Abstract
The murine Igh locus has a 3′ regulatory region (3′ RR) containing four enhancers (hs3A, hs1,2, hs3B, and hs4) at DNase I-hypersensitive sites. The 3′ RR exerts long-range effects on class switch recombination (CSR) to several isotypes through its control of germ line transcription. By measuring levels of acetylated histones H3 and H4 and of dimethylated H3 (K4) with chromatin immunoprecipitation assays, we found that early in B-cell development, chromatin encompassing the enhancers of the 3′ RR began to attain stepwise modifications typical of an open conformation. The hs4 enhancer was associated with active chromatin initially in pro- and pre-B cells and then together with hs3A, hs1,2, and hs3B in B and plasma cells. Histone modifications were similar in resting splenic B cells and in splenic B cells induced by lipopolysaccharide to undergo CSR. From the pro-B-cell stage onward, the ∼11-kb region immediately downstream of hs4 displayed H3 and H4 modifications indicative of open chromatin. This region contained newly identified DNase I-hypersensitive sites and several CTCF target sites, some of which were occupied in vivo in a developmentally regulated manner. The open chromatin environment of the extended 3′ RR in mature B cells was flanked by regions associated with dimethylated K9 of histone H3. Together, these data suggest that 3′ RR elements are located within a specific chromatin subdomain that contains CTCF binding sites and developmentally regulated modules.
The immunoglobulin heavy chain locus (Igh) (see Fig. 1) is arguably the most complex mammalian locus. B-cell development is marked by the progressive expression of Igh genes (via VDJ recombination, somatic hypermutation, and class switch recombination [CSR]). These regulated processes, some involving substantial DNA rearrangements and deletions, are required for the generation of antibody diversity (reviewed in references 47 and 57). Spanning ∼3 Mb, the murine Igh locus contains a limited number of enhancers, including the intronic enhancer (Eμ), which has been implicated in V to DJ joining and μ chain expression (reviewed in reference 47), and a complex 3′ regulatory region (3′ RR) located immediately downstream of Cα, the last constant region gene. The human Igh locus contains two 3′ RRs, each located downstream of one of the duplicated Cα genes (9, 49, 62).
FIG. 1.
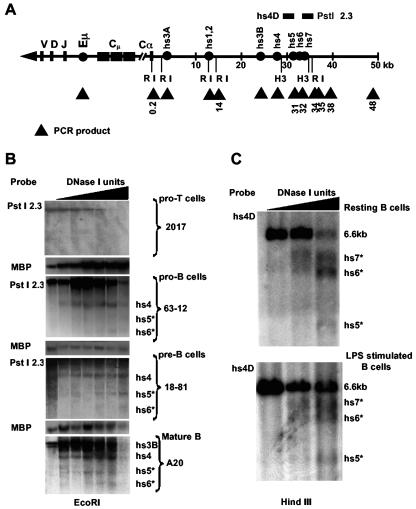
DNase I hypersensitive sites in the region downstream of hs4. (A) Schematic map of the murine Igh locus shows V, D, J, and constant region genes (solid bars) and enhancers and DNase I-hypersensitive sites (solid circles). The murine Igh locus is located on chromosome 12 and comprises hundreds of variable (V) genes, 16 diversity (D) genes, and 4 joining (J) genes spanning more than 3 Mb. Between Cμ and Cα, there are additional constant region genes: δ, γ3, γ1, γ2b, γ2a, and ɛ. The numbers represent the position, in kilobases, along the insert in BAC199M11, which begins downstream of the last exon of the Cα gene. EcoRI (RI) restriction enzyme sites are depicted, as are HindIII (H3) sites used for analysis of DNase I hypersensitivity. The horizontal bars above the line in the vicinity of hs4 represent the PstI-2.3 and hs4D probes used in this study. The PstI-2.3 probe detects a ∼22-kb RI fragment (BALB/c), while hs4D detects a ∼6.6-kb HindIII fragment (C57BL/6). Triangles are targets used for ChIP analysis. (B) EcoRI digestion of DNase I-treated nuclei from the 63-12 pro-B cell line, the 18-81 pre-B cell line, the A20 mature B cell line, and the 2017 pro-T cell line. The ∼22-kb fragment detected by PstI-2.3 includes hs4 as well as two novel DNase I-hypersensitive sites, hs5 and hs6 (*), located ∼4.5 and 3.5 kb, respectively, upstream of the distal EcoRI site. (C) HindIII digestion of DNase I-treated nuclei from resting and LPS-stimulated B cells revealed an additional hs4 downstream DNase I-hypersensitive site, hs7 (*).
The murine 3′ RR spans ∼28 kb downstream of Cα and comprises four enhancers, each associated with DNase I hypersensitivity (hs3A, hs1,2, hs3B, and hs4) (reviewed in reference 33). hs3A and hs3B are located at the termini of a 25-kb inverted repeat flanking hs1,2 (7, 66), and these three enhancers acquire DNase I hypersensitivity at later stages of B-cell development (66). The hs4 enhancer is the only 3′ enhancer with activity and DNase I hypersensitivity throughout B-cell development (19, 48), although no role for hs4 or any of the 3′ enhancers in early B-cell development has been shown.
Targeted deletions of the 3′ RR enhancers have shown that hs3A and hs1,2 are individually dispensable for B-cell development and Igh expression (46), while the combination of hs3B and hs4 is critical for the process of CSR (63). The 3′ RR has also been proposed to regulate Igh expression in plasma cells (23, 40, 48, 63, 70). In accord with earlier suggestions that it functions as a locus control region (43), the 3′ RR shows synergistic activity and position-independent regulation when tested in transgenic model systems (8, 33, 59). However, copy-number-dependent expression has not been observed (8). In addition to a function within the Igh locus, the 3′ RR is likely to be responsible for dysregulation of c-myc expression resulting from certain Igh::c-myc translocations in mouse plasmacytoma cells as well as in human Burkitt's lymphoma and multiple myeloma cells (30, 36, 43).
The unique, almost acrobatic, alterations in genomic context that occur within the extended Igh locus, and the multiple associated epigenetic modifications of the locus, do not have any apparent influence on or from the adjacent non-Igh genes. The nearest non-Igh gene is hole (55), which is located beginning ∼30 kb downstream of hs4 (68) and is followed by crip1, crip2, and mta1 (87). The transcriptional orientation of these genes is opposite to that of the Igh genes, their expression pattern does not follow the enhancer pattern of activity, and they are not subject to the DNA rearrangement and mutation processes characteristic of the Igh locus. It is therefore likely that the Igh locus is maintained within a chromatin context that, while conducive to promoting various Igh locus-specific events, can nevertheless limit its cis and trans factors from facilitating such events elsewhere in the genome. The 3′ RR is a prime candidate for exerting this complex role.
It has been shown that all known vertebrate chromatin insulators contain ∼50-bp target DNA sequences that, while varying in nucleotide sequence, are specific recognition sites of the multivalent 11 Zn finger domain of the highly conserved nuclear factor CTCF (3, 33, 35, 58). Regulation of several complex loci has been suggested to be directed by in vivo-regulated CTCF-driven chromatin insulators conducting a complex orchestra of functional communications between promoters and silencers-enhancers (reviewed in reference 77). The best-studied examples of insulator functions include CTCF target sites permanently bound by the protein at the constitutive DNase I-hypersensitive sites flanking the β-globin loci of avian (3), murine (5), and human (80) origin, as well as methylation-sensitive CTCF sites of the imprinting control region of the Igf2/h19 locus (31, 32, 61).
The ability of enhancers to control long-range events (estimated to be ∼40 to 150 kb for CSR) most likely requires the relaxation of the chromatin architecture in their vicinity, in part through posttranslational modifications of histones (reviewed in references 27 and 76). Measures of active chromatin have included histone H3 and H4 acetylation (AcH3 and AcH4) as well as histone H3 K4 methylation (di-me K4 H3). In contrast, histone H3 K9 methylation (di-me K9 H3) has been associated with heterochromatin. DNase I sensitivity has also been used to assess open chromatin environments (25, 44) and to identify potential cis regulators.
Recent studies have shown that certain histone modifications appear to be required for proper B-cell development via their effects on VDJ assembly (4, 38, 77). Beginning early in B-cell development at the pro-B-cell stage, the association of di-me K9 with Vh sequences is no longer evident (29), and a segment primed for D-J recombination (inclusive of the DH through the Cμ region and halting before Cδ) is associated with AcH3, AcH4, and histone H3 K79 methylation and is flanked by peaks of di-me K4 H3 (10, 15, 51, 56, 77). The open chromatin domain appears to be associated with chromatin remodeling complexes, as indicated by binding to BRG1 (51). Upon progression to the pre-B-cell stage, the region of acetylated histones extends 5′ to include proximal and distal subgroups of VH genes that differ in their interleukin-7 dependence (10, 11, 28) and their readiness for use in V-DJ recombination. Other histone modifications have been described for later stages of B-cell development when somatic hypermutation (83) and CSR (54, 65) occur. However, no extensive studies of histone modifications of the 3′ RR have been reported during these processes.
To begin to understand at the epigenetic level how the 3′ RR may play a significant role in limiting DNA rearrangements and expression to the Igh locus, we performed chromatin immunoprecipitation (ChIP) analyses of transitions in histone acetylation and lysine methylation and DNase I sensitivity assays during B-cell development and activation. We found strong evidence for additional cis elements downstream of hs4 based on the identification of an extended region of open chromatin downstream of hs4, in which additional DNase I-hypersensitive sites were located. These assays revealed the presence of regulated subdomains of chromatin activation in the 3′ RR and downstream sequences. A region of open chromatin appeared to terminate ∼11 kb downstream of hs4, suggestive of a potential 3′ terminus of the Igh locus. A screen of the newly extended 3′ RR in B cells with electrophoretic mobility shift assays (EMSA) identified new CTCF target sequences, and CTCF-specific ChIP assays showed in vivo occupancy.
MATERIALS AND METHODS
Cell lines.
