, reaction mixture lacking nuclear extract; lane 2, reaction mixture lacking antibody. (E) Nuclear extract (20 μg) from HUVECs or HeLa cells was analyzed by Western blotting with antibodies against specific AP-1 components or tubulin as a control.
To determine if the AP-1 motif mediates complex formation, EMSA analysis was conducted with the labeled AP-1(wt) oligonucleotide, an AP-1 motif-mutated oligonucleotide [AP-1(mut-1)], and an oligonucleotide in which a sequence of no known function downstream of the AP-1 motif was mutated [AP-1(mut-2)]. Mutation of the AP-1 motif [AP-1(mut-1) probe], but not the downstream sequence [AP-1(mut-2) probe], abolished complex formation (Fig. 5C). Competitive binding assays were conducted with a 100-fold excess of the unlabeled AP-1(wt) oligonucleotide or the GATA oligonucleotide. The AP-1(wt) oligonucleotide abrogated binding of the HUVEC factors to the AP-1(wt) and AP-1(mut-2) probes, whereas the GATA oligonucleotide had no effect (Fig. 5C). These results strongly indicate that AP-1 or a highly related factor binds the conserved AP-1 motif of the NOTCH4 promoter in vitro.
AP-1 exists in multiple heterodimeric configurations, which can have different biochemical and biological activities (2, 7, 14, 18-20, 26, 30, 43, 44, 54, 55, 68, 78, 80, 90, 110). For example, tethering AP-1 subunits via a flexible peptide linker revealed that c-Jun-Fra-2, but not c-Jun-Fra-1 or c-Jun-c-Fos, inhibited growth arrest of immortalized fibroblasts (4). The slightly different mobility of the HUVEC and HeLa AP-1 complexes suggested that they contain distinct AP-1 components or that the components differ in posttranslational modifications. To define the subunit composition of HUVEC versus HeLa AP-1 complexes that form on the NOTCH4 promoter, extracts were preincubated with antibodies against multiple Fos and Jun family members, and DNA binding was measured by EMSA. HUVEC complexes were supershifted or inhibited by antibodies reacting with multiple Fos species (All-Fos), Fra-1, multiple Jun species (All-Jun), c-Jun [H-79 and c-Jun(N)], and JunD (Fig. 5D, top). HeLa complexes were supershifted or inhibited by antibodies reacting with All-Fos, All-Jun, JunD, and JunB (Fig. 5D, bottom). The greatest difference in the AP-1 complexes is that the HUVEC AP-1 complex preferentially contained the Fos family member Fra-1. A second major difference between HUVEC and HeLa AP-1 complexes is that the HUVEC complexes contained c-Jun almost exclusively, whereas the HeLa complexes contained JunB predominantly.
As HUVEC and HeLa AP-1 complexes that assemble on the NOTCH4 promoter in vitro differ in composition, it is attractive to propose that the differential composition constitutes a mechanism that confers or contributes to endothelial cell specificity of NOTCH4 transcription. AP-1 integrates information via diverse cellular signals (20, 88), including signals initiated by FGF-2 (66, 67), a vascular angiogenic remodeling factor. FGF-2 signaling activates mitogen-activated protein kinase and extracellular signal-regulated kinase, which stimulate AP-1 activity via regulation of the synthesis and activity of AP-1 components (66, 67).
The different compositions of the HUVEC and HeLa AP-1 complexes might result from the cell-type-specific expression or activation of AP-1 components. To determine the basis of the distinct AP-1 complexes, the relative levels of AP-1 components in HUVEC and HeLa nuclear extracts were measured by Western blotting (Fig. 5E). Multiple Fos species were detected with the All-Fos antibody in both extracts, consistent with the ability of this antibody to supershift AP-1 complexes formed with both extracts. By contrast, Fra-1 levels were high in HUVEC extracts but were nearly undetectable in HeLa extracts. The inability of the anti-Fra-1 antibody to supershift or inhibit the HeLa AP-1 complex (Fig. 5D) can therefore be explained by low levels of Fra-1 in HeLa cells. The All-Jun antibody detected a major species in HUVEC extracts and multiple low-level components in HeLa extracts. HUVEC extracts contained very little JunB compared to HeLa extracts, consistent with the supershift results. JunD and tubulin were detected in the two extracts at nearly identical levels. These results indicate that the differential expression of AP-1 subunits in HUVECs versus HeLa cells gives rise to cell-type-specific complexes that assemble on the conserved AP-1 motif of the NOTCH4 promoter in vitro.
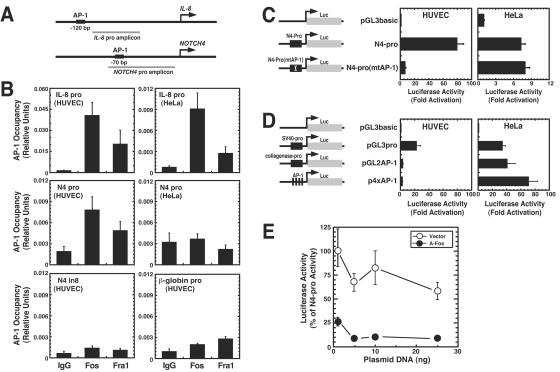
Despite the utility of EMSA in identifying prospective trans-acting factors, DNA recognition motifs in cells are often inaccessible. Quantitative ChIP analysis was used to determine whether AP-1 complexes occupy the NOTCH4 promoter in HUVECs and HeLa cells. Multiple AP-1 antibodies were used to test for AP-1 occupancy in HUVECs at the promoter of the established AP-1 target gene IL-8 (Fig. 6A). The only antibodies that yielded specific signals were the All-Fos and Fra-1 antibodies (Fig. 6B). Under conditions in which Fos and Fra-1 occupied the IL-8 promoter, occupancy was also detected at the NOTCH4 promoter in HUVECs but not in HeLa cells. By contrast, no occupancy was detected at NOTCH4 intron 8 (N4-in8) or at the β-globin promoter. As expected from the results of Fig. 3B, which show that only a small percentage of the HUVECs express NOTCH4 primary transcripts, the All-Fos and Fra-1 enrichment levels were relatively low.
FIG. 6.
Endothelial cell-specific occupancy of the NOTCH4 promoter by AP-1 and preferential requirement of the AP-1 motif for promoter activity in endothelial cells. (A) Diagram of the IL-8 promoter and NOTCH4 promoter amplicons used in the ChIP assay. The amplicon either includes or is very close to the conserved AP-1 motif. (B) Quantitative ChIP analysis of AP-1 occupancy at the IL-8 promoter and NOTCH4 promoter in HUVECs and HeLa cells (mean ± SEM; at least three independent experiments). Anti-Fos antibody against multiple Fos components and anti-Fra-1 antibody were used in the ChIP assay. Rabbit IgG was used as a control. AP-1 occupancy was also analyzed at intron 8 of NOTCH4 and the β-globin promoter as negative controls. pro, promoter; in, intron. (C) The conserved AP-1 motif on the NOTCH4 promoter is required for endothelial cell-specific transcription. HUVECs and HeLa cells were transiently transfected with the following luciferase reporter constructs: pGL3basic, pGL3basic containing NOTCH4 promoter (N4-pro), or pGL3basic containing the NOTCH4 promoter with a mutated AP-1 motif [N4-pro(mtAP-1)] (mean ± SEM; at least three independent experiments). (D) HUVECs and HeLa cells were transiently transfected with the following luciferase reporter constructs: pGL3basic, pGL3basic containing the SV40 promoter (pGL3pro), and pGL2basic containing either the collagenase promoter (pGL2AP-1) or four synthetic AP-1 motifs (p4xAP-1). Luciferase activity was normalized by the protein content of the lysates. Relative luciferase activities are shown as fold activation, with the activity of pGL3basic designated as 1.0 (mean ± SEM; at least three independent experiments). (E) HUVECs were transiently cotransfected with the N4-pro reporter construct and increasing amounts of the blank vector pCMV500 or an expression vector encoding the dominant-negative AP-1 molecule A-Fos. Relative luciferase activities, normalized by the protein content of the lysates, are shown as the percentage of the activity obtained with the N4-pro construct (mean ± SEM; at least three independent experiments).
