Abstract
Myotonic dystrophy protein kinase (DMPK) is a Ser/Thr-type protein kinase with unknown function, originally identified as the product of the gene that is mutated by triplet repeat expansion in patients with myotonic dystrophy type 1 (DM1). Alternative splicing of DMPK transcripts results in multiple protein isoforms carrying distinct C termini. Here, we demonstrate by expressing individual DMPKs in various cell types, including C2C12 and DMPK−/− myoblast cells, that unique sequence arrangements in these tails control the specificity of anchoring into intracellular membranes. Mouse DMPK A and C were found to associate specifically with either the endoplasmic reticulum (ER) or the mitochondrial outer membrane, whereas the corresponding human DMPK A and C proteins both localized to mitochondria. Expression of mouse and human DMPK A—but not C—isoforms in mammalian cells caused clustering of ER or mitochondria. Membrane association of DMPK isoforms was resistant to alkaline conditions, and mutagenesis analysis showed that proper anchoring was differentially dependent on basic residues flanking putative transmembrane domains, demonstrating that DMPK tails form unique tail anchors. This work identifies DMPK as the first kinase in the class of tail-anchored proteins, with a possible role in organelle distribution and dynamics.
Myotonic dystrophy protein kinase (DMPK) was discovered more than a decade ago as the product of the gene that is altered by (CTG)n repeat expansion in patients with myotonic dystrophy type 1 (DM1) (6, 13, 31). Study of DMPK has thus far been aimed primarily at its normal physiological role, because the coding information of the DMPK gene remains unaltered in DM1 patients and disease-causing effects of mutation appear to act primarily at the RNA rather than at the protein level (28, 40). Bioinformatic, biochemical, and cell biological studies demonstrated that DMPK is an evolutionarily new protein found only in skeletal, cardiac, and smooth muscle and epithelial cells in mammals (reference 45 and our unpublished data).
The protein is related to Rho-kinase type protein kinases (14, 26) and belongs to the family of AGC-kinases, with myotonic dystrophy kinase-related Cdc42-binding kinase (MRCKα/β) (29) and human p160ROCK, rat ROKα, Caenorhabditis elegans LET-502, and murine citron Rho-interacting kinase (54) as its closest homologues. These kinases modulate the actin cytoskeleton by regulating myosin phosphatase activity or by directly phosphorylating the myosin regulatory light chain, thereby affecting stress fiber formation, smooth muscle contraction, and cytokinesis (2, 11, 39).
Recent findings revealed that DMPK may participate in a variety of cellular processes. As potential substrates for DMPK, phospholemman (34), the β-subunit of DHPR (51), MKBP (49), CUGBP/hNab50 (41), and the myosin phosphatase targeting subunit 1 (MYPT1) (35, 54) have been identified. The latter finding could point to a role for DMPK in cytoskeletal movement or intracellular transport dynamics, similar to the function of ROCK/Rho-kinase/ROK and MRCK in reorganization of the actin-based cytoskeleton, as effectors of RhoA and Cdc42, respectively. However, the precise site(s) of action and the relationship between DMPK expression and regulation of myosin phosphatase activity via MYPT phosphorylation (35) remain to be established.
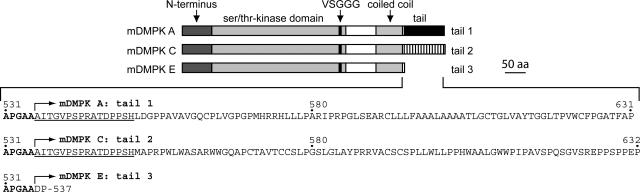
By prediction from gene structure data and by direct mRNA expression studies, it was shown, by us and others, that multiple DMPK isoforms arise in both humans and mice by alternative splicing (15, 20, 52). DMPK isoforms have a leucine-rich N terminus, a serine/threonine-type protein kinase domain, and a coiled-coil region in common but vary in the presence or absence of a VSGGG amino acid sequence and in the nature of the C terminus. Recently, the VSGGG motif was found to be involved in mDMPK autophosphorylation and to be a determinant of in-gel migration behavior and perhaps folding configuration (54). The nature of the C terminus depends on the mode of splicing, leading to expression of DMPK isoforms with long tails predominant in the heart, skeletal muscle, and brain and DMPKs with a 2-amino-acid tail in smooth muscle (15, 20) (Fig. 1).
FIG. 1.
Mouse DMPK splice isoforms have distinct C termini. The domain organization and C-terminal amino acid sequences of mDMPK splice isoforms A, C, and E are shown. The N terminus, catalytic kinase domain, VSGGG segment, coiled-coil domain, and tail segments are indicated. Amino acids common to all mDMPK isoforms are shown in bold, and residues identical in tail 1 and tail 2 are underlined.
Previous studies have localized hDMPK to the neuromuscular junction in skeletal muscle and intercalated disks in cardiac muscle (30, 38). However, in this work, mixtures of coexpressed instead of individual DMPK isoforms were studied, because antibodies were used that recognize epitopes shared between the different coexpressed DMPK isoforms and with other members of the family of Rho kinases (26).
With the ultimate goal of understanding the biological function of DMPK, we focused in this study on a comparison between the subcellular localization of individual DMPK isoforms of mice and humans. We report that different DMPK splice forms either are associated with the endoplasmic reticulum (ER) or mitochondria or are cytosolic and that orthologous splice isoforms of humans and mice do not behave similarly. Use of confocal laser scanning microscopy and immunoelectron microscopy (immuno EM) in combination with transient transfection and mutation analysis of individual DMPK cDNAs in various cell types revealed new information on the sequence and structure of targeting signals required for localization to the mitochondrial outer membrane (MOM) or the ER. Furthermore, we report that anchoring of human DMPK A into the MOM affects mitochondrial distribution whereas mouse DMPK A clusters the ER. Based on our observations, we classify DMPK as the first kinase member of a special class of membrane proteins, termed tail-anchored (TA) proteins. DMPK binding to membranes, with a specificity that is divergent between mice and humans, could be a mechanism for regulation of organellar fusion and transport and mitochondrial physiology.
MATERIALS AND METHODS
Cell culture and DNA transfection.
COS-1 cells were cultured and transfected using the DEAE-dextran procedure as described previously (15). Neuro-2A (N2A), NIH 3T3, HeLa, and C2C12 cells were grown to subconfluence in Dulbecco minimal essential medium supplemented with 10% fetal calf serum and maintained at 37°C under a 5% CO2 atmosphere. N2A, NIH 3T3, HeLa, and C2C12 cells were transiently transfected with various expression plasmids by using Lipofectamine (Invitrogen) as specified by the manufacturer and maintained in culture for an additional 24 h prior to analysis. Alternatively, we used adenovirus vector-based DNA transduction for expression of single mDMPK isoforms in immortalized DMPK−/− myoblasts, which were grown at 33°C on Matrigel (BD Biosciences)-coated dishes in Dulbecco minimal essential medium supplemented with 20% fetal calf serum, 60 μg of gentamicin per ml, and 10 U of gamma interferon per ml. This myogenic cell line was derived from DMPK−/− knockout mice harboring the H-2Kb-tsA58 allele as described previously (33).
Plasmid and adenovirus vectors for DMPK expression and site-directed mutagenesis.
HA-tagged mDMPK A, C, and E cDNAs were generated in pSG8ΔEco expression vectors (15). Various pEYFP/CFP-DMPK expression vectors and mutation and deletion constructs were obtained by cloning the appropriate PCR fragments into restriction sites of pEYFP-C1 or pECFP-C1 (Clontech) (see online supplemental information for details). All expression plasmids contained the proper mouse or human DMPK 3′ untranslated region. The sequences of all fragments obtained by PCR were verified by DNA sequencing. The ER was visualized using plasmid vector pEGFP-ER, which encodes enhanced green fluorescent protein (EGFP) with a calreticulin-derived ER targeting signal at its N terminus and a KDEL ER retention signal at its C terminus (Clontech). To mark mitochondria for live-cell imaging (see below), cells were labeled with a mitochondrial selective probe, Rhodamine 123 (Molecular Probes), at a final concentration of 10 μg/ml. The Golgi was visualized by transfection of plasmid pEGFP-Golgi, which encodes an EGFP C-terminally fused to the cytoplasmic, transmembrane, and stalk regions of human N-acetylglucosaminyl transferase I, a known Golgi marker protein (47). E1/E3-deleted serotype 5 adenovirus vectors encoding yellow fluorescent protein (YFP)-mDMPK isoforms A, C, or E under the control of a cytomegalovirus immediate-early promoter were generated using the AdEasy Vector System (17) as described previously (9).
Immunofluorescence microscopy.