RAG2-deficient Abelson virus-transformed murine pro-B cells (63-12) (71), a gift from K. Calame, Columbia University, New York, N.Y.; 3-1 cells (BALB/c, pre-B cells) (1); 18-81 cells (BALB/c, pre-B cells; μ) (72); A20 cells (BALB/c, B-cell lymphoma; γ, κ) (34); and Jurkat T cells were maintained in RPMI 1640 medium (BioWhittaker, Walkersville, Md.) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Gemini Bio-Products, Woodland, Calif.), 50 μM 2-mercaptoethanol, and 1% penicillin-streptomycin (Cellgro). Mouse erythroleukemia (MEL) cells (84), a gift from A. Skoultchi, Albert Einstein College of Medicine, Bronx, N.Y., and MPC11 cells (BALB/c; myeloma; γ2b, κ) (64) were maintained in Dulbecco's modification of Eagle’s medium (Cellgro) supplemented with 10% FBS and 1% penicillin-streptomycin. All cells were grown at 37°C in an atmosphere of 5% CO2.
Primary B cells.
Splenic B cells were isolated from spleens of 6- to 8-week-old female BALB/c mice (The Jackson Laboratory, Bar Harbor, Maine) as previously described (21). Briefly, spleens were homogenized to release cells, red blood cells were lysed, T cells were depleted (by using anti-T-cell antigen cocktail: anti-Thy-1.2 monoclonal antibody, anti-CD8α monoclonal antibody, and complement treatment), and CD43+ cells were removed by negative selection using anti-CD43 magnetic beads (Miltenyi Biotec, Auburn, Calif.). The resulting CD43− B220+ CD3ɛ− population of resting mature B cells was >95% CD43− B220+ as determined by fluorescence-activated cell sorting (FACS) analysis. This isolation procedure was performed four independent times. B-cell stimulation using bacterial lipopolysaccharide (LPS) was performed as previously described (21) except that 20 μg of LPS (437625; CalBiochem, San Diego, Calif.)/ml was used to stimulate the cells for 48 h. Class switching was assessed by reverse transcription-PCR of Iγ1, Iγ2b, and Iγ3 germ line transcripts, as previously described (53, 69), with GAPDH (glyceraldehyde-3-phosphate dehydrogenase) as a control (2). B cells used for DNase I hypersensitivity assays were isolated from the spleens of C57BL/6 mice after removal of T cells by use of the mouse B-cell isolation kit (catalog no. 130-090-862; Miltenyi). The resulting population was >99% B220+ by FACS.
Bone marrow pro-B cells were isolated from the femurs and tibias of 6- to 8-week-old female BALB/c RAG1−/− mice (The Jackson Laboratory) as previously described (10), with minor modifications. After bone marrow cells were recovered, a 5-min lysis of red blood cells was performed. Additionally, prior to positive selection by anti-CD19 magnetic beads, the cells were preblocked for 15 min on ice with 0.2 μg of rat anti-mouse FcγIII/II receptor antibody (BD Pharmingen) per 106 cells. Recovered cells were 99% B220+ CD19+ MAC1−, as determined by FACS analysis. Primary splenic B cells and bone marrow-derived pro-B cells were maintained in RPMI for <1 h prior to formaldehyde cross-linking for ChIP assays.
DNase I hypersensitivity assay.
The DNase I hypersensitivity assay was performed as described previously (19). Briefly, cell nuclei isolated by Dounce homogenization (2.5 × 107 cells per treatment condition) were treated with increasing doses of DNase I (Sigma) for 30 s to 10 min at room temperature or at 37°C. DNA was isolated by use of a DNA blood midi kit (QIAGEN, Valencia, Calif.). DNase I-treated samples from the pro-T-cell line 2017 were a gift from R. Sen, National Institute of Child Health and Human Development, National Institutes of Health (NIH), Baltimore, Md. (10). Ten to fifteen micrograms of DNA was digested for 24 h with EcoRI (New England Laboratories) or with HindIII and separated on a 0.7% agarose gel in 1× Tris-acetate-EDTA buffer. The DNA was transferred to a Hybond-N+ membrane (Amersham Pharmacia Biotech) by the Southern technique (per the manufacturer's procedure and reference 74). Southern blots were prehybridized for 2 h in hybridization solution (50% formamide, 1 M NaCl, 1% sodium dodecyl sulfate [SDS], 5 mM Tris-HCl [pH 7.4], 0.1 mg of denatured salmon sperm DNA/ml) at 42°C. Southern blots were hybridized with myelin basic protein (78) and PstI-2.3 (23) (EcoRI digests) or with a probe extending from 28382 to 30156 (BAC199M11; GenBank accession no. AF450245) (HindIII digests) at a final concentration of 1 × 106 to 2 × 106 cpm/ml for 16 to 24 h at 42°C. Blots were washed two times with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at room temperature and one or two times in 2× SSC-1% SDS at 65°C and subjected to autoradiography at −80°C for various lengths of time with KODAK Biomax MS film.
ChIPs.
The method of Kuo and Allis (37) was adapted from a protocol from Upstate (Lake Placid, N.Y.) as follows. Cells (108) were cross-linked in 225 ml of medium by the addition of 22.5 ml of fixation solution (50 mM HEPES, pH 8; 1.0 mM EDTA, 100 mM NaCl, 0.5 mM EGTA, 4% formaldehyde) for 5 to 10 min at room temperature or at 37°C, as optimized for each cell line and cell population used (18-81 cells, 10 min at 37°C; 3-1 cells, 10 min at 37°C; A20 cells, 5 min at room temperature; MPC11 cells, 10 min at 37°C; MEL cells, 10 min at 37°C; RAG1−/− pro-B cells, 10 min at 37°C; and mouse splenic B cells, 10 min at room temperature). Fixation was stopped by adding glycine to a final concentration of 0.125 M and incubating at 4°C for 10 min. Protease inhibitors (P-8340; Sigma) were added to all buffers immediately before use (final concentration of 1 ml per 109 cells). Cells were washed twice with cold phosphate-buffered saline and lysed at a concentration of 4 × 107 cells/ml in lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl, pH 8.1) in the presence of 10 mM sodium butyrate (Upstate [U.B.]) for 10 min. DNA fragments between 200 and 1,000 bp were generated by sonication using a cell type-specific optimized number (four to nine) of 30-s continuous pulses provided by a Sonic Dismembrator (Fisher Scientific model 500) at 55% amplitude at 4°C. Cell debris was removed by centrifugation at 14,000 rpm with an Eppendorf 5417C centrifuge for 10 min, and chromatin from ∼0.5 × 107 cells was used per pull-down. Immunoprecipitations (IP) were performed at 4°C. Chromatin samples were diluted in a total volume of 400 μl of IP buffer (1% Triton X, 0.1% Na-deoxycholate, 0.14 M NaCl in DNase- and RNase-free water) (Gibco) and precleared with 80 μl of salmon sperm DNA-protein A-agarose 50% slurry (Upstate) for 2 to 4 h. Immunoprecipitations were done overnight using 4 μg of one of the following antibodies: normal rabbit immunoglobulin G (IgG) (U.B. catalog no. 12-370), anti-acetylated histone H3 recognizing the K9 and K14 diacetylated tail (U.B. catalog no. 06-599), anti-acetylated histone H4 recognizing a K3, K6, K10, and K14 tetra-acetylated tail (U.B. catalog no. 06-866), anti-dimethyl K4 of histone H3 (U.B. catalog no. 07-030), anti-dimethyl K9 of histone H3 (U.B. catalog no. 07-212 or Abcam catalog no. ab1772) or anti-CTCF (U.B. catalog no. 06-917, lot no. 06-917). For semiquantitative PCR analysis, a water control without antibody addition was also performed. The salmon sperm DNA-protein A-agarose slurry (60 μl) was added for a 4-h immunoprecipitation. Beads were pelleted at 3,000 rpm for 2 min. The supernatant from the IgG sample was recovered and used as the “input” sample. Immunoprecipitations were washed two to three times in wash buffer I (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.1, 150 mM NaCl), two to three times in wash buffer II (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.1, 500 mM NaCl), two to three times in wash buffer III (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris-HCl, pH 8.1), and three to four times in Tris-EDTA. Complexes were eluted twice by using 250 μl of IP elution buffer (0.1 M NaHCO3, 1% SDS). The eluates were treated with RNase A (0.2 mg/ml) at 55°C for 1 h. Twenty microliters of 5 M NaCl were added, and the cross-links were reversed by an overnight incubation at 65°C. Samples were treated with proteinase K (0.2 mg/ml) for 2 h at 45°C, and DNA was isolated by 1× phenol-chloroform-isoamyl alcohol extraction, 1× chloroform-isoamyl alcohol extraction, and overnight ethanol precipitation. DNA was dissolved in 50 μl of DNase- and RNase-free water. The DNA samples were analyzed by semiquantitative PCR or real-time quantitative PCR. Differential amplification efficiency of PCR primers was accommodated by use of standard curves and input DNA to normalize all primer pairs in every experiment. Chromatin for CTCF ChIP analysis was prepared as described elsewhere (79).
PCR analysis.
Table 1 lists the primers used for PCR analysis. New primers were designed based on the BAC199M11 (125-kb insert) sequence from 129S1/SvImJ mice (accession no. AF450245). Numerical assignments of the primer pairs correspond with the location (in kilobases) of their amplified product in BAC199M11, i.e., from immediately downstream of the Cα membrane (87) (see Fig. 1). For semiquantitative PCR, 1 μl of input and 5 μl of the pull-down samples were used and subjected to a 1:3 serial dilution. Targets were amplified by using 20 pmol of each primer and Taq polymerase (Roche) for 32 cycles in a ThermoHybaid PCR machine. PCR products were separated in a 2% agarose gel. All primer pairs were subjected to an annealing temperature optimization process and verified to produce a single product. The identity of all targets was verified by sequencing (Albert Einstein College of Medicine sequencing facility). Samples were also analyzed quantitatively by real-time PCR. Standard curves of SYBR green fluorescence signals obtained from diluted input DNA from each cell line and primary B-cell source were used to quantify the amount of target DNA in the immunoprecipitated samples. Triplicate reactions using SYBR Green Master Mix (Applied Biosystems) were analyzed in an ABI PRISM 7700HT in the 384-well format. Each reaction contained 4 μl of the SYBR green mix, 2 μl of a 1.2 μM concentration of each primer, and 2 μl of input or pull-down DNA samples (1:100 dilution of the input samples or 1:3 to 1:5 dilutions of the experimental samples). To ensure that a single product was amplified, PCR products were initially analyzed by electrophoresis on agarose gels, and the dissociation curve for each PCR product was routinely examined. Real-time data were quantitated according to an established protocol (Essentials of Real Time PCR, User Bulletin 2 for the ABI Prism 7700 Sequence Detection System, The Relative Standard Curve Method and Creating Standard Curves with Genomic DNA or Plasmid DNA Templates for Use in Quantitative PCR [ABI, Foster City, Calif.]) and then plotted as relative enrichment (acetylation, methylation, or CTCF binding), which is defined as the amount of target DNA in the experimental sample divided by the amount of target DNA in the input sample.
