Abstract
Objective
The combination of phenomic data from electronic health records (EHR) and clinical data repositories with dense biological data has enabled genomic and pharmacogenomic discovery, a first step toward precision medicine. Computational methods for the identification of clinical phenotypes from EHR data will advance our understanding of disease risk and drug response, and support the practice of precision medicine on a national scale.
Methods
Based on our experience within three national research networks, we summarize the broad approaches to clinical phenotyping and highlight the important role of these networks in the progression of high-throughput phenotyping and precision medicine. We provide supporting literature in the form of a non-systematic review.
Results
The practice of clinical phenotyping is evolving to meet the growing demand for scalable, portable, and data driven methods and tools. The resources required for traditional phenotyping algorithms from expert defined rules are significant. In contrast, machine learning approaches that rely on data patterns will require fewer clinical domain experts and resources.
Conclusions
Machine learning approaches that generate phenotype definitions from patient features and clinical profiles will result in truly computational phenotypes, derived from data rather than experts. Research networks and phenotype developers should cooperate to develop methods, collaboration platforms, and data standards that will enable computational phenotyping and truly modernize biomedical research and precision medicine.
Keywords: Machine learning, Clinical phenotyping, Electronic health records, Networked research, Precision medicine
1. Introduction
The national Precision Medicine Initiative aims to enroll one million members in a national cohort that will integrate data from biospecimens, sensor and mobile technologies, and health-care, largely from electronic health record (EHR) data, to advance biomedical discovery and improve health [1]. The realization of this vision will require efficient and effective methods to convert data from EHRs into specific and reliable phenotype characterizations that can be used to predict an individual’s risk of disease or response to drug therapy.
Phenotypes are the measurable biological, behavioral and clinical markers of a condition or disease. The process of deriving research-grade phenotypes from clinical data using computer-executable algorithms is called computational phenotyping (phenotyping for short) [2]. Phenotyping includes a range of approaches from finding a phenotype using expert-derived rules and those phenotypes emerging from novel computational methods that potentially represent new clinical entities. The widespread adoption of EHRs will increase the reliance on phenotyping for a number of activities, including genomic studies of disease and drug response, clinical predictive modeling, pragmatic clinical trials, and healthcare quality measurement. Current methods face bottlenecks for development, implementation, sharability, and the ability to derive novel, not-foreseen findings. We provide a survey of the approaches to computational phenotyping and challenges experienced by several national research networks with which we are affiliated, and provide supporting literature in the form of a non-systematic review. The aim of this paper is to provide a summary of the approaches and tools that clinical research networks are using to realize the scale of high-throughput computational phenotyping. Based on the common challenges faced by these networks, we suggest cultural change and resources that will be needed to support computational phenotyping on a grand scale and advance data-driven precision medicine research.
2. National networks and phenotyping activity
A number of national research and public health surveillance networks have leveraged data from EHRs for defining conditions and risk. The Electronic Medical Records & Genomics (eMERGE) Network, formed in 2007 and arguably the pioneer of computational phenotyping, has investigated more than 40 phenotypes for genomic studies using algorithms that combine billing codes, medication data, laboratory and test results, and natural language processing of clinical notes [3,4]. Sites from the Pharmacogenomics Research Network (PGRN) have used EHR data to identify genetic predictors of drug-response phenotypes across multiple sites [5–7]. The Mini-Sentinel surveillance initiative, funded by the U.S. Food and Drug Administration, uses phenotype algorithms to define conditions from administrative data from 18 national health plans to identify adverse drug outcomes [8–15]. In addition, provider networks use computational phenotyping to identify patients with particular conditions for health services or population-level research. These include the Health Care Systems Research Network, formerly known as the HMO Research Network, and the Observational Medical Outcomes Partnership (OMOP) [16], now part of the Observational Health Data Sciences and Informatics (OHDSI) collaborative [17].
A number of disease-specific research networks and multi-site registries have developed and validated EHR-based phenotype definitions for specific conditions [18,19]. In the National Institutes of Health’s Health Care Systems Research Collaboratory, a number of multi-site pragmatic clinical trial demonstration projects are using computable phenotypes for cohort identification, development of interventions, and study outcomes [20–22] More recently, the Patient Centered Outcomes Research Institute funded the National Patient-Centered Clinical Research Network (PCORnet) to conduct comparative effectiveness studies across 13 Clinical Data Research Networks and 21 Patient Powered Research Networks [23]. Partnering institutions are expected to support up to 200 queries in the next few years, signifying the imminent need for high-throughput and reproducible phenotyping methods.