For immunofluorescence microscopy, cells were grown and transfected on glass coverslips. The cells were fixed in phosphate-buffered saline containing 2% formaldehyde ∼24 h after transfection and permeabilized in phosphate-buffered saline containing 0.2% NP-40. Samples were processed for immunofluorescence microscopy using standard procedures. A rabbit anti-cytochrome c oxidase antibody was used to visualize mitochondria. Mouse monoclonal antibody (mAb) 12CA5 was used to detect the HA epitope tag, and mAb 414 was used as a nuclear envelope (NE) marker (57). Images of fixed cells were obtained with a Bio-Rad MRC1024 confocal laser-scanning microscope equipped with an argon/krypton laser, using a 60× 1.4 NA oil objective and LaserSharp2000 acquisition software. Images were further processed with Adobe Photoshop 7.0.
Live-cell imaging.
N2A cells were grown and transfected with pEYFP-mDMPK A or pECFP-mDMPK C together with pEGFP-ER or were labeled with Rhodamine 123 in 35-mm glass-bottom dishes (Willco Wells BV). They were washed twice in OptiMEM-I (Life Technologies), and live cells were imaged while placed on a temperature-controlled stage of a Zeiss LSM510-Meta confocal microscope (running software release 3.2) using the appropriate argon laser lines and a 63× 1.4 NA oil objective. In the case of overlapping GFP and cyan fluorescent protein signals, the metadetector in conjunction with the linear unmixing option in the software was used to generate the final images.
Immuno-EM.
N2A cells were grown and transfected with plasmids YFP-mDMPK A, C or E. They were fixed for 2 h at room temperature in 100 mM sodium phosphate buffer (pH 7.0) containing 1% formaldehyde and 0.1% glutaraldehyde. Next, they were pelleted in 10% gelatin and postfixed in 1% formaldehyde for another 24 h. Ultrathin cryosections were prepared on a Leica EMFCS, incubated with polyclonal antiserum raised against EGFP, and then incubated with protein A complexed to 10-nm-diameter gold particles by standard procedures (12). Sections were observed in a JEOL 1010 electron microscope operating at 80 kV.
SDS-PAGE and Western blotting.
Transfected COS-1 cells were lysed in RIPA buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 1 mM phenylmethylsulfonyl fluoride, 10 mM NaF) and centrifuged for 10 min at 14,000 × g. Cell lysates were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) (8 or 12% polyacrylamide) and transferred to nitrocellulose membranes (Amersham Pharmacia Biotech) for immunodetection using B79, a DMPK-specific antibody (15), or anti-EGFP, followed by horseradish peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories). Immunodetection was performed by enhanced chemiluminescence and exposure to film (Kodak X-OMAT AR).
Membrane isolation and extraction.
N2A cells were grown in 100-mm-diameter culture dishes and transfected with pSG8ΔEco-mDMPK expression plasmids. They were scraped into ice-cold 10% TES buffer (20 mM Tris-HCl [pH 7.5], 1 mM EDTA, 100 mM NaCl) plus 1 mM phenylmethylsulfonyl fluoride and a protease inhibitor cocktail (Roche), pelleted by centrifugation for 10 min at 3,000 × g, resuspended in 10% TES buffer, and incubated on ice for 15 min. Cell lysates were made using a glass-glass Dounce homogenizer and were cleared from intact cells and nuclei by centrifugation for 10 min at 3,000 × g. An input sample was collected from this supernatant for analysis by SDS-PAGE. Total membranes were collected using a micro-ultracentrifuge (Sorvall RC M150 GX) at 100,000 × g for 30 min in a S55S rotor, and the supernatant fraction was collected (designated S1) for SDS-PAGE analysis. Membranes obtained were extracted on ice in (i) PBS, (ii) 1 M NaCl in PBS, (iii) 100 mM Na2CO3 (pH 11), or (iv) 1% Triton X-100 in PBS and centrifuged for 30 min at 100,000 × g. Subsequent pellet (P) and supernatant (S2) fractions were analyzed by SDS-PAGE followed by Western blotting using B79.
Supplemental material.
Supplemental material is available online. Information includes supplemental Materials and Methods, legends to supplemental figures, and expression characterization of DMPK mutants used in this work (Fig. S1), NE immunolocalization of YFP-mDMPK A mutants (Fig. S2), real-time analysis of YFP-mDMPK A associated with the ER (movies Fig. S3A and S3B), real-time analysis of CFP-mDMPK C associated with mitochondria (movie Fig. S4), comparative analysis of mouse and human DMPK C-termini (Fig. S5; Table S1), and expression of hDMPK A and C in C2C12 cells (Fig. S6).
RESULTS
The DMPK gene specifies different gene products.
DMPK splice forms A and C carry distinct C termini, here referred to as tail 1 and tail 2, respectively, which are 96 and 95 amino acids in length and are identical only in the N-terminal 16 amino acids (encoded by exon 13) (Fig. 1) (for details, see reference 15). The short isoform DMPK E is distinctly different and carries a short, truncated tail of 2 amino acids (designated tail 3). We previously reported that the splice modes leading to this C-terminal variation have been conserved between mice and humans and we determined the subcellular distribution of DMPK isoforms (54). Since this distribution was originally studied in ectopic host cells and since neither the conditions nor the exact mechanism for this targeted accumulation are well understood, we decided to focus first on the localization of individual splice products in authentic host cells. C2C12 myoblasts and immortalized myoblasts derived from DMPK−/− knockout mice harboring the H-2Kb-tsA58 transgene (33) were used for this purpose.
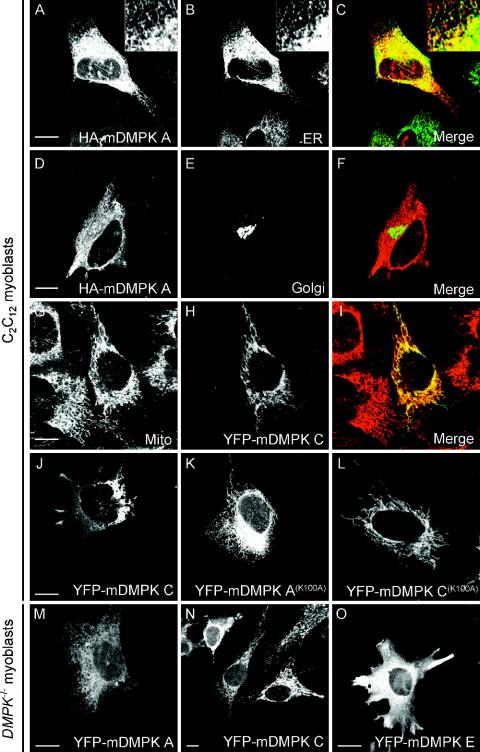
To analyze the specific distribution behavior of single isoforms, DMPKs were expressed as HA- or YFP-tagged versions from plasmid or adenovirus vectors and their localization was compared to that of various organellar markers by immunofluorescence microscopy. As shown in Fig. 2A and B, reticular structures revealed by staining of HA-tagged mDMPK A in C2C12 cells overlapped with the ER network stained by transient expression of the GFP-ER marker, containing the calreticulin ER targeting signal and KDEL ER retention signal. Since mDMPK A was confined solely to ER contiguous membranes (Fig. 2C), we tested colocalization with MAb414, a nuclear envelope (NE) marker, directed against the nuclear pore complex (57). Also, this marker colocalized (see Fig. S2A to C in the supplemental material) showing that mDMPK A decorates all membranes of the ER, since the NE is a subdomain of, and continuous with, the ER (53). The intensely stained area observed close to the nucleus did not show any appreciable colocalization with a GFP-Golgi marker containing the transmembrane and stalk region of human N-acetylglucosaminyl transferase I (47), suggesting that tail 1 is specific for ER and NE membranes and not for membranes in the Golgi apparatus (Fig. 2D to F; also see Fig. S2A to C in the supplemental material).
FIG. 2.
Mouse DMPK A and C localize to the ER or mitochondria. To assign subcellular DMPK location, confocal images were taken of fluorescent products in C2C12 myoblast cells 24 h after cotransfection with HA-tagged or YFP-tagged mDMPK isoforms A and C and various organellar markers. (A to C) HA-mDMPK A colocalized with a GFP-ER marker containing the calreticulin ER-targeting signal and the KDEL ER retention signal. The insert shows an enlargement of the peripheral ER network. (D to F) Fluorescent visualization of cotransfected HA-mDMPK A (D) and a GFP-Golgi (E) marker shows that mDMPK A is not targeted to the Golgi. (G to I) YFP-mDMPK C localized to mitochondria, as shown by staining with an anti-cytochrome c oxidase antibody. (J) Strong overexpression of YFP-mDMPK C leads to a cytosolic location in addition to perinuclear mitochondrial clustering. (K and L) Localization of kinase-dead mutants YFP-mDMPK A(K100A) and C(K100A) to the ER and mitochondria, respectively. (M and N) Adenovirus gene transfer of YFP-mDMPK A and C into DMPK−/− myoblasts result in ER and mitochondrial localization, respectively. (O) Expression of YFP-mDMPK E in DMPK−/− myoblasts gives a cytosolic distribution. Bars, 10 μm.
