Abstract
Cas9 proteins are RNA-guided endonucleases that protect bacteria from viral infection. These endonucleases have been creatively repurposed as programmable molecular scalpels for surgical manipulation of DNA. Now, two papers in Cell identify viral proteins that suppress the Cas9 endonucleases, and show that these proteins may function like molecular sheaths for the Cas9 scalpel.
To call it a ‘revolution’ might be cliché. In the short span of a decade, research aimed at determining the biological role of DNA repeats called CRISPRs (clusters of interspaced short palindromic repeats) has gone from scientific obscurity to mainstream celebrity. CRISPR repeats are common in bacterial and archaeal genomes, and in 2007 Barrangou et al showed that CRISPRs were part of an adaptive immune system that protects bacteria from viral infection (Barrangou et al., 2007). The discovery of an adaptive immune system in bacteria united a group of scientists around the common goal of understanding the molecular mechanisms of these immune systems — a line of work that quickly lead to an unexpected “revolution” in genome editing technologies that may cure genetic diseases.
Bacteria and archaea acquire immunity by integrating short fragments of foreign (e.g. viral) DNA into CRISPR loci in their own genome. CRISPR loci provide a molecular memory of previous encounters with foreign DNA and CRISPR transcripts are processed into short CRISPR RNAs (crRNAs) that guide protective nucleases to foreign targets for cleavage. Cas9 is one of these crRNA-guided nucleases and the discovery that this bacterial protein cleaves both strands of a complementary DNA target led to the creative repurposing of these enzymes as programmable molecular scalpels capable of precise genome surgery in a variety of different cell types and organisms, including humans (Barrangou and Doudna, 2016). These genome editing technologies are rapidly moving toward clinical applications, and now two different papers in this issue of Cell (pg. XXX and YYY) identified proteins that may improve the safety of these enzymes by functioning like molecular sheaths for the Cas9 scalpel.
Presumably, bacterial immune systems like CRISPRs evolved in response to antagonistic interactions with molecular parasites like phage, where the competing selfish interests of viral replication and host fitness often create a dynamic landscape of selective pressures that drive evolution and genetic innovation. In 1973, the evolutionary biologist Leigh Van Valen famously compared this dynamic evolutionary landscape to Alice's predicament in Lewis Carroll's fantasy novel “Through the Looking Glass”. An exasperated Alice complains to the Red Queen that she is exhausted from running, only to find she is still beneath the same tree under which she had started. Van Valen's metaphor provides a conceptual framework for understanding the constant ‘arms race’ between co-evolving species that must perpetually adapt and proliferate; not merely to gain reproductive advantage, but to simply survive (Van Valen, 1973).
CRISPR-mediated adaptive immune systems represent a formidable barrier to viral predation and — consistent with the expectations of a biological ‘arms race’ — viruses have evolved ‘anti-CRISPR’ proteins that suppress these immune systems. However, much in the same way that Cas9 wasn't “discovered” by scientists looking for a way to precisely edit genomes, anti-CRISPRs were not discovered by scientists looking for a way to suppress CRISPR-mediated immune systems. In 2010, Joe Bondy-Denomy was an inquisitive graduate student in Alan Davidson's laboratory at the University of Toronto looking for new phenotypes in Pseudomonas aeruginosa (an environmentally ubiquitous and medically relevant gram-negative bacterium) that occur as a consequence of viral infection. Some viruses, known as temperate phage, integrate into the bacterial genome upon infection and occasionally these lysogens (strains of bacteria that contain an integrated viral genome) display new phenotypes, which for pathogens are often associated with virulence or antibiotic resistance.
