Abstract
Autophagy is a fundamental cellular process used for the turnover and recycling of cytosolic components and damaged organelles. Originally characterized as a response to cellular stress, it now is well established that autophagy also is used as a defensive mechanism to combat the infection of host cells by intracellular pathogens. However, although this defensive strategy does limit the proliferation of most pathogens within their host cells, successful pathogens have evolved countermeasures that subvert or circumvent the autophagic response. In this review, we discuss the mechanisms used by a number of these pathogens to escape autophagy, with a particular focus on Salmonella enterica serovar Typhimurium, which has been the most extensively studied example. We also discuss the consequences of bacterial autophagy for the broader innate immune response.
Keywords: Autophagy, Xenophagy, Salmonella, FAK, Akt, Interferon
Abbreviations used in this paper: DAG, diacylglycerol; FAK, focal adhesion kinase; IFN, interferon; LC3, microtubule-associated light chain 3; mTORC1, mechanistic target of rapamycin 1; SCV, Salmonella-containing vacuole; TLR, Toll-like receptor; T3SS, type III secretion system
Summary.
Many intracellular pathogens induce an autophagic response upon entering host cells. However, successful pathogens have evolved mechanisms for avoiding or suppressing autophagy. Here we discuss some of these mechanisms, with a special emphasis on the enteric pathogen Salmonella enterica serovar Typhimurium.
Macroautophagy (referred to hereafter as autophagy) is an evolutionarily conserved, essential process in which cellular components such as cytosolic protein aggregates or damaged organelles are delivered to lysosomes for degradation and recycling of the resulting metabolites.1 Although autophagy occurs at a low rate under homeostatic conditions, it is amplified in response to cellular stresses including nutrient deprivation, hypoxia, energy loss, or infection with microbial agents. The capture of cytosolic components during autophagy is achieved through formation of double-membraned structures called phagophores, which encircle and ultimately enclose the captured cargo to form double-membraned autophagosomes.1 Autophagosomes themselves are not degradative, but subsequent fusion of autophagosomes with existing lysosomes delivers the enclosed cargo for degradation. A large body of work over the past 20 years has identified more than 30 proteins (Atg proteins) that mediate the various stages of autophagy including initiation, cargo recognition, phagophore formation, and autophagosome/lysosome fusion.2, 3 Importantly, autophagy can be either nonselective (eg, the capture of random bits of cytosol during nutrient starvation) or selective (as in the turnover of damaged mitochondria, referred to as mitophagy).
In addition to its role in homeostatic maintenance, it now is clear that cells also use autophagy as an innate immune mechanism for the clearance of intracellular pathogens, where it is referred to as xenophagy. Many invasive bacterial pathogens including Salmonella, Shigella, Listeria, Legionella, Mycobacterium, Franciscella, and group A streptococcus trigger an autophagic response in host cells, which at least partially restricts their intracellular growth. However, as successful pathogens, these organisms have evolved sophisticated mechanisms that either suppress autophagy entirely or prevent their recognition and capture by the autophagic machinery (for reviews see Huang and Brumell,4 Sorbara and Girardin,5 and Kohler and Roy6).
General Features of Autophagy
Autophagy is initiated in response to a variety of upstream signals, most commonly some form of cellular stress. These can include nutrient starvation, hypoxia, damage to cellular membranes, accumulation of protein aggregates, or microbial infection. Upon initiation, phagophores begin to form from specialized regions of the endoplasmic reticulum.7 These early phagophores often adopt a horseshoe shape, referred to as omegasomes.7 As early phagophores mature, a group of ubiquitin-like proteins, the Atg8 family composed of microtubule-associated light chain 3 (LC3) and gamma-aminobutyric acid receptor associated proteins, undergoes a processing event in which they are conjugated to phosphatidylethanolamine and become anchored into the membrane of the growing phagophore.8 These proteins function as receptors for autophagic cargo and are critical for selective autophagy. Of note, conversion of LC3 (LC3-I) to its lipid-conjugated form (LC3-II) often is used as a marker of autophagy progression.1 Cargos that are targeted for autophagic degradation typically are marked by ubiquitylation (see later). These ubiquitylated cargos are recognized by a family of adaptor proteins that includes p62/SQSTM1, NDP52, optineurin, NBR1, and TAX1BP1, which then link them to the nascent phagophore through interactions with LC3 or gamma-aminobutyric acid receptor associated proteins.9 The double-membraned phagophore then expands around the captured cargo, eventually closing to form a mature autophagosome. Mature autophagosomes then fuse with lysosomes to form a hybrid organelle, the autophagolysosome, in which the captured contents are degraded and recycled for use elsewhere in the cell.