In contrast, YFP-mDMPK C in C2C12 cells was completely colocalized with the mitochondrial marker cytochrome c oxidase but not with the other organellar markers (Fig. 2G to I and data not shown). We noticed that in some cells weak YFP-mDMPK C staining also appeared at cytosolic locations. In these cases, mitochondria often appeared clustered in the perinuclear region (Fig. 2J) (see below).
To test whether the ER and mitochondrial localization of mDMPK A and C was uniquely dependent on the sequence arrangement in their tails and independent of intrinsic kinase activity, we tagged kinase-dead variants of mDMPK isoforms. The YFP-mDMPK A(K100A) and C(K100A) distributions at ER and mitochondria, respectively, were identical to those of their kinase-active counterparts (Fig. 2K and L).
Next, we tested if partitioning of DMPKs occurred similarly in myoblast host cells that were rendered completely DMPK deficient (19). Although these myoblasts lacked the normal mixture of endogenous DMPK isoforms completely, we expected them to provide an otherwise fully authentic host environment, closely mimicking the situation in vivo. When individual YFP-tagged mDMPK isoforms A or C were expressed by adenovirus gene delivery, the typical and selective targeting to either ER or mitochondria was observed (Fig. 2M and N). Finally, we also tested other cell types with a zero or very low level of endogenous DMPK gene expression and hence “ectopic” host infrastructure, such as N2A and fibroblast (NIH 3T3) cells, and obtained identical results (data not shown). Since all these cell types were of mouse origin, we also studied mDMPKs in human HeLa cells and again found distributions at exact cognate locations (see below). In all cell types tested, the third shorter isoform, mDMPK E, adopted a diffuse cytosolic distribution (Fig. 2O and data not shown). Based on these findings, we studied other aspects of DMPK location mainly in one cell type, choosing N2A as the most amendable cell line for live cell microscopy, video imaging, and transfection analysis.
The ER and mitochondria are dynamic structures, which continuously move and rearrange shape by transport and fusion and fission events (10, 53). To evaluate the role or fate of mDMPK A and C in these events, we monitored the localization of YFP- or CFP-tagged mDMPK isoforms over time in transiently transfected N2A cells and made parallel comparisons to the behavior of otherwise stained organellar markers. Alterations of the ER network in YFP-mDMPK A-transfected cells were observed by collecting 2-s-interval time frames (see Fig. S3A DMPK-A.mov in the supplemental material) and were similar to alterations observed with a GFP-ER marker (see Fig. S3B GFP-ER.mov in the supplemental material). Images of CFP-mDMPK C, collected with a 10-s interval, reproduced the typical mobility of mitochondria (see Fig. S4 DMPK-C.mov in the supplemental material), as was observed in colocalization with Rhodamine 123-labeled mitochondria. Live-cell imaging thus corroborated the association of mDMPK with ER and mitochondria, each having their own typical dynamics.
Ultrastructural localization of mDMPK isoforms.
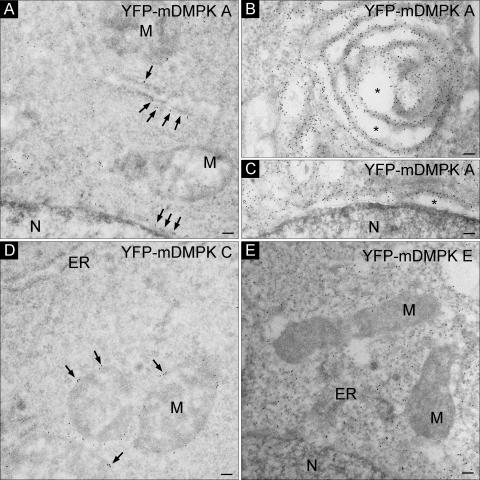
Next, we resolved the localization of mDMPK isoforms at the ultrastructural level by performing immuno-EM using an anti-GFP antiserum on YFP-mDMPK-transfected cells. This analysis showed that mDMPK A was localized exclusively (>95% of the gold particles) to the cytosolic side of the ER and NE (Fig. 3A). The mitochondria (Fig. 3A) and Golgi complex (not shown) were unlabeled. High levels of expression of ER-resident proteins can lead to the formation of stacked ER membrane structures, collectively called organized smooth ER (OSER), as was recently demonstrated for the TA protein sequence of cytochrome b5, Sec61β, and Sec61γ (48). Also in multiple mDMPK A-overexpressing cells, but never with other mDMPK isoforms, the formation of comparable parallel or circular membrane stacks was detected (Fig. 3B). The newly induced membrane structures, as well as the NE, appeared dilated and typical for strongly expressing cells (asterisks Fig. 3B and C).
FIG. 3.
Mouse DMPK A and C localize to ER membranes or the MOM. Immuno-EM of YFP-mDMPKs A, C, or E in transiently transfected N2A cells visualized with an anti-GFP antibody followed by incubation with protein A complexed to 10-nm gold particles is shown (A). YFP-mDMPK A localized to ER and NE structures. (B and C) Overexpression caused abnormal ER membrane stacks (B) next to dilated ER and NE structures (B and C, asterisks). (D and E) YFP-mDMPK C localized exclusively to the MOM (D), whereas a cytosolic localization was observed for YFP-mDMPK E (E). M, mitochondrion; N, nucleus. Bars, 100 nm.
The mDMPK C isoform was specifically associated with mitochondria. Over 95% of the gold label was found at the MOM and not at the ER/NE (Fig. 3D and data not shown). For mDMPK E, no association with particular (membrane) structures was observed (Fig. 3E). Mouse DMPK E signal could be observed only in cells fixed with a strong cross-linker (i.e., glutaraldehyde) in the fixation mixture. Combined, these data show that isoforms lacking a C-terminal tail are freely diffusible cytosolic proteins whereas the presence of a tail is involved in ER or MOM association.
Mouse DMPK A and C are tail-anchored kinases.
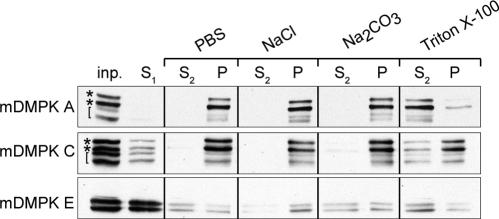
Cell fractionation followed by chemical extraction procedures was used to study the mode of mDMPK membrane association. To assess binding, different buffer and solvent conditions were used to extract mDMPK from the total membrane fraction. As shown in Fig. 4, association of mDMPK A and C with membranes, i.e., their presence in the pellet fraction, was resistant to high salt conditions and alkaline sodium carbonate, indicating that these proteins are integrated into their target membranes. Only Triton X-100 extraction was able to release mDMPK A and C from the membrane fraction, shifting it into the soluble fraction. Previous studies have shown that mDMPK A, C, and E display doublet bands on immunoblots associated with autophosphorylation (Fig. 4, asterisks) (15, 54). In addition, we observed that certain lysis and buffer conditions induced nonenzymatic clipping of mDMPK with a long C terminus (Fig. 4, brackets; compare mDMPK A and C with mDMPK E), as occurred here also. Under the conditions used, the majority of mDMPK E was present in the first supernatant fraction (S1), confirming that its microscopic classification as a cytosolic protein was correct.
FIG. 4.
Mouse DMPK A and C are tail-anchored proteins. Western blots of pellet (P) and supernatant (S1 and S2) fractions of N2A cells transfected with mDMPK A, C, or E after phosphate-buffered saline (PBS), high-salt, alkaline Na2CO3, or Triton X-100 extraction of 100,000 × g pellets. N2A cells were grown, transfected, and fractionated into supernatant and pellet fractions before extraction as described in Materials and Methods. DMPK was visualized with an anti-DMPK antibody. inp., input; S1/S2 and P, supernatant and pellet fractions after 100,000 × g centrifugation. Asterisks mark doublet DMPK signals, and brackets indicate C-terminally truncated mDMPK products as a result of nonenzymatic breakdown (see the text).