Although the aforementioned research networks have unique objectives and constraints, they share common challenges related to the use of clinical data for research, including heterogeneous EHR systems, a lack of standardized data, concerns about data completeness and inherent biases, and variation in medical diagnosis, procedures, treatments, and data documentation across providers, organizations, and regions. In response, several networks have published methodological guides for data quality assurance [24–29].
3. Evolution of phenotyping methods
Research networks by their very nature require scalable approaches that can be implemented quickly with reproducible performance characteristics in multiple settings and information systems. There are several broad classes of methods to computational phenotyping that are continuously improving.
The use of expert-defined rules is most widely adopted method for phenotyping, and this approach was used for the early phenotypes developed from the eMERGE network, such as type 2 diabetes [30] and cataracts [31]. This approach begins with the manual development of an algorithm – often using Boolean logic, scoring thresholds, or a decision tree – based on domain expertise. The logic is then iteratively enhanced through validation and chart review on EHR data. Advantages of this approach are that it yields human-interpretable algorithms, which can be portable to other sites [32], and the number of charts needed to review to train/validate an algorithm can be lower. However, the effort and time for developing the algorithms can be significant, requiring clinical and informatics knowledge, and this approach cannot be used to identify phenotypes not first envisioned by a researcher.
Machine learning methods rely on data patterns to develop the phenotype definitions, and can reduce the effort required from clinical domain experts. Supervised learning aims to construct classifiers to differentiate cases (positive for the phenotype) and controls (negative for the phenotype). The high level steps involve (1) characterizing patients as feature vectors, (2) determining the class label (case vs. control) for each patient, (3) building and optimizing the classifier. Typically the number of charts reviewed is higher than required for rule-based algorithms, a time-consuming task requiring domain experts. Chen et al. explored active learning as a more efficient labeling process, demonstrating reduction in the number of cases needed [33]. However, machine learning classification models can be difficult to interpret, require significant training data, and may not transfer well to other sites, as a model may learn features that are unique to an institution (e.g., physician name, local note type, or clinical unit). Yu et al. extracted clinical features from publicly-available knowledge sources to develop more “interpretable” machine learning algorithms that performed as well as or better than expert-derived algorithms [34].
Unsupervised learning provides approaches to cluster EHR data into patient groups corresponding to phenotypes or subtypes. Unsupervised learning does not require expert labels, which tremendously reduces the time needed for manual chart review. However, the validation of the resulting phenotypic groups is challenging, as no clear ground truth on those groups are given. While these methods require very large volumes of training data, they do not carry costs of manually labeling individuals as cases or controls. Various tensor factorization methods have been developed for unsupervised phenotyping [35–37]. Deep learning is another approach which has successfully identified patterns in clinical data representing distinct phenotypes [38].
Because important relevant clinical data is included in narrative clinical notes rather than structured data elements or standardized coding systems, natural language processing methods can be used to extract phenotypes from clinical notes [39,40] and to process data for more advanced machine learning techniques. Phenotype definitions including general purpose natural language processing (NLP) tools [41–43] have accelerated the widespread use of NLP, which is an important component of some complex phenotypes [44].
4. Toward a future of higher throughput phenotyping
The planned Precision Medicine Initiative study will require higher-throughput, more easily shared computational approaches than have been demonstrated to date. Scalable precision medicine will require clinical phenotypes that can be rapidly developed, executed in high volume, and easily adapted to new sites with high algorithm reliability (Fig. 1).
Fig. 1.
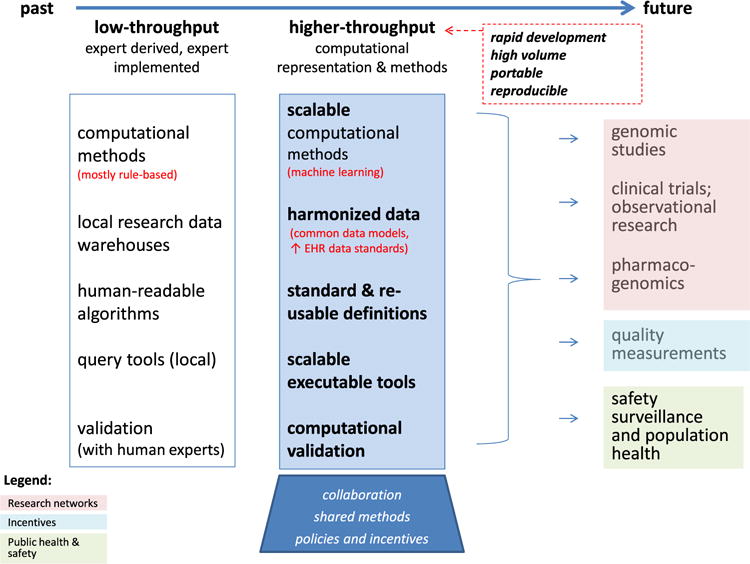
The evolution of computational phenotyping methods and the key biomedical applications that needs high-throughput phenotyping.
