Abstract
CCAAT/enhancer-binding protein alpha (C/EBPα) has been previously considered a strong inhibitor of cell proliferation which uses multiple pathways to cause growth arrest. In this paper, we describe a new function of C/EBPα, which is an acceleration of cell proliferation. This new function of C/EBPα is created in proliferating livers by protein phosphatase 2A-mediated dephosphorylation of C/EBPα at Ser193. The Ser193-dephosphorylated C/EBPα interacts with retinoblastoma protein (Rb) independently on E2Fs and sequesters Rb, leading to a reduction of E2F-Rb repressors and to acceleration of proliferation. This new function of C/EBPα requires Rb, since the dephosphorylated C/EBPα does not promote proliferation in Rb-negative cells. We also show that a balance of Rb and Ser193-dephosphorylated C/EBPα determines if the cells are growth arrested or have an increased rate of proliferation. Consistently with these findings, a significant portion of Rb is sequestered into Rb-C/EBPα complexes in proliferating livers, and E2F-Rb complexes are not detectable in these livers. Our data demonstrate a new pathway by which the phosphorylation-dependent switch of biological functions of C/EBPα promotes liver proliferation.
Partial hepatectomy (PH) in animals is a classical model for the investigations of biochemical pathways that control liver proliferation after surgeries (4). A commonly useful model of liver regeneration includes a removal of 70% of liver. The remaining liver proliferates and restores its original size (4, 6, 10, 12). The initiation of cell cycle in regenerating livers has been intensively investigated, and the major pathways of the initiation have been elucidated (4, 6). In addition to this step, liver proliferation requires removing a negative control, which is supported by certain liver-specific proteins and by retinoblastoma protein (Rb) family proteins (pocket proteins). These pocket proteins display their growth-inhibitory activities via interaction with E2F transcription factors and through the formation of complexes that occupy promoters and repress expression of S-phase and mitotis-specific genes (3). Quiescence in the liver is also supported by growth-inhibitory activity of a transcription factor, CCAAT/enhancer-binding protein alpha (C/EBPα). C/EBPα is a strong inhibitor of proliferation of cultured cells, and it is also required for the inhibition of liver growth. Ablation of C/EBPα in animals leads to an increased rate of proliferation in the liver and in cultured hepatocytes derived from C/EBPα−/− livers (5, 21, 23, 24). Although C/EBPα is a transcription factor, C/EBPα displays its inhibitory activity through interactions with cell cycle proteins such as cyclin-dependent kinase 2 (cdk2), cdk4, Rb, E2F, and Brm (9, 13, 15, 24-27). A number of recent papers revealed that the ability of C/EBPα to cause growth arrest is regulated on several levels. In addition to transcriptional regulation of C/EBPα mRNA and regulation of protein stability (20), certain cellular transduction pathways are able to activate or inhibit activities of C/EBPα without changing the protein levels of C/EBPα. Ross et al. have recently found that the extracellular signal-regulated kinases 1 and 2 inhibit C/EBPα-mediated differentiation of granulocytes through phosphorylation of C/EBPα at Ser21 (17). The authors demonstrated that this pathway regulates C/EBPα activities in a tissue-specific manner; it operates only in myeloid cells but does not affect C/EBPα activities in other cells such as adipocytes (17). We have recently found a pathway that blocks growth-inhibitory activity of C/EBPα in hepatoma cells and in liver tumors. The activation of phosphatidylinositol 3-kinase (PI3K)/Akt in liver tumors leads to accumulation of protein phosphatase 2A (PP2A) in nuclei where PP2A dephosphorylates C/EBPα on Ser193 and blocks its growth-inhibitory activity (25). This PI3K/Akt-mediated block of C/EBPα inhibition leads to the lack of negative control of proliferation in liver and to development of tumors (25). These examples clearly demonstrate that certain signal transduction pathways regulate C/EBPα activities on the level of posttranslational modifications.
In this paper, we identified a new function of C/EBPα: acceleration of proliferation, which is opposite to that previously described for this protein. We found that the choice of a biological function of C/EBPα is achieved by phosphorylation-dephosphorylation of a single Ser193 residue within the C/EBPα growth-inhibitory region. Phosphorylated C/EBPα binds to cdk2 and Brm and inhibits proliferation, while Ser193-dephosphorylated C/EBPα accelerates proliferation via sequestering Rb. These findings show that phosphorylation-dependent switch of biological activities of C/EBPα not only eliminates C/EBPα-mediated negative control of proliferation but also neutralizes growth-inhibitory activity of Rb through sequestering Rb from E2F-Rb complex repressors. This phosphorylation-dependent neutralization of two negative regulators of liver proliferation, C/EBPα and Rb, contributes to liver proliferation after surgical resections and to development of liver tumors.
MATERIALS AND METHODS
Materials and plasmids.
Antibodies (Abs) to C/EBPα (14AA and N19), cdk4 (C-22), cdk2 (M2), Brm, and Rb (C-15) were purchased from Santa Cruz Biotechnology. Expression vectors for wild-type (WT) mouse C/EBPα and mutations were described in our previous paper (25). A short growth-inhibitory region of human C/EBPα (hGIR) was generated by fusing a synthetic DNA oligomer (amino acids [aa] 182 to 207) to AUG codon on the 5′ end and cloning it into the KpnI and NotI sites of a pcDNA6/myc-His expression vector.
Human liver tumor samples and liver regeneration.
Human liver samples were obtained as part of an institutional review board-approved protocol, where tumor and normal sections were collected from resected samples. A detailed characterization of these tumors was presented in our previous paper (25).
Transient transfection assay.
Analysis of C/EBPα growth arrest was performed with 3T3-L1 cells, HT1080 cells, and Rb-deficient SAOS2 and C33A cells with pAdTrack-C/EBPα mutants. In experiments with pAdTrack-C/EBPα, the inhibition-acceleration of cell growth was calculated by counting the number of cells appearing green under fluorescence microscopy in each colony. This procedure is described in detail in previously published papers (9, 25-27). The present study presents data of three to four independent experiments. From 200 to 300 colonies were examined in each experiment.
BrdU uptake.
SAOS2 cells were transfected with pAdTrack-C/EBPα constructs. Control cells were transfected with an empty pAdTrack vector. Twenty-four hours later, bromodeoxyuridine (BrdU) was added for 1 h, and cells were fixed and stained with monoclonal Abs to BrdU. DAPI (4′,6′-diamidino-2-phenylindole) staining was performed to visualize nontransfected cells. The manuscript presents data obtained from three independent experiments. The percentage of BrdU-positive cells is shown as a summary of these experiments.
Generation of C/EBPα recombinant adenoviruses.
pAdTrack-C/EBPα constructs and pAdEasy-1 vector were cotransformed into BJ 5183 cells. Recombinant virus DNAs were selected from the kanamycin plates. The recombinant adenoviruses were packaged and produced in HEK 293 cells. Purified high-titer virus supplies were used in culture cell infections as previously described (7).
Protein isolation and Western blotting.
Nuclear extracts (NEs) were isolated as described in previous papers (22-24). Briefly, livers were homogenized in low-salt buffer and centrifuged at 12,000 rpm for 10 min at 4°C. Nuclei (in pellet form) were incubated for 30 min on ice with high-salt buffer B (20 mM Tris-HCl [pH 7.5], 25% sucrose, 0.42 M NaCl, 5 mM dithiothreitol, 2 mM MgCl2, 0.2 mM EDTA). After centrifugation, the supernatant (NE) was frozen and kept at −80°C. Proteins (30 to 50 μg) were loaded onto an 8 to 16% polyacrylamide gel, transferred onto the membrane, and probed with antibodies to C/EBPα, cdk2, cdk4, Rb, E2F4, or Brm. To verify protein loading, each filter was reprobed with β-actin and then stained with Coomassie blue.
Two-dimensional (2D) gel Western blotting.
C/EBPα was immunoprecipitated from transfected cells with specific Abs (N19; Santa Cruz Biotechnology). Immunoprecipitates (IPs) were separated by 2D gel electrophoresis with Protean IEF (Bio-Rad), and C/EBPα was transferred on the membrane and probed with rabbit Abs to C/EBPα (14AA; Santa Cruz Biotechnology).
Gel shift with C/EBPα probe.
Conditions for a gel shift assay with a bZIP probe are described in our earlier papers (22-23). Briefly, DNA oligomer containing a C/EBP consensus site within the C3 promoter (bZIP) was labeled by Klenow and [α-32P]dCTP. A total of 5 to 10 μg of NEs from livers were incubated with the probe in the binding buffer containing 25 mM Tris-HCl, 100 mM KCl, 5 mM dithiothreitol, 10% glycerol, 2 mM MgCl, 2 μg of poly(dI-dC) nonspecific competitor-reaction mixture, and 50,000 cpm of the probe. DNA-protein complexes were separated by nondenaturing 5 to 6% polyacrylamide gel electrophoresis in 0.5× Tris-borate-EDTA buffer. After electrophoresis, the gel was dried and exposed to X-ray film.