We tested whether the preferential activity of the NOTCH4 promoter in transfection assays in HUVECs versus HeLa cells requires the AP-1 motif. Mutation of the AP-1 motif nearly abolished the strong reporter activity conferred by the NOTCH4 promoter in HUVECs (Fig. 6C). As mutation of the AP-1 motif did not affect the low activity of the promoter in HeLa cells (Fig. 6C), the HUVEC AP-1 complexes are uniquely able to activate the NOTCH4 promoter. Taken together with the selective AP-1 occupancy of endogenous NOTCH4 chromatin in HUVECs (Fig. 6B), these results indicate that the unique ability of a cell-type-specific AP-1 complex to access or to form a stable complex with the NOTCH4 chromatin template contributes to or confers endothelial cell-specific NOTCH4 transcription. Since antibodies against other AP-1 components were not efficacious in the ChIP assay, one cannot rule out the possibility that other AP-1 components also occupy the NOTCH4 locus.
Since the HUVEC and HeLa AP-1 complexes have distinct compositions, HUVEC AP-1 might have an intrinsically greater transactivation capacity, which could explain the results shown in Fig. 6C. We tested this by measuring the activity of two AP-1 reporter constructs in HUVECs and HeLa cells. These constructs, containing either the collagenase promoter with a single AP-1 motif or four synthetic AP-1 motifs, are known to be activated only by AP-1. The AP-1 reporters had considerably higher activities in HeLa cells, whereas pGL3pro had high activities both HeLa cells and HUVECs (Fig. 6D). This result indicates that AP-1 complexes are not more efficacious in HUVECs than HeLa cells in activating transcription through AP-1 motifs. A model in which AP-1 functionally interacts with other components of the NOTCH4 promoter can therefore explain the preferential requirement of the NOTCH4 promoter AP-1 motif, and this interaction occurs in HUVECs but not HeLa cells.
As endogenous Fos species occupy the endogenous NOTCH4 promoter in HUVECs but not HeLa cells and as the AP-1 motif is selectively required for promoter activity in HUVECs but not HeLa cells, it is highly likely that endogenous AP-1 confers high-level NOTCH4 promoter activity in HUVECs. To further test this, we asked whether expression of a dominant-negative molecule that antagonizes endogenous AP-1, A-Fos (71), affects NOTCH4 promoter activity in HUVEC cells. Transfecting increasing amounts of A-Fos expression vector strongly reduced promoter activity, whereas the blank vector pCMV500 had little effect (Fig. 6E). This result solidifies a role for endogenous AP-1 in conferring activation through the AP-1 motif of the NOTCH4 promoter.
Endothelial cell growth supplement reprograms the NOTCH4 gene in HeLa cells from a repressed to a transcriptionally active state.
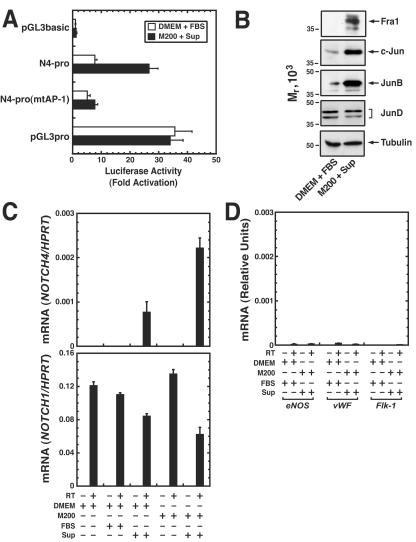
HUVECs are cultured in media containing a low-serum growth supplement with FGF-2, epidermal growth factor, hydrocortisone, and heparin. FGF-2, an important regulator of vascular angiogenic remodeling, activates AP-1-responsive transcriptional elements and AP-1 DNA binding activity in cultured murine corneal cells and a calvarial osteoblast cell line (66, 67). We reasoned that the signaling molecules in the supplement might induce components of the HUVEC-specific AP-1 complexes both in endothelial cells and in nonendothelial cells. We tested this by measuring NOTCH4 promoter activity and the expression of AP-1 components in HeLa cells cultured for 24 h with the supplement. The supplement increased the activity of the N4-pro reporter construct ∼3-fold without affecting the activity of the N4-pro(mtAP-1) reporter containing a mutated AP-1 motif or the SV40 promoter-containing reporter pGL3pro (Fig. 7A). Fra-1, c-Jun, and JunB expression were strongly induced by the supplement, whereas low-level JunD expression was unchanged (Fig. 7B). Thus, signal-dependent reconfiguration of the expression profile of AP-1 components, yielding a profile that resembles the HUVEC profile, is associated with elevated NOTCH4 promoter activity.
FIG. 7.
Endothelial cell growth supplement induces AP-1 components and reprograms endogenous NOTCH4 from a repressed to a transcriptionally active state in HeLa cells. (A) HeLa cells were transiently transfected with the following luciferase reporter constructs: pGL3basic, pGL3basic containing NOTCH4 promoter (N4-pro), pGL3basic containing the NOTCH4 promoter with a mutated AP-1 motif [N4-pro(mtAP-1)], or pGL3basic containing the SV40 promoter (pGL3pro). Transfected HeLa cells were cultured in normal medium (DMEM plus 10% FBS) or in the endothelial medium (Medium 200 plus endothelial cell supplement) for 24 h before harvest. Luciferase activity was normalized by the protein content of the lysates. Relative luciferase activities are shown as fold activation, with the activity of pGL3basic designated as 1.0 (mean ± SEM; three independent experiments). (B) Nuclear extract (20 μg) from HeLa cells cultured in normal medium or endothelial medium was analyzed by Western blotting with antibodies against specific AP-1 components or tubulin as a control. (C) Quantitative real-time RT-PCR analysis of NOTCH4 and NOTCH1 mRNA expression in HeLa cells cultured in DMEM or Medium 200 with or without 10% FBS or the endothelial cell supplement. NOTCH4 and NOTCH1 transcripts were also measured in HUVECs cultured in endothelial cell medium. The levels of NOTCH4 and NOTCH1 mRNA were normalized by HPRT mRNA. The NOTCH4/HPRT or NOTCH1/HPRT mRNA ratios in HUVEC cells were designated as 1.0. The graph shows the expression pattern of NOTCH4 (top) and NOTCH1 (bottom) in HeLa cells (mean ± SEM; three independent experiments). (D) HeLa cells and HUVECs were cultured as described in panel C. eNOS, vWF, and Flk-1 mRNA levels were normalized by HPRT mRNA levels, and the transcript ratios in HUVECs were designated as 1.0. The graph shows the expression patterns of eNOS, vWF, and Flk-1 in HeLa cells (mean ± SEM; three independent experiments).