Another frequent outcome of lysogeny is that the integrated virus will block subsequent infections by related phages, a phenomenon known as “superinfection exclusion”. However, Joe found a few lysogens with the opposite phenotype. In other words, bacterial strains formerly resistant to infection by a particular virus suddenly became sensitive to infection after they had been lysogenized (Bondy-Denomy et al., 2013). What could explain this unexpected result? Joe and Alan showed that viral resistance in the original strains was due to CRISPR-mediated immunity and hypothesized that the lysogens contained new viral gene(s) responsible for suppressing the CRISPR-mediated immune system. But not all lysogens suppressed the CRISPR immune system; so to guide their search for these enigmatic suppressors they aligned a family of related viral genomes and searched for differences that correlated with the phage sensitive phenotype. This comparative genomic analysis revealed a diverse set of small open reading frames (ORFs) between two conserved genes involved in viral assembly (i.e. head morphogenesis). To determine if these genes were responsible for suppression of the CRISPR system they cloned and overexpressed 17 of the genes and showed that 5 of them resulted in a phage sensitive phenotype (i.e. functioned to suppress the immune system). They called these suppressors “anti-CRISPRs” (Acrs), and in a series of follow-up experiments they showed that these anti-CRISPR proteins are mechanistically diverse and that different anti-CRISPRs target different components of the type I-F CRISPR system in P. aeruginosa (Bondy-Denomy et al., 2015; Bondy-Denomy et al., 2013). Many, but not all of the anti-CRISPRs they cloned suppressed the type I-F CRISPR system in P. aeruginosa; so one logical extension of this study was to test these anti-CRISPRs on other immune systems. There are six main types and 19 different subtypes of the CRISPR-Cas immune systems and many of the anti-CRISPRs that had no phenotype on the type I-F immune system were shown to target the Type I-E system (Pawluk et al., 2014).
The original anti-CRISPRs were small and contained no conserved sequence motifs that could be used to identify other anti-CRISPRs. To expand the search, Pawluk et al went looking for a genetic landmark that could be used as a proxy for finding anti-CRISPRs (Pawluk et al., 2016). They identified a conserved gene with a helix-turn-helix (HTH) motif that was downstream of the known anti-CRISPR genes, but absent in related phages lacking anti-CRISPRs. Using this anti-CRISPR associated (aca) gene to query the database they identified five additional anti-CRISPRs with broad distribution across the phylum Proteobacteria. Now in this issue of Cell (pg XXX), Pawluk et al further expand the anti-CRISPR hunt by looking for novel anti-CRISPRs in other phage families that might target other CRISPR-Cas systems. Using the Aca sequences as powerful fiducial markers; they identify a putative acr gene found in mobile genetic elements (MGEs) associated with microbes that contain a Type II-C CRISPR-Cas system (i.e. AcrIIC). Then, in a sort of bioinformatic ping-pong, they switched from searching for Aca proteins (ping), to looking for AcrIIC homologues (pong). This approach identifies new proteins related to the original AcrIIC protein, some of which are located adjacent to new Aca proteins. These new Aca proteins now “serve” (ping) as fresh starting points to query for new Acr proteins. Focusing on the mechanism of suppression, these authors go on to show that the AcrIIC proteins bind directly to the Cas9 protein from Neisseria meningitidis (NmeCas9). NmeCas9 has been repurposed for targeted genome engineering in human cells (Hou et al., 2013), and the authors show that the AcrIIC proteins block NmeCas9 from binding to the crRNA-guided target and without DNA binding there is no cleavage.
In a complementary study by Rauch et al (pg XXX), these authors implement a creative new approach for finding novel anti-CRISPRs (Rauch et al). Previous work has shown that CRISPR loci sometimes contain spacers complementary to locations in their own genome, which should result in autoimmunity (i.e. crRNA-guided targeting of the bacterial genome) (Stern and Sorek, 2010). However, Rauch et al hypothesized that bacterial lysogens (strains containing an integrated prophage) might contain anti-CRISPRs that block the autoimmune reaction. To find these inhibitors, they searched cas9-containing genomes for co-existence of a spacer and a complimentary target. This analysis led to the discovery of four unique anti-CRISPRs that inhibit the Type II-A CRISPR-Cas9 systems (i.e. acrIIA1, acrIIA2, acrIIA3, and acrIIA4). Like Pawluk et al, Rauch and colleagues do not miss the opportunity to demonstrate how these anti-CRISPRs might be useful for controlling Cas9 activity. They show that two of these proteins can be used to block crRNA-guided DNA binding in bacterial cells and that these same AcrIIA proteins also block Cas9-mediated target DNA cleavage in human cells.