The observation that mDMPK A and C are strongly associated with membranes via their C termini places them in a protein family classified as C-terminally anchored or TA proteins, a recently discovered group of proteins which anchor preferentially in membranes of the ER or MOM (reference 4 and references therein). This notion prompted us to analyze in more detail the amino acid composition of tails 1 and 2, with emphasis on their overall hydrophobicity and positions of known relevant amino acid residues. Within tail 1, a long stretch of hydrophobic amino acids is present between positions 593 and 631. Computer analysis predicted the presence of a putative transmembrane domain (TMD) between positions 593 and 616 (Fig. 5A). Tail 2 is more hydrophilic, and computer analysis predicted a low probability for the presence of a TMD (Fig. 5A). We did identify a stretch of amino acids of moderate hydrophobicity, excluding any charged amino acids, between positions 597 and 616. Next, C termini of TA proteins known to bind membranes of the ER or MOM were compared with mDMPK tail sequences (Fig. 5B). One evident similarity between mDMPK tails and ER/MOM targeting sequences of other TA proteins is that basic residues (i.e., lysine and arginine) are highly prevalent in the regions flanking the 17- to 23-residue long hydrophobic stretches. Since no significant other similarity emerged from this comparison, these results suggest that amino acid content as well as amino acid distribution specifies mDMPK anchoring.
FIG. 5.
Mouse DMPK tail 1 and 2 anchors have different sequence arrangements and hydrophobicity profiles. (A) Hydrophobicity plots of mDMPK tails 1 and 2. The average hydrophobicity of the assigned TMD (25) is indicated. (B) Comparison of DMPK tail 1 and 2 sequences with ER and MOM targeting sequences of known tail-anchored proteins. Putative TMDs are boxed, and basic residues are shown in bold. Amino acid numbering for mouse and human DMPK isoforms is indicated (see also Fig. 1A).
A hydrophobic domain in mDMPK tail 1 is responsible for ER/NE targeting.
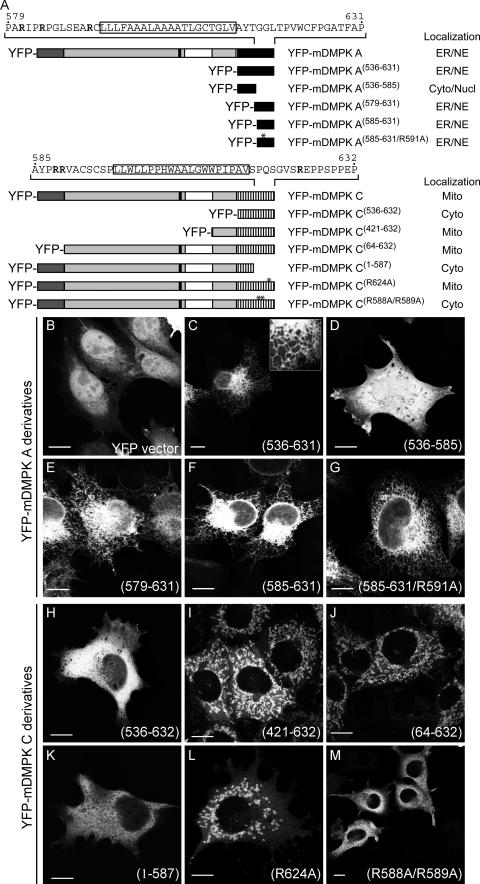
Previous studies using mutagenesis approaches have shown that the nature of the TMD and positioning of flanking lysine and arginine residues determine ER or MOM localization of TA proteins (5, 18, 21). Therefore, we examined which particular amino acid residues in tails 1 and 2 were responsible for ER and MOM targeting and whether basic residues contributed to the targeting specificity of mDMPK. Deletion mutants of mDMPK A and C or the separate tails were fused to YFP, and the subcellular localization of the chimeric proteins was examined during transient expression in N2A cells (Fig. 6A). Proper expression of YFP fusions was assessed via immunoblotting (see Fig. S1 in the supplemental material). Tail 1 alone was sufficient to target YFP, normally mainly a cytosolic/nuclear protein (Fig. 6B), to the ER (Fig. 6C) and the NE (see Fig. S2D to F in the supplemental material). Analysis of additional mutants showed that all information was present in amino acids 579 to 631 (Fig. 6D and E; see Fig. S2G to I in the supplemental material). This confirmed that, as predicted from comparison with other TA proteins, the ER targeting signal is contained within the ultimate C-terminal part of tail 1. Involvement of basic residues was investigated by further truncating tail 1 to amino acids 585 to 631, thereby removing R581 and R584 but leaving R591 in place. Fusion protein YFP-mDMPK A(585-631) still localized to ER networks (Fig. 6F) as well as to the NE (see Fig. S2J to L in the supplemental material), indicating that R581 and R584 are not required for ER targeting. Also, an additional R591A mutation did not affect the ER/NE association (Fig. 6G; see Fig. S2M to O in the supplemental material). This supports the idea that positively charged amino acids are not required for proper ER targeting of tail 1. Hence, the ER targeting signal for mDMPK A is confined within the ultimate 40 amino acids of tail 1, encompassing a putative TMD.
FIG.6.
Involvement of basic residues in tail anchors in localization of mDMPK A and C. (A) Schematic representation of mDMPK A and C deletion constructs and their subcellular localization: ER, NE (nuclear envelope), Cyto (cytosol), Nucl (nucleus), Mito (mitochondrial). (B to M) Confocal images of fluorescent YFP-tagged DMPK A or C deletion mutants in N2A cells. Transfected N2A cells expressing unmodified YFP were used as the control (B). ER localization of mDMPK A deletion constructs was found for tail 1 alone (YFP-mDMPK A(536-631)) (C) and YFP-mDMPK A(579-631) (E), YFP-mDMPK A(585-631) (F), and YFP-mDMPK A(585-631/R591A) (G), but not for YFP-mDMPK A(536-585) (D). Tail 2 of mDMPK C alone did not target to mitochondria (YFP-mDMPK C(536-632)) (H), whereas YFP-mDMPK C(421-632) (I) and YFP-mDMPK C(64-632) (J) yielded a mitochondrial localization. The C-terminal 45 amino acids are essential for mitochondrial localization (YFP-mDMPK C(1-587) (K) and also the basic residues therein, since expression of YFP-mDMPK C(R624A) results in rounded and fragmented mitochondria (L) whereas YFP-mDMPK C(R588A/R589A) localizes to the cytosol (M). Bars, 10 μm.
Mitochondrial targeting by mDMPK tail 2 requires basic residues flanking a putative TMD and presence of the coiled-coil domain.
We also examined requirements for mDMPK C anchoring in more detail. Unlike for mDMPK A, just the tail region of mDMPK C appeared insufficient for targeting YFP to mitochondria, since a YFP-mDMPK C(536-632) fusion protein was found in the cytosol and to some extent also in the nucleus (Fig. 6H). When the coiled-coil domain was included, as in fusion protein YFP-mDMPK C(421-632), mitochondrial localization was nearly completely restored (Fig. 6I). To determine whether a leucine zipper motif in the mDMPK N terminus participated in the binding of DMPK C to mitochondria, this domain was deleted. Its absence did not affect normal mitochondrial distribution (Fig. 6J). Although this does not necessarily exclude any targeting role for the motif, it makes a direct role in mitochondrial targeting unlikely. In contrast, the C-terminal 45-amino-acid domain plays a dominant role, since removal of the polypeptide stretch between positions 588 and 632 resulted in a cytosolic localization (Fig. 6K). Thus, the targeting signal of mDMPK C must be contained within the C-terminal ∼45 amino acids, similar to the situation in mDMPK A. When R624 positioned at the C-terminal side of the putative TMD in tail 2 was mutated to alanine, a small but significant fraction of the YFP-tagged protein shifted to a location in the cytosol, and the mitochondria to which the majority of protein still bound appeared rounded and fragmented (Fig. 6L). This suggests that R624 plays an important role in determining targeting specificity for the MOM (see also below). Finally, introduction of mutations R588A and R589A completely abolished mitochondrial localization (Fig. 6M), indicating that positively charged residues preceding the TMD are important in the targeting signal of tail 2. Taken together, our results demonstrate that MOM targeting by mDMPK tail 2 requires the presence of the coiled-coil domain as well as critical basic residues flanking the putative TMD in the 45-amino-acid tail domain.
The human DMPK A orthologue associates with mitochondria instead of the ER and causes mitochondrial clustering.