We tested whether the supplement-mediated induction of AP-1 components is associated with changes in expression of the endogenous, inactive NOTCH4 gene in HeLa cells. HeLa cells were derived from an epidermoid carcinoma of the cervix and have epithelial cell properties (34). HeLa cells might lack multiple factors required for NOTCH4 transcription, or transcriptional repression might result from a deficiency of specific AP-1 components, such as those expressed in HUVECs. Quantitative RT-PCR revealed that culturing HeLa cells in medium containing the growth supplement for 24 h under conditions that induce AP-1 components and increase NOTCH4 promoter activity in transient transfections (Fig. 7B) was sufficient to reprogram the endogenous NOTCH4 gene from a repressed state to a transcriptionally active state (Fig. 7C). However, the NOTCH4 expression level was considerably lower than in HUVECs. The supplement-mediated induction of NOTCH4 expression was confirmed by quantitative RT-PCR analysis with an independent primer set (NOTCH4-2), was maximal after 24 h of treatment, and persisted for at least 6 days in the continued presence of the supplement (data not shown). NOTCH1 was expressed in HeLa cells and was not induced by the endothelial cell supplement (Fig. 7C). To determine whether the endothelial cell supplement induces expression of other endothelial cell-specific genes, we measured the expression of genes encoding endothelial cell nitric oxide synthase (eNOS), von Willebrand factor (vWF), and the type 2 receptor for vascular endothelial cell growth factor (Flk-1) (Fig. 7D). The supplement had little if any effect on the expression of these genes. This result indicates that signaling molecules in the supplement have the capacity to induce AP-1 components that are expressed in HUVECs and to selectively activate endogenous NOTCH4 transcription in HeLa cells.
AP-1-dependent high-level activity of NOTCH4 promoter in vascular endothelium in vivo.
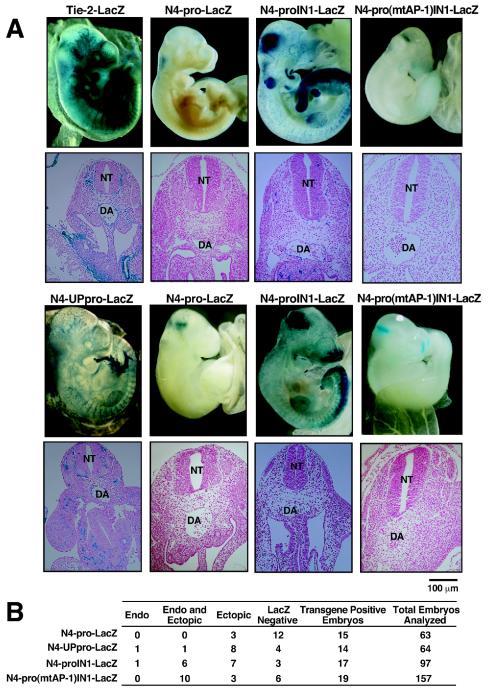
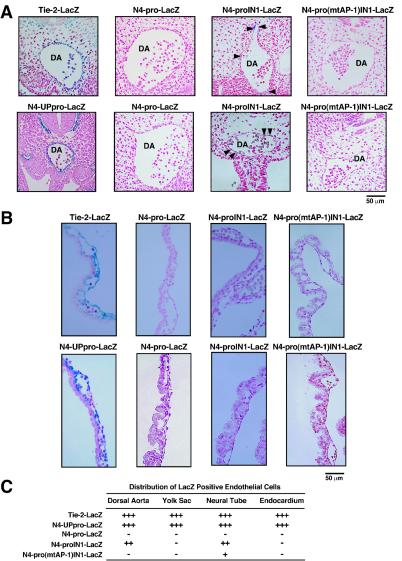
To test whether the NOTCH4 promoter AP-1 motif is important for endothelial cell-specific transcription in vivo, NOTCH4 promoter-lacZ fusion constructs were assayed in F0 E10.5 transgenic mouse embryos (Fig. 8). A construct containing the endothelial cell-specific Tie-2 enhancer and promoter (Tie-2-LacZ) (81) was used as a control. As expected, analysis of whole mount embryos revealed that Tie-2-LacZ was expressed in the vascular endothelium (Fig. 8A). By contrast, the NOTCH4 promoter alone (N4-pro-LacZ) was almost completely inactive (Fig. 8A), although 3 of 15 transgene-positive embryos revealed ectopic staining (Fig. 8B). Inclusion of intron 1 in the N4-pro-LacZ construct (N4-proIN1-LacZ) resulted in endothelial cell expression in 7 of 17 embryos with a similar frequency of ectopic expression (Fig. 8). Mutation of the NOTCH4 promoter AP-1 motif in the presence of intron 1 [N4-pro(mtAP-1)IN1-LacZ] strongly reduced LacZ expression, although careful analysis revealed very low level expression in subregions of the vascular endothelium and at ectopic sites in 10 of 19 transgene-positive embryos (Fig. 8). Inclusion of an additional ∼4 kb of sequence upstream of the NOTCH4 promoter (N4-UPpro-LacZ) conferred high-level expression in vascular endothelium but only at a low frequency (2 of 14 transgene-positive embryos) (Fig. 8). Extensive attempts (analysis of >150 embryos) to analyze an additional construct containing both the upstream sequences and intron 1 were unsuccessful, as the stable integration of this construct in chromosomal DNA could not be reliably established (data not shown).
FIG. 8.
Requirement of NOTCH4 promoter AP-1 motif for high-level promoter activity in E10.5 transgenic mouse embryos. (A) Two representative pictures of E10.5 transgenic mouse embryos (top) and transverse embryo sections (bottom) containing the N4-pro-LacZ, N4-proIN1-LacZ, and N4-pro(mtAP-1)IN1-LacZ transgenes are shown. One representative picture of Tie-2-LacZ and N4-UPpro-LacZ transgenic embryos is shown. DA, dorsal aorta; NT, neural tube. (B) The table summarizes the total numbers of embryos analyzed, the number of embryos that genotyped positive for the lacZ transgenes, and the number of embryos that stained positive or negative for LacZ.
To investigate the vascular expression pattern of NOTCH4 promoter-lacZ constructs in detail, sections of E10.5 embryos were analyzed for LacZ expression in endothelial cells of the dorsal aorta (Fig. 9A), yolk sac (Fig. 9B), neural tube (data not shown), and the endocardium (data not shown). The Tie-2-LacZ and N4-UPpro-LacZ constructs were expressed in endothelial cells of all four anatomical regions (Fig. 9C). The N4-proIN1-LacZ construct was expressed in endothelial cells of the dorsal aorta and the neural tube (Fig. 9C). By contrast, mutation of the AP-1 motif [N4-pro(mtAP-1)IN1-LacZ] abolished expression in endothelial cells of the dorsal aorta and reduced expression in endothelial cells of the neural tube (Fig. 9C).
FIG. 9.
NOTCH4 transgene expression in the vasculature of E10.5 mouse embryos. (A) Expression of NOTCH4 promoter-lacZ constructs in vascular endothelium. The photomicrographs show embryo sections, which reveal the dorsal aorta (DA) of E10.5 transgenic mouse embryos with the indicated transgenes. The arrowheads indicate endothelial cells with LacZ positivity. (B) Expression of NOTCH4 promoter-lacZ constructs in sections of E10.5 transgenic mouse embryo yolk sac. (C) The table summarizes a quantitative analysis of the distribution of LacZ-positive endothelial cells in the dorsal aorta, neural tube, endocardium, and the yolk sac. +++, all endothelial cells were LacZ positive; ++, intermediate number of LacZ-positive endothelial cells; +, rare LacZ-positive endothelial cells; −, LacZ-negative endothelial cells.