The vision of rapid, portable phenotyping implies that multiple providers and applications can reuse computational methods and definitional logic, enhanced by accessible repositories for phenotyping logic and methods. While individual research networks undoubtedly have infrastructure for sharing, national repositories for definitions and methods will enable cooperation across networks. The Phenotype Knowledge Base (PheKB) is one such knowledge resource for phenotyping methods, which hosts algorithms from eMERGE, PCORnet, PGRN, and other sites and networks [45]. Many algorithms on PheKB have been validated by multiple sites [28].
Several tools have been developed to simplify “EHR-wide” analyses. These include groupings of billing codes into meaningful phenotypes, such as done by the Agency for Healthcare Research and Quality’s Clinical Classifications Software [46,47] or the phenome-wide association study (PheWAS) tools [3,48–50]. PheWAS approaches have been used for a number of genomics and clinical studies, including recent studying leveraging text. Data driven approaches leveraging PheWAS have also elucidated sub-classifications of phenotypes, such as for rheumatoid arthritis and autism [51]. Other resources, such as mapping medications to their indications or adverse effects [52,53], offer aids to building algorithms. Mo et al. offer desiderata for computable representations of EHR-driven phenotype algorithms [54].
The implementation of phenotypes (in multisite clinical or research networks) can be accelerated by using harmonized clinical data. One approach is the use of a common data model (CDM) to create a common set of definitions, formats, and allowable values for data elements from heterogeneous EHRs. A number of different CDMs are used by different research networks for different purposes, including the OMOP CDM [55] adopted by OHDSI [17], the Mini-sentinel data model [56], the PCORnet CDM [57]. Other clinical data models include the Health Level Seven (HL7) virtual medical record (vMR) model for clinical decision support [58,59], OpenEHR Archetypes [60,61], and the HL7 Detailed Clinical Models [62,63]. An important consideration is that most CDMs are limited to the types of data they include, and unstructured or ad hoc structured data types are often excluded. Thus, many of the phenotype algorithms executed within eMERGE and potentially as a part of precision medicine are not fully addressable via existing CDMs. Novel combinations that facilitate the use of a CDM in tandem with custom applications of NLP or other cohort selection tools will be needed.
For rapid authoring, the Phenotype Execution Modeling Architecture (PhEMA) project is a phenotyping algorithm authoring and execution platform [64] designed to streamline the process for developing computer-readable, rule-based phenotyping algorithms that can be shared as executable representations between different sites. By creating a common representation model for phenotypes (such as the Quality Data Model from the National Quality Forum), PhEMA could enable a common language to be used against a variety of CDMs and EHR systems [65]. Other tests of computable phenotyping approaches have leveraged the Konstanz Information Miner (KNIME) [66,67] or Drools [68]. The recent desiderata recommend use of a common data model, and the use standardized terminologies and ontologies, and facilitate reuse of value sets [54].
5. Discussion
This survey represents the opinions of authors based upon our experience with computational phenotyping within several national research networks, and does not represent a systematic review or national consensus. However, the research networks we represent are leveraging deep multidisciplinary expertise and collective resources to develop new methods and tools for computational phenotyping across heterogeneous organizations, data systems, and populations. These methods promise to significantly advance the identification of disease cohorts and the quantification of an individual’s risk for disease or drug toxicity, and provide path for rapid translational of research findings in healthcare delivery.