TABLE 1.
PCR primers used in ChIP analysis
| Locus | Label on graphsa | Gene | Primer | Upstream primer sequence | Reference | Useb |
|---|---|---|---|---|---|---|
| CAD | CAD | CAD | Upstream | CACCTTTTCCCCCGTTTT | 51 | SQ and RTM |
| Downstream | CAAGCCAGAGCCCTAAGC | |||||
| β-globin | β-globin | β-Globin gene | Upstream | GCCTTGCCTGTTCCTGCTC | 11 | SQ and RTM |
| Downstream | CAGACCATAAACTGTATTTTTCTTATTGAGCCC | |||||
| Globin hs3 | Globin hs3 | Upstream | TGTTTCCCTGATGAGGATTCAATGG | 18 | SQ | |
| Downstream | CCCACACATGGTCATCTATCTGAGC | |||||
| Myc | Myc | Myc (N site) | Upstream | AAGGAAGCATCTTCCCAGAAC | RTM | |
| Downstream | AAGTGTGCCCTCTACTGGCCA | |||||
| IgH | Eμ | Eμ | Upstream | GGAATGGGAGTGAGGCTCTCTC | 10 | SQ |
| Downstream | GCTGCAGGTGTTCCGGTTCTGATCGGCC | |||||
| Eμ | Eμ | Upstream | TGGCAGGAAGCAGGTCAT | 51 | RTM | |
| Downstream | GGACTTTCGGTTTGGTGG | |||||
| 0.2 | 0.2 | Upstream | GCCCTATATTGGTAACTGAACCGTG | SQ and RTM | ||
| Downstream | GCTATGATCAAAACCAGTCATCGG | |||||
| 1.6 | hs3A | Upstream | GGGTAGGGCAGGGATGCTCACAT | 22 | SQ and RTM | |
| Downstream | GCTCTGGTTTGGGGCACCTGTGC | |||||
| 11.9 | hs1,2 | Upstream | CACACAGCTGCCGAGTTACTCA | SQ | ||
| Downstream | CATCCTTGCCCATCTCCTGTCA | |||||
| 12.0 | hs1,2 | Upstream | AGCATAGGCCACTGGGACTGG | RTM | ||
| Downstream | CTCTCACTTCCCTGGGGTGTT | |||||
| 14 | 14 | Upstream | CTTACAGCCCCACATGGCTATAGC | SQ and RTM | ||
| Downstream | CTGACAGAGACAGACAGCGTTCC | |||||
| 24.5 | hs3B | Upstream | TGGTTTGGGGCACCTGTGCTGAG | 22 | SQ and RTM | |
| Downstream | GGGTAGGGCAGGGATGTTCACAT | |||||
| 27.7 | hs4 | Upstream | CCATGGGACTGAAACTCAGGGAACCAGAAC | SQ and RTM | ||
| Downstream | CTCTGTGACTCGTCCTTAGG | |||||
| 30.9 | hs5 or 31 | Upstream | GCTTGCCTTCTGGGCTAGAAC | SQ and RTM | ||
| Downstream | AGCAGCCAGTGCTCTTAACCC | |||||
| 32.1 | hs6 or 32 | Upstream | GTTGAGTCTGAAGATACACGCG | SQ and RTM | ||
| Downstream | TCGCAGCTATTCCCAACTACG | |||||
| 34.3 | hs7 or 34 | Upstream | AATACGTTCCTAGGGCCTAACGATACT | SQ and RTM | ||
| Downstream | TGGAGACGCCACAGGTGTATC | |||||
| 35.1 | 35 | Upstream | AATACGTTCCTAGGGCCTAACGATACT | SQ and RTM | ||
| Downstream | ATGCCCTTCCCCATCGTAACT | |||||
| 38.1 | 38 | Upstream | CCTGTGGACACATGGTTACGC | SQ and RTM | ||
| Downstream | GTTGGCAAGAGGACTCACTACGTC | |||||
| 48.5 | 48 | Upstream | GGTGGGAAGGAGTGGAGCTAC | SQ and RTM | ||
| Downstream | AATGTGGAGATGCGAGATTGCCC |
EMSA analysis for CTCF binding sites.
Consecutive overlapping DNA fragments of ∼250 bp were amplified and end labeled by PCR for the ∼6-kb region beginning with hs4 and extending downstream. EMSA using murine DNA as a template was performed by using recombinant CTCF, as previously described (16). Binding reactions were carried out in the presence of poly(dI-dC), poly(dG)-poly(dC), and the 44-mer double-stranded FpV oligonucleotide containing overlapping Sp1/Egr1-binding sites.
Colony assay for insulator activity.
Originally formulated by Zhong and Krangel (86), the colony assay for insulator activity was carried out as recently described (20). Constructs provided by M. Krangel, Duke University, were the basis for analysis of insulator activity in stable transfection assays. These constructs contained various constellations of the T-cell-specific Vδ promoter, the Εα/δ enhancer, and the known BEAD-1 insulator (86), together with a NeoR reporter gene, followed by the scs′ Drosophila insulator. Fragments to be analyzed (hs5, 30516 to 31500; and CTCF/hs7, 32289 to 33879) were inserted in both orientations between the promoter and the upstream enhancer at unique SalI sites (numbers represent the base number in BAC199M11 [AF25045]). DNA was introduced into Jurkat T cells by electroporation, and cells were cultured for 48 h in RPMI containing 10% FBS and then plated in 2% soft agar containing the same medium together with Jurkat-conditioned medium, l-glutamine and 1 mg of G418/ml. Plates were incubated for ∼3 weeks, after which NeoR colonies were counted.
RESULTS
DNase I hypersensitivity downstream of hs4.
The hs4 enhancer was previously detected as a 15-kb fragment after DNase I digestion by genomic Southern analysis using a probe for the 5′ end of the 22-kb EcoRI fragment in which it was located, and no additional 3′ sites were detected (19, 43). However, a probe (PstI-2.3), which hybridizes to the 3′ end of the EcoRI fragment (Fig. 1), detected additional weak DNase I-hypersensitive sites downstream of hs4 in pro-B (63-12), pre-B (18-81), and mature B (A20) cells (and also in the MPC11 plasma cell line [data not shown]) but not in a pro-T-cell line (2017). A probe for myelin basic protein was used as a negative control, and hs3B and hs4 in A20 (and MPC11) and hs4 in the other B-cell lines were positive measures for the assay. The most prominent of the hs4 downstream DNase I-hypersensitive sites (now called hs5 and hs6) were located ∼3.5 and 4.5 kb downstream of hs4. Using HindIII (and BglII [data not shown]) digests and different probes, we confirmed the presence of these sites and identified an additional downstream site (hs7) in both resting and LPS-activated splenic B cells (Fig. 1). These data are indicative of a potential downstream extension of the 3′ RR.
Histone acetylation of the Igh 3′ RR during B-cell development.
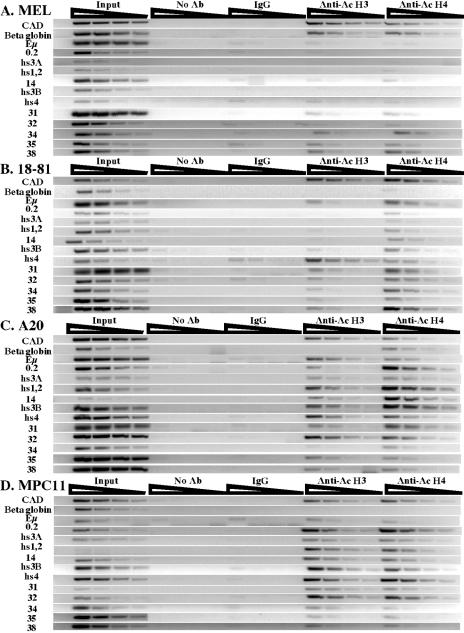
The presence of acetylated histones has been correlated with active chromatin domains (27). To determine whether there were changes in histone acetylation of the Igh 3′ RR and in the region containing additional downstream DNase I-hypersensitive sites during B-cell development, ChIP assays were performed on an MEL cell line, the 18-81 pre-B-cell line, a mature B-cell line (A20), and a plasma cell line (MPC11). Acetylated histone H3 and H4 (AcH3 and AcH4) were targeted as the immunoprecipitation epitopes, and analysis of enhancers of the 3′ RR and regions of additional DNase I hypersensitivity was done by semiquantitative PCR. Neither Eμ nor any of the 3′ RR sequences was associated with AcH3 or AcH4 in the MEL non-B-cell line (Fig. 2A). Eμ and hs4 were both associated with AcH3 in the pre-B-cell, B-cell, and plasma cell stages (Fig. 2B, C, and D). hs3A, hs1,2, and hs3B gained this association with AcH3 at the mature B-cell stage (Fig. 2C) and maintained this association in the plasma cell stage (Fig. 2D).
FIG. 2.
Developmental analysis of AcH3 and AcH4 association at the 3′ RR. ChIP assays were performed on chromatin from the MEL cell line (A), 18-81 pre-B cell line (B), A20 mature cell line (C), and MPC11 plasma cell line (D). No antibody and IgG serve as experimental controls. While β-globin and CAD (the gene encoding carbamoyl-phosphate synthetase II [EC 6.3.5.5], aspartate transcarbamylase [EC 2.1.3.2], and dihydroorotase [EC 3.5.2.3]) serve as positive controls, Eμ is used as a negative control in the MEL cell line. CAD and the β-globin gene are used as a positive and a negative control for histone acetylation, respectively, in B cells. The primers used are indicated next to relevant gels. Shown are serial (1:3) dilutions of input and IP samples for the semiquantitative PCR analysis. The signal for hs1,2 in the input of MPC11 is absent due to a technical error.