The transgenic analysis indicated that a transgene containing the NOTCH4 promoter alone is insufficient to confer expression in vascular endothelium in E10.5 transgenic mouse embryos. However, either intron 1 or sequences upstream of the promoter conferred promoter activity in vascular endothelium. Intron 1 and the upstream sequences therefore have intrinsic enhancer activity in vivo, which was not apparent in transient transfections in HUVECs and HeLa cells (Fig. 4). The strongest expression in vascular endothelium was achieved with the N4-UPpro-LacZ construct. However, based on the low frequency (14%) of N4-UPpro-LacZ expression in transgenic embryos, this construct could not be used for additional mechanistic analysis. By contrast, N4-proIN1-LacZ had a considerably higher frequency of expression (41%), making additional mechanistic analysis tractable. Mutation of the AP-1 motif in the N4-proIN1-LacZ construct strongly reduced expression in vascular endothelium, even though intron 1 was present, indicating that AP-1 cooperates with intron 1 to confer expression in vivo.
Our results, showing that the NOTCH4 promoter has cell-type-specific activity in cultured cells but is insufficient to recapitulate this activity in transgenic mice, are reminiscent of results obtained from analyses of other tissue-specific genes, such as the β-globin genes. Extensive studies with β-globin locus transgenes in mice have revealed transcriptional silencing and ectopic expression with constructs containing only the β-globin promoters (48a). Subsequent efforts identified the β-globin locus control region, which overcomes chromosome position effects and confers copy number-dependent expression in transgenic mice (24a, 29a). Importantly, the β-globin promoters, analogous to the NOTCH4 promoter, bind factors that confer cell-type-specific transcription in cultured cells. The locus control region establishes an additional level of regulation, which allows promoter activity to be manifested at ectopic chromosomal sites in mice. Mechanistically, these results showing that isolated promoters with intrinsic cell type specificity determinants need additional regulatory sequences in vivo can be explained by a requirement for chromatin modifying or remodeling activities conferred by the additional sequences, e.g., by intron 1 and upstream sequences of the NOTCH4 locus. Such activities can be crucial for counteracting chromatin-mediated repression of promoter-only constructs integrated at ectopic chromosomal sites.
An AP-1-Notch4 axis for angiogenic vascular remodeling and oncogenesis?
Targeted deletions in mice have established an important role of Fra-1 (83), JunB (82), and Notch4 (47) in vascular development. fra-1-null mice die at E10 to E10.5 due to severe reduction in vascular endothelial cells and defective placental vascularization (83). Large vessels develop in the chorionic plate but fail to undergo vascular remodeling. Similarly, junB-null mice die at E8.5 to E10 due to defective vascularization of extraembryonic tissues, including defective vascularization of the placental labyrinth (82). Given the sequence homology of AP-1 subunits, the scope of these vascular phenotypes might be restricted due to functional redundancies in specific cell types. Functional redundancies among AP-1 family members have been revealed by gene targeting experiments in mice, including the knock in of junB into c-jun-null mice, which rescues defects in liver and cardiac development (74). Moreover, the knock in of fra-1 into c-fos-null mice rescues bone development and light-induced photoreceptor apoptosis (24). In the case of Fra-1 and c-Fos, this is not an absolute redundancy, however, as Fra-1 does not rescue defective transcriptional responses in c-fos-null fibroblasts (24).
Our analysis has clearly shown that AP-1 preferentially activates the NOTCH4 promoter in HUVECs versus HeLa cells (Fig. 6); the conserved AP-1 motif of the NOTCH4 promoter, in the context of the N4-proIN1-LacZ construct, is crucial for promoter activity in transgenic mouse embryos (Fig. 8). Thus, it is attractive to consider the possibility that vascular phenotypes associated with fra-1 and junB knockouts might result, in part, from defective regulation of NOTCH4 transcription. It is unlikely, however, that such phenotypes in the Fra-1- and JunB-null mice can be explained solely by defective Notch4 transcription, since Notch4-null mice do not have detectable vascular phenotypes (29, 47). Almost certainly, AP-1 regulates additional genes besides Notch4 that are dysregulated in the Fra-1 and JunB knockout mice and that mediate angiogenic remodeling. Given the collective requirement of Notch1 and Notch4 for angiogenic remodeling during mouse embryogenesis (29, 47), we propose that AP-1-dependent Notch4 transcription is required to establish an AP-1-Notch4 vascular angiogenic pathway.
Based on our finding that the endothelial growth supplement reprograms the NOTCH4 gene in HeLa cells from a repressed to a transcriptionally active state, it is attractive to propose that aberrant signaling in nonendothelial cells, for example in breast cancer cells, ectopically activates NOTCH4 expression. This mode of establishing ectopic Notch4 signaling would have important consequences for the control of cellular proliferation and differentiation, since many examples exist in which experimental strategies that deregulate Notch signaling are oncogenic (3, 6, 10, 23, 76, 101). Intriguingly, NIC-1 represses expression of the endogenous AP-1 target genes IL-8 and MMP-1 and represses AP-1-mediated transactivation in transfection assays (15, 16). Thus, activation of Notch1 signaling might suppress AP-1-mediated induction of NOTCH4 expression, establishing a crucial molecular link between Notch1 and Notch4 signaling. The studies described herein, dissecting the mechanism underlying endothelial cell-specificity of NOTCH4 transcription, have laid the groundwork for identifying the factors and signals required to reprogram the repressed NOTCH4 gene and for testing the validity of our model that such factors contribute to oncogenesis via NOTCH4 deregulation.
Acknowledgments
We thank Louis Lurie and Soumen Pal for providing critical comments on the manuscript.
This work was funded by grants from the National Institutes of Health (DK50107) to E.H.B. E.H.B. is a Romnes Faculty Scholar and a Shaw Scientist. J.W. is an American Heart Association predoctoral fellow.
REFERENCES
- 1.Ando, K., S. Kanazawa, T. Tetsuka, S. Ohta, X. Jiang, T. Tada, M. Kobayashi, N. Matsui, and T. Okamoto. 2003. Induction of Notch signaling by tumor necrosis factor in rheumatoid synovial fibroblasts. Oncogene 22:7796-7803. [DOI] [PubMed] [Google Scholar]
- 2.Angel, P., M. Imagawa, R. Chiu, B. Stein, R. J. Imbra, H. J. Rahmsdorf, C. Jonat, P. Herrlich, and M. Karin. 1987. Phorbol ester-inducible genes contain a common cis element recognized by a TPA-modulated trans-acting factor. Cell 49:729-739. [DOI] [PubMed] [Google Scholar]
- 3.Aster, J. C., L. Xu, F. G. Karnell, V. Patriub, J. C. Pui, and W. S. Pear. 2000. Essential roles for ankyrin repeat and transactivation domains in induction of T-cell leukemia by Notch1. Mol. Cell. Biol. 20:7505-7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bakiri, L., K. Matsuo, M. Wisniewska, E. F. Wagner, and M. Yaniv. 2002. Promoter specificity and biological activity of tethered AP-1 dimers. Mol. Cell. Biol. 22:4952-4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bavendiek, U., P. Libby, M. Kilbride, R. Reynolds, N. Mackman, and U. Schonbeck. 2002. Induction of tissue factor expression in human endothelial cells by CD40 ligand is mediated via activator protein 1, nuclear factor κB, and Egr1. J. Biol. Chem. 277:25032-25039. [DOI] [PubMed] [Google Scholar]
- 6.Beverly, L. J., and A. J. Capobianco. 2003. Perturbation of Ikaros isoform selection by MLV integration is a cooperative event in Notch(IC)-induced T cell leukemogenesis. Cancer Cell 3:551-564. [DOI] [PubMed] [Google Scholar]
- 7.Bohmann, D., T. J. Bos, A. Admon, T. Nishimura, P. K. Vogt, and R. Tjian. 1987. Human proto-oncogene c-jun encodes a DNA binding protein with structural and functional properties of transcription factor AP-1. Science 238:1386-1392. [DOI] [PubMed] [Google Scholar]
- 8.Boyd, K. E., and P. J. Farnham. 1999. Coexamination of site-specific transcription factor binding and promoter activity in living cells. Mol. Cell. Biol. 19:8393-8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bresnick, E. H., H. Im, and K. D. Johnson. 2003. Long-range acetylation patterns in the genome, p. 260-264. In D. Cooper (ed.), Nature encyclopedia of the human genome. Nature Publishing Group, Macmillan Publishers Ltd., Basingstoke, United Kingdom.