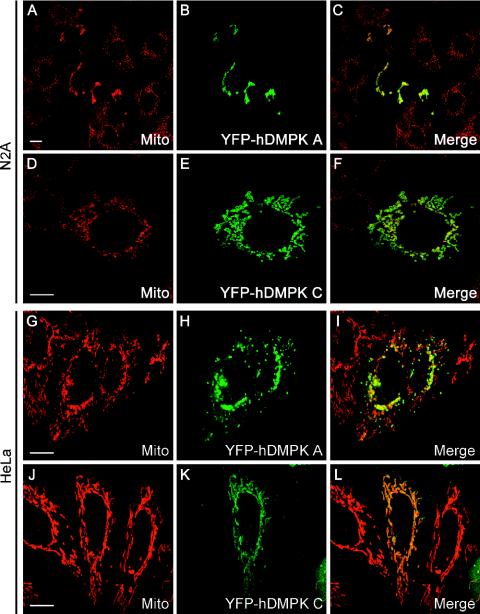
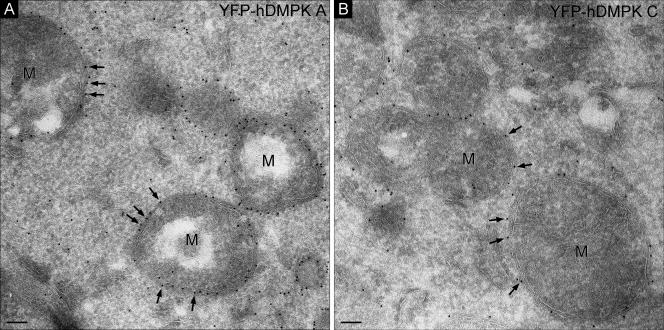
Although the amino acid sequences of DMPK isoforms are highly homologous between humans and mice (15), we found it conspicuous that the percent sequence identity and similarity for the C termini fell far below the overall values for the whole tail region (see Table S1 in the supplemental material). Between the two species, strong differences exist mainly in the presence and arrangement of essential basic residues, in particular for tail 1. The positioning of basic residues in the human sequence suggested that hDMPK A would localize to mitochondria and not to the ER, like mDMPK A (see Fig. S5 in the supplemental material). To test this hypothesis, we compared the localization of YFP-hDMPK A and C with that of their mouse orthologues (Fig. 7). In our analyses, we included HeLa cells in order to exclude cross-species effects caused by membrane composition in combination with features in the DMPK sequences. As anticipated, we observed that not only hDMPK C but now also hDMPK A localized to mitochondria in both N2A and HeLa cells (Fig. 7). Again, the subcellular localization of mDMPK A and C was independent of the species origin of the recipient cell line (data not shown). Immuno-EM confirmed that both hDMPK isoforms were indeed localized exclusively (>95% of the gold label) at the MOM (Fig. 8).
FIG. 7.
Human DMPK A and C localize to mitochondria and alter mitochondrial distribution. Confocal fluorescence microscopy analysis of YFP-hDMPK A and C in transiently transfected N2A (A to F) and HeLa (G to L) cells. YFP-hDMPK A localized at mitochondria, stained by an anti-cytochrome c oxidase marker, which accumulated in the nuclear periphery. YFP-hDMPK C also localized to mitochondria (D to F and J to L) but did not show the accumulation near the nucleus as observed for hDMPK A (B and H). Note that mitochondria devoid of hDMPK A remain unclustered (H). Bars, 10 μm.
FIG. 8.
Human DMPK A and C both localize to the MOM. Immuno-EM of YFP-hDMPK A (A) and C (B) in transfected N2A cells show localization of human DMPK isoforms to the MOM. Procedures for transfection, immunogold labeling, and imaging are as described in the legend to Fig. 3 and in Materials and Methods. M, mitochondrion. Bars, 100 nm.
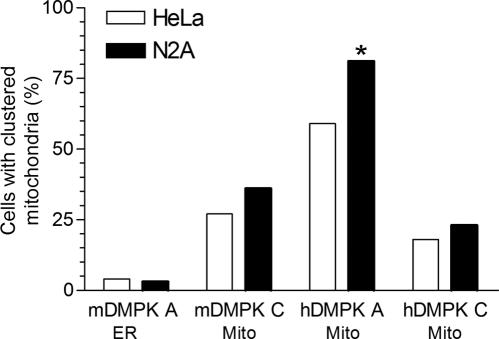
Even more strikingly, we observed a dramatic effect of hDMPK A expression on the morphology and distribution of mitochondria throughout the cytoplasm. In the majority of YFP-hDMPK A-transfected cells, mitochondrial clustering occurred in the perinuclear region (Fig. 7A to C and G to I and Fig. 9). Importantly, this was found exclusively for hDMPK A-decorated mitochondria and was also observed in C2C12 myoblasts (Fig. 7I; see Fig. S6 in the supplemental material). Clustering of mitochondria was not seen in cells in which the mDMPK A isoform was expressed, although here aggregated ER structures were frequently observed (Fig. 3B and 9). Moreover, comparison of cells with high and low hDMPK A expression revealed that mitochondrial aggregation occurred seemingly independently of the expression level (data not shown). These observations suggest that the mode of hDMPK A binding to the target organelle rather than overexpression per se is involved in the organellar redistribution. Further analyses also revealed that many transiently transfected cells showed extensive blebbing and a high incidence of cell death. This could be a consequence of the anomalous distribution of mitochondria, associated with as yet unknown effects on cell physiology. It is of note here that relatively mild mitochondrial clustering was observed in cells expressing human or mouse DMPK C (Fig. 7D to F and J to L and Fig. 9), but increased cell death was never observed in these cells. Taken together, these data show that DMPK isoforms have a localized function and that mitochondrial isoforms can differentially influence mitochondrial characteristics.
FIG. 9.
Human DMPK A promotes mitochondrial clustering at perinuclear sites. The fraction of N2A and HeLa cells containing clustered mitochondria was determined after transient transfection with vectors encoding mouse or human DMPK isoforms A and C. Mitochondria were regarded as clustered when virtually all cytochrome c oxidase-positive mitochondria aggregated near the nucleus. The extent of clustering induced by hDMPK A in N2A cells is probably underestimated because many transfected cells showed an apoptosis-like appearance and therefore were excluded from the assay (asterisk). A minimum of 100 cells was analyzed for each cell type and each DMPK expression vector. Subcellular localization of DMPK isoforms is indicated.
DISCUSSION
In the present study, we analyzed parameters involved in the subcellular distribution of protein products of DMPK, the gene mutated in DM1 (16). We demonstrate that distinct mouse DMPK splice isoforms specifically associate with either the ER membrane or the MOM, guided by unique sequence information in their C-terminal tail. Human DMPK forms show no divergence in localization and target to mitochondria only. To our knowledge, DMPK is the first protein kinase that can be classified into the recently established group of TA proteins (reference 4 and references therein).
DMPK is a tail-anchored serine/threonine protein kinase.
TA proteins constitute a group of proteins that specifically insert into intracellular membranes, using a single membrane-spanning region located close to the COOH terminus (reference 4 and references therein). Similar to what was found for the location preference of most tail-anchored proteins (4, 55), mDMPK localization is confined to either membranes of the ER or NE (for tail 1) or to the MOM (for tail 2). Since no appreciable colocalization with Golgi markers or the plasma membrane was observed for mDMPK A, we conclude that the protein, after translation and insertion into the ER membrane, is not further sorted into the vesicular transport route. How this is regulated, via lipids or through active retention, is at present unknown.
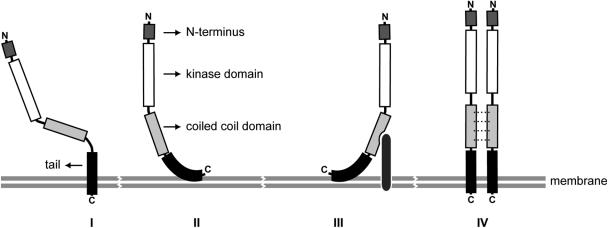
Anchoring of mDMPK A and C into their target membranes must be strong, since they cannot be liberated by high-salt or alkaline sodium carbonate treatment. Since the membrane anchors of mDMPK A and C are positioned within the last 40 C-terminal amino acids, we consider it most likely that membrane insertion occurs posttranslationally in an Nout-Cin orientation. Exactly how mDMPK is anchored in the membrane is not clear, however, and hypothetical models in which mDMPK tails span the membrane or attach to the lipid bilayer are still open (Fig. 10, modes I and II). Further mechanistic studies are therefore required to decide between possible modes of membrane association.
FIG. 10.
Hypothetical modes of membrane association of DMPK isoforms. A schematic representation of the putative association of DMPK isoforms with their target membrane is shown. C-terminal DMPK anchors can either pass the membrane entirely (mode I) or give close surface binding to the outer leaflet of the lipid bilayer (mode II). A requirement of the coiled-coil domain for proper MOM localization of mDMPK C suggests that either heterologous protein interactions may stabilize mDMPK localization at the membrane (mode III) or multimerization of DMPK via the coiled-coil domain may promote binding specificity and/or strength (mode IV). Possible intramolecular effects of proximity of the coiled-coil segment on folding of the hydrophobic domain can also be envisaged but are not included in the figure.
Structural features of ER- and MOM-targeting signals of DMPK isoforms.