The number of national initiatives and activities focused on the use of clinical data is promising, but effective realization of precision medicine on a grand scale will require higher-throughput computational phenotyping. Machine learning approaches have great potential to transform our understanding of disease by allowing phenotypes to be defined by what patients present with, rather than by what research experts know or believe. This will result in truly computational phenotypes, derived from data rather than experts. We might never get to full confidence in the output of a purely machine-generated phenotype, but a goal instead could be to get to a well characterized phenotype, complete with a confidence level or interval, based on the data availability and quality, and the specific use case where it can be applied. New tools will facilitate faster development and implementation of computational phenotyping and lead to more nuanced understanding of “phenotypes.” [69] The latter is a primary goal of precision medicine, to achieve a deeper understanding of diseases and drug response that allow tailoring of therapy toward better health on the basis of clinical and molecular data. In the future, this goal will be enhanced with synergy and harmonization with phenotypes and phenotypic traits defined the biology community [70].
In parallel, national incentives for quality measurement and reporting are driving the development of standardized approaches for deriving research-grade phenotypes from EHR data to advance precision medicine. The future alignment between research phenotyping and quality improvement efforts will enable further efficiencies for both domains. This should include libraries of phenotype definitions, annotated by use case and data required, that can easily be discovered by implementers working in genomic research, precision medicine, or healthcare quality measurement. The next step – and the greater challenge – is to build a culture and supporting infrastructure to share the knowledge and tools that can advance all these efforts. Clinical research networks and phenotype developers should cooperate to develop methods, collaboration platforms, and data standards that will enable high-throughput phenotyping to be implemented across millions of individuals, for a spectrum of use cases from personalized medicine to drug safety surveillance and population health. Successful synergy between clinical research networks, providers, and national initiatives will truly modernize computational phenotyping, biomedical research, precision medicine, and health outcomes.
Acknowledgments
The authors are affiliated with the following research networks, although the statements contained in this article represent the opinions of the authors and were not vetted by any of the networks or their sponsoring organizations. This publication was supported in part by funding from grants: U54 AT007748-02, R01 GM105688, R01 HS023077, R01 GM103859, R01 LM010685, R01 MH105384, U01 HG004603, U01 HG006388, U01 HG007253, and P50 GM115305 from the National Institutes of Health; 1R18HS023921 (AHRQ); the National Science Foundation (award IIS-1418511 and CCF-1533768 and NSF 13-543); and the Patient Centered Outcomes Research Institute (CDRN-1306-04737 and Coordinating Center P122013-499A). Dr. Sun was also supported in part by the Children’s Healthcare of Atlanta, CDC I-SMILE project, Google Faculty Award, AWS Research Award, Microsoft Azure Research Award and UCB.
The views presented here are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health, the National Science Foundation, PCORI, or any of the funding sources and sponsors mentioned above.
References
- 1.Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;372(9):793–5. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pathak J, Kho AN, Denny JC. Electronic health records-driven phenotyping: challenges, recent advances, and perspectives. J Am Med Inform Assoc. 2013;20(e2):e206–11. doi: 10.1136/amiajnl-2013-002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Denny JC, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013;31(12):1102–10. doi: 10.1038/nbt.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crawford DC, et al. eMERGEing progress in genomics-the first seven years. Front Genet. 2014;5:184. doi: 10.3389/fgene.2014.00184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naidoo D, et al. A polymorphism in HLA-G modifies statin benefit in asthma. Pharmacogenomics J. 2015;15(3):272–7. doi: 10.1038/tpj.2014.