E2F gel shift.
Conditions for the E2F gel shift assay are described in our previous papers (9, 24). Briefly, E2F probes (see Fig. 5) were labeled with dCTP in the reaction mixture with Klenow enzyme. E2F probes were incubated with NEs (5 to 10 μg) in binding reaction mixtures that had a composition similar to that described above for C/EBPα. Salmon DNA was used as the competitor in the E2F binding reaction mixtures. Binding reaction mixtures were separated by a native 5% polyacrylamide gel.
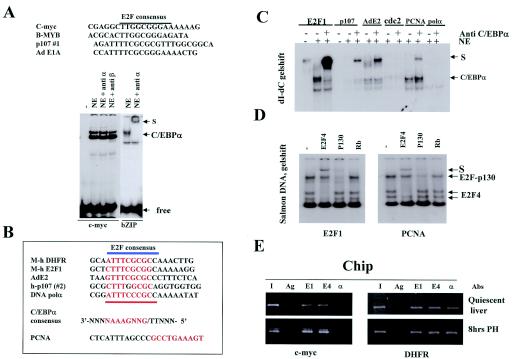
FIG. 5.
C/EBPα binds directly to certain E2F dependent promoters. (A) Analysis of E2F consensuses that lack the C/EBPα binding site. (Top) Nucleotide sequences of E2F consensuses that lack a putative binding site for C/EBPα. (Bottom) Gel shift showing that the c-myc promoter does not interact directly with C/EBPα. DNA probes were incubated with HT1-IPTG extracts containing overexpressed C/EBPα and with a poly(dI-dC) competitor which inhibits E2F binding (9). Abs to C/EBPα and to C/EBPβ were added to the binding reaction mixtures before the addition of probe. A gel shift assay with bZIP is shown as a positive control for C/EBPα binding. (B) Nucleotide sequences of E2F promoters that contain a potential C/EBPα binding site (marked by red letters) within E2F consensuses. (C) Gel shift assay with poly(dI-dC) competitor which specifically inhibits E2F binding. C/EBPα overexpressed in HT1 cells (9, 22) was incubated with E2F probes (shown on the bottom) and analyzed by native 5% gel electrophoresis. Abs to C/EBPα were added to the binding reaction mixtures. Positions of C/EBPα and the supershift (S) are shown on the right. (D) Gel shift assay with probes containing E2F consensuses from E2F1 and PCNA promoters under conditions specific for E2F binding (salmon DNA as a competitor). Abs to E2F4, p130, and Rb were added before the addition of the probe. Positions of E2F-Rb complexes are shown on the right. (E) C/EBPα occupies the DHFR promoter in proliferating livers. Chromatin solutions were prepared from quiescent livers and from livers at 8 h after PH. E2F1, E2F4, and C/EBPα were precipitated from the solutions by specific Abs. The IPs were examined by PCR with primers specific to c-myc (left) and the DHFR (right) promoters. I, input; Ag, mock-agarose control.
Coimmunoprecipitation (Co-IP) and GST pull-down.
C/EBPα was immunoprecipitated from NEs with polyclonal Abs (14AA; Santa Cruz), and the presence of Rb, Brm, E2F4, cdk4, or cdk2 in C/EBPα IPs were examined by Western blotting with monoclonal Abs to the proteins mentioned. Glutathione S-transferase (GST)-C/EBPα constructs were generated, and a GST pull-down assay was performed as described previously (24-27).
Chromatin immunoprecipitation.
The chromatin immunoprecipitation (ChIP) assay was performed with cultured cells and with liver tissues as described in a previous paper (9). The chromatin solution was sonicated to obtain DNA fragments of 500 to 1,000 bp in length. Abs against C/EBPα (14AA), E2F1 (sc-193), and E2F4 (purchased from Santa Cruz) were added to each aliquot of chromatin and incubated overnight. Blocked protein A-agarose (Sigma) was added and incubated for 30 to 40 min at room temperature. After the cross-linking proteins were removed, DNA was precipitated and used for PCR with primers specific for E2F-dependent promoters. The sequences of the primers for c-myc and dihydrofolate reductase (DHFR) promoters were published in a previous paper (9). PCR mixtures were amplified for 1 cycle of 95°C for 5 min, annealing temperature for primers for 5 min, and at 72°C for 2 min. PCR mixtures were then amplified for 34 cycles of 95°C for 1 min, annealing temperature for 2 min, and 72°C for 1.5 min. PCR products were separated by 1.5% agarose gel electrophoresis.
RESULTS
Dephosphorylation of C/EBPα at Ser193 in proliferating livers creates a new activity of C/EBPα, which is the acceleration of cell proliferation.
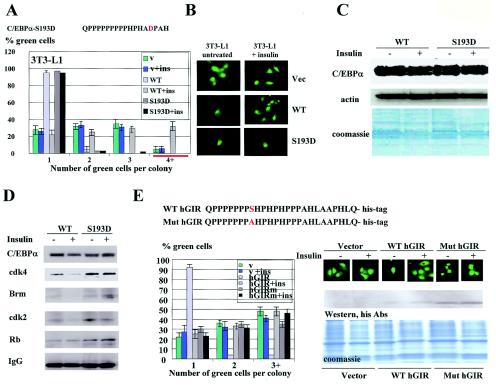
The growth-inhibitory function of C/EBPα in quiescent livers is mediated via direct inhibition of cdk's and through E2F repression (9, 13, 15, 25-27). In proliferating livers, C/EBPα activity is eliminated by dephosphorylation of Ser193, which blocks the interactions of C/EBPα with cdk2 and Brm (25). A typical picture of C/EBPα isoforms in quiescent and proliferating livers is shown in Fig. 1A, left). Although the dephosphorylated C/EBPα does not interact with cdk's and Brm, it still possessed two active regions: the Rb-interacting region and the DNA binding domain, both of which might affect cell proliferation (Fig. 1A). Therefore, we investigated the effects of these regions (activities) on cell proliferation when the growth-inhibitory region of C/EBPα was neutralized by dephosphorylation. Our previous studies (27) identified a signal transduction pathway, PI3K/Akt/PP2A, which dephosphorylates Ser193 in cultured cells and creates a C/EBPα molecule identical to that in proliferating livers (Fig. 1C). To investigate the activities of dephosphorylated C/EBPα, we initially performed experiments with 3T3-L1 cells, since the PI3K-Akt pathway (which triggers dephosphorylation of Ser193) is not active in these cells but may be induced by insulin signaling (8, 9, 16, 17, 25). 3T3-L1 cells were transfected with empty pAdTrack plasmid and with pAdTrack-WT-C/EBPα plasmid (which expresses green fluorescent protein and C/EBPα). Cell proliferation in green-fluorescing cells treated or untreated with insulin was examined. Results of these studies are shown in Fig. 1B to D. Western blotting with ph-S473-Akt Abs showed that insulin activates the PI3K/Akt pathway in 3T3-L1 cells (Fig. 1B). 2D gel electrophoresis of C/EBPα revealed that insulin dephosphorylates C/EBPα on Ser193, but protein levels of C/EBPα are not altered (Fig. 1C and D). Examination of cell numbers in each colony showed that the activation of PI3K/Akt by insulin led to an accelerated rate of growth in cells transfected with WT C/EBPα, while untreated control cells were arrested by C/EBPα (Fig. 1D). These data are consistent with observations obtained with a C/EBPα-S193A mutant, which mimics dephosphorylated state of C/EBPα and also accelerates cell proliferation (25).
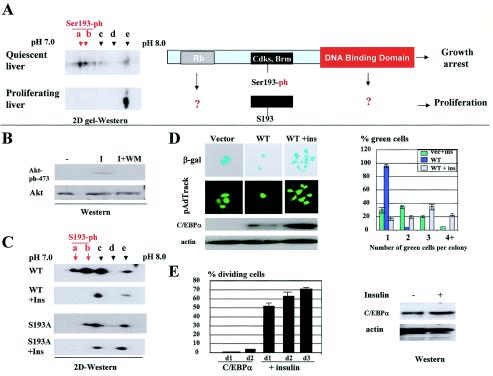
FIG. 1.