To determine whether acetylation was associated only with enhancers of the 3′ RR, we examined a segment 1 kb upstream of hs3A, termed “0.2,” and another segment 2 kb downstream of hs1,2, termed “14,” both numbered based on their location along BAC199M11 (see Materials and Methods and Table 1 for details). Like hs3A, hs1,2, and hs3B, fragments 0.2 and 14 were also associated with AcH3 in A20 and MPC11 cells. The associations observed with AcH3 were similarly detected with AcH4. We conclude that the 3′ RR acquires association with AcH3 (and AcH4) in a developmentally regulated manner, first hs4 in pre-B-cell lines, which then extends to include hs3A, hs1,2, and hs3B in B cells (together with 0.2 and 14), while the intronic enhancer appears to be acetylated at all stages of B-cell development tested.
Sequences downstream of hs4 in which DNase I-hypersensitive sites were detected were also associated with AcH3 and AcH4 in B-cell lines. In 18-81, the region from “31” to “38” showed less association with AcH3 than with AcH4, while this segment showed comparable associations with both acetylated histones in A20 and MPC11 cells. In MEL cells, however, the lack of association of the 3′ RR with AcH3 and AcH4 extended several kilobases downstream of hs4.
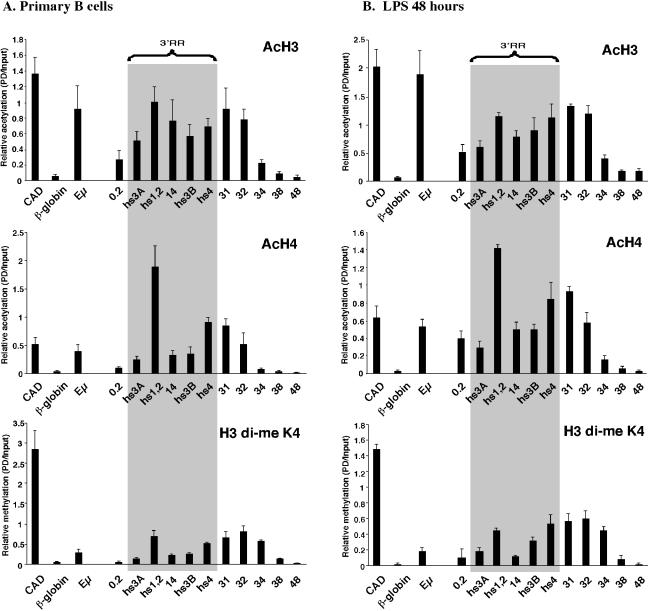
Histone acetylation of the 3′ RR and additional downstream sequences in primary B cells.
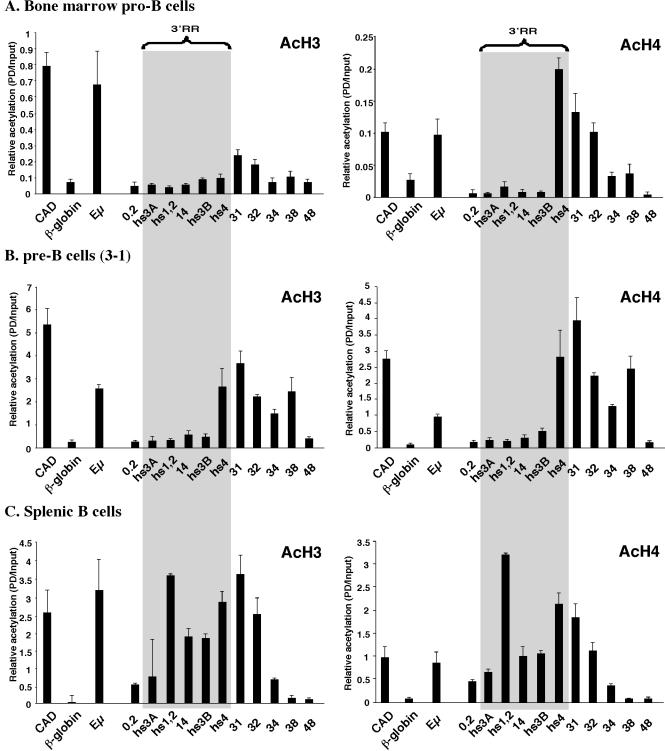
We used real-time PCR analysis to extend these observations to pro-B cells isolated from the bone marrow of RAG1−/− BALB/c mice (CD19+ B220+ Mac1−; 99% purity) (50) (Fig. 3A). In pro-B cells, hs4 was associated with AcH4 but not with AcH3 (with a level comparable to that of the β-globin negative control). This epigenetic state is in contrast to that observed in 18-81 (Fig. 2) and 3-1 (Fig. 3B) pre-B cells, where hs4 was associated with both AcH4 and AcH3. The histone acetylation profile of splenic B cells (CD43− B220+; ≥95% purity) (Fig. 3C) was similar to that of A20 cells (Fig. 2) in that the entire 3′ RR extending from 0.2 through hs4 was associated with AcH3 and AcH4. Similar observations from ChIP analysis by two different methods on cells representing a similar stage of B-cell development (e.g., for 3-1 and 18-81 pre-B-cell lines and A20 and normal splenic B cells) support the use of cell lines in this study and reinforce our findings of developmentally associated histone modifications.
FIG. 3.
Association of AcH3 and AcH4 at the 3′ end of the Igh locus in mouse primary pro-B-cell and splenic B-cell populations and a pre-B-cell line. Shown are real-time PCR analyses of a ChIP done with anti-AcH3 and with anti-AcH4 on pro-B cells isolated from the bone marrow of a RAG1-deficient mouse (A), the 3-1 pre-B-cell line (B), and splenic B cells (C). The primers used are indicated below the histogram bars. Data are plotted as a numerical comparison of signal in immunoprecipitated samples to that detected in the input, as described in Materials and Methods. A gray swath represents the 3′ RR extending from hs3A to hs4.
To determine where downstream of hs4 the open chromatin environment ended, additional sequences were analyzed for association with AcH3 and AcH4 (Fig. 2 and 3). AcH3 and AcH4 were detected at sequences 4 and 5 kb downstream of hs4 (primer sets 31 and 32) in bone marrow pro-B cells, pre-B cells, and splenic B cells. In pre-B-cell lines, acetylation extended another 6 kb downstream (primer sets 34 and 38). There was no detectable acetylation in any of these cell lines at a region that lay an additional 10 kb downstream (primer set 48). These data further support the conclusion that the cis elements of the 3′ RR undergo a stepwise association with epigenetic modifications associated with open chromatin, beginning with hs4 downstream sequences in pro-B cells and progressively moving 5′ to include hs4 in pre-B cells and hs3A, hs1,2, and hs3B in B cells. The epigenetic extension of acetylated histones at the 3′ end, coupled with the presence of DNase I-hypersensitive sites in this segment, further suggests the possibility that the 3′ RR extends downstream of hs4.
Histone H3 methylation at the 3′ end of the Igh locus.
Methylated histones have been found to demarcate regions of active and inactive chromatin. Histone H3 K4 methylation (di-me K4 H3) has been associated with open or active chromatin, in some cases comprising a domain spanning an entire locus, e.g., the chicken β-globin locus, and in other cases focused only on cis regulators, e.g., the mouse β-globin locus (5, 42). In contrast, histone H3 K9 methylation (di-me K9 H3) is a mark of heterochromatin (reviewed in reference 14) and has been localized to boundaries of open chromatin (20, 41, 51). To determine whether H3 methylation delineates different chromatin regions at the extended 3′ RR, ChIP assays targeting these epitopes were performed.
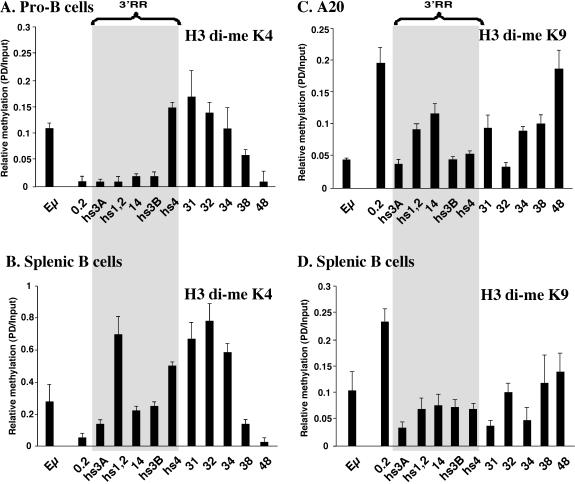
ChIP analysis of the bone marrow-derived pro-B cells (Fig. 4A) revealed that, like AcH4, di-me K4 H3 was associated with Eμ as well as with hs4 and its downstream flanking sequences (within ∼11 kb) but not with enhancers hs3A, hs1,2, or hs3B. The presence of di-me K4 H3 at Eμ in normal pro-B cells was in contrast to its absence in a 63-12 pro-B-cell line (51). A similar difference has been reported for the histone acetylation status of the Cμ-Cδ intergenic region (10) and may represent a slight difference in the developmental stage represented by these two sources of pro-B cells. Results from mouse splenic B cells (Fig. 4B) revealed that di-me K4 H3 was additionally associated with hs3A, hs1,2, and hs3B. Association with di-me K4 H3 is markedly less in splenic B cells at the flanks of the extended 3′ RR, e.g., at 0.2/hs3A and also at 38/48. Hence, the pattern of association of the 3′ RR with di-me K4 H3 is similar to that observed with AcH4.
FIG. 4.
Reciprocal association of di-me K4 H3 and di-me K9 H3 at sequences flanking the extended 3′ RR and real-time PCR analysis of di-me K4 H3 and di-me K9 H3 modifications. Shown is the enrichment of di-me K4 H3 in pro-B cells (A) and in splenic B cells (B) and the enrichment of di-me K9 H3 in A20 cells (C) and in splenocytes (D).
The pattern of association of the 3′ RR with di-me K9 H3, as assayed in the A20 B-cell line (Fig. 4C) and in splenic B cells (Fig. 4D), differed from that observed with di-me K4 H3. While the entire 3′ RR was associated with both forms of methylated histones, the upstream flank of the 3′ RR, i.e., 0.2, showed a significant enhancement of association with di-me K9 H3 over that seen with di-me K4 H3. In A20 B cells, the downstream “48” region showed a similar high level of association. An increase in K9 methylation was also detected in this downstream region in splenic B cells, although it was not as pronounced and appeared to extend over a longer interval. No activity has been ascribed to either the 0.2 or 48 regions at any stage of development in this lineage.