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Driest SL, et al. Genome-wide association study of serum creatinine levels during vancomycin therapy. PLoS One. 2015;10(6):e0127791. doi: 10.1371/journal.pone.0127791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karol SE, et al. Genetics of glucocorticoid-associated osteonecrosis in children with acute lymphoblastic leukemia. Blood. 2015 doi: 10.1182/blood-2015-05-643601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Platt R, et al. The U.S. Food and Drug Administration’s Mini-Sentinel program: status and direction. Pharmacoepidemiol Drug Saf. 2012;21(Suppl. 1):1–8. doi: 10.1002/pds.2343. [DOI] [PubMed] [Google Scholar]
- 9.Cutrona SL, et al. Validation of acute myocardial infarction in the food and drug administration’s mini-sentinel program. Pharmacoepidemiol Drug Saf. 2013;22(1):40–54. doi: 10.1002/pds.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lo Re V, 3rd, et al. Validity of diagnostic codes to identify cases of severe acute liver injury in the US food and drug administration’s mini-sentinel distributed database. Pharmacoepidemiol Drug Saf. 2013;22(8):861–72. doi: 10.1002/pds.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toh S, et al. Rapid assessment of cardiovascular risk among users of smoking cessation drugs within the US food and drug administration’s mini-sentinel program. JAMA Intern Med. 2013;173(9):817–9. doi: 10.1001/jamainternmed.2013.3004. [DOI] [PubMed] [Google Scholar]
- 12.Walsh KE, et al. Validation of anaphylaxis in the food and drug administration’s mini-sentinel. Pharmacoepidemiol Drug Saf. 2013;22(11):1205–13. doi: 10.1002/pds.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McClure DL, et al. Mini-sentinel methods: framework for assessment of positive results from signal refinement. Pharmacoepidemiol Drug Saf. 2014;23(1):3–8. doi: 10.1002/pds.3547. [DOI] [PubMed] [Google Scholar]
- 14.Psaty BM, Breckenridge AM. Mini-Sentinel and regulatory science–big data rendered fit and functional. N Engl J Med. 2014;370(23):2165–7. doi: 10.1056/NEJMp1401664. [DOI] [PubMed] [Google Scholar]
- 15.Winiecki S, et al. Complementary use of passive surveillance and mini-sentinel to better characterize hemolysis after immune globulin. Transfusion. 2015;55(Suppl. 2):S28–35. doi: 10.1111/trf.13116. [DOI] [PubMed] [Google Scholar]
- 16.OHSDI. OMOP common data model. 2015 Available from: http://www.ohdsi.org/data-standardization/the-common-data-model/. (accessed: 17.05.16).
- 17.Hripcsak G, et al. Observational health data sciences and informatics (ohdsi): opportunities for observational researchers. Stud Health Technol Inform. 2015;216:574–8. [PMC free article] [PubMed] [Google Scholar]
- 18.Nichols GA, et al. Construction of a multisite data link using electronic health records for the identification, surveillance, prevention, and management of diabetes mellitus: the SUPREME-DM project. Prev Chronic Dis. 2012;9:E110. doi: 10.5888/pcd9.110311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prieto-Centurion V, et al. Multicenter study comparing case definitions used to identify patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2014;190(9):989–95. doi: 10.1164/rccm.201406-1166OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richesson RL, et al. Electronic health records based phenotyping in next-generation clinical trials: a perspective from the NIH health care systems collaboratory. J Am Med Inform Assoc. 2013;20(e2):e226–31. doi: 10.1136/amiajnl-2013-001926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collaboratory., N.H.C.S.R. Rethinking clinical trials In: A living textbook of pragmatic clinical trials. Duke University; Durham: 2015. [Google Scholar]
- 22.Rusincovitch SA. Practical development and implementation of EHR phenotypes. presented on NIH collaboratory grand rounds; November 15; 2013; Available from: https://www.nihcollaboratory.org/Pages/Grand-Rounds-11-15-13.aspx. [Google Scholar]
- 23.Fleurence RL, et al. Launching PCORnet: a national patient-centered clinical research network. J Am Med Inform Assoc. 2014;21(4):578–82. doi: 10.1136/amiajnl-2014-002747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zozus MN, et al. Assessing Data Quality for Healthcare Systems Data Used in Clinical Research (Version 1.0) An NIH Health Care Systems Research Collaboratory Phenotypes, Data Standards, and Data Quality Core White Paper. 2013 Available from: https://sites.duke.edu/rethinkingclinicaltrials/tag/data-quality/. (accessed: 17.05.16).