Together, these two papers build on a foundation of work that has repeatedly shown that the interfaces of genetic conflict are hot spots for biological and biotechnological innovation. In Van Valen's original paper outlining his conception of the Red Queen Hypothesis, he notes that his biological observations are directly analogous to Newton's third law of motion (Reminder: “For every action there is always an equal and opposite reaction”). Bacteria have evolved sophisticated CRISPR-based immune systems in responses to phage predation, and phages evolved anti-CRISPRs that neutralize these immune systems. But this tit-for-tat will not stalemate at a standoff between CRISRPs and anti-CRSIPRs, so keep an eye out for the anti-anti-CRISPR.
Figure. An evolutionary tit-for-tat between host CRISPRs and viral anti-CRISPRs.
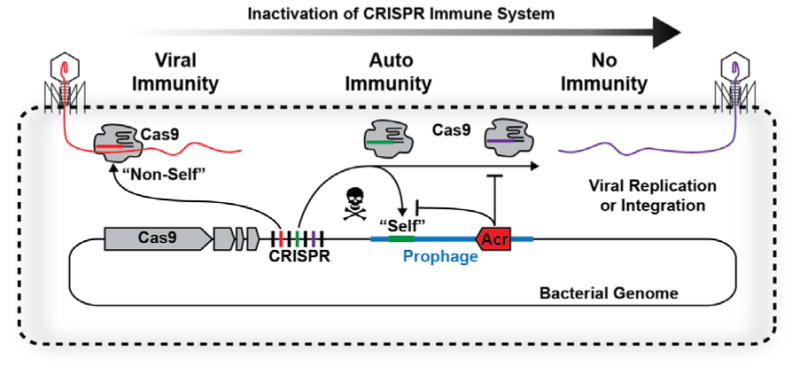
Schematic representation of a virus infecting a bacterial cell. The cell contains an active type II CRISPR system—typified by Cas9—and a CRISPR locus with a spacer (red) complementary to a target in the viral genome (red). However, in some cases a phage (a virus that infect bacteria) is able to integrate the bacterial genome. The integrated phage is now called a prophage (blue) and the cell is referred to as a lysogen. If the host CRISPR locus contains a spacer (green) that targets the prophage, then this may elicit an autoimmune reaction that results in the degradation of the host genome. Some (pro)phages encode anti-CRISPR (Acr, red arrow) proteins that blocks Cas9 cleavage, preventing CRISPR-induced autoimmunity. In this case, the host and the prophage have a shared interest in preventing Cas9 degradation, but the trade-off is that Acr-mediated blocking of the immune systems also make the cell more susceptible infection by subsequent viruses.
References
- Barrangou R, Doudna JA. Applications of CRISPR technologies in research and beyond. Nature Biotechnology. 2016;34:933–941. doi: 10.1038/nbt.3659. [DOI] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Bondy-Denomy J, Garcia B, Strum S, Du M, Rollins MF, Hidalgo-Reyes Y, Wiedenheft B, Maxwell KL, Davidson AR. Multiple mechanisms for CRISPR-Cas inhibition by anti-CRISPR proteins. Nature. 2015;526:136–139. doi: 10.1038/nature15254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondy-Denomy J, Pawluk A, Maxwell KL, Davidson AR. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature. 2013;493:429–U181. doi: 10.1038/nature11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou ZG, Zhang Y, Propson NE, Howden SE, Chu LF, Sontheimer EJ, Thomson JA. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:15644–15649. doi: 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawluk A, Bondy-Denomy J, Cheung VHW, Maxwell KL, Davidson AR. A New Group of Phage Anti-CRISPR Genes Inhibits the Type I-E CRISPR-Cas System of Pseudomonas aeruginosa. Mbio. 2014;5 doi: 10.1128/mBio.00896-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawluk A, Staals RHJ, Taylor C, Watson BNJ, Saha S, Fineran PC, Maxwell KL, Davidson aAR. Inactivation of CRISPR-Cas systems by anti-CRISPR proteins in diverse bacterial species. Nature Microbiology. 2016 doi: 10.1038/nmicrobiol.2016.85. [DOI] [PubMed] [Google Scholar]
- Stern A, Sorek R. The phage-host arms race: shaping the evolution of microbes. Bioessays. 2010;33:43–51. doi: 10.1002/bies.201000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Valen L. A new evolutionary law. Evol Theor. 1973;1:1–30. [Google Scholar]