Little is known about the molecular machinery and steps involved in partitioning of TA proteins into the ER or MOM, except that conventional import pathways do not seem to apply (3, 4). A specific role for the signal recognition particle for targeting of TA proteins to the ER was reported (1), but detailed insight in the mechanism of membrane insertion is currently lacking. Data from various studies suggest that shared structural elements in C termini reflect the only critical features implicated in membrane selectivity. To discriminate between various mechanistic hypotheses, we started here by making a comparison between elements in the tail sequences of DMPK A and C and other members of the group of TA proteins. Length and hydrophobicity of the TMD, together with flanking basic amino acids, have been proposed as the most crucial determinants of membrane selectivity (5, 18, 21, 24). Positively charged amino acids preceding the TMD may be essential for ER localization, as demonstrated for VAMP-1A, VAMP-2, and VAMP-8 (22). We observed here, however, that an mDMPK A mutant lacking all three basic residues preceding the TMD remained associated with ER and NE membranes. We can exclude compensatory replacement effects of amino acids placed immediately upstream of the TMD in the tail since all basic residues derived from the C-terminus of YFP and a short stretch of amino acids specified by the multiple-cloning site in the vector were removed (see the supplemental information). Previous reports showed that a reduction in the overall positive charge C-terminally of the TMD results in higher affinity for ER membranes (18, 24). The fact that amino acids following the TMD in the mDMPK A tail are all neutral is consistent with this observation and may determine binding strength. Taking these data together, we conclude that it is the strong hydrophobic character of the mDMPK A TMD (average hydrophobicity of 2.1) that constitutes the ER and NE targeting signal. Our findings are compatible with other studies showing that artificially engineered stretches of hydrophobic amino acids of different lengths can be sufficient for ER targeting (56, 58).
Clearly, for the MOM targeting of mDMPK C, more elaborate structural information is required, as can be inferred from the cytosolic localization of YFP-mDMPK C(1-587) and YFP-mDMPK C(536-632). Although the coiled-coil domain by itself confers no targeting properties, as is evident from the cytosolic localization of mDMPK E, the amino acid information in this domain appeared a critical component for correct MOM association of mDMPK C. This could indicate that, apart from providing the appropriate sequence context, the coiled-coil domain could also play a role in protein-protein interactions that determine the specificity and strength of MOM binding (Fig. 10, hypothetical binding modes III and IV). Alternatively, close proximity of coil segments could exert intramolecular effects on the tail conformation necessary for determining MOM anchoring specificity. Whether we can draw any structure-function analogy to proteins with a similar arrangement of membrane-proximal coils and membrane-inserted domains, like the SNARE proteins, remains a question for future study (32). Previous work indicated that basic residues flanking a TMD dictate MOM targeting of TA proteins (4, 21). Consistent with this observation, we report here that mutations R588A and R589A render mDMPK C a cytosolic protein. Remarkably, reducing the basic amino acid content of the very C-terminal end of the tail, by mutating Arg at position 624 to Ala, did not abolish the specificity of the MOM signal. This observation and the finding that its C-terminal end has a net negative charge gives mDMPK C a unique position in the group of MOM anchored proteins.
Interestingly, the sequence arrangements that differ between human and mouse DMPK A tails confer differential properties to the protein, in that hDMPK A is targeted to the MOM whereas mDMPK A associates with the ER. This finding was unanticipated, since there is a remarkable conservation of gene structure and alternative splice sites between the two DMPK genes (15). Perhaps preference for MOM anchoring of hDMPK A is determined by the distinct positioning of basic residues within its tail. Neutral amino acids in the C-terminal stretch of mDMPK A are replaced by basic residues in hDMPK A (see Fig. S5 in the supplement material). More probably, the Arg at position 600 in the human protein breaks up the hydrophobic stretch that was identified as the minimal ER-targeting signal in the mouse form. As a consequence, the resulting tail has a TMD with a lower hydrophobicity, probably too low to promote ER binding. Indeed, an R600A mutation in the human tail 1 sequence resulted in an ER-localized protein, but deletion of the final 10 C-terminal residues, including 3 arginines, did not alter mitochondrial localization (data not shown). Variation in the guidance motifs in tail anchors may provide an adaptive mechanism for matching intracellular location to function during evolution. On the one hand, we should therefore consider the possibility that the difference in location preference rendered the functions of mouse and human DMPK. A orthologues completely divergent. On the other hand, it is conceivable that for proper function DMPK needs to be positioned close to sites of ER-MOM contact, which can be specified by association with either the ER or MOM side of the membranous interphase. No such speculation about an evolutionary shift in function for the DMPK C isoforms is needed, since both DMPK C isoforms show an identical distribution at the MOM in both human and mouse cells.
Under conditions of high expression, human and mouse DMPK C both adapted a cytosolic localization. This suggests that the MOM interaction of DMPK C can become saturated, as was also shown for VAMP-1B (27). It is important to note here that such an effect was never observed for hDMPK A. Our interpretation of this finding is that, besides the stronger hydrophobicity of the TMD, the net positive charge of +3 of the C-terminal region of the hDMPK A tail, as opposed to the net negative charge found in human and mouse DMPK C, contributes to its more efficient anchoring into the MOM. Species differences in lipid composition of the MOM may influence the requirements of the targeting signal, as shown for the TOM5 protein in yeast and mammalian cells (18). Our observation that the specificity of location is retained between human and mouse DMPK isoforms in cultured cells of human and mouse origin implies, however, that species effects of membrane lipid composition on the location of tail-anchored proteins are of no or only minor concern when compared between mammals.
Perinuclear clustering of mitochondria and OSER formation by DMPK A.
An interesting and important difference between DMPK isoforms from mice and humans was that hDMPK A preferentially clusters mitochondria near nuclei while mDMPK A can form OSER structures. Perinuclear clustering and aggregation of mitochondria has also been observed in cultured cells when expressing proteins like mitofusin 1 and 2, as well as with hFIS1 (42, 44, 59), OMP25 (36), or OPA1 (46). Except for OMP25, these proteins are involved in fusion or fission of the mitochondrial membrane, contain a coiled-coil domain, and have targeting properties in common with TA proteins (42, 43, 59). One possible explanation for the observed aggregation effects could be that cytosolic exposure of the coiled-coil domain of hDMPK A induces complex formation and brings together opposing mitochondrial membranes, similar to what was reported recently (23). Interactions between coiled-coil regions contribute to DMPK multimerization (8; R. E. M. A. van Herpen et al., unpublished data), and it is therefore likely that these segments just upstream of the tails play an active role in the clustering behavior. We can virtually rule out low-affinity dimer-binding effects of the YFP-tag since this moiety was present in both constructs but mitochondrial aggregation ability differed profoundly between hDMPK C and A isoforms (compare Fig. 7K and H). To explain why the effect was much weaker with DMPK C, we will need more information about the topology of membrane insertion for each of the DMPK variants. Current evidence also indicates that low-affinity protein interactions at the cytosolic side of the ER membrane are sufficient to promote OSER formation (48). Coiled-coil regions of mDMPK A may therefore have contributed to the effects seen at the ER membrane as well.
Perinuclear clustering of mitochondria can play diverse roles, including the initiation of apoptosis (50) or local modulation of intracellular Ca2+ transients, as observed in parotid acinar cells (7). Fission and fusion behavior and movement of mitochondria are dependent on the integrity of the cytoskeletal network, and we know that DMPK may play a role in actomyosin dynamics by controlling myosin phosphatase activity via MYPT1 phosphorylation (2, 35, 54). Thus, a functional link between DMPK activity on the cytosolic face of the MOM and interaction with the actin cytoskeleton could well affect mitochondrial distribution. Whether similar associations also could play a role in OSER formation is not known.
Results presented here, combined with preliminary findings in a previous study (54), now suggest a model where distinct locations by insertion into ER or MOM membranes via unique tail anchors determine DMPK's functional selectivity of activity. Organelles themselves appear to serve as organizing platforms for DMPK function and its subsequent role in cell signaling. Elucidation of possible reciprocal effects of multimerization and kinase activity on organellar membrane dynamics ultimately will be essential for understanding the function of DMPK. Parallel study of DMPK isoforms from different species may be helpful in these studies.
Finally, it is important to note that our findings were confirmed by results obtained with various types of cells, including skeletal muscle-derived myoblasts which, before and after fusion into myotubes, provide the normal host environment for DMPK A and C. Strict control of isoform ratios of C-tailed DMPKs and their levels and location may very well be important for the well-being and viability of muscle cells and other cell types in which the DMPK gene is expressed. In view of the cytopathological effects reported here, this may be especially true for the human DMPK A and C gene products. Just recently, our group has published results showing that cumulative distress of overexpression of hDMPK in a transgenic-mouse model translates into increased muscle workload intolerance, hypertrophic cardiomyopathy with dysrhythmia, myotonic myopathy, and hypotension (37). Further research is thus warranted to see if an imbalance in the mitochondrial targeting and the associated aggregation behavior demonstrated here could have contributed to these DM1-like traits.
Supplementary Material
Acknowledgments
We are grateful to J. van Deursen (Mayo Clinic, Rochester, Minn.) for providing monoclonal antibody mAb414.