- 25.PCORI, Building PCOR Value and Integrity with Data Quality and Transparency Standards Michael G. Kahn, MD, PhD, Principal Investigator. Improving Methods for Conducting Patient-Centered Outcomes Research Award 2013 [Google Scholar]
- 26.Kahn MG, et al. A pragmatic framework for single-site and multisite data quality assessment in electronic health record-based clinical research. Med Care. 2012;(Suppl. 50):S21–9. doi: 10.1097/MLR.0b013e318257dd67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown JS, Kahn M, Toh S. Data quality assessment for comparative effectiveness research in distributed data networks. Med Care. 2013;51(8 Suppl. 3):S22–9. doi: 10.1097/MLR.0b013e31829b1e2c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newton KM, et al. Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network. J Am Med Inform Assoc. 2013;20:e147–54. doi: 10.1136/amiajnl-2012-000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Denny JC. Chapter 13: Mining Electronic Health Records in the Genomics Era. PloS Comput Biol. 2012;8(12) doi: 10.1371/journal.pcbi.1002823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kho AN, et al. Use of diverse electronic medical record systems to identify genetic risk for type 2 diabetes within a genome-wide association study. J Am Med Inform Assoc. 2012;19(2):212–8. doi: 10.1136/amiajnl-2011-000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peissig PL, et al. Importance of multi-modal approaches to effectively identify cataract cases from electronic health records. J Am Med Inform Assoc. 2012;19(2):225–34. doi: 10.1136/amiajnl-2011-000456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Denny JC, et al. Variants near FOXE1 are associated with hypothyroidism and other thyroid conditions: using electronic medical records for genome- and phenome-wide studies. Am J Hum Genet. 2011;89(4):529–42. doi: 10.1016/j.ajhg.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Y, et al. Applying active learning to high-throughput phenotyping algorithms for electronic health records data. J Am Med Inform Assoc. 2013;20(e2):e253–9. doi: 10.1136/amiajnl-2013-001945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu S, et al. Toward high-throughput phenotyping: unbiased automated feature extraction and selection from knowledge sources. J Am Med Inform Assoc. 2015;22(5):993–1000. doi: 10.1093/jamia/ocv034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ho JC, et al. Limestone: high-throughput candidate phenotype generation via tensor factorization. J Biomed Inform. 2014;52:199–211. doi: 10.1016/j.jbi.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ho JC, Joydeep G, Sun J. Marble: high-throughput phenotyping from electronic health records via sparse nonnegative tensor factorization. 20th ACM SIGKDD international conference on knowledge discovery and data mining. 2014;14 KDD. [Google Scholar]
- 37.Wang Y, et al. Rubik: Knowledge Guided Tensor Factorization and Completion for Health Data Analytics. 21th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. 2015;15 doi: 10.1145/2783258.2783395. KDD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lasko TA, Denny JC, Levy MA. Computational phenotype discovery using unsupervised feature learning over noisy, sparse, and irregular clinical data. PLoS One. 2013;8(6):e66341. doi: 10.1371/journal.pone.0066341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doan S, et al. Integrating existing natural language processing tools for medication extraction from discharge summaries. J Am Med Inform Assoc. 2010;17(5):528–31. doi: 10.1136/jamia.2010.003855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Byrd RJ, et al. Automatic identification of heart failure diagnostic criteria, using text analysis of clinical notes from electronic health records. Int J Med Inform. 2014;83(12):983–92. doi: 10.1016/j.ijmedinf.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Savova GK, et al. Mayo clinical text analysis and knowledge extraction system (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc. 2010;17(5):507–13. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aronson AR, Lang FM. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc. 2010;17(3):229–36. doi: 10.1136/jamia.2009.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meystre S, Haug PJ. Evaluation of medical problem extraction from electronic clinical documents using MetaMap transfer (MMTx) Stud Health Technol Inform. 2005;116:823–8. [PubMed] [Google Scholar]
- 44.Wei WQ, Denny JC. Extracting research-quality phenotypes from electronic health records to support precision medicine. Genome Med. 2015;7(1):41. doi: 10.1186/s13073-015-0166-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.University., V. PheKB. 2012 Available from: http://www.phekb.org/. (accessed: 17.05.16).
- 46.AHRQ. Clinical classifications software (ICD-9-CM) summary and download-redirect. 2012 Available from: http://www.ahrq.gov/research/data/hcup/ccs.html. (accessed: 17.05.16).
- 47.Kozma CM. Clinical classifications software for identification of codes. Manag Care Interface. 2006;19(3):37–8. [PubMed] [Google Scholar]
- 48.Carroll RJ, Bastarache L, Denny JC. R PheWAS: data analysis and plotting tools for phenome-wide association studies in the R environment. Bioinformatics. 2014;30(16):2375–6. doi: 10.1093/bioinformatics/btu197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pendergrass SA, et al. Visually integrating and exploring high throughput phenome-wide association study (PheWAS) results using PheWAS-view. BioData Min. 2012;5(1):5. doi: 10.1186/1756-0381-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Denny JC, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26(9):1205–10. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Doshi-Velez F, Ge Y, Kohane I. Comorbidity clusters in autism spectrum disorders: an electronic health record time-series analysis. Pediatrics. 2014;133(1):e54–63. doi: 10.1542/peds.2013-0819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tatonetti NP, et al. Data-driven prediction of drug effects and interactions. Sci Transl Med. 2012;4(125):125ra31. doi: 10.1126/scitranslmed.3003377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wei WQ, et al. Development and evaluation of an ensemble resource linking medications to their indications. J Am Med Inform Assoc. 2013;20(5):954–61. doi: 10.1136/amiajnl-2012-001431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mo H, et al. Desiderata for computable representations of electronic health records-driven phenotype algorithms. J Am Med Inform Assoc. 2015 Nov 22;(6):1220–30. doi: 10.1093/jamia/ocv112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.OHDSI. Observational Health Data Sciences and Informatics. Available from: http://www.ohdsi.org/data-standardization/the-common-data-model/. (accessed: 17.05.6).