- 10.Capobianco, A. J., P. Zagouras, C. M. Blaumueller, S. Artavanis-Tsakonas, and J. M. Bishop. 1997. Neoplastic transformation by truncated alleles of human NOTCH1/TAN1 and NOTCH2. Mol. Cell. Biol. 17:6265-6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan, Y., J. E. Fish, C. D'Abreo, S. Lin, G. B. Robb, A.-M. Teichert, F. Karantzoulis-Fegaras, A. Keightley, B. M. Steer, and P. A. Marsden. 2004. The cell-specific expression of endothelial nitric-oxide synthase: a role for DNA methylation. J. Biol. Chem. 279:35087-35100. [DOI] [PubMed] [Google Scholar]
- 12.Cheutin, T., A. J. McNairn, T. Jenuwein, D. M. Gilbert, P. B. Singh, and T. Misteli. 2003. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science 299:721-725. [DOI] [PubMed] [Google Scholar]
- 13.Chi, J. T., H. Y. Chang, G. Haraldsen, F. L. Jahnsen, O. G. Troyanskaya, D. S. Chang, Z. Wang, S. G. Rockson, M. van de Rijn, D. Botstein, and P. O. Brown. 2003. Endothelial cell diversity revealed by global expression profiling. Proc. Natl. Acad. Sci. USA 100:10623-10628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiu, R., P. Angel, and M. Karin. 1989. Jun-B differs in its biological properties from, and is a negative regulator of, c-Jun. Cell 59:979-986. [DOI] [PubMed] [Google Scholar]
- 15.Chu, J., and E. H. Bresnick. 2004. Evidence that C promoter-binding factor is required for Notch-1-mediated repression of activator protein-1. J. Biol. Chem. 279:12337-12345. [DOI] [PubMed] [Google Scholar]
- 16.Chu, J., S. Jeffries, J. E. Norton, A. J. Capobianco, and E. H. Bresnick. 2002. Repression of activator protein-1-mediated transcriptional activation by the Notch-1 intracellular domain. J. Biol. Chem. 277:7587-7597. [DOI] [PubMed] [Google Scholar]
- 17.Chung, J. H., M. Whiteley, and G. Felsenfeld. 1993. A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell 74:505-514. [DOI] [PubMed] [Google Scholar]
- 18.Cohen, D. R., and T. Curran. 1988. fra-1: a serum-inducible, cellular immediate-early gene that encodes a Fos-related antigen. Mol. Cell. Biol. 8:2063-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng, T., and M. Karin. 1993. JunB differs from c-Jun in its DNA binding and dimerization domains, and represses c-Jun by formation of inactive heterodimers. Genes Dev. 7:479-490. [DOI] [PubMed] [Google Scholar]
- 20.Eferl, R., and E. F. Wagner. 2003. AP1: A double-edged sword in tumorigenesis. Nat. Rev. Cancer 3:859-868. [DOI] [PubMed] [Google Scholar]
- 21.Elefant, F., Y. Su, S. A. Liebhaber, and N. E. Cooke. 2000. Patterns of histone acetylation suggest dual pathways for gene activation by a bifunctional locus control region. EMBO J. 19:6814-6822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evans, T., and G. Felsenfeld. 1989. The erythroid-specific transcription factor Eryf1: a new finger protein. Cell 58:877-885. [DOI] [PubMed] [Google Scholar]
- 23.Fitzgerald, K., A. Harrington, and P. Leder. 2000. Ras pathway signals are required for notch-mediated oncogenesis. Oncogene 19:4191-4198. [DOI] [PubMed] [Google Scholar]
- 24.Fleischmann, A., F. Hafezi, C. Elliott, C. E. Reme, U. Ruther, and E. F. Wagner. 2000. Fra-1 replaces c-Fos-dependent functions in mice. Genes Dev. 14:2695-2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24a.Forrester, W. C., S. Takegawa, T. Papayannopoulou, G. Stamatoyannopoulos, and M. Groudine. 1987. Evidence for a locus activation region: the formation of developmentally stable hypersensitive sites in globin-expressing hybrids. Nucleic Acids Res. 15:10159-10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forsberg, E. C., K. M. Downs, H. M. Christensen, H. Im, P. A. Nuzzi, and E. H. Bresnick. 2000. Developmentally dynamic histone acetylation pattern of a tissue-specific chromatin domain. Proc. Natl. Acad. Sci. USA 97:14494-14499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Franza, B. R., F. J. Rauscher, S. F. Josephs, and T. Curran. 1988. The Fos complex and Fos-related antigens recognize sequence elements that contain AP-1 binding sites. Science 239:1150-1153. [DOI] [PubMed] [Google Scholar]
- 27.Fryer, C. J., E. Lamar, I. Turbachova, C. Kintner, and K. A. Jones. 2002. Mastermind mediates chromatin-specific transcription and turnover of the Notch enhancer complex. Genes Dev. 16:1397-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grass, J. A., M. E. Boyer, S. Paul, J. Wu, M. J. Weiss, and E. H. Bresnick. 2003. GATA-1-dependent transcriptional repression of GATA-2 via disruption of positive autoregulation and domain-wide chromatin remodeling. Proc. Natl. Acad. Sci. USA 100:8811-8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28a.Gribnau, J., K. Diderich, S. Pruzina, R. Calzolari, and P. Fraser. 2000. Intergenic transcription and developmental remodeling of chromatin subdomains in the human beta-globin locus. Mol. Cell 5:377-386. [DOI] [PubMed] [Google Scholar]
- 29.Gridley, T. 2001. Notch signaling during vascular development. Proc. Natl. Acad. Sci. USA 98:5377-5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29a.Grosveld, F., G. B. van Assendelft, D. R. Greaves, and G. Kollias. 1987. Position-independent, high-level expression of the human beta-globin gene in transgenic mice. Cell 51:975-985. [DOI] [PubMed] [Google Scholar]
- 30.Hirai, S. I., R. P. Ryseck, F. Mechta, R. Bravo, and M. Yaniv. 1989. Characterization of JunD: a new member of the jun proto-oncogene family. EMBO J. 8:1433-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ho, Y., F. Elefant, N. Cooke, and S. Liebhaber. 2002. A defined locus control region determinant links chromatin domain acetylation with long-range gene activation. Mol. Cell 9:291-302. [DOI] [PubMed] [Google Scholar]
- 32.Hsieh, J. J., T. Henkel, P. Salmon, E. Robey, M. G. Peterson, and S. D. Hayward. 1996. Truncated mammalian Notch1 activates CBF1/RBPJk-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol. Cell. Biol. 16:952-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsieh, J. J., S. Zhou, L. Chen, D. B. Young, and S. D. Hayward. 1999. CIR, a corepressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc. Natl. Acad. Sci. USA 96:23-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsu, S. H., B. Z. Schacter, N. L. Delaney, T. B. Miller, V. A. McKusick, R. H. Kennett, J. G. Bodmer, D. Young, and W. F. Bodmer. 1976. Genetic characteristics of the HeLa cell. Science 191:392-394. [DOI] [PubMed] [Google Scholar]