Dephosphorylation of C/EBPα at Ser193 alters biological functions of C/EBPα. (A) Proliferating livers express Ser193-dephosphorylated C/EBPα. (Left) Examination of C/EBPα by 2D gel Western blotting in quiescent and proliferating livers 8 h after PH. Positions of Ser193-phosphorylated isoforms (a and b) are shown by red arrowheads. (Right) Regions of C/EBPα that display distinct biological activities of C/EBPα. The Rb-interacting region (aa 50 to 95) is shown by a grey box. The growth-inhibitory region, which interacts with cdk2, cdk4, and Brm, is shown by a black box. The DNA binding domain is shown by a red box. This paper examines the role of the Rb-interacting region and DNA binding domain in the ability of dephosphorylated C/EBPα to accelerate cell proliferation (indicated by question marks). (B) Activation of PI3K/Akt pathway by insulin in 3T3-L1 cells. Western blotting with Abs to ph-Akt and total Akt was performed with cytoplasmic proteins isolated from 3T3-L1 cells treated with insulin (I) and with insulin-wortmannin (I+WM). (C) 2D gel electrophoresis of C/EBPα isolated from 3T3-L1 cells treated with insulin. The WT or the C/EBPα-S193A mutant were transfected into 3T3-L1 cells, cells were treated with 100 nM insulin, and C/EBPα was examined by 2D gel electrophoresis. Positions of five isoforms (a to e) of C/EBPα are shown on the top. Isoforms a and b are generated by phosphorylation of Ser193, since they are not detectable in the C/EBPα-S193A mutant. (D) Ser193-dephosphorylated C/EBPα accelerates cell proliferation. Colony formation assays of 3T3-L1 cells transfected with C/EBPα and treated with insulin are shown. (Top) Cotransfections of C/EBPα with β-gal were performed as previously described (25, 27). (Bottom) Transfections of 3T3-L1 cells with pAdTrack-C/EBPα. Colonies are shown on day 3 after transfections. Bar graphs (right) represent a summary of three experiments for pAdTrack transfections. Western blotting shows protein levels of C/EBPα in extracts isolated from experimental plates. (E) Overexpression of C/EBPα does not cause cell death. Insulin was added to cells arrested by C/EBPα (2 days after transfections with C/EBPα), and cell growth was monitored on days 1, 2, and 3 after addition of insulin. (Left) Bar graphs present a summary of three independent experiments. (Right) Western blotting shows the levels of C/EBPα in protein extracts isolated from the experimental plates.
Since our growth arrest-growth promotion assay included the visualization of green-fluorescing cells, we performed control studies to ensure that the appearance of single green cells was the result of growth arrest and was not related to the cell death. For this goal, we determined if cells arrested by C/EBPα might proliferate after insulin-mediated release of C/EBPα growth-inhibitory activity. 3T3-L1 cells transfected with WT C/EBPα were kept on plates for 2 days, and then insulin was added to the plates. Figure 1E shows that C/EBPα-arrested cells were viable and started proliferation again if C/EBPα growth-inhibitory activity was blocked by insulin. Western blotting of C/EBPα with protein extracts from experimental plates revealed that the insulin-mediated release of C/EBPα growth arrest did not involve alterations in protein levels of C/EBPα (Fig. 1E, right).
Taken together, these studies revealed that PI3K/Akt-dependent dephosphorylation of C/EBPα at Ser193 creates a new function of C/EBPα, which is the acceleration of cell proliferation. We next performed a careful examination of molecular pathways by which dephosphorylated C/EBPα accelerates proliferation.
Disruption of E2F-Rb complexes by Ser193-dephosphorylated C/EBPα correlates with accelerated proliferation.
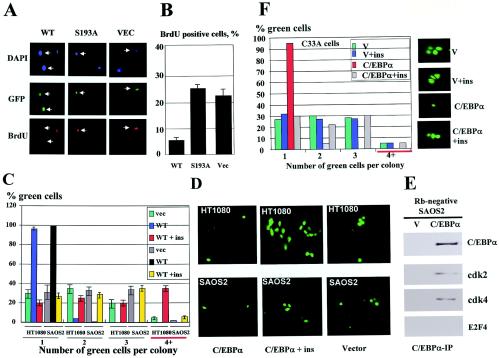
Since the Ser193-dephosphorylated C/EBPα contains an intact Rb-interacting region (Fig. 1A), we examined the hypothesis that this interaction might be involved in growth promotion activities of the Ser193-dephosphorylated C/EBPα. To test this hypothesis, we first analyzed E2F-Rb complexes in cells expressing C/EBPα under conditions that dephosphorylate C/EBPα at Ser193 and block the growth-inhibitory activity of C/EBPα. Since transient experiments have a limited efficiency of transfection, we utilized a previously described stable clonal line, HT1, in which C/EBPα is under Lac repressor control (22). Incorporation of Abs into the gel shift assay with an E2F probe showed that untreated cells contained abundant E2F-Rb complexes and minor E2F-p130 complexes (Fig. 2A). Under standard conditions, isopropyl-ā-d-thiogalactopyranoside (IPTG)-mediated induction of C/EBPα led to a significant reduction of E2F-Rb complex and to the formation of a growth-inhibitory C/EBPα-Rb-E2F-Brm complex, which was observed on the top of the gel (Fig. 2B). This complex was previously detected in these cells by high-performance liquid chromatography-based size exclusion chromatography (9). As shown in Fig. 2C, insulin-mediated dephosphorylation of C/EBPα blocked the formation of the growth-inhibitory C/EBPα-Rb-E2F4-Brm complexes and led to the disruption of E2F-Rb complexes. Colony formation assay of untreated cells and cells treated with insulin showed that the disruption of E2F-Rb complexes by dephosphorylated C/EBPα correlated with acceleration of proliferation in HT1 cells (Fig. 2D). The acceleration of proliferation by insulin-mediated dephosphorylation of C/EBPα was specific, since insulin did not affect proliferation of cells transfected with empty pAdTrack vector. Examination of C/EBPα in protein extracts isolated from experimental plates revealed that the treatment of cells with the insulin did not affect protein levels of C/EBPα (Fig. 2D). Taken together, the studies of Rb-E2F complexes in HT1 cells supported the hypothesis that Ser193-dephosphorylated C/EBPα accelerates proliferation through disruption of E2F-Rb complexes. Since Ser193-dephosphorylated C/EBPα contains the intact region for the interaction with Rb, these observations suggest that the direct interaction of the dephosphorylated C/EBPα with Rb causes the disruption of the E2F-Rb complexes. To further examine this hypothesis and to elucidate precise molecular mechanisms, we performed a number of experiments with cultured cells and livers.
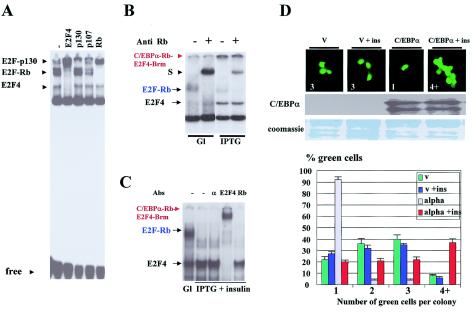
FIG. 2.
Ser193-dephosphorylated C/EBPα sequesters Rb from Rb-E2F complexes. (A) Composition of E2F complexes in stable clonal line HT1. NEs of untreated HT1 were incubated with a DNA probe containing E2F consensus from the DHFR promoter. Abs to E2F4, p130, p107, and Rb (shown on the top) were added to the binding reaction mixtures. Positions of E2F complexes are shown on the left. (B) Ectopic expression of C/EBPα reduces E2F-Rb complexes and leads to the appearance of a high-molecular-weight growth-inhibitory C/EBPα-Rb-E2F4-Brm complex. HT1 cells were treated with glucose (Gl) and IPTG (C/EBPα inducer), and NEs were isolated and examined by gel shift assay with aE2F probe. Abs to Rb (shown on the top) were incorporated into the binding reaction mixtures. Positions of E2F-Rb complexes are shown on the left. (C) Ser193-dephosphorylated C/EBPα disrupts E2F-Rb complexes but fails to form the growth-inhibitory C/EBPα-Rb-E2F4-Brm complex. C/EBPα was induced in HT1 cells by IPTG, and cells were treated with insulin to dephosphorylate C/EBPα. Abs (shown on the top) were added to the binding reaction mixtures before the addition of probe. (D) The disruption of E2F-Rb complexes by dephosphorylated C/EBPα correlates with acceleration of proliferation. Empty vector and pAdTrack-C/EBPα were transfected into HT1080 cells. Cells were treated with insulin and examined for the formation of colonies. (Top) A typical picture of cell colonies. After the visualization of green-fluorescing cells, total protein extracts were isolated and used for Western blotting analysis with Abs to C/EBPα (bottom). The membrane was stained with Coomassie blue to verify protein loading. Bar graphs indicate a summary of three independent experiments.