- 56.Mini-Sentinel. Distributed database and common data model. 2015 Available from: http://mini-sentinel.org/data_activities/distributed_db_and_data/details.aspx?ID=105. (accessed: 17 May 2016).
- 57.PCORnet PCORnet Common Data Model (CDM) Why, what and how? 2015 Available from: http://www.pcornet.org/pcornet-common-data-model/. (accessed: 17.05.16).
- 58.Ebrahiminia V, Yasini M, Lamy JB. Mapping ASTI patient’s therapeutic-data model to virtual medical record: can VMR represent therapeutic data elements used by ASTI in clinical guideline implementations? AMIA Annu Symp Proc. 2013;2013:372–8. [PMC free article] [PubMed] [Google Scholar]
- 59.HL7. Virtual medical record for clinical decision support (vMR-CDS) standards. 2015 Available from: http://wiki.hl7.org/index.php?title=HL7_CDS_Standards#Virtual_Medical_Recordfor_Clinical_Decision_Support_.28vMR-CDS.29_Standards. (accessed: 17.05.16).
- 60.Garde S, et al. Expressing clinical data sets with openEHR archetypes: a solid basis for ubiquitous computing. Int J Med Inform. 2007;76(Suppl 3):S334–41. doi: 10.1016/j.ijmedinf.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 61.Martinez-Costa C, Menarguez-Tortosa M, Fernandez-Breis JT. An approach for the semantic interoperability of ISO EN 13606 and OpenEHR archetypes. J Biomed Inform. doi: 10.1016/j.jbi.2010.05.013. [DOI] [PubMed] [Google Scholar]
- 62.Goossen W, Goossen-Baremans A, van der Zel M. Detailed clinical models: a review. Healthc Inform Res. 2010;16(4):201–14. doi: 10.4258/hir.2010.16.4.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jiang G, et al. Harmonization of detailed clinical models with clinical study data standards. Methods Inf Med. 2014;54(1) doi: 10.3414/ME13-02-0019. [DOI] [PubMed] [Google Scholar]
- 64.Jiang G, et al. A standards-based semantic metadata repository to support ehr-driven phenotype authoring and execution. Stud Health Technol Inform. 2015;216:1098. [PMC free article] [PubMed] [Google Scholar]
- 65.Li D, et al. Modeling and executing electronic health records driven phenotyping algorithms using the NQF quality data model and JBoss(R) drools engine. AMIA Annu Symp Proc. 2012;2012:532–41. [PMC free article] [PubMed] [Google Scholar]
- 66.Mazanetz MP, et al. Drug discovery applications for KNIME: an open source data mining platform. Curr Top Med Chem. 2012;12(18):1965–79. doi: 10.2174/156802612804910331. [DOI] [PubMed] [Google Scholar]
- 67.Mo H, et al. A prototype for executable and portable electronic clinical quality measures using the KNIME analytics platform. AMIA Jt Summits Transl Sci Proc. 2015;2015:127–31. [PMC free article] [PubMed] [Google Scholar]
- 68.Peterson KJ, Pathak J. Scalable and high-throughput execution of clinical quality measures from electronic health records using mapreduce and the JBoss(R) drools engine. AMIA Annu Symp Proc. 2014;2014:1864–73. [PMC free article] [PubMed] [Google Scholar]
- 69.Hripcsak G, Albers DJ. Next-generation phenotyping of electronic health records. J Am Med Inform Assoc. 2013;20(1):117–21. doi: 10.1136/amiajnl-2012-001145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Deans AR, et al. Finding our way through phenotypes. PloS Biol. 2015;13(1) doi: 10.1371/journal.pbio.1002033. [DOI] [PMC free article] [PubMed] [Google Scholar]