- 35.Im, H., J. A. Grass, K. D. Johnson, M. E. Boyer, J. Wu, and E. H. Bresnick. 2004. Measurement of protein-DNA interactions in vivo by chromatin immunoprecipitation. Methods Mol. Biol. 284:129-146. [DOI] [PubMed] [Google Scholar]
- 36.Iso, T., Y. Hamamori, and L. Kedes. 2003. Notch signaling in vascular development. Arterioscler. Thromb. Vasc. Biol. 23:543-553. [DOI] [PubMed] [Google Scholar]
- 37.Jarriault, S., C. Brou, F. Logeat, E. H. Schroeter, R. Kopan, and A. Israel. 1995. Signalling downstream of mammalian Notch. Nature 377:355-358. [DOI] [PubMed] [Google Scholar]
- 38.Jeffries, S., D. J. Robbins, and A. J. Capobianco. 2002. Characterization of a high-molecular-weight Notch complex in the nucleus of Notchic-transformed RKE cells and in a human T-cell leukemia cell line. Mol. Cell. Biol. 22:3927-3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson, K. D., and E. H. Bresnick. 2002. Dissecting long-range transcriptional mechanisms by chromatin immunoprecipitation. Methods 26:27-36. [DOI] [PubMed] [Google Scholar]
- 40.Johnson, K. D., J. D. Grass, M. E. Boyer, C. M. Kiekhaefer, G. A. Blobel, M. J. Weiss, and E. H. Bresnick. 2002. Cooperative activities of hematopoietic regulators recruit RNA polymerase II to a tissue-specific chromatin domain. Proc. Natl. Acad. Sci. USA 99:11760-11765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawana, M., M. E. Lee, E. E. Quertermous, and T. Quertermous. 1995. Cooperative interaction of GATA-2 and AP1 regulates transcription of the endothelin-1 gene. Mol. Cell. Biol. 15:4225-4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kendall, E., C. A. Sargent, and R. D. Campbell. 1990. Human major histocompatibility complex contains a new cluster of genes between the HLA-D and complement C4 loci. Nucleic Acids Res. 18:7251-7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kerppola, T. K., and T. Curran. 1993. Selective DNA bending by a variety of bZIP proteins. Mol. Cell. Biol. 13:5479-5489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kerppola, T. K., D. Luk, and T. Curran. 1993. Fos is a preferential target of glucocorticoid receptor inhibition of AP-1 activity in vitro. Mol. Cell. Biol. 13:3782-3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kiekhaefer, C. M., J. A. Grass, K. D. Johnson, M. E. Boyer, and E. H. Bresnick. 2002. Hematopoietic activators establish an overlapping pattern of histone acetylation and methylation within a tissue-specific chromatin domain. Proc. Natl. Acad. Sci. USA 99:14309-14314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45a.Kimura, H., K. Sugaya, and P. R. Cook. 2002. The transcription cycle of RNA polymerase II in living cells. J. Cell Biol. 159:777-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ko, L. J., and J. D. Engel. 1993. DNA-binding specificities of the GATA transcription factor family. Mol. Cell. Biol. 13:4011-4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krebs, L. T., Y. Xue, C. R. Norton, J. R. Shutter, M. Maguire, J. P. Sundberg, G. Gallahan, V. Closson, J. Kitajewski, R. Callahan, G. H. Smitch, K. L. Stark, and T. Gridley. 2000. Notch signaling is essential for vascular morphogenesis in mice. Genes Dev. 14:1343-1352. [PMC free article] [PubMed] [Google Scholar]
- 48.Kurooka, H., and T. Honjo. 2000. Functional interaction between the mouse notch1 intracellular region and histone acetyltransferases PCAF and GCN5. J. Biol. Chem. 275:17211-17220. [DOI] [PubMed] [Google Scholar]
- 48a.Lacy, E., S. Roberts, E. P. Evans, M. D. Burtenshaw, and F. D. Costantini. 1983. A foreign beta-globin gene in transgenic mice: integration at abnormal chromosomal positions and expression in inappropriate tissues. Cell 34:343-358. [DOI] [PubMed] [Google Scholar]
- 49.Lam, L. T., and E. H. Bresnick. 1995. Evidence for distinct DNA binding forms of the erythroid-specific transcription factor NF-E2. Biochemistry 34:16347-16358. [DOI] [PubMed] [Google Scholar]
- 50.Lam, L. T., and E. H. Bresnick. 1998. Identity of the beta-globin locus control region binding protein HS2NF5 as the mammalian homolog of the notch-regulated transcription factor suppressor of hairless. J. Biol. Chem. 273:24223-24231. [DOI] [PubMed] [Google Scholar]
- 51.Lardelli, M., J. Dahlstrand, and U. Lendahl. 1994. The novel Notch homolog mouse Notch3 lacks specific epidermal growth factor-repeats and is expressed in proliferating neuroepithelium. Mech. Dev. 46:123-136. [DOI] [PubMed] [Google Scholar]
- 52.Lawson, N. D., N. Scheer, V. N. Pham, C.-H. Kim, A. B. Chitnis, J. A. Campos-Ortega, and B. M. Weinstein. 2001. Notch signaling is required for arterial-venous differentiation during embryonic vascular development. Development 128:3675-3683. [DOI] [PubMed] [Google Scholar]
- 53.Lee, D. Y., J. J. Hayes, D. Pruss, and A. P. Wolffe. 1993. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell 72:73-84. [DOI] [PubMed] [Google Scholar]
- 54.Lee, W., A. Haslinger, M. Karin, and R. Tjian. 1987. Activation of transcription by two factors that bind promoter and enhancer sequences of the human metallothionein gene and SV40. Nature 325:368-372. [DOI] [PubMed] [Google Scholar]
- 55.Lee, W., P. Mitchell, and R. Tjian. 1987. Purified transcription factor AP-1 interacts with TPA-inducible enhancer elements. Cell 49:741-752. [DOI] [PubMed] [Google Scholar]
- 56.Leong, K. G., X. Hu, L. Li, M. Noseda, B. Larrivee, C. Hull, L. Hood, F. Wong, and A. Karsan. 2002. Activated Notch4 inhibits angiogenesis: role of β1-integrin activation. Mol. Cell. Biol. 22:2830-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liang, G., J. C. Y. Lin, V. Wei, C. Yoo, J. C. Cheng, C. T. Nguyen, D. J. Weisenberger, G. Egger, D. Takai, F. A. Gonzales, and P. A. Jones. 2004. Distinct localization of histone H3 acetylation and H3-K4 to the transcription start sites in the human genome. Proc. Natl. Acad. Sci. USA 101:7357-7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Litt, M. D., M. Simpson, M. Gaszner, C. D. Allis, and G. Felsenfeld. 2001. Correlation between histone lysine methylation and developmental changes at the chicken beta-globin locus. Science 293:2453-2455. [DOI] [PubMed] [Google Scholar]
- 59.Liu, Z.-J., T. Shirakawa, Y. Li, A. Soma, M. Oka, G. P. Dotto, R. M. Fairman, O. C. Velazquez, and M. Herlyn. 2003. Regulation of Notch1 and Dll4 by vascular endothelial growth factor in arterial endothelial cells: implications for modulating arteriogenesis and angiogenesis. Mol. Cell. Biol. 23:14-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.MacKenzie, F., P. Duriez, B. Larrivee, L. Chang, I. Pollet, F. Wong, C. Yip, and A. Karsan. 2004. Notch4-induced inhibition of endothelial sprouting requires the ankyrin repeats and involves signaling through RBP-Jκ. Blood 104:1760-1768. [DOI] [PubMed] [Google Scholar]