We conclude that the cis elements of the 3′ RR acquire association with both di-me K4 H3 and acetylated histones in a stepwise manner in primary B cells (summarized in Table 2). This epigenetic modification extends to include sequences as much as ∼11 kb downstream of the last known element, hs4, in pro-B cells and mature B cells. Furthermore, at the mature B-cell stage, there is an inverse correlation between the association with di-me K4 H3 and di-me K9 H3 at the 5′ and 3′ ends of the extended 3′ RR of the Igh locus, indicative of an epigenetic domain for the 3′ RR.
TABLE 2.
Summary of AcH4, di-me K4 H3, and AcH3 modifications at the 3′ end of the Igh locus during B-cell development
| Histone modification | hs3A, hs1,2, and hs3Ba
|
hs4
|
hs5, hs6, and hs7
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pro | Pre | Mat | Plas | Pro | Pre | Mat | Plas | Pro | Pre | Mat | Plas | |
| AcH4 | − | +/− | + | + | + | + | + | + | + | + | + | + |
| H3 di-me K4 | − | − | + | + | + | + | + | + | + | + | + | + |
| AcH3 | − | − | + | + | − | + | + | + | + | + | + | + |
Pro, pro-B cells; Pre, pre-B cells; Mat, mature B cells; Plas, plasma cells. +, significant enhancement of the relevant modification; −, no association was detected in ChIP assays.
Histone modifications of the Igh regulatory elements after B-cell stimulation.
The 3′ RR cis elements play an essential role in the regulation of germ line transcription of multiple isotypes prior to CSR (63). We therefore wanted to determine whether changes in epigenetic modifications at the 3′ RR accompany this process. LPS induces the entry of splenic B cells into the G1 phase of the cell cycle as well as CSR to γ2b and γ3 (reviewed in reference 75); both isotypes are included in those regulated by the 3′ RR. ChIP assays were performed on splenic B cells treated with LPS for 48 h (Fig. 5B), and results were compared to the 0-h time point (Fig. 5A; resting B cells were isolated in an experiment independent from that described in Fig. 3C and 4B and D). CSR was monitored by the appearance of γ3 and γ2b germ line transcripts (as described in reference 69). Interestingly, no significant alterations in the pattern of histone acetylation or of di-me K4 H3 at the 3′ RR were observed. However, the levels of AcH3 and AcH4 associated with 3′ enhancers, especially AcH4 at hs1,2, decreased relative to controls (CAD and Eμ) after LPS stimulation.
FIG. 5.
Histone H3 and H4 acetylation as well as di-me K4 H3 at the Igh 3′ RR in cells undergoing CSR. ChIP assays with antibodies to AcH3, AcH4, and di-me K4 H3 were performed on freshly isolated splenic B cells (A) or after LPS stimulation for 48 h (B).
CTCF binding sites downstream of hs4.
CTCF is the first example of a multivalent nuclear factor capable of specific recognition of various DNA sites with no single consensus sequence (reviewed in references 17 and 58). A growing number of different CTCF target sites in various genomes have been implicated in a variety of regulatory functions, including promoter repression and activation. Moreover, all vertebrate chromatin insulators identified so far interact with CTCF, including the insulator in the IGF2/H19 locus that governs the imprinting control region (24, 31). Because the 3′ RR region associated with acetylated histones and methylated histone H4 extends downstream of hs4, we wanted to determine whether CTCF binding could be detected in this region. In addition, it is known that CTCF is associated with DNase I-hypersensitive sites (16), and we have detected additional hypersensitive sites downstream of hs4.
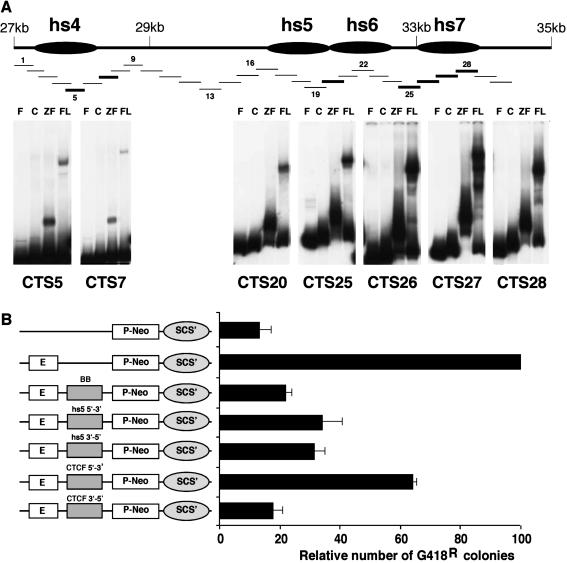
We therefore carried out EMSA on 30 overlapping DNA fragments from the region extending from hs4 through hs7 (Fig. 6A). The selected fragments were of sufficient size for detection of CTCF binding. EMSA analysis detected seven CTCF binding sites in this segment, associated with hs4 (CTS5 and CTS7) and hs5 and hs6 (CTS20), with four strong sites (CTS25 to CTS28) located in the vicinity of hs7 (∼33.7 with respect to BAC199M11) (Fig. 1 and 6A). Using a stable transfection assay (86), we examined selected 3′ RR fragments containing CTCF binding sites for insulator activity. The number of G418R colonies in Jurkat cells is augmented by enhancer-promoter interaction (Fig. 6, compare the results with P-Neo alone to those obtained upon addition of the enhancer). While experiments with this insulator system have shown that the introduction of irrelevant DNA segments had no inhibitory effects on NeoR expression (20, 86), the introduction of BB (the BEAD-1 insulator) between the enhancer and the promoter substantially diminished the expression of the NeoR gene, as previously reported (86). Both hs5 and the region downstream of hs6 containing four strong CTCF binding sites also had substantial insulator activity (Fig. 6B). The latter fragment showed some evidence of orientation-dependent insulator activity.
FIG. 6.
Identification and analysis of CTCF sites in the region downstream of hs4. (A) In-scale schematic of the 3′ RR showing 30 consecutive overlapping DNA fragments (fragment numbers intermittently indicated) used for EMSA. EMSA identified CTCF binding sites associated with hs4, hs5, and the region downstream of hs6, i.e., hs7. F, free DNA probe; C, control TnT reaction with luciferase template; ZF, 11 zinc finger region of CTCF; FL, full-length CTCF. (B) Assessment of insulator activity of hs5 and the region containing strong CTCF binding sites (hs7) (see Materials and Methods for exact fragments tested) by using a stable transfection assay that measures NeoR colonies. The number of colonies detected with the construct containing both the T-cell receptor enhancer and the promoter ranged from ∼130 to 180 in different experiments. We set this number at 100 and normalized all the other data accordingly. BB identifies the BEAD-1 insulator and is used as a positive control.
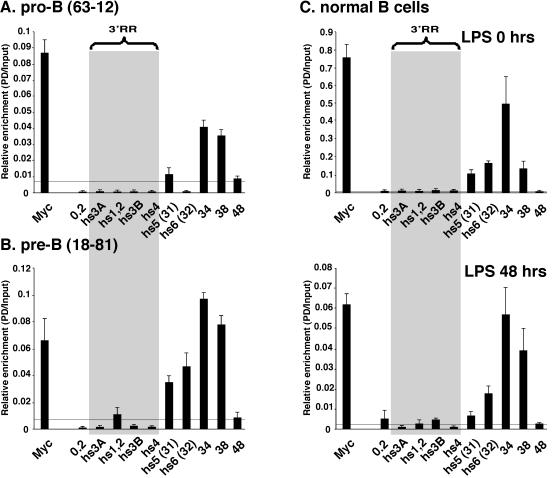
ChIP assays were used to assay the interaction of CTCF with 3′ RR sequences in cells representing different stages of B-cell differentiation (Fig. 7). In 63-12 pro-B cells, we detected binding not only to the 34 segment, which contains four strong CTCF binding sites, but also to the 38 region ∼4 kb downstream. In 18-81 pre-B cells, in addition to the 34 and 38 regions, the hs5 and hs6 region also showed CTCF binding. In resting splenic B cells, association with 34 was most prominent, with additional smaller interactions at hs5, h6, and 38 (Fig. 7). After LPS activation, the 38 region became more strongly associated with CTCF.
FIG. 7.
ChIP analysis of CTCF binding in 3′ RR in 63-12 pro-B cells (A), 18-81 pre-B cells (B), and resting and LPS-stimulated normal B cells (C). The horizontal line above the x axis represents the highest level of signal detected with normal IgG.
DISCUSSION
We have shown that the known cis regulatory elements of the 3′ RR undergo stepwise opening during B-cell development (summarized in Table 2). Although in MEL non-B cells none of the 3′ enhancers was associated with acetylated histones, in pro-B cells, hs4, the most distal enhancer, was associated with AcH4 (but not with AcH3) and with di-me K4 H3. Progression to the pre-B-cell stage was characterized by the additional acquisition of AcH3 at the hs4 enhancer. Mature B cells appear to extend the association with AcH3, AcH4, and di-me K4 H3 to include the entire 3′ RR region. This pattern of histone modifications was similar in plasma cells. Due to the large number of cells required for ChIP, we analyzed only primary pro-B and splenic B cells and relied on cell lines to complete the timeline of differentiation. Our data support the conclusion that cell lines can provide adequate systems to investigate the epigenetic changes of the Igh locus, as also used by others (10, 28, 51, 83).
The association with histone modifications characteristic of open chromatin environments extended downstream of hs4 to include ∼11 kb of sequence in which additional DNase I hs sites were detected in all B-cell lines and sources tested. This segment downstream of hs4 is associated with AcH3, AcH4, and di-me K4 H3 from the pro-B-cell stage onwards, thus distinguishing its regulation from hs4, which is not associated with AcH3 in pro-B cells. Previous experiments have addressed the possibility that additional enhancers are located downstream of hs4. Some lines of investigation suggested that hs4 was the most distal 3′ enhancer. For example, while NeoR replacement, individually, of hs3A (46) and hs1,2 (12) had strong deleterious effects on CSR, insertion of the NeoR gene 2 kb downstream of hs4 resulted in no detectable phenotype (45). This observation suggested that a boundary for CSR was located within close proximity to hs4. Targeted deletion of hs3B and hs4 together (63), in fact, provided a phenotype comparable to that provided by upstream NeoR insertions, consistent with this prediction. However, the phenotype observed when the NeoR gene was retained in place of the combination of hs3B and hs4 was stronger than that observed when these two enhancers were cleanly deleted (63), suggesting that the NeoR gene was interfering with the function of additional regulators downstream of hs4 that contributed to CSR and/or IgM expression levels. Our identification of these 3′ DNase I-hypersensitive sites in the context of an extended open chromatin region predicts potential functional roles for these elements in Igh processes.