This study was supported by the Prinses Beatrix Fonds and the Stichting Spieren voor Spieren, the American Muscular Dystrophy Association, and the Association Française contre les Myopathies.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abell, B. M., M. R. Pool, O. Schlenker, I. Sinning, and S. High. 2004. Signal recognition particle mediates post-translational targeting in eukaryotes. EMBO J. 23:2755-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amano, M., Y. Fukata, and K. Kaibuchi. 2000. Regulation and functions of Rho-associated kinase. Exp. Cell Res. 261:44-51. [DOI] [PubMed] [Google Scholar]
- 3.Borgese, N., S. Brambillasca, P. Soffientini, M. Yabal, and M. Makarow. 2003. Biogenesis of tail-anchored proteins. Biochem. Soc. Trans. 31:1238-1242. [DOI] [PubMed] [Google Scholar]
- 4.Borgese, N., S. Colombo, and E. Pedrazzini. 2003. The tale of tail-anchored proteins: coming from the cytosol and looking for a membrane. J. Cell Biol. 161:1013-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borgese, N., I. Gazzoni, M. Barberi, S. Colombo, and E. Pedrazzini. 2001. Targeting of a tail-anchored protein to endoplasmic reticulum and mitochondrial outer membrane by independent but competing pathways. Mol. Biol. Cell 12:2482-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brook, J. D., M. E. McCurrach, H. G. Harley, A. J. Buckler, D. Church, H. Aburatani, K. Hunter, V. P. Stanton, J.-P. Thirion, T. Hudson, R. Sohn, B. Zemelman, R. G. Snell, S. A. Rundle, S. Crow, J. Davies, P. Shelbourne, J. Buxton, C. Jones, V. Juvonen, K. Johnson, P. S. Harper, D. J. Shaw, and D. E. Housman. 1992. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell 68:799-808. [DOI] [PubMed] [Google Scholar]
- 7.Bruce, J. I. E., D. R. Giovannucci, G. Blinder, T. J. Shuttleworth, and D. I. Yule. 2004. Modulation of [Ca2+]i signaling dynamics and metabolism by perinuclear mitochondria in mouse parotid acinar cells. J. Biol. Chem. 279:12909-12917. [DOI] [PubMed] [Google Scholar]
- 8.Bush, E. W., S. M. Helmke, R. A. Birnbaum, and M. B. Perryman. 2000. Myotonic dystrophy protein kinase domains mediate localization, oligomerization, novel catalytic activity, and autoinhibition. Biochemistry 39:8480-8490. [DOI] [PubMed] [Google Scholar]
- 9.Chartier, C., E. Degryse, M. Gantzer, A. Dieterle, A. Pavirani, and M. Mehtali. 1996. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J. Virol. 70:4805-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins, T. J., M. J. Berridge, P. Lipp, and M. D. Bootman. 2002. Mitochondria are morphologically and functionally heterogeneous within cells. EMBO J. 21:1616-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cunto, F. D., S. Imarisio, P. Camera, C. Boitani, F. Altruda, and L. Silengo. 2002. Essential role of citron kinase in cytokinesis of spermatogenic precursors. J. Cell Sci. 115:4819-4826. [DOI] [PubMed] [Google Scholar]
- 12.Fransen, J. A., L. A. Ginsel, H. P. Hauri, E. Sterchi, and J. Blok. 1985. Immuno-electronmicroscopical localization of a microvillus membrane disaccharidase in the human small-intestinal epithelium with monoclonal antibodies. Eur. J. Cell Biol. 38:6-15. [PubMed] [Google Scholar]
- 13.Fu, Y.-H., A. Pizzuti, R. G. Fenwick, Jr., J. King, S. Rajnarayan, P. W. Dunne, J. Dubel, G. A. Nasser, T. Ashizawa, P. de Jong, B. Wieringa, R. G. Korneluk, M. B. Perryman, H. F. Epstein, and C. T. Caskey. 1992. An unstable triplet repeat in a gene related to myotonic muscular dystrophy. Science 255:1256-1258. [DOI] [PubMed] [Google Scholar]
- 14.Groenen, P., and B. Wieringa. 1998. Expanding complexity in myotonic dystrophy. Bioessays 20:901-912. [DOI] [PubMed] [Google Scholar]
- 15.Groenen, P. J., D. G. Wansink, M. Coerwinkel, W. van den Broek, G. Jansen, and B. Wieringa. 2000. Constitutive and regulated modes of splicing produce six major myotonic dystrophy protein kinase (DMPK) isoforms with distinct properties. Hum. Mol. Genet. 9:605-616. [DOI] [PubMed] [Google Scholar]
- 16.Harper, P. S. 2001. Myotonic dystrophy. The W. B. Saunders Co., London, United Kingdom.
- 17.He, T. C., S. Zhou, L. T. da Costa, J. Yu, K. W. Kinzler, and B. Vogelstein. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA 95:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horie, C., H. Suzuki, M. Sakaguchi, and K. Mihara. 2002. Characterization of signal that directs C-tail-anchored proteins to mammalian mitochondrial outer membrane. Mol. Biol. Cell 13:1615-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jansen, G., P. J. Groenen, D. Bachner, P. H. Jap, M. Coerwinkel, F. Oerlemans, W. van den Broek, B. Gohlsch, D. Pette, J. J. Plomp, P. C. Molenaar, M. G. Nederhoff, C. J. van Echteld, M. Dekker, A. Berns, H. Hameister, and B. Wieringa. 1996. Abnormal myotonic dystrophy protein kinase levels produce only mild myopathy in mice. Nat. Genet. 13:316-324. [DOI] [PubMed] [Google Scholar]
- 20.Jansen, G., M. Mahadevan, C. Amemiya, N. Wormskamp, B. Segers, W. Hendriks, K. O'Hoy, S. Baird, L. Sabourin, G. Lennon, P. L. Jap, D. Iles, M. Coerwinkel, M. Hofker, A. V. Carrano, P. J. de Jong, R. G. Korneluk, and B. Wieringa. 1992. Characterization of the myotonic dystrophy region predicts multiple protein isoform-encoding mRNAs. Nat. Genet. 1:261-266. [DOI] [PubMed] [Google Scholar]
- 21.Kaufmann, T., S. Schlipf, J. Sanz, K. Neubert, R. Stein, and C. Borner. 2003. Characterization of the signal that directs Bcl-xL, but not Bcl-2, to the mitochondrial outer membrane. J. Cell Biol. 160:53-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, P. K., C. Hollerbach, W. S. Trimble, B. Leber, and D. W. Andrews. 1999. Identification of the endoplasmic reticulum targeting signal in vesicle-associated membrane proteins. J. Biol. Chem. 274:36876-36882. [DOI] [PubMed] [Google Scholar]
- 23.Koshiba, T., S. A. Detmer, J. T. Kaiser, H. Chen, J. M. McCaffery, and D. C. Chan. 2004. Structural basis of mitochondrial tethering by mitofusin complexes. Science 305:858-862. [DOI] [PubMed] [Google Scholar]
- 24.Kuroda, R., T. Ikenoue, M. Honsho, S. Tsujimoto, J. Y. Mitoma, and A. Ito. 1998. Charged amino acids at the carboxyl-terminal portions determine the intracellular locations of two isoforms of cytochrome b5. J. Biol. Chem. 273:31097-31102. [DOI] [PubMed] [Google Scholar]
- 25.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 26.Lam, L. T., Y. C. Pham, T. M. Nguyen, and G. E. Morris. 2000. Characterization of a monoclonal antibody panel shows that the myotonic dystrophy protein kinase, DMPK, is expressed almost exclusively in muscle and heart. Hum. Mol. Genet. 9:2167-2173. [DOI] [PubMed] [Google Scholar]
- 27.Lan, L., S. Isenmann, and B. W. Wattenberg. 2000. Targeting and insertion of C-terminally anchored proteins to the mitochondrial outer membrane is specific and saturable but does not strictly require ATP or molecular chaperones. Biochem. J. 349:611-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.La Spada, A. R., R. I. Richards, and B. Wieringa. 2004. Dynamic mutations on the move in Banff. Nat. Genet. 36:667-670. [DOI] [PubMed] [Google Scholar]
- 29.Leung, T., X. Q. Chen, I. Tan, E. Manser, and L. Lim. 1998. Myotonic dystrophy kinase-related Cdc42-binding kinase acts as a Cdc42 effector in promoting cytoskeletal reorganization. Mol. Cell. Biol. 18:130-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maeda, M., C. S. Taft, E. W. Bush, E. Holder, W. M. Bailey, H. Neville, M. B. Perryman, and R. D. Bies. 1995. Identification, tissue-specific expression, and subcellular localization of the 80- and 71-kDa forms of myotonic dystrophy kinase protein. J. Biol. Chem. 270:20246-20249. [DOI] [PubMed] [Google Scholar]