- 61.Matsunami, N., Y. Hamaguchi, Y. Yamamoto, K. Kuze, K. Kangawa, H. Matsuo, M. Kawaichi, and T. Honjo. 1989. A protein binding to the Jk recombination sequence of immunoglobulin genes contains a sequence related to the integrase motif. Nature 342:934-937. [DOI] [PubMed] [Google Scholar]
- 62.Merika, M., and S. H. Orkin. 1993. DNA-binding specificity of GATA family transcription factors. Mol. Cell. Biol. 13:3999-4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Minami, T., and W. C. Aird. 2001. Thrombin stimulation of the vascular cell adhesion molecule-1 promoter in endothelial cells is mediated by tandem nuclear factor kappa-B and GATA motifs. J. Biol. Chem. 276:47632-47641. [DOI] [PubMed] [Google Scholar]
- 64.Minami, T., J. A. Kuivenhoven, V. Evans, T. Kodama, R. D. Rosenberg, and W. C. Aird. 2003. Ets motifs are necessary for endothelial cell-specific expression of a 723-bp Tie-2 promoter/enhancer in Hprt targeted transgenic mice. Arterioscler. Thromb. Vasc. Biol. 23:2041-2047. [DOI] [PubMed] [Google Scholar]
- 65.Minami, T., T. Murakami, K. Horiuchi, M. Miura, T. Noguchi, J. Miyazaki, T. Hamakubo, W. C. Aird, and T. Kodama. 2004. Interaction between Hex and GATA transcription factors in vascular endothelial cells inhibits flk-1/KDR-mediated vascular endothelial growth factor signaling. J. Biol. Chem. 279:20626-20635. [DOI] [PubMed] [Google Scholar]
- 66.Mohan, R., J. Sivak, P. Ashton, L. A. Russo, B. Q. Pham, N. Kasahara, M. B. Raizman, and M. E. Fini. 2000. Curcuminoids inhibit the angiogenic response stimulated by fibroblast growth factor-2, including expression of matrix metalloproteinase gelatinase B. J. Biol. Chem. 275:10405-10412. [DOI] [PubMed] [Google Scholar]
- 67.Newberry, E. P., D. Willis, T. Latifi, J. M. Boudreaux, and D. A. Towler. 1997. Fibroblast growth factor receptor signaling activates the human interstitial collagenase promoter via the bipartite Ets-AP1 element. Mol. Endocrinol. 11:1129-1144. [DOI] [PubMed] [Google Scholar]
- 68.Nishina, H., H. Sato, T. Suzuki, M. Sato, and H. Iba. 1990. Isolation and characterization of fra-2, an additional member of the fos gene family. Proc. Natl. Acad. Sci. USA 87:3619-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nofziger, D., A. Miyamoto, K. M. Lyons, and G. Weinmaster. 1999. Notch signaling imposes two distinct blocks in the differentiation of C2C12 myoblasts. Development 126:1689-1702. [DOI] [PubMed] [Google Scholar]
- 70.Noma, K., C. D. Allis, and S. I. Grewal. 2001. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science 293:1150-1155. [DOI] [PubMed] [Google Scholar]
- 71.Olive, M., D. Krylov, D. R. Echlin, K. Gardner, E. Taparowsky, and C. Vinson. 1997. A dominant negative to activation protein-1 (AP1) that abolishes DNA binding and inhibits oncogenesis. J. Biol. Chem. 272:18586-18594. [DOI] [PubMed] [Google Scholar]
- 72.Onodera, K., S. Takahashi, S. Nishimura, J. Ohta, H. Motohashi, K. Yomogida, N. Hayashi, J. D. Engel, and M. Yamamoto. 1997. GATA-1 transcription is controlled by distinct regulatory mechanisms during primitive and definitive erythropoiesis. Proc. Natl. Acad. Sci. USA 94:4487-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72a.Osborne, C. S., L. Chakalova, L., K. E. Brown, D. Carter, A. Horton, E. Debrand, B. Goyenechea, J. A. Mitchell, S. Lopes, W. Reik, and P. Fraser. 2004. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet. 36:1065-1071. [DOI] [PubMed] [Google Scholar]
- 73.Pal, S., A. B. Cantor, K. D. Johnson, T. Moran, M. E. Boyer, S. H. Orkin, and E. H. Bresnick. 2004. Coregulator-dependent facilitation of chromatin occupancy by GATA-1. Proc. Natl. Acad. Sci. USA 101:980-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Passegue, E., W. Jochum, A. Behrens, R. Ricci, and E. F. Wagner. 2002. JunB can substitute for Jun in mouse development and cell proliferation. Nat. Genet. 30:158-166. [DOI] [PubMed] [Google Scholar]
- 75.Patterson, C., Y. Wu, M.-E. Lee, J. D. DeVault, M. S. Runge, and E. Haber. 1997. Nuclear protein interactions with the human KDR/flk-1 promoter in vitro. J. Biol. Chem. 272:8410-8416. [DOI] [PubMed] [Google Scholar]
- 76.Pear, W. S., J. C. Aster, M. L. Scott, R. P. Hasserjian, B. Soffer, J. Sklar, and D. Baltimore. 1996. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J. Exp. Med. 183:2283-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Prioleau, M. N., P. Nony, M. Simpson, and G. Felsenfeld. 1999. An insulator element and condensed chromatin region separate the chicken beta-globin locus from an independently regulated erythroid-specific folate receptor gene. EMBO J. 18:4035-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rausscher, F. J., L. C. Sambucetti, T. Curran, R. J. Distel, and B. M. Spiegelman. 1988. Common DNA binding site for FOS protein complexes and transcription factor AP-1. Cell 52:471-480. [DOI] [PubMed] [Google Scholar]
- 79.Rossant, J., and L. Howard. 2002. Signaling pathways in vascular development. Annu. Rev. Cell Dev. Biol. 18:541-573. [DOI] [PubMed] [Google Scholar]
- 80.Sassone-Corsi, P., W. W. Lamph, M. Kamps, and I. M. Verma. 1988. Fos-associated cellular p39 is related to nuclear transcription factor AP-1. Cell 54:553-560. [DOI] [PubMed] [Google Scholar]
- 81.Schlaeger, T. M., S. Bartunkova, J. A. Lawitts, G. Teichmann, W. Risau, U. Duetsch, and T. N. Sato. 1997. Uniform vascular endothelial-cell-specific gene expression in both embryonic and adult transgenic mice. Proc. Natl. Acad. Sci. USA 94:3058-3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schorpp-Kistner, M., Z. Q. Wang, P. Angel, and E. F. Wagner. 1999. JunB is essential for mammalian placentation. EMBO J. 18:934-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schreiber, M., Z.-Q. Wang, W. Jochum, I. Fetka, C. Elliott, and E. F. Wagner. 2000. Placental vascularisation requires the AP-1 component Fra1. Development 127:4937-4948. [DOI] [PubMed] [Google Scholar]
- 84.Schroeter, E. H., J. A. Kisslinger, and R. Kopan. 1998. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature 393:382-386. [DOI] [PubMed] [Google Scholar]
- 85.Schubeler, D., C. Francastel, D. M. Cimbora, A. Reik, D. I. Martin, and M. Groudine. 2000. Nuclear localization and histone acetylation: a pathway for chromatin opening and transcriptional activation of the human beta-globin locus. Genes Dev. 14:940-950. [PMC free article] [PubMed] [Google Scholar]