While preliminary experiments testing hs5 and hs6 in reporter constructs have not detected enhancer activity in a plasma cell line (F. E. Garrett, unpublished observations), these two segments are positioned in a region with potential for insulator function between the Igh locus and the next non-Igh genes (hole, 33 kb downstream from hs6 [55, 69]) and other genes farther downstream (87), none of which is expressed in a B-cell-specific manner. Insulators are DNA elements that protect genes from inappropriate neighboring regulatory signals by enhancer blocking activity and/or by delineating chromatin domains (82). All vertebrate insulators tested so far have been associated with CTCF binding sites (5, 20, 82, 86). Conversely, CTCF binding sites identified by ChIP experiments have been associated with insulator activity in in vitro assays (52). Nonetheless, it should be noted that some CTCF sites are known to be involved in transcriptional activation in vivo, even when scored as insulators in vitro (81, 85).
Semisystematic screening of the ∼7-kb region extending 3′ from hs4 for CTCF binding sites identified individual CTCF binding sites in the vicinity of hs4 (CTS5 and CTS7) and hs5 and hs6 (CTS20) together with a cluster of four very strong sites (CTS25 to CTS28) in which hs7 was located. CTCF binding sites associated with hs5, hs6, and hs7 are occupied in vivo, as assessed in cell lines representing various stages of B-cell development and normal splenic B cells. We propose that the 3′ RR thus extends several kilobases beyond hs4. Our previous studies revealed the presence of a constitutively undermethylated region (19) that we now know to be located in the vicinity of the mapped CTCF binding sites, thereby rendering these sites broadly accessible to CTCF binding. Although the role of each individual CTCF site in the 3′ RR remains to be elucidated, the association of these sites with changes in histone modification and in vitro insulator activity suggests an important role in the overall regulation of the locus. Furthermore, high levels of di-me K9 H3 are evident at the 5′ end of the 3′ RR, and di-me K9 H3 is also associated with the 3′ end of this regulatory region, although in a less pronounced manner. Together, these data suggest that the entire extended 3′ RR is located within a chromatin subdomain.
This study documents for the first time that the murine 3′ RR cis elements are subject to association with histone acetylation and histone H3 K4 methylation. This association appears to evolve in a developmentally regulated stepwise progression, impacting subsets of the extended 3′ RR (i.e., first the region containing hs5, hs6, and hs7; then hs4; and lastly hs3A, hs1,2, and hs3B). This progression is unlike the collective association of enhancers of the β-globin locus control region with acetylated histones (41). As the association with AcH3 is acquired at the 3′ RR elements after AcH4 and di-me K4 H3 are detected, and its pattern correlates with in vitro enhancer activity, we suggest that AcH3 serves as an index of enhancer function. Accordingly, the human Igh hs4 enhancer is associated with AcH3 only in B cells, while it is associated with AcH4 in both B and T cells (68). A preferred association of AcH3 with active chromatin has also been observed at the human β-globin locus (67).
The 3′ enhancers hs3B and hs4 have been implicated in the control of CSR through their regulation of germ line transcripts of multiple isotypes (63). Analysis by ChIP of epigenetic modifications at the 3′ RR after CSR induction by LPS did not reveal substantial changes in the pattern of di-me K4 H3, AcH3, or AcH4 modifications, although a small decrease in the levels of AcH3 and AcH4 relative to Eμ was observed at some of the 3′ RR cis elements, especially hs1,2. While modest changes in the 3′ RR could reflect the relatively limited percentage of cells that undergo CSR, more substantial increases in histone acetylation at germ line transcript promoters and switch regions have been detected after similar exposure to LPS (39, 54; F. E. Garrett, unpublished observations). The measurement of acetylated histones at the 3′ RR could be a summation resulting from masking of the target epitopes used in the ChIP assays by the recruitment of trans-acting factors to the 3′ RR, recruitment of histone deacetylases to the 3′ RR, a partial shift in histone acetyltransferase activity from the 3′ RR to the germ line transcript promoters and/or switch sequences, or an average of differential histone modifications detected by antibodies with broad specificity. We consider it more likely, however, that epigenetic modifications of the 3′ RR, while necessary, are not sufficient for its activity in CSR. In fact, 3′ RR sequences have also been implicated in high levels of Igh expression in plasma cells (23, 40, 48); yet the profile of histone acetylation in the MPC11 plasma cell line is similar to that seen in B cells. 3′ RR activity is likely to depend on interactions with specific transcription factors and other factors that are upregulated or released upon stimulation for CSR and that may similarly be part of a plasma cell transcription factor profile. The detectable increase in CTCF binding to the 38 region upon LPS stimulation is perhaps one example. Various steps in CSR, e.g., targeting of AID and/or other proteins to the S regions to initiate CSR, and/or the role of H2AX in facilitating long-range inter-switch region recombination during CSR (65), may also depend on the 3′ RR cis elements.
A functional role for 3′ RR cis elements early in B-cell differentiation has not yet been detected. Given the temporal parallel in histone modifications at both the 5′ (10, 28, 29) and 3′ (this study) regions of the Igh locus in early stages of B-cell development, it is possible that to ensure proper B-cell development, epigenetic changes and/or open chromatin domains are simultaneously required on both ends of the locus. Constellations of chromatin loops have been recently proposed as part of the active chromatin hub model for regulation of complex loci (13, 60). Experimental proof of the importance of both ends of the Igh locus will require localization of DNase I-hypersensitive sites as well as mapping of CTCF binding sites at the 5′ end of the locus. Our previous studies have documented a physical interaction between Vh and 3′ RR sequences (6). The finding that Pax5 is a transcription factor that has regulatory activity at the 3′ RR elements (73) and appears to have a key regulatory role in targeting distal V genes for V-DJ recombination in pro-B cells (26) leads us to suggest a possible role for Pax5 in Igh locus regulation. We propose that Pax5 binding at the 3′ RR can influence the open chromatin region at the 5′ end of the locus, potentially via DNA looping. This hypothesis is further supported by our observations that Pax5 has the potential to mediate interaction with both histone acetyltransferase complexes and SWI/SNF chromatin remodeling complexes (2). Alternatively, a major role in DNA looping could be played by CTCF, which could bring 5′ and 3′ insulators together as a main loop and foster regulatory subloops through its interaction with tissue-specific DNase I-hypersensitive sites and trans-acting, tissue-specific factors like Pax5.
Acknowledgments
This work was supported by NIH grant AI13509 (B.K.B.), NIH training grant T32 CA09173 (A.V.E.), an Albert Einstein College of Medicine Cancer Center Core grant P30CA13330, NIH grant AI30653 (L.A.E.), and NIH intramural funding (V.V.L.). F.E.G. was supported by NCI 5 F31 CA76942, a UNCF/MERCK Graduate Science Research Dissertation fellowship, and a National Medical Fellowship in Academic Medicine for Minority Medical Students. P.F. was a 2003 summer graduate student with V.V.L.
We thank Nasrin Ashouian, Alexa Price-Whelan, Huafeng Xie, and Nydiaris Hernandez for technical assistance and all members of the Lobanenkov and Birshtein laboratories for helpful discussions. We also thank Matthew Scharff, Ari Melnick, Ziqiang Li, Herbert Morse III, Svetlana Pack, and Dmitry Fyodorov for critical review of the manuscript. Our appreciation is extended to Kathryn Calame, Ranjan Sen, Arthur Skoultchi, Michael Krangel, and Anton Krumm for kind gifts of reagents and to Dipanjan Chowdhury, Brian Dynlacht, Katrina Morshead, Caroline Woo, Joseph Locker, Ari Melnick, Tomas Stopka, and Wendy Gombert for helpful advice.