C/EBPα-S193A mutant sequesters Rb into C/EBPα-Rb complexes.
Since the C/EBPα-S193A mutant mimics dephosphorylated state of C/EBPα, this mutant was used for the examination of the interactions of C/EBPα with Rb by Co-IP and GST pull-down assays. The experiments with C/EBPα mutants showed that WT C/EBPα and two other C/EBPα mutants (182 and 184) (25) interacted with cdk2, cdk4, Rb, and Brm. On the contrary, C/EBPα-S193A mutant interacted strongly with Rb and much more weakly with cdk4, but no interactions of the C/EBPα-S193A mutant with cdk2 and Brm were observed (Fig. 3A and B). We next performed a careful examination of the amounts of Rb in C/EBPα-S193A-Rb complexes and the amounts of free Rb in cells transfected with the C/EBPα-S193A mutant. Figure 3C shows a protocol for these experiments. 3T3-L1 cells were infected with adenoviruses driving the expression of WT C/EBPα and the C/EBPα-S193A mutant. In these experiments, we obtained an efficiency of transfections of approximately 92 to 95%. C/EBPα was immunoprecipitated from transfected cells, and Rb was examined in supernatants and in C/EBPα IPs. The major portion of Rb (80 to 85%) was associated with C/EBPα, while free Rb represented only 15 to 20% of the total Rb (Fig. 3D). In the case of WT C/EBPα, Rb was observed in C/EBPα IPs as part of a complex with C/EBPα, E2F4, and Brm (growth-inhibitory complex) (25), while the C/EBPα-S193A mutant failed to form this complex but sequestered Rb (Fig. 3E). These data were consistent with results of gel shift assays showing the disruption of Rb-E2F complexes by dephosphorylated C/EBPα (Fig. 2B and C). Thus, these studies suggested that the sequestering Rb might be a major pathway by which the C/EBPα-S193A mutant and Ser193-dephosphorylated C/EBPα cause the acceleration of cell growth.
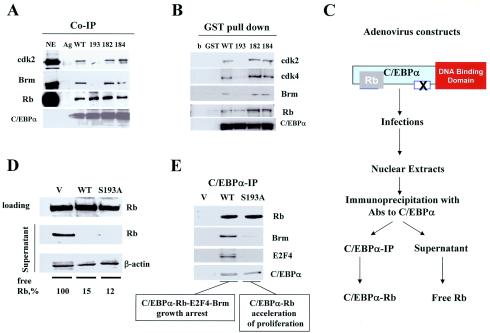
FIG. 3.
C/EBPα-S193A mutant sequesters Rb. (A) Co-IP studies. WT C/EBPα and C/EBPα mutants S193A, 182, and 184 (25) were transfected into 3T3-L1 cells. C/EBPα was immunoprecipitated from transfected cells; cdk2, cdk4, Rb, and Brm levels were determined in C/EBPα IPs by Western blotting with specific Abs. (B) GST pull-down assay. GST-C/EBPα constructs (shown on the top) were incubated with NEs from 3T3-L1 cells, washed with PBS, and examined by Western blotting with Abs to cdk2, cdk4, Brm, Rb, and C/EBPα. (C) A diagram showing the strategy for the experiment presented in panels D and E. Adenovirus-based WT C/EBPα and the C/EBPα-S193A mutant were infected into HT1080 cells (transfection efficiency, 92 to 95%). C/EBPα was immunoprecipitated from NEs, and Rb in supernatants was examined. (D) C/EBPα-S193A mutant sequesters Rb. The levels of Rb were determined in supernatants by Western blotting and calculated as ratios to β-actin and then as a percentage of Rb in cells transfected with the empty vector. A representative result of three experiments is shown. (E) WT C/EBPα forms growth-inhibitory C/EBPα-Rb-E2F4-Brm complexes, while the C/EBPα-S193A mutant interacts only with Rb. C/EBPα IPs were examined by Western blotting with Abs to Rb, E2F4, Brm, and C/EBPα.
Ser193 is a key residue which is required for the switch of biological activities of C/EBPα.
Our experimental design for specific dephosphorylation of C/EBPα at Ser193 included treatment of cells with insulin. Insulin signaling affects many biological pathways, which might indirectly change biological functions of CEBPα (2, 11, 18, 19). Therefore, we examined if dephosphorylation of Ser193 residue of C/EBPα was a key step in phosphorylation-dependent alterations of biological functions of C/EBPα. For this goal, we mutated Ser193 to aspartate (S193D) to mimic the phosphorylation state of C/EBPα and to potentially remove the C/EBPα-S193D molecule from insulin/PI3K/Akt/PP2A-mediated control. Figure 4A to C shows that the C/EBPα-S193D mutant strongly inhibited proliferation of 3T3-L1 cells and that the activation of the PI3K/Akt pathway by insulin failed to block the inhibitory activity of this constitutively active C/EBPα and to create a C/EBPα molecule that accelerated proliferation. Measurements of C/EBPα levels in experimental plates revealed that C/EBPα was expressed at the same levels in insulin-treated and untreated cells (Fig. 4C). We next examined the protein-protein interactions of the constitutively active C/EBPα-S193D mutant in 3T3-L1 cells. The WT and the C/EBPα-S193D mutant were immunoprecipitated from 3T3-L1 cells, and cdk2, cdk4, Rb, and Brm were examined in these IPs by Western blotting with specific Abs. Figure 4D shows that the insulin/PI3K/Akt pathway failed to block the interaction of the C/EBPα-S193D mutant with cdk's and Brm, while the interactions of WT C/EBPα with cdk2 and Brm were completely blocked in insulin-treated cells. Interestingly, the interaction of the dephosphorylated C/EBPα with cdk4 was reduced only to 30 to 50% but was easily detectable (Fig. 4D). Taken together, these studies confirmed that S193 is a key residue of C/EBPα, dephosphorylation of which changes growth-inhibitory activity of C/EBPα to the ability to accelerate proliferation.
FIG. 4.
Ser193 is a key residue which is required for the switch of biological functions of C/EBPα. (A) The C/EBPα-S193D mutant is resistant to insulin signaling. 3T3-L1 cells were transfected with empty pAdTrack vector, WT C/EBPα, and C/EBPα-S193D. Insulin was added at the time of transfection, and colony formation was examined at day 3 after transfections. Bar graphs represent a summary of three independent experiments. (B) A typical picture of green-fluorescing cells at day 3 after transfections. (C) Protein levels of C/EBPα determined by Western blotting (top) with extracts isolated from experimental plates. The membrane was reprobed with Abs to β-actin and stained with Coomassie blue (bottom). (D) Insulin does not block the interactions of the constitutively active C/EBPα-S193D mutant with cdk's and Brm. C/EBPα was immunoprecipitated from cells transfected with WT C/EBPα and the C/EBPα-S193D mutant; cdk2, cdk4, Brm, and Rb were examined in C/EBPα IPs. IgG, heavy IgG chains. (E) Ser190 in the growth-inhibitory region of human C/EBPα is required for the insulin-mediated regulation of growth-inhibitory activity of the human C/EBPα. (Left, top) Amino acid sequences of growth-inhibitory regions of human WT C/EBPα and Ser190A mutant linked to the His tag and cloned into the pAdTrack plasmid. Empty pAdTrack vector and these C/EBPα plasmids were transfected into 3T3-L1 cells, and formation of cell colonies was examined at day 3. (Left, bottom) The bar graphs show a summary of three independent experiments. (Right, top) Typical picture of colonies in insulin-treated and untreated cells. (Right, bottom) Western blotting demonstrates protein levels of hGIR determined by immunostaining with Abs to the His tag.
Both phosphorylation of Ser193 and the S193D mutation bring a negative charge to the growth-inhibitory region of C/EBPα, suggesting that the addition of the negative charge to C/EBPα is a key step in the activation of the inhibitory activity of C/EBPα. This suggestion is consistent with our studies of the hGIR of human C/EBPα. Although both mouse and human regions are proline rich, they have slightly different sequences surrounding Ser193 (Ser190 in human C/EBPα) (Fig. 4A and E). Despite this difference, growth-inhibitory activity of hGIR is also regulated by phosphorylation-dephosphorylation of Ser190. Our data demonstrate that hGIR alone inhibited cell proliferation and that its inhibitory activity could be blocked by insulin signaling (Fig. 4E). This insulin-dependent regulation required Ser190, since the mutation of Ser190 to Ala abolished the insulin-mediated block of inhibitory activity of hGIR (Fig. 4E). In these experiments, we monitored the expression of hGIR with Abs to the His tag. As one can see in Fig. 4E (bottom), the expression levels of hGIR were not affected by insulin. Taken together, these studies demonstrate that the activation of the PI3K/Akt/PP2A pathway creates a new biological activity of C/EBPα through dephosphorylation of a single Ser193 residue.