- 86.Sekinger, E. A., and D. S. Gross. 2001. Silenced chromatin is permissive to activator binding and PIC recruitment. Cell 105:403-414. [DOI] [PubMed] [Google Scholar]
- 87.Selkoe, D., and R. Kopan. 2003. Notch and Presenilin: regulated intramembrane proteolysis links development and degeneration. Annu. Rev. Neurosci. 26:565-597. [DOI] [PubMed] [Google Scholar]
- 88.Shaulian, E., and M. Karin. 2002. AP-1 as a regulator of cell life and death. Nat. Cell Biol. 4:131-136. [DOI] [PubMed] [Google Scholar]
- 89.Shie, J.-L., G. Wu, J. Wu, F.-F. Liu, R. J. Laham, P. Oettgen, and J. Li. 2004. RTEF1, a novel transcriptional stimulator of vascular endothelial growth factor in hypoxic endothelial cells. J. Biol. Chem. 279:25010-25016. [DOI] [PubMed] [Google Scholar]
- 90.Suzuki, T., H. Okuno, T. Yoshida, T. Endo, H. Nishina, and H. Iba. 1991. Difference in transcriptional regulatory function between c-Fos and Fra-2. Nucleic Acids Res. 19:5537-5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Swiatek, P. J., C. E. Lindsell, F. F. del Amo, G. Weinmaster, and T. Gridley. 1994. Notch1 is essential for postimplantation development in mice. Genes Dev. 8:707-719. [DOI] [PubMed] [Google Scholar]
- 92.Taylor, K. L., A. M. Henderson, and C. C. Hughes. 2002. Notch activation during endothelial cell network formation in vitro targets the basic HLH transcription factor HESR-1 and downregulates VEGFR-2/KDR expression. Microvasc. Res. 64:372-383. [DOI] [PubMed] [Google Scholar]
- 93.Tsai, S. F., D. I. Martin, L. I. Zon, A. D. D'Andrea, G. G. Wong, and S. H. Orkin. 1989. Cloning of cDNA for the major DNA-binding protein of the erythroid lineage through expression in mammalian cells. Nature 339:446-451. [DOI] [PubMed] [Google Scholar]
- 94.Tse, C., T. Sera, A. P. Wolffe, and J. C. Hansen. 1998. Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol. Cell. Biol. 18:4629-4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Uyttendaele, H., V. Closson, G. Wu, F. Roux, G. Weinmaster, and J. Kitajewski. 2000. Notch4 and Jagged-1 induce microvessel differentiation of rat brain endothelial cells. Microvasc. Res. 60:91-103. [DOI] [PubMed] [Google Scholar]
- 96.Uyttendaele, H., J. Ho, J. Rossant, and J. Kitajewski. 2001. Vascular patterning defects associated with expression of activated Notch4 in embryonic endothelium. Proc. Natl. Acad. Sci. USA 98:5643-5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Uyttendaele, H., G. Marazzi, G. Wu, Q. Yan, D. Sassoon, and J. Kitajewski. 1996. Notch4/int-3, a mammary proto-oncogene, is an endothelial cell-specific mammalian Notch gene. Development 122:2251-2259. [DOI] [PubMed] [Google Scholar]
- 98.Vettese-Dadey, M., P. A. Grant, T. R. Hebbes, C. Crane-Robinson, C. D. Allis, and J. L. Workman. 1996. Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro. EMBO J. 15:2508-2518. [PMC free article] [PubMed] [Google Scholar]
- 99.Villa, N., L. Walker, C. E. Lindsell, J. Gasson, M. L. Iruela-Arispe, and G. Weinmaster. 2001. Vascular expression of Notch pathway receptors and ligands is restricted to arterial vessels. Mech. Dev. 108:161-164. [DOI] [PubMed] [Google Scholar]
- 100.Wallberg, A. E., K. Pedersen, U. Lendahl, and R. G. Roeder. 2002. p300 and PCAF act cooperatively to mediate transcriptional activation from chromatin templates by notch intracellular domains in vitro. Mol. Cell. Biol. 22:7812-7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Weijzen, S., P. Rizzo, M. Braid, R. Vaishnav, S. M. Jonkheer, A. Zlobin, B. A. Osborne, S. Gottipati, J. C. Aster, W. C. Hahn, M. Rudolf, K. Siziopikou, W. M. Kast, and L. Miele. 2002. Activation of Notch-1 signaling maintains the neoplastic phenotype in human Ras-transformed cells. Nat. Med. 8:979-986. [DOI] [PubMed] [Google Scholar]
- 102.Weinmaster, G., V. J. Roberts, and G. Lemke. 1991. A homolog of Drosophila Notch expressed during mammalian development. Development 113:199-205. [DOI] [PubMed] [Google Scholar]
- 103.Weinmaster, G., V. J. Roberts, and G. Lemke. 1992. Notch2: a second mammalian Notch gene. Development 116:931-941. [DOI] [PubMed] [Google Scholar]
- 104.Wu, Y., M. Moser, V. L. Bautch, and C. Patterson. 2003. HoxB5 is an upstream transcriptional switch for differentiation of the vascular endothelium from precursor cells. Mol. Cell. Biol. 23:5680-5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xiong, J.-W., A. Leahy, H.-H. Lee, and H. Stuhlmann. 1999. Vezf1: A Zn finger transcription factor restricted to endothelial cells and their precursors. Dev. Biol. 206:123-141. [DOI] [PubMed] [Google Scholar]
- 106.Xu, J. C., and E. Goldberg. 2000. Identification of a novel testis-specific leucine-rich protein in humans and mice. Biol. Reprod. 62:1278-1284. [DOI] [PubMed] [Google Scholar]
- 107.Xu, L., C. K. Glass, and M. G. Rosenfeld. 1999. Coactivator and corepressor complexes in nuclear receptor function. Curr. Opin. Genet. Dev. 9:140-147. [DOI] [PubMed] [Google Scholar]
- 108.Yamashita, K., D. J. Discher, J. Hu, N. H. Bishopric, and K. A. Webster. 2001. Molecular regulation of the endothelin-1 gene by hypoxia. J. Biol. Chem. 276:12645-12653. [DOI] [PubMed] [Google Scholar]
- 109.Yancopoulos, G., S. Davis, N. W. Gale, J. S. Rudge, S. J. Wiegand, and J. Holash. 2000. Vascular-specific growth factors and blood vessel formation. Nature 407:242-248. [DOI] [PubMed] [Google Scholar]
- 110.Yoshioka, K., T. Deng, M. Cavigelli, and M. Karin. 1995. Antitumor promotion by phenolic antioxidants: inhibition of AP-1 activity through induction of Fra expression. Proc. Natl. Acad. Sci. USA 92:4972-4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zaichuk, T. A., E. H. Shroff, R. Emmanuel, S. Filleur, T. Nelius, and O. Volpert. 2004. Nuclear factor of activated T-cells balances angiogenesis activation and inhibition. J. Exp. Med. 199:1513-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhou, S., M. Fujimuro, J. J. Hsieh, L. Chen, A. Miyamoto, G. Weinmaster, and S. D. Hayward. 2000. SKIP, a CBF1-associated protein, interacts with the ankyrin repeat domain of NotchIC to facilitate NotchIC function. Mol. Cell. Biol. 20:2400-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhou, S., and S. D. Hayward. 2001. Nuclear localization of CBF1 is regulated by interactions with the SMRT corepressor complex. Mol. Cell. Biol. 21:6222-6232. [DOI] [PMC free article] [PubMed] [Google Scholar]