REFERENCES
- 1.Alt, F., N. Rosenberg, S. Lewis, E. Thomas, and D. Baltimore. 1981. Organization and reorganization of immunoglobulin genes in A-MuLV-transformed cells: rearrangement of heavy but not light chain genes. Cell 27:381-390. [DOI] [PubMed] [Google Scholar]
- 2.Barlev, N. A., A. V. Emelyanov, P. Castagnino, P. Zegerman, A. J. Bannister, M. A. Sepulveda, F. Robert, L. Tora, T. Kouzarides, B. K. Birshtein, and S. L. Berger. 2003. A novel human Ada2 homologue functions with Gcn5 or Brg1 to coactivate transcription. Mol. Cell. Biol. 23:6944-6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell, A. C., A. G. West, and G. Felsenfeld. 1999. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98:387-396. [DOI] [PubMed] [Google Scholar]
- 4.Bradney, C., M. Hjelmeland, Y. Komatsu, M. Yoshida, T. P. Yao, and Y. Zhuang. 2003. Regulation of E2A activities by histone acetyltransferases in B lymphocyte development. J. Biol. Chem. 278:2370-2376. [DOI] [PubMed] [Google Scholar]
- 5.Bulger, M., D. Schubeler, M. A. Bender, J. Hamilton, C. M. Farrell, R. C. Hardison, and M. Groudine. 2003. A complex chromatin landscape revealed by patterns of nuclease sensitivity and histone modification within the mouse β-globin locus. Mol. Cell. Biol. 23:5234-5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calvo, C.-F., S. L. Giannini, N. Martinez, and B. K. Birshtein. 1991. DNA sequences 3′ of the IgH chain cluster rearrange in mouse B cell lines. J. Immunol. 146:1353-1360. [PubMed] [Google Scholar]
- 7.Chauveau, C., and M. Cogne. 1996. Palindromic structure of the IgH 3′ locus control region. Nat. Genet. 14:15-16. [DOI] [PubMed] [Google Scholar]
- 8.Chauveau, C., E. A. Jansson, S. Muller, M. Cogne, and S. Pettersson. 1999. Cutting edge: Ig heavy chain 3′ HS1-4 directs correct spatial position-independent expression of a linked transgene to B lineage cells. J. Immunol. 163:4637-4641. [PubMed] [Google Scholar]
- 9.Chen, C., and B. K. Birshtein. 1997. Virtually identical enhancers containing a segment of homology to murine 3′IgH-E(hs1,2) lie downstream of human Ig C alpha 1 and C alpha 2 genes. J. Immunol. 159:1310-1318. [PubMed] [Google Scholar]
- 10.Chowdhury, D., and R. Sen. 2001. Stepwise activation of the immunoglobulin mu heavy chain gene locus. EMBO J. 20:6394-6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chowdhury, D., and R. Sen. 2003. Transient IL-7/IL-7R signaling provides a mechanism for feedback inhibition of immunoglobulin heavy chain gene rearrangements. Immunity 18:229-241. [DOI] [PubMed] [Google Scholar]
- 12.Cogne, M., R. Lansford, A. Bottaro, J. Zhang, J. Gorman, F. Young, H. L. Cheng, and F. W. Alt. 1994. A class switch control region at the 3′ end of the immunoglobulin heavy chain locus. Cell 77:737-747. [DOI] [PubMed] [Google Scholar]
- 13.de Laat, W., and F. Grosveld. 2003. Spatial organization of gene expression: the active chromatin hub. Chromosome Res. 11:447-459. [DOI] [PubMed] [Google Scholar]
- 14.Felsenfeld, G., and M. Groudine. 2003. Controlling the double helix. Nature 421:448-453. [DOI] [PubMed] [Google Scholar]
- 15.Fernandez, L. A., M. Winkler, and R. Grosschedl. 2001. Matrix attachment region-dependent function of the immunoglobulin μ enhancer involves histone acetylation at a distance without changes in enhancer occupancy. Mol. Cell. Biol. 21:196-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Filippova, G. N., S. Fagerlie, E. M. Klenova, C. Myers, Y. Dehner, G. Goodwin, P. E. Neiman, S. J. Collins, and V. V. Lobanenkov. 1996. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol. Cell. Biol. 16:2802-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Filippova, G. N., C. F. Qi, J. E. Ulmer, J. M. Moore, M. D. Ward, Y. J. Hu, D. I. Loukinov, E. M. Pugacheva, E. M. Klenova, P. E. Grundy, A. P. Feinberg, A. M. Cleton-Jansen, E. W. Moerland, C. J. Cornelisse, H. Suzuki, A. Komiya, A. Lindblom, F. Dorion-Bonnet, P. E. Neiman, H. C. Morse III, S. J. Collins, and V. V. Lobanenkov. 2002. Tumor-associated zinc finger mutations in the CTCF transcription factor selectively alter its DNA-binding specificity. Cancer Res. 62:48-52. [PubMed] [Google Scholar]
- 18.Forsberg, E. C., K. M. Downs, H. M. Christensen, H. Im, P. A. Nuzzi, and E. H. Bresnick. 2000. Developmentally dynamic histone acetylation pattern of a tissue-specific chromatin domain. Proc. Natl. Acad. Sci. USA 97:14494-14499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giannini, S. L., M. Singh, C.-F. Calvo, G. Ding, and B. K. Birshtein. 1993. DNA regions flanking the mouse Ig 3′α enhancer are differentially methylated and DNAse I hypersensitive during B cell differentiation. J. Immunol. 150:1772-1780. [PubMed] [Google Scholar]
- 20.Gombert, W. M., S. D. Farris, E. D. Rubio, K. M. Morey-Rosler, W. H. Schubach, and A. Krumm. 2003. The c-myc insulator element and matrix attachment regions define the c-myc chromosomal domain. Mol. Cell. Biol. 23:9338-9348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gordon, S. J., S. Saleque, and B. K. Birshtein. 2003. Yin Yang 1 is a lipopolysaccharide-inducible activator of the murine 3′ Igh enhancer, hs3. J. Immunol. 170:5549-5557. [DOI] [PubMed] [Google Scholar]
- 22.Greenbaum, S., and Y. Zhuang. 2002. Identification of E2A target genes in B lymphocyte development by using a gene tagging-based chromatin immunoprecipitation system. Proc. Natl. Acad. Sci. USA 99:15030-15035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gregor, P. D., and S. L. Morrison. 1986. Myeloma mutant with a novel 3′ flanking region: loss of normal sequence and insertion of repetitive elements leads to decreased transcription but normal processing of the alpha heavy-chain gene products. Mol. Cell. Biol. 6:1903-1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hark, A. T., C. J. Schoenherr, D. J. Katz, R. S. Ingram, J. M. Levorse, and S. M. Tilghman. 2000. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405:486-489. [DOI] [PubMed] [Google Scholar]
- 25.Hebbes, T. R., A. L. Clayton, A. W. Thorne, and C. Crane-Robinson. 1994. Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken beta-globin chromosomal domain. EMBO J. 13:1823-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hesslein, D. G., D. L. Pflugh, D. Chowdhury, A. L. Bothwell, R. Sen, and D. G. Schatz. 2003. Pax5 is required for recombination of transcribed, acetylated, 5′ IgH V gene segments. Genes Dev. 17:37-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 28.Johnson, K., C. Angelin-Duclos, S. Park, and K. L. Calame. 2003. Changes in histone acetylation are associated with differences in accessibility of VH gene segments to V-DJ recombination during B-cell ontogeny and development. Mol. Cell. Biol. 23:2438-2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson, K., D. L. Pflugh, D. Yu, D. G. Hesslein, K. I. Lin, A. L. Bothwell, A. Thomas-Tikhonenko, D. G. Schatz, and K. Calame. 2004. B cell-specific loss of histone 3 lysine 9 methylation in the V(H) locus depends on Pax5. Nat. Immunol. 5:853-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanda, K., H. M. Hu, L. Zhang, J. Grandchamps, and L. M. Boxer. 2000. NF-kappa B activity is required for the deregulation of c-myc expression by the immunoglobulin heavy chain enhancer. J. Biol. Chem. 275:32338-32346. [DOI] [PubMed] [Google Scholar]
- 31.Kanduri, C., V. Pant, D. Loukinov, E. Pugacheva, C. F. Qi, A. Wolffe, R. Ohlsson, and V. V. Lobanenkov. 2000. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr. Biol. 10:853-856. [DOI] [PubMed] [Google Scholar]
- 32.Kanduri, M., C. Kanduri, P. Mariano, A. A. Vostrov, W. Quitschke, V. Lobanenkov, and R. Ohlsson. 2002. Multiple nucleosome positioning sites regulate the CTCF-mediated insulator function of the H19 imprinting control region. Mol. Cell. Biol. 22:3339-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khamlichi, A. A., E. Pinaud, C. Decourt, C. Chauveau, and M. Cogne. 2000. The 3′ IgH regulatory region: a complex structure in search of a function. Adv. Immunol. 75:317-345. [DOI] [PubMed] [Google Scholar]
- 34.Kim, K. J., C. Kanellopoulos-Langevin, R. M. Merwin, D. H. Sachs, and R. Asofsky. 1979. Establishment and characterization of BALB/c lymphoma lines with B cell properties. J. Immunol. 122:549-554. [PubMed] [Google Scholar]
- 35.Klenova, E. M., H. C. Morse III, R. Ohlsson, and V. V. Lobanenkov. 2002. The novel BORIS + CTCF gene family is uniquely involved in the epigenetics of normal biology and cancer. Semin. Cancer Biol. 12:399-414. [DOI] [PubMed] [Google Scholar]
- 36.Kuehl, W. M., and P. L. Bergsagel. 2002. Multiple myeloma: evolving genetic events and host interactions. Nat. Rev. Cancer 2:175-187. [DOI] [PubMed] [Google Scholar]
- 37.Kuo, M. H., and C. D. Allis. 1999. In vivo cross-linking and immunoprecipitation for studying dynamic protein:DNA associations in a chromatin environment. Methods 19:425-433. [DOI] [PubMed] [Google Scholar]
- 38.Lee, S. C., A. Bottaro, and R. A. Insel. 2003. Activation of terminal B cell differentiation by inhibition of histone deacetylation. Mol. Immunol. 39:923-932. [DOI] [PubMed] [Google Scholar]
- 39.Li, Z., Z. Luo, and M. D. Scharff. 2004. Differential regulation of histone acetylation and generation of mutations in switch regions is associated with Ig class switching. Proc. Natl. Acad. Sci. USA 101:15428-15433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lieberson, R., J. Ong, X. Shi, and L. A. Eckhardt. 1995. Immunoglobulin gene transcription ceases upon deletion of a distant enhancer. EMBO J. 14:6229-6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Litt, M. D., M. Simpson, M. Gaszner, C. D. Allis, and G. Felsenfeld. 2001. Correlation between histone lysine methylation and developmental changes at the chicken beta-globin locus. Science 293:2453-2455. [DOI] [PubMed] [Google Scholar]
- 42.Litt, M. D., M. Simpson, F. Recillas-Targa, M. N. Prioleau, and G. Felsenfeld. 2001. Transitions in histone acetylation reveal boundaries of three separately regulated neighboring loci. EMBO J. 20:2224-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Madisen, L., and M. Groudine. 1994. Identification of a locus control region in the immunoglobulin heavy-chain locus that deregulates c-myc expression in plasmacytoma and Burkitt's lymphoma cells. Genes Dev. 8:2212-2226. [DOI] [PubMed] [Google Scholar]