C/EBPα binds directly to certain E2F-dependent promoters.
We next examined if transcriptional activity of C/EBPα might play a role in the acceleration of growth. Although previous studies failed to connect the transcriptional activity of C/EBPα with the ability of C/EBPα to control the cell cycle, we considered this possibility because C/EBPα binds directly to certain E2F consensuses, such as one within the DHFR promoter (9). Since E2F targets are critical regulators of cell cycle progression, this interaction may contribute to the ability of C/EBPα to accelerate growth after dephosphorylation on Ser193. We first determined if other known E2F-dependent promoters could be directly bound by C/EBPα. Point mutational analysis of the E2F consensus within the DHFR promoter showed that CGCGAAAT nucleotides within this promoter were required for the interaction with C/EBPα (9). Examination of known E2F binding sites for the presence of a similar sequence divided E2F promoters into two groups. One group (the c-myc group) did not contain this consensus and did not interact with C/EBPα (Fig. 5A) (9). A second group of E2F-dependent promoters contained sequences that potentially might interact with C/EBPα (Fig. 5B). Gel shift-supershift analysis with the dI-dC competitor (a specific inhibitor of E2F, which does not affect binding of C/EBPα) showed that C/EBPα did bind to E2F binding sites within DHFR, E2F1, Ad2, p107, and the PCNA promoter (Fig. 3C), while the interaction of C/EBPα with DNA polymerase α and cdc2 promoters was very weak or not detectable. The binding of C/EBPα to E2F consensuses did not appear to involve E2Fs, since E2F activity is completely inhibited by the concentrations of dI-dC that were used in these experiments (2 μg/10 μl), and incorporation of Abs to E2F4 and E2F1 did not detect C/EBPα-DNA complexes (data not shown). A parallel gel shift assay with salmon DNA competitor (which does not block E2F binding) confirmed that these sequences interacted with E2F4 and E2F-Rb complexes (Fig. 5D shows data for E2F1 and PCNA promoters). The interaction of C/EBPα with E2F consensuses was specific, since cold bZIP, DHFR, and AdE2 oligomers competed for the interaction (data not shown). Thus, this analysis revealed that C/EBPα directly interacts with E2F consensuses of certain E2F-dependent promoters and suggests that dephosphorylated C/EBPα may accelerate growth through direct regulation of E2F-dependent promoters.
To further determine if the direct binding of C/EBPα to E2F-dependent promoters might be involved in the regulation of liver proliferation, we examined by ChIP assay the occupation of these promoters by C/EBPα in livers that proliferate after PH. We have previously found that C/EBPα also interacts with E2F-dependent promoters in quiescent livers through the association with E2F, as the component of the C/EBPα-Rb-E2F4-Brm complex (9). To distinguish the direct and indirect interactions of C/EBPα with E2F-dependent promoters, the ChIP assay was performed with the c-myc promoter (C/EBPα does not bind directly) and with the DHFR promoter to which C/EBPα could bind directly or as the complex with E2Fs. Chromatin solutions were isolated from quiescent livers and from livers 8 h after PH; C/EBPα, E2F1, and E2F4 were immunoprecipitated; and the presence of c-myc and DHFR promoters in IPs was examined by PCR with specific primers (9). As can be seen in Fig. 5E, C/EBPα occupied the DHFR promoter in quiescent young livers through direct interaction, since it was not detectable on the c-myc promoter. Although PH dephosphorylated C/EBPα at Ser193 (Fig. 1A), the dephosphorylated C/EBPα was also present on the DHFR promoter at 8 h after PH. The interaction of C/EBPα with DHFR promoter is specific, since C/EBPα was not detectable in the mock (agarose) control and on the c-myc promoter to which it did not bind directly. The occupation of the DHFR promoter by C/EBPα in proliferating livers suggests that the direct binding of C/EBPα to E2F-dependent promoters might contribute to the ability of dephosphorylated C/EBPα to accelerate cell proliferation.
The acceleration of cell proliferation by dephosphorylated C/EBPα requires functional Rb.
Since both direct binding of C/EBPα to E2F-dependent promoters and sequestering Rb might be involved in acceleration of proliferation, we next examined a possible contribution of these interactions to growth promotion activity of dephosphorylated C/EBPα and the C/EBPα-S193A mutant. To distinguish these pathways, we utilized two cell lines, SAOS2 and C33A, in which functional Rb is not expressed and therefore where cell growth cannot be affected through a depletion of Rb. WT C/EBPα and the C/EBPα-S193A mutant were transfected into SAOS2 cells, and proliferation was examined by measuring BrdU uptake. As can be seen in Fig. 6A and B, the percentage of SAOS2 cells synthesizing DNA in cells transfected with empty vector was similar to that in cells expressing C/EBPα-S193A mutant. The failure of the C/EBPα-S193A mutant to increase DNA replication in SAOS2 cells suggested that Rb is required for the ability of the S193A mutant to accelerate cell proliferation. To further examine this suggestion, we performed a colony formation assay with Rb-negative SAOS2 cells transfected with WT C/EBPα and treated with insulin. Since SAOS2 cells are human cells, we used Rb-positive human fibrosarcoma HT1080 cells as the control. We have previously shown that C/EBPα inhibits proliferation of these cells (22). Figure 6C and D shows a summary of these studies. Consistent with data obtained with mouse 3T3-L1 cells, the treatment of Rb-positive HT1080 cells with insulin not only blocked growth-inhibitory activity of C/EBPα, but also increased proliferation of cells expressing C/EBPα. On the contrary, no significant difference was observed in the proliferation of SAOS2 cells transfected with vector and in the proliferation of cells transfected with C/EBPα and treated with insulin. In untreated SAOS2 cells, WT C/EBPα inhibited proliferation, perhaps through inhibition of cdk2 and cdk4, since these kinases are associated with C/EBPα (Fig. 6E). To confirm the role of Rb in the acceleration of proliferation, we performed similar experiments with a second cell line, C33A, in which growth-inhibitory activity of Rb was blocked (13). Figure 6F shows that insulin-mediated dephosphorylation of C/EBPα in these Rb-deficient cells also did not lead to acceleration of proliferation. Thus, the studies with two Rb-deficient cell lines demonstrated that Rb is required for the dephosphorylated C/EBPα and for C/EBPα-S193A mutant to accelerate cell proliferation.
FIG. 6.
The ability of Ser193-dephosphorylated C/EBPα to accelerate proliferation requires functional Rb. (A) C/EBPα-S193A mutant does not promote proliferation in Rb-deficient SAOS2 cells. BrdU uptake was examined in SAOS2 cells transfected with WT C/EBPα and with the C/EBPα-S193A mutant. A typical picture of immunostaining of SAOS2 cells is shown. DAPI staining shows all cells on the field. Green fluorescent protein represents cells transfected with pAdTrack-C/EBPα or with empty pAdTrack vectors. (B) A summary of four independent experiments with BrdU uptake. A total of 100 to 150 cells were used for calculations in each experiment. (C) Ser193-dephosphorylated C/EBPα does not accelerate proliferation in Rb-deficient SAOS2 cells. A colony formation assay in Rb-positive HT1080 and Rb-negative SAOS2 cells transfected with WT C/EBPα and treated with insulin is shown. Bar graphs represent a summary of three independent experiments. Bar graphs labeled by 4+ show the percentage of colonies containing ≥4 cells per colony. (D) Colony formation assay in HT1080 (Rb-positive) and SAOS2 (Rb-negative) cells. Typical pictures are shown for cells transfected with empty vector and for cells transfected with pAdTrack-WT-C/EBPα, treated or untreated with insulin. Insulin-mediated dephosphorylation of C/EBPα in Rb-deficient cells does not accelerate proliferation. (E) WT C/EBPα inhibits proliferation of Rb-negative SAOS2 cells by interacting with cdk's. C/EBPα and empty vector were transfected into Rb-deficient SAOS2 cells and precipitated with specific Abs. cdk2, cdk4, and E2F4 were examined in C/EBPα IPs. E2F4 does not bind to C/EBPα in cells lacking Rb (9). (F) Ser193-dephosphorylated C/EBPα does not accelerate proliferation of C33A cells that lack a functional Rb pathway. The experiment was performed as described above. Bar graphs (left) represent a summary of three experiments. Typical pictures of green-fluorescing cells are shown on the right.