- 31.Mahadevan, M. S., C. Amemiya, G. Jansen, L. Sabourin, S. Baird, C. E. Neville, N. Wormskamp, B. Segers, M. Batzer, J. Lamerdin, et al. 1993. Structure and genomic sequence of the myotonic dystrophy (DM kinase) gene. Hum. Mol. Genet. 2:299-304. [DOI] [PubMed] [Google Scholar]
- 32.Melia, T. J., T. Weber, J. A. McNew, L. E. Fisher, R. J. Johnston, F. Parlati, L. K. Mahal, T. H. Sollner, and J. E. Rothman. 2002. Regulation of membrane fusion by the membrane-proximal coil of the t-SNARE during zippering of SNAREpins. J. Cell Biol. 158:929-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morgan, J. E., J. R. Beauchamp, C. N. Pagel, M. Peckham, P. Ataliotis, P. S. Jat, M. D. Noble, K. Farmer, and T. A. Partridge. 1994. Myogenic cell lines derived from transgenic mice carrying a thermolabile T antigen: a model system for the derivation of tissue-specific and mutation-specific cell lines. Dev. Biol. 162:486-498. [DOI] [PubMed] [Google Scholar]
- 34.Mounsey, J. P., J. E. John III, S. M. Helmke, E. W. Bush, J. Gilbert, A. D. Roses, M. B. Perryman, L. R. Jones, and J. R. Moorman. 2000. Phospholemman is a substrate for myotonic dystrophy protein kinase. J. Biol. Chem. 275:23362-23367. [DOI] [PubMed] [Google Scholar]
- 35.Muranyi, A., R. Zhang, F. Liu, K. Hirano, M. Ito, H. F. Epstein, and D. J. Hartshorne. 2001. Myotonic dystrophy protein kinase phosphorylates the myosin phosphatase targeting subunit and inhibits myosin phosphatase activity. FEBS Lett. 493:80-84. [DOI] [PubMed] [Google Scholar]
- 36.Nemoto, Y., and P. De Camilli. 1999. Recruitment of an alternatively spliced form of synaptojanin 2 to mitochondria by the interaction with the PDZ domain of a mitochondrial outer membrane protein. EMBO J. 18:2991-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Cochlain, D. F., C. Perez-Terzic, S. Reyes, G. C. Kane, A. Behfar, D. M. Hodgson, J. A. Strommen, X. K. Liu, W. Van Den Broek, D. G. Wansink, B. Wieringa, and A. Terzic. 2004. Transgenic overexpression of human DMPK accumulates into hypertrophic cardiomyopathy, myotonic myopathy and hypotension traits of myotonic dystrophy. Hum. Mol. Genet. 13:2505-2518. [DOI] [PubMed] [Google Scholar]
- 38.Pham, Y. C., N. Man, L. T. Lam, and G. E. Morris. 1998. Localization of myotonic dystrophy protein kinase in human and rabbit tissues using a new panel of monoclonal antibodies. Hum. Mol. Genet. 7:1957-1965. [DOI] [PubMed] [Google Scholar]
- 39.Piekny, A. J., and P. E. Mains. 2002. Rho-binding kinase (LET-502) and myosin phosphatase (MEL-11) regulate cytokinesis in the early Caenorhabditis elegans embryo. J. Cell Sci. 115:2271-2282. [DOI] [PubMed] [Google Scholar]
- 40.Ranum, L. P., and J. W. Day. 2004. Myotonic dystrophy: RNA pathogenesis comes into focus. Am. J. Hum. Genet. 74:793-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roberts, R., N. A. Timchenko, J. W. Miller, S. Reddy, C. T. Caskey, M. S. Swanson, and L. T. Timchenko. 1997. Altered phosphorylation and intracellular distribution of a (CUG)n triplet repeat RNA-binding protein in patients with myotonic dystrophy and in myotonin protein kinase knockout mice. Proc. Natl. Acad. Sci. USA 94:13221-13226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rojo, M., F. Legros, D. Chateau, and A. Lombes. 2002. Membrane topology and mitochondrial targeting of mitofusins, ubiquitous mammalian homologs of the transmembrane GTPase Fzo. J. Cell Sci. 115:1663-1674. [DOI] [PubMed] [Google Scholar]
- 43.Rube, D. A., and A. M. van der Bliek. 2004. Mitochondrial morphology is dynamic and varied. Mol. Cell. Biochem. 256-257:331-339. [DOI] [PubMed] [Google Scholar]
- 44.Santel, A., and M. T. Fuller. 2001. Control of mitochondrial morphology by a human mitofusin. J. Cell Sci. 114:867-874. [DOI] [PubMed] [Google Scholar]
- 45.Sarkar, P. S., J. Han, and S. Reddy. 2004. In situ hybridization analysis of Dmpk mRNA in adult mouse tissues. Neuromuscul. Disord. 14:497-506. [DOI] [PubMed] [Google Scholar]
- 46.Satoh, M., T. Hamamoto, N. Seo, Y. Kagawa, and H. Endo. 2003. Differential sublocalization of the dynamin-related protein OPA1 isoforms in mitochondria. Biochem. Biophys. Res. Commun. 300:482-493. [DOI] [PubMed] [Google Scholar]
- 47.Shima, D. T., K. Haldar, R. Pepperkok, R. Watson, and G. Warren. 1997. Partitioning of the Golgi apparatus during mitosis in living HeLa cells. J. Cell Biol. 137:1211-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Snapp, E. L., R. S. Hegde, M. Francolini, F. Lombardo, S. Colombo, E. Pedrazzini, N. Borgese, and J. Lippincott-Schwartz. 2003. Formation of stacked ER cisternae by low affinity protein interactions. J. Cell Biol. 163:257-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suzuki, A., Y. Sugiyama, Y. Hayashi, N. Nyu-i, M. Yoshida, I. Nonaka, S. Ishiura, K. Arahata, and S. Ohno. 1998. MKBP, a novel member of the small heat shock protein family, binds and activates the myotonic dystrophy protein kinase. J. Cell Biol. 140:1113-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takada, S., Y. Shirakata, N. Kaneniwa, and K. Koike. 1999. Association of hepatitis B virus X protein with mitochondria causes mitochondrial aggregation at the nuclear periphery, leading to cell death. Oncogene 18:6965-6973. [DOI] [PubMed] [Google Scholar]
- 51.Timchenko, L., W. Nastainczyk, T. Schneider, B. Patel, F. Hofmann, and C. T. Caskey. 1995. Full-length myotonin protein kinase (72 kDa) displays serine kinase activity. Proc. Natl. Acad. Sci. USA 92:5366-5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tiscornia, G., and M. S. Mahadevan. 2000. Myotonic dystrophy: the role of the CUG triplet repeats in splicing of a novel DMPK exon and altered cytoplasmic DMPK mRNA isoform ratios. Mol. Cell 5:959-967. [DOI] [PubMed] [Google Scholar]
- 53.Voeltz, G. K., M. M. Rolls, and T. A. Rapoport. 2002. Structural organization of the endoplasmic reticulum. EMBO Rep. 3:944-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wansink, D. G., R. E. M. A. van Herpen, M. M. Coerwinkel-Driessen, P. J. T. A. Groenen, B. A. Hemmings, and B. Wieringa. 2003. Alternative splicing controls myotonic dystrophy protein kinase structure, enzymatic activity, and subcellular localization. Mol. Cell. Biol. 23:5489-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wattenberg, B., and T. Lithgow. 2001. Targeting of C-terminal (tail)-anchored proteins: understanding how cytoplasmic activities are anchored to intracellular membranes. Traffic 2:66-71. [DOI] [PubMed] [Google Scholar]
- 56.Whitley, P., E. Grahn, U. Kutay, T. A. Rapoport, and G. von Heijne. 1996. A 12-residue-long polyleucine tail is sufficient to anchor synaptobrevin to the endoplasmic reticulum membrane. J. Biol. Chem. 271:7583-7586. [DOI] [PubMed] [Google Scholar]
- 57.Wu, X., L. H. Kasper, R. T. Mantcheva, G. T. Mantchev, M. J. Springett, and J. M. A. van Deursen. 2001. Disruption of the FG nucleoporin NUP98 causes selective changes in nuclear pore complex stoichiometry and function. Proc. Natl. Acad. Sci. USA 98:3191-3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang, M., J. Ellenberg, J. S. Bonifacino, and A. M. Weissman. 1997. The transmembrane domain of a carboxyl-terminal anchored protein determines localization to the endoplasmic reticulum. J. Biol. Chem. 272:1970-1975. [DOI] [PubMed] [Google Scholar]
- 59.Yoon, Y., E. W. Krueger, B. J. Oswald, and M. A. McNiven. 2003. The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Mol. Cell. Biol. 23:5409-5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.