- 44.Maes, J., L. P. O'Neill, P. Cavelier, B. M. Turner, F. Rougeon, and M. Goodhardt. 2001. Chromatin remodeling at the Ig loci prior to V(D)J recombination. J. Immunol. 167:866-874. [DOI] [PubMed] [Google Scholar]
- 45.Manis, J. P., J. S. Michaelson, B. K. Birshtein, and F. W. Alt. 2003. Elucidation of a downstream boundary of the 3′ IgH regulatory region. Mol. Immunol. 39:753-760. [DOI] [PubMed] [Google Scholar]
- 46.Manis, J. P., N. van der Stoep, M. Tian, R. Ferrini, L. Davidson, A. Bottaro, and F. W. Alt. 1998. Class switching in B cells lacking 3′ immunoglobulin heavy chain enhancers. J. Exp. Med. 188:1421-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Max, E. E. 2003. Immunoglobulins: molecular genetics, p. 107-158. In W. E. Paul (ed.), Fundamental immunology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 48.Michaelson, J. S., S. L. Giannini, and B. K. Birshtein. 1995. Identification of 3′ alpha-hs4, a novel Ig heavy chain enhancer element regulated at multiple stages of B cell differentiation. Nucleic Acids Res. 23:975-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mills, F. C., N. Harindranath, M. Mitchell, and E. E. Max. 1997. Enhancer complexes located downstream of both human immunoglobulin Ca genes. J. Exp. Med. 186:845-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mombaerts, P., J. Iacomini, R. S. Johnson, K. Herrup, S. Tonegawa, and V. E. Papaioannou. 1992. RAG-1-deficient mice have no mature B and T lymphocytes. Cell 68:869-877. [DOI] [PubMed] [Google Scholar]
- 51.Morshead, K. B., D. N. Ciccone, S. D. Taverna, C. D. Allis, and M. A. Oettinger. 2003. Antigen receptor loci poised for V(D)J rearrangement are broadly associated with BRG1 and flanked by peaks of histone H3 dimethylated at lysine 4. Proc. Natl. Acad. Sci. USA 100:11577-11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mukhopadhyay, R., W.-Q. Yu, J. Whitehead, J.-W. Xu, M. Lezcano, S. D. Pack, C. Kanduri, M. Kanduri, V. Ginjala, A. A. Vostrov, W. Quitschke, I. Chernukhin, E. Klenova, V. Lobanenkov, and R. Ohlsson. 2004. The binding sites for the chromatin insulator protein CTCF map to DNA methylation-free domains genomewide. Genome Res. 14:1594-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muramatsu, M., K. Kinoshita, S. Fagarasan, S. Yamada, Y. Shinkai, and T. Honjo. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102:553-563. [DOI] [PubMed] [Google Scholar]
- 54.Nambu, Y., M. Sugai, H. Gonda, C. G. Lee, T. Katakai, Y. Agata, Y. Yokota, and A. Shimizu. 2003. Transcription-coupled events associating with immunoglobulin switch region chromatin. Science 302:2137-2140. [DOI] [PubMed] [Google Scholar]
- 55.Nesset, A. L., and D. M. Bader. 2002. Hole is a novel gene product expressed in the developing heart and brain. Mech. Dev. 117:347-350. [DOI] [PubMed] [Google Scholar]
- 56.Ng, H. H., D. N. Ciccone, K. B. Morshead, M. A. Oettinger, and K. Struhl. 2003. Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: a potential mechanism for position-effect variegation. Proc. Natl. Acad. Sci. USA 100:1820-1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nossal, G. J. 2003. The double helix and immunology. Nature 421:440-444. [DOI] [PubMed] [Google Scholar]
- 58.Ohlsson, R., R. Renkawitz, and V. Lobanenkov. 2001. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 17:520-527. [DOI] [PubMed] [Google Scholar]
- 59.Ong, J., S. Stevens, R. G. Roeder, and L. A. Eckhardt. 1998. 3′ IgH enhancer elements shift synergistic interactions during B cell development. J. Immunol. 160:4896-4903. [PubMed] [Google Scholar]
- 60.Palstra, R. J., B. Tolhuis, E. Splinter, R. Nijmeijer, F. Grosveld, and W. de Laat. 2003. The beta-globin nuclear compartment in development and erythroid differentiation. Nat. Genet. 35:190-194. [DOI] [PubMed] [Google Scholar]
- 61.Pant, V., S. Kurukuti, E. Pugacheva, S. Shamsuddin, P. Mariano, R. Renkawitz, E. Klenova, V. Lobanenkov, and R. Ohlsson. 2004. Mutation of a single CTCF target site within the H19 imprinting control region leads to loss of Igf2 imprinting and complex patterns of de novo methylation upon maternal inheritance. Mol. Cell. Biol. 24:3497-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pinaud, E., C. Aupetit, C. Chauveau, and M. Cogne. 1997. Identification of a homolog of the C alpha 3′/hs3 enhancer and of an allelic variant of the 3′IgH/hs1,2 enhancer downstream of the human immunoglobulin alpha 1 gene. Eur. J. Immunol. 27:2981-2985. [DOI] [PubMed] [Google Scholar]
- 63.Pinaud, E., A. A. Khamlichi, C. Le Morvan, M. Drouet, V. Nalesso, M. Le Bert, and M. Cogne. 2001. Localization of the 3′ IgH locus elements that effect long-distance regulation of class switch recombination. Immunity 15:187-199. [DOI] [PubMed] [Google Scholar]
- 64.Potter, M. 1967. The plasma cell tumors and myeloma proteins of mice, vol. 2. Academic Press Inc., New York, N.Y.
- 65.Reina-San-Martin, B., S. Difilippantonio, L. Hanitsch, R. F. Masilamani, A. Nussenzweig, and M. C. Nussenzweig. 2003. H2AX is required for recombination between immunoglobulin switch regions but not for intra-switch region recombination or somatic hypermutation. J. Exp. Med. 197:1767-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saleque, S., M. Singh, R. D. Little, S. L. Giannini, J. S. Michaelson, and B. K. Birshtein. 1997. Dyad symmetry within the mouse 3′ IgH regulatory region includes two virtually identical enhancers (C alpha3′E and hs3). J. Immunol. 158:4780-4787. [PubMed] [Google Scholar]
- 67.Schubeler, D., C. Francastel, D. M. Cimbora, A. Reik, D. I. Martin, and M. Groudine. 2000. Nuclear localization and histone acetylation: a pathway for chromatin opening and transcriptional activation of the human beta-globin locus. Genes Dev. 14:940-950. [PMC free article] [PubMed] [Google Scholar]
- 68.Sepulveda, M. A., A. V. Emelyanov, and B. K. Birshtein. 2004. NF-kappaB and Oct-2 synergize to activate the human 3′ Igh hs4 enhancer in B cells. J. Immunol. 172:1054-1064. [DOI] [PubMed] [Google Scholar]
- 69.Sepulveda, M. A., F. E. Garrett, A. Price-Whelan, and B. K. Birshtein. 2005.. Comparative analysis of human and mouse 3′ Igh regulatory regions identifies distinctive structural features. Mol. Immunol. 42:605-615. [DOI] [PubMed]
- 70.Shi, X., and L. A. Eckhardt. 2001. Deletional analyses reveal an essential role for the hs3b/hs4 IgH 3′ enhancer pair in an Ig-secreting but not an earlier-stage B cell line. Int. Immunol. 13:1003-1012. [DOI] [PubMed] [Google Scholar]
- 71.Shinkai, Y., G. Rathbun, K. P. Lam, E. M. Oltz, V. Stewart, M. Mendelsohn, J. Charron, M. Datta, F. Young, A. M. Stall, et al. 1992. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell 68:855-867. [DOI] [PubMed] [Google Scholar]
- 72.Siden, E. J., D. Baltimore, D. Clark, and N. E. Rosenberg. 1979. Immunoglobulin synthesis by lymphoid cells transformed in vitro by Abelson murine leukemia virus. Cell 16:389-396. [DOI] [PubMed] [Google Scholar]
- 73.Singh, M., and B. K. Birshtein. 1993. NF-HB (BSAP) is a repressor of the murine immunoglobulin heavy-chain 3′ α enhancer at early stages of B-cell differentiation. Mol. Cell. Biol. 13:3611-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Southern, E. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 75.Stavnezer, J. 2000. Molecular processes that regulate class switching. Curr. Top. Microbiol. Immunol. 245:127-168. [DOI] [PubMed] [Google Scholar]
- 76.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 77.Su, I. H., A. Basavaraj, A. N. Krutchinsky, O. Hobert, A. Ullrich, B. T. Chait, and A. Tarakhovsky. 2003. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat. Immunol. 4:124-131. [DOI] [PubMed] [Google Scholar]
- 78.Takahashi, N., A. Roach, D. B. Teplow, S. B. Prusiner, and L. Hood. 1985. Cloning and characterization of the myelin basic protein gene from mouse: one gene can encode both 14 kd and 18.5 kd MBPs by alternate use of exons. Cell 42:139-148. [DOI] [PubMed] [Google Scholar]
- 79.Takahashi, Y., J. B. Rayman, and B. D. Dynlacht. 2000. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 14:804-816. [PMC free article] [PubMed] [Google Scholar]
- 80.Tanimoto, K., A. Sugiura, A. Omori, G. Felsenfeld, J. D. Engel, and A. Fukamizu. 2003. Human β-globin locus control region HS5 contains CTCF- and developmental stage-dependent enhancer-blocking activity in erythroid cells. Mol. Cell. Biol. 23:8946-8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vostrov, A. A., and W. W. Quitschke. 1997. The zinc finger protein CTCF binds to the APBbeta domain of the amyloid beta-protein precursor promoter. Evidence for a role in transcriptional activation. J. Biol. Chem. 272:33353-33359. [DOI] [PubMed] [Google Scholar]
- 82.West, A. G., M. Gaszner, and G. Felsenfeld. 2002. Insulators: many functions, many mechanisms. Genes Dev. 16:271-288. [DOI] [PubMed] [Google Scholar]
- 83.Woo, C. J., A. Martin, and M. D. Scharff. 2003. Induction of somatic hypermutation is associated with modifications in immunoglobulin variable region chromatin. Immunity 19:479-489. [DOI] [PubMed] [Google Scholar]
- 84.Yamasaki, H., I. B. Weinstein, E. Fibach, R. A. Rifkind, and P. Marks. 1979. Tumor promoter-induced adhesion of the DS19 clone of murine erythroleukemia cells. Cancer Res. 39:1989-1994. [PubMed] [Google Scholar]
- 85.Yang, Y., W. W. Quitschke, A. A. Vostrov, and G. J. Brewer. 1999. CTCF is essential for up-regulating expression from the amyloid precursor protein promoter during differentiation of primary hippocampal neurons. J. Neurochem. 73:2286-2298. [DOI] [PubMed] [Google Scholar]
- 86.Zhong, X. P., and M. S. Krangel. 1997. An enhancer-blocking element between alpha and delta gene segments within the human T cell receptor alpha/delta locus. Proc. Natl. Acad. Sci. USA 94:5219-5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhou, J., N. Ashouian, M. Delepine, F. Matsuda, C. Chevillard, R. Riblet, C. L. Schildkraut, and B. K. Birshtein. 2002. The origin of a developmentally regulated Igh replicon is located near the border of regulatory domains for Igh replication and expression. Proc. Natl. Acad. Sci. USA 99:13693-13698. [DOI] [PMC free article] [PubMed] [Google Scholar]