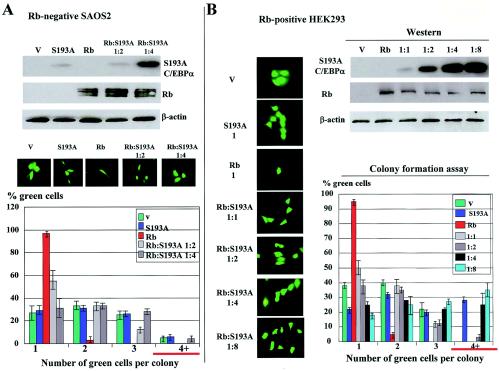
Ser-193-dephosphorylated C/EBPα competes with Rb for the regulation of cell cycle division.
We next examined if Rb was a primary target within pathway that leads to acceleration of proliferation. To address this issue, we asked if expression of Rb in Rb-deficient cells would restore the ability of the S193A mutant to accelerate proliferation. Rb-deficient SAOS2 cells were transfected with the S193A mutant and with Rb at Rb-S193A ratios of 1:2 and 1:4. Western blotting was performed with proteins isolated from experimental plates to monitor levels of transfected proteins. These data are shown in Fig. 7A. As can be seen, the expression of Rb and S193A proteins correlated with the amount of plasmid DNA transfected into the cells. The results of the colony formation assay are shown in Fig. 7A (bottom). As can be seen, the S193A mutant did not accelerate proliferation of these cells, while Rb inhibited proliferation of SAOS2 cells. Cotransfections of Rb and the S193A mutant revealed that S193A eliminated Rb-mediated growth arrest. These data suggested that Rb is a direct target within the pathway by which the C/EBPα-S193A accelerates proliferation. To further investigate this pathway, we performed similar experiments with Rb-positive 3T3-L1 and HEK293 cells. Since the C/EBPα-S193A mutant accelerated proliferation of these cells, one would expect that the ratio of Rb and C/EBPα-S193A would determine whether cells were arrested or had an increased rate of proliferation. Results of these studies with HEK293 cells are shown in Fig. 7B. Western blotting showed that Rb and the S193A mutant were expressed at expected levels. As expected, Rb alone inhibited proliferation of more than 95% of transfected cells, while the C/EBPα-S193A mutant alone accelerated proliferation of these cells. If Rb and the C/EBPα-S193A mutant cotransfected together, cell growth arrest-acceleration would depend on the ratios of these proteins. Under 1:1 and 1:2 ratios, cells proliferated at a rate close to that of control cells transfected with empty vector. If the C/EBPα-S193A mutant was expressed at levels four- to eightfold higher than Rb, the proliferation of these cells was accelerated. The acceleration of proliferation under these ratios seemed to be mediated through sequestering endogenous Rb, since under these ratios the C/EBPα-S193A mutant did not cause acceleration in Rb-deficient SAOS2 cells (Fig. 7A). We also performed experiments in which Rb expression was inhibited by small interfering RNA (siRNA) in Rb-positive HEK293 and HT1080 cells. The C/EBPα-S193A mutant was transfected into these cells expressing very low levels of Rb. Under our experimental conditions, the inhibition of Rb in these cells was sufficient to accelerate cell proliferation. The sequestering of remaining Rb by the C/EBPα-S193A mutant had a minor effect above the acceleration of proliferation mediated by siRNA inhibition of Rb (data not shown). Since both siRNA and the C/EBPα-S193A mutant targeted Rb, the acceleration of cell proliferation by siRNA-mediated inhibition of Rb supports the hypothesis that the C/EBPα-S193A mutant accelerates proliferation of 3T3-L1 and HT1080 cells by neutralizing the inhibitory activity of Rb. Thus, these investigations demonstrated that Rb is the primary target within the pathway by which Ser193-dephosphorylated C/EBPα accelerates cell proliferation.
FIG. 7.
The C/EBPα-S193A mutant and Rb compete for the regulation of cell growth. (A) Rb-mediated growth arrest of SAOS2 cells is inhibited by the Ser193-dephosphorylated C/EBPα. (Top) Protein levels of transfected Rb and C/EBPα-S193A proteins determined by Western blotting. β-actin shows a reprobe of the filter with Abs to β-actin. (Middle) Typical picture of cells. (Bottom) Bar graphs represent a summary of three experiments for colony growth assay. (B) A ratio of Rb and C/EBPα-S193A determines if cells are growth arrested or have the increased rate of proliferation. Rb and the C/EBPα-S193A mutant were cotransfected into Rb-positive HEK293 cells under different ratios, and cell proliferation was examined by a colony formation assay. (Left) Typical picture of cells. Western blotting indicates protein levels of Rb and C/EBPα-S193A mutant determined in extracts isolated from experimental plates (endogenous Rb is detectable after longer exposures). Bar graphs (right) represent a summary of three independent experiments.
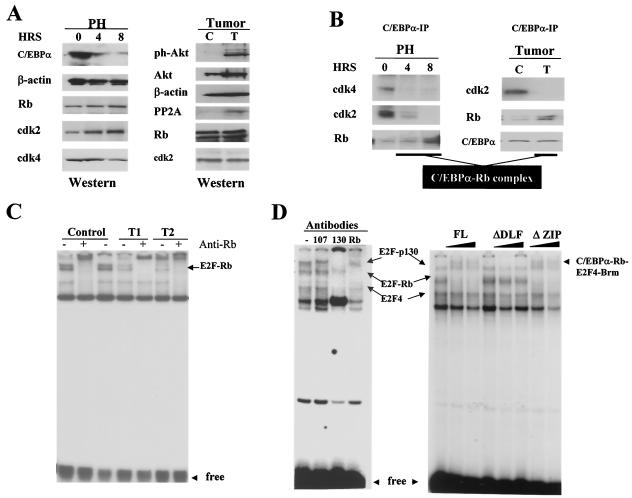
Dephosphorylated C/EBPα sequesters Rb in proliferating livers.
Given that dephosphorylated C/EBPα accelerates proliferation in cultured cells by sequestering Rb, we examined if this activity of C/EBPα was involved in the promotion of liver proliferation after surgical resection. We first examined C/EBPα-Rb interactions in mouse livers proliferating after PH. In this system, C/EBPα protein levels were reduced after surgery, and remaining C/EBPα was dephosphorylated on Ser193 (Fig. 1A) (25). Co-IP studies with NEs from regenerating livers showed that although the expression of cdk2 and cdk4 was not reduced after PH (Fig. 8A), C/EBPα-cdk2-cdk4 complexes were not detectable after PH. However, the amounts of Rb in C/EBPα complexes were not changed despite the reduction of C/EBPα levels (Fig. 8B). We observed a slight increase of Rb in C/EBPα IPs, which was within the variability of the Co-IP assay. Since C/EBPα is also dephosphorylated at Ser190 in human tumors (25), we next examined if Ser190-dephosphorylated C/EBPα sequesters Rb in human tumors. Western blotting with Abs to ph-Akt shows that PI3K/Akt was activated in human tumors and that PP2A (which removes a phosphate from Ser193) was increased in the nuclei of tumor sections (Fig. 8A). Examination of cdk2 and Rb in C/EBPα IPs showed that cdk2 was no longer associated with C/EBPα in tumor samples, while amounts of Rb were not changed in C/EBPα IPs from the tumors. Total levels of cdk2 and Rb were not altered in tumors. These data are consistent with observations obtained with mouse livers and indicate that PI3K/Akt/PP2A pathway blocks the growth-inhibitory activity of C/EBPα but does not change its ability to interact with Rb and sequester Rb from Rb-E2F complexes. Thus, Co-IP studies show that Ser193- or Ser190-dephosphorylated C/EBPα interacts with Rb in proliferating livers and suggest that the Ser193-dephosphorylated C/EBPα may sequester a portion of Rb from E2F-Rb complexes-repressors.
FIG. 8.
Phosphorylation-dependent switch of biological functions of C/EBPα in proliferating livers. (A) Protein levels of C/EBPα and cell cycle proteins after PH and in liver tumors. (Left) Western blotting was performed with NEs isolated at 4 and 8 h after PH with Abs to C/EBPα, Rb, cdk2, and cdk4. The membrane was reprobed with Abs to β-actin. (Right) Western blotting shows protein levels of Akt, ph-Akt, PP2A, cdk2, and Rb in NEs from control (C) and tumor (T) sections of the same liver. (B) C/EBPα-cdk complexes are reduced in proliferating livers, while C/EBPα-Rb complexes are abundant. (Left) C/EBPα was precipitated from NEs isolated from regenerating livers and amounts of cdk2, cdk4, and Rb were determined within C/EBPα IPs. (Right) C/EBPα was immunoprecipitated from control and tumor sections of the liver, and cdk2 and Rb were determined in C/EBPα IPs by Western blotting with specific Abs. (C) E2F-Rb complexes are reduced in liver tumors that express dephosphorylated C/EBPα. An E2F gel shift assay was performed with NEs from two tumor samples (T1 and T2) and from a control section. Abs to Rb were incorporated into the binding reaction mixtures (shown on the top). (D) Composition of E2F complexes in NEs from mouse livers (left). Abs to p107, p130, and Rb were added to the binding reactions containing liver NEs and a DNA probe with c-myc E2F consensus. Rb-interacting region (DLF) (24) of C/EBPα is required to sequester Rb from the E2F-Rb complexes. Increasing amounts of purified FL C/EBPα and C/EBPα deletion mutants ΔDLF and ΔZIP were preincubated with NEs from mouse livers and added to the E2F binding reaction mixture with the E2F probe. Positions of E2F-Rb complexes and C/EBPα-Rb-E2F4-Brm complexes are shown by arrows.
To test if the sequestering Rb affects Rb-E2F complexes in proliferating livers, E2F gel shift-supershift assays were performed with NEs isolated from two tumor samples and from control sections. The reproducible result of these studies is shown in Fig. 8C. In control liver samples, Rb-E2F complexes were abundant and represented the majority of the E2F-Rb family complexes. Although the composition of E2F-Rb complexes was not changed in tumor samples, the amounts of Rb-E2F complexes were significantly reduced. The reduction of the E2F-Rb complexes in proliferation livers correlated with the abundant C/EBPα-Rb complexes (Fig. 8B) and was consistent with the hypothesis that dephosphorylated C/EBPα sequesters Rb from E2F complexes.
N-terminal region of C/EBPα is required for sequestering Rb from E2F complexes.
We next examined if the direct interaction of Ser193-dephosphorylated C/EBPα with Rb is involved in the disruption of E2F-Rb complexes observed in livers. To address this issue, we performed experiments with the NEs isolated from mouse liver. Examination of the composition of E2F complexes in the mouse liver by gel shift assay demonstrated that quiescent livers expressed E2F-p130 and E2F-Rb complexes (Fig. 8D). We then examined effects of full-length (FL) C/EBPα and two C/EBPα deletion mutants on the E2F complexes. Liver NEs were incubated with an excess of C/EBPα proteins and then examined by gel shift assay with the c-myc E2F probe, to which C/EBPα does not bind directly. Two mutants were used in these studies: C/EBPα-ΔDLF, lacking the Rb-interacting region, and C/EBPα-ΔZIP, lacking the C-terminal zipper region but containing the Rb-interacting and growth-inhibitory regions (24). Figure 8D shows that the incubation of FL C/EBPα with NEs completely disrupted the Rb-E2F complex and partially disrupted the E2F-p130 complex, while the ΔDLF protein (Rb-interacting region deleted) did not affect E2F-Rb complexes. Similar to HT1 cells, the disruption of E2F-Rb complexes by FL C/EBPα was accompanied by the appearance of a high-molecular-weight C/EBPα-Rb-E2F4-Brm complex (Fig. 8D). It is interesting that the addition of C/EBPα-ΔZIP (which lacks the C-terminal zipper region) disrupted E2F-Rb complexes and also formed the new high-molecular-weight complex. Taken together, these results show that the Rb-interacting region is required for the sequestration of Rb from E2F-Rb complexes.
DISCUSSION
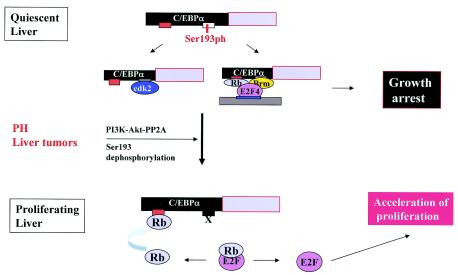
Data presented in this paper describe a new function of C/EBPα, which is generated in proliferating livers by dephosphorylation of C/EBPα and which supports liver proliferation. Previous studies showed that C/EBPα is a strong inhibitor of liver proliferation, suggesting that C/EBPα should interfere with liver proliferation after surgical resections and in tumor samples; however, livers proliferate and express high levels of C/EBPα (6, 25). This suggested that the liver might initiate pathways to reorganize activities of key regulators of cell cycle progression, such as C/EBPα, to promote proliferation. Recent publications revealed that one of these pathways might be the phosphorylation of C/EBPα (17, 25). Previous studies demonstrated that mechanisms of C/EBPα-mediated growth arrest are complicated and involve interactions with several cell cycle proteins (9, 13, 14, 15, 23, 24, 26, 27). Our recent findings show that these interactions are tightly regulated in cells and might be used as pathways to inhibit cell proliferation, as well as serve as pathways to accelerate cell growth. In this paper, we investigated molecular mechanisms by which Ser193-dephosphorylated C/EBPα accelerates cell proliferation. Discovery of this new biological function of C/EBPα is quite surprising, since it is opposite to the well-characterized growth-inhibitory activity of C/EBPα. On the basis of our previous data, we considered two pathways by which Ser193-dephosphorylated C/EBPα might increase rates of proliferation: (i) direct binding to and activation of E2F-dependent promoters and (ii) sequestering Rb from E2F-Rb complexes-repressors. Both these pathways target a key event of cell cycle progression: activation of S-phase and mitotic-specific genes (3). Data obtained with a variety of cultured cells and with the mouse livers suggest that sequestering Rb is a major mechanism by which Ser193-dephosphorylated C/EBPα accelerates proliferation. Because Ser193-dephosphorylated C/EBPα is created by the PI3K/Akt/PP2A pathway in mouse livers after PH and in human tumors (25), we examined this mechanism with mouse (3T3-L1) and human (HT1080) cells, as well as with Rb-deficient SAOS2 and C33A cells. Experimental data from all of these cells show that Rb is a primary target of Ser193-dephosphorylated C/EBPα. A hypothetical pathway by which dephosphorylated C/EBPα sequesters Rb and accelerates cell proliferation is shown in Fig. 9. In the quiescent liver and in cultured cells that have reduced PI3K/Akt/PP2A activity, C/EBPα inhibits proliferation through two pathways: inhibition of cdk's and E2F repression. The activation of the PI3K/Akt/PP2A pathway in proliferating livers and in cultured cells eliminates growth-inhibitory activity of C/EBPα by dephosphorylation of Ser193. Moreover, the remaining interactions with Rb lead to sequestering Rb from E2F-Rb repressor complexes and to the neutralization of Rb-mediated repression of the cell cycle progression. Thus, our model suggests that the elimination of growth-inhibitory activity of Rb by Ser193-dephosphorylated C/EBPα causes the acceleration of cell proliferation.
FIG. 9.
A hypothetical pathway by which Ser193-dephosphorylated C/EBPα accelerates liver proliferation. In quiescent livers, C/EBPα arrests proliferation through inhibition of cdk2 and E2F transcription. PH and liver tumors activate the PI3K/Akt/PP2A pathway, which dephosphorylates Ser193 and blocks the interactions of C/EBPα with cdk2 and Brm. The Ser193-dephosphorylated C/EBPα interacts with Rb and sequesters Rb from the E2F-Rb complexes, leading to accelerated proliferation.
The interaction of C/EBPα with Rb was discovered in 1996 in work carried out in Lee's laboratory (1) and was later confirmed by other studies (14, 15, 24). These investigations demonstrated that Rb increases the binding of C/EBPα to promoters and the activation of C/EBPα-dependent genes (1, 14). In the light of these publications, the association of C/EBPα with Rb in proliferating livers (Fig. 8A) suggests that the ability of C/EBPα to activate liver-specific genes is not decreased after PH, despite the reduction of total protein levels of C/EBPα. Our unpublished observations confirm this suggestion. In this paper, we focused our studies on the detailed examination of mechanisms by which the Ser193-dephosphorylated C/EBPα accelerates cell proliferation. This new activity of C/EBPα is tightly regulated in cultured cells and in the liver by signal transduction pathways such as insulin/PI3K/Akt/PP2A, which is activated after PH (25). We also found that a molar ratio of Ser193-dephosphorylated C/EBPα and Rb is crucial for cell fate. Data from Rb-negative and Rb-positive cells showed that expression of Ser193-dephosphorylated C/EBPα over Rb levels blocks Rb-mediated growth arrest, while overexpression of Rb above the levels of Ser193-dephosphorylated C/EBPα eliminates growth promotion activity of the Ser193-dephosphorylated C/EBPα. In addition to the interactions with Rb, dephosphorylated C/EBPα is able to bind to DNA and activate promoters (25). Experiments in Rb-deficient SAOS2 cells clearly showed that Rb is required for the growth promotion activity of Ser193-dephosphorylated C/EBPα. Although these observations favor the hypothesis that dephosphorylated C/EBPα accelerates proliferation via sequestering Rb, they do not rule out the possibility that the direct interaction of C/EBPα with E2F-dependent promoters of the cell cycle genes also contributes to the promotion of proliferation. In fact, examination of E2F-dependent promoters in the livers showed that the Ser193-dephosphorylated C/EBPα occupies the DHFR promoter in proliferating livers. However, the contribution of this pathway is not clear. Further experiments are necessary to elucidate this issue. Co-IP data showing that a portion of Rb is associated with C/EBPα in proliferating livers and observations in cultured cells strongly suggest that Ser193-dephosphorylated C/EBPα promotes proliferation by sequestering a portion of Rb from E2F-Rb complexes-repressors. Since Rb and C/EBPα are key proteins that maintain liver quiescence, our data provide a new mechanism by which proliferating livers neutralize growth-inhibitory activity of both C/EBPα and Rb.
Acknowledgments
This work was supported by National Institutes of Health grants AG20752, CA100070, and GM55188 (N.A.T.).
We thank Xiurong She for the assistance with Western blotting experiments.
REFERENCES
- 1.Chen, P.-L., D. J. Riley, Y. Chen, and W.-H. Lee. 1996. Retinoblastoma protein positively regulates terminal adipocyte differentiation through direct interaction with C/EBPs. Genes Dev. 10:2794-2804. [DOI] [PubMed] [Google Scholar]
- 2.Duong, D. T., M. E. Waltner-Law, R. Sears, L. Sealy, and D. K. Granner. 2002. Insulin inhibits hepatocellular glucose production by utilizing liver-enriched transcriptional inhibitory protein to disrupt the association of CREB-binding protein and RNA polymerase II with the phosphoenolpyruvate carboxykinase gene promoter. J. Biol. Chem. 277:32234-32242. [DOI] [PubMed] [Google Scholar]
- 3.Dyson, N. 1998. The regulation of E2F by RB-family proteins. Genes Dev. 12:2245-2262. [DOI] [PubMed] [Google Scholar]
- 4.Fausto, N. 2000. Liver regeneration. J. Hepatol. 32:19-31. [DOI] [PubMed] [Google Scholar]
- 5.Flodby, P. C., H. Barlow, L. Kalefjord, L. Ahrlund-Richer, and K. G. Xanthopolous. 1996. Increased hepatic cell proliferation and lung abnormalities in mice deficient in CCAAT/enhancer binding protein α. J. Biol. Chem. 271:24753-24760. [DOI] [PubMed] [Google Scholar]
- 6.Greenbaum, L. E., D. E. Cressman, B. A. Haber, and R. Taub. 1995. Coexistence of C/EBPα, β, growth-induced proteins and DNA synthesis in hepatocytes during liver regeneration. J. Clin. Investig. 96:1351-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He, T.-C., S. Zhou, L. T. Costa, J. Yu, K. W. Kinzler, and B. Vogelstein. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA 95:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hemati, N., S. E. Ross, R. L Erickson, G. E. Groblewski, and O. A. MacDougald. 1997. Signaling pathway through which insulin regulates CCAAT/enhancer binding protein α (C/EBPα) phosphorylation and gene expression in 3T3-L1 adipocytes. J. Biol. Chem. 272:25913-25919. [DOI] [PubMed] [Google Scholar]
- 9.Iakova, P., S. S. Awad, and N. A. Timchenko. 2003. Aging reduces proliferative capacities of liver by switching pathways of C/EBPα growth arrest. Cell 113:495-506. [DOI] [PubMed] [Google Scholar]
- 10.Kurumiya, Y., K. Nozawa, K. Sakaguchi, M. Nagino, Y. Nimura, and S. Yoshida. 2000. Differential suppression of liver-specific genes in regenerating rat liver induced by extended hepatectomy. J. Hepatol. 32:636-644. [DOI] [PubMed] [Google Scholar]
- 11.Lawlor, M. A., and D. R. Alessi. 2001. PBK/Akt: a key mediator of cell proliferation, survival and insulin response. J. Cell Sci. 114:2903-2910. [DOI] [PubMed] [Google Scholar]
- 12.Mitchell, C., and H. Gilgenkrantz. 2003. Transcriptional profiling of liver regeneration: new approaches to an old trick! J. Hepatol. 38:847-849. [DOI] [PubMed] [Google Scholar]
- 13.Müller, C., C. F. Calkhoven, X. Sha, and A. Leutz. 2004. The CCAAT enhancer-binding protein α (C/EBPα) requires a SWI/SNF complex for proliferation arrest. J. Biol. Chem. 279:7353-7358. [DOI] [PubMed] [Google Scholar]
- 14.Pederson, T. A., E. Kowenz-Leutz, A. Leutz, and C. Nerlov. 2001. Cooperation between C/EBPα, TBP/TFIIB and SWI/SNF recruiting domains is required for adipocyte differentiation. Genes Dev. 15:3208-3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Porse, B. T., T. A. Pederson, X. Xu, B. Lindbergh, U. M. Wewer, L. Fris-Hansen, and C. Nerlov. 2001. E2F repression by C/EBPα is required for adipogenesis and granulopoiesis in vivo. Cell 107:247-258. [DOI] [PubMed] [Google Scholar]
- 16.Ross, S. E., R. L. Erickson, N. Hemati, and O. A. MacDougald. 1999. Glycogen synthase kinase 3 is an insulin-regulated C/EBPα kinase. Mol. Cell. Biol. 19:8433-8441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ross, S. E., H. S. Radomska, B. Wu, P. Zhang, J. N. Winnay, L. Baijnok, W. S. Wright, F. Schaufele, D. G. Tenen, and O. A. MacDougald. 2004. Phosphorylation of C/EBPα inhibits granulopoiesis. Mol. Cell. Biol. 24:675-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shamji, A. F., P. Nghiem, and S. L. Schreiber. 2003. Integration of growth factor and nutrient signaling: implications for cancer biology. Mol. Cell 12:271-280. [DOI] [PubMed] [Google Scholar]
- 19.Shi, D., Y.-R. Deng, S.-L. Liu, Y.-D. Zhang, and L. Wei. 2003. Redox stress regulates cell proliferation and apoptosis of human hepatoma through Akt protein phosphorylation. FEBS Lett. 542:60-64. [DOI] [PubMed] [Google Scholar]
- 20.Shim, M., and R. C. Smart. 2003. Lithium stabilizes the CCAAT/enhancer-binding protein α (C/EBPα) through a glycogen synthase kinase 3 (GSK3)-independent pathway involving direct inhibition of proteasomal activity. J. Biol. Chem. 278:19674-19681. [DOI] [PubMed] [Google Scholar]
- 21.Soriano, H. E., D. C. Kang, M. J. Finegold, M. J. Hicks, N.-D. Wang, W. Harrison, and G. J. Darlington. 1998. Lack of C/EBPα gene expression results in increased DNA synthesis and in an increased frequency of immortalization of freshly isolated rat hepatocytes. Hepatology 27:392-401. [DOI] [PubMed] [Google Scholar]
- 22.Timchenko, N. A., M. Wilde, M. Nakanishi, J. R. Smith, and G. J. Darlington. 1996. CCAAT/enhancer-binding protein α (C/EBPα) inhibits cell proliferation through the p21 (WAF-/CIP-1/SDI-1) protein. Genes Dev. 10:804-815. [DOI] [PubMed] [Google Scholar]
- 23.Timchenko, N. A., T. E. Harris, M. Wilde, T. A. Bilyeu, B. L. Burgess-Beusse, M. J. Finegold, and G. J. Darlington. 1997. CCAAT/enhancer binding protein α regulates p21 protein and hepatocyte proliferation in newborn mice. Mol. Cell. Biol. 17:7353-7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Timchenko, N. A., M. Wilde, and G. J. Darlington. 1999. C/EBP alpha regulates formation of S-phase-specific E2F-p107 complexes in livers of newborn mice. Mol. Cell. Biol. 19:2936-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang, G.-L., P. Iakova, M. Wilde, S. Awad, and N. A. Timchenko. 2004. Liver tumors escape negative control of proliferation via PI3K/Akt-mediated block of C/EBPα growth inhibitory activity. Genes Dev. 18:912-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang, H., T. Goode, P. Iakova, J. Albrecht, and N. A. Timchenko. 2002. C/EBPα triggers proteasome-dependent degradation of cdk4 during growth arrest. EMBO J. 21:930-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang, H., P. Iakova, M. Wilde, A. Welm, T. Goode, W. J. Roesler, and N. A. Timchenko. 2001. C/EBPα arrests cell proliferation through direct inhibition of cdk2 and cdk4. Mol. Cell 8:817-828. [DOI] [PubMed] [Google Scholar]