Recognition of Intracellular Bacteria by the Autophagic Machinery
Different intracellular bacterial pathogens adopt distinct lifestyles within their host cells. Although some (Salmonella, Mycobacterium, and Legionella) are enclosed in a host-derived membrane (vacuole) that they modify into a replicative compartment, others (Shigella, Listeria, group A streptococcus) escape the vacuole and replicate in the nutrient-rich environment of the cytosol. Bacteria that are free in the cytosol are recognized by host cells and rapidly become ubiquitylated, although the specific ubiquitin ligases and the type of ubiquitin linkage may differ from one pathogen to another.10 In the case of S typhimurium, ubiquitylation appears largely to be mediated by a specific E3 ligase, leucine-rich repeat and sterile alpha motif-containing 1 (LRSAM1).11 Similar to other ubiquitylated cargos, bacteria are recognized by the adaptor proteins NDP52, optineurin, p62/SQSTM1, NBR1, and TAX1BP1, which then bring them together with nascent phagophores.12, 13, 14, 15, 16
Perhaps not surprisingly, successful pathogens have evolved mechanisms for avoiding recognition by the autophagic machinery. For example, the Shigella flexneri effector protein VirA inhibits the activity of Rab1, a small guanosine triphosphatase required for early phagophore formation from the endoplasmic reticulum.17 S flexneri also expresses a surface protein, IcsA (also known as VirG), which is a ligand for the autophagy protein Atg5, which is part of the LC3 conjugation machinery and is present on the phagophore membrane. However, a second protein, IcsB, binds with high affinity to IcsA and blocks its interaction with Atg5, thereby inhibiting phagophore recruitment.18 Another cytosolic pathogen, Listeria monocytogenes, escapes autophagic recognition by coating itself in a dense actin network, through the activity of the secreted protein ActA.19 In a particularly elegant mechanism, Legionella pneumophila secretes a protein, RavZ, that irreversibly cleaves the amide bond linking LC3 to phosphatidylethanolamine, thereby blocking the ability of phagophores to recognize ubiquitylated cargo.20 Other species, including adherent-invasive Escherichia coli,21 Mycobacterium marinum,22 Chlamydia trachomatis,23, 24 and Yersinia pestis25 appear to be recognized effectively by host cells and captured in autophagosomes, but avoid killing by blocking the fusion of autophagosomes with lysosomes. In most cases, the mechanisms used by these pathogens to block fusion remain unknown (Table 1). However, many bacterial effectors target the regulatory machinery that controls endomembrane transport and fusion, and it is likely that at least some of these disrupt the function of Rabs, Arfs, SNAREs, or other machinery required for autophagosome maturation and fusion.
Table 1.
Mechanisms Used by Bacteria to Suppress or Evade Autophagy
| Pathogen | Secreted factor(s) | Host target | References |
|---|---|---|---|
| S flexneri | VirA | Rab1 | 17 |
| S flexneri | IcsA/B | Atg5 | 18 |
| L monocytogenes | ActA | Actin | 19 |
| L pneumophila | RavZ | LC3-II | 20 |
| Adherent/invasive E coli | Unknown | Unknown | 21 |
| M marinum | Esx-1 secretion system | Unknown | 22 |
| C trachomatis | Unknown | Unknown | 23, 24 |
| Y pestis | Unknown | Unknown | 25 |
Manipulation of Autophagy by Salmonella enterica Serovar Typhimurium
Perhaps the best understood example of bacterial interplay with the host autophagic machinery is S enterica serovar Typhimurium (S typhimurium). Pathogenic Salmonella strains express 2 type III secretion systems (T3SSs) encoded within distinct pathogenicity islands, Salmonella pathogenicity island-1 and -2.26 These syringe-like protein complexes (T3SS1 and T3SS2) insert into host cell membranes and deliver distinct sets of effector proteins into the cytosol. Expression of the Salmonella pathogenicity island-1 T3SS and its translocated effectors is induced by environmental conditions in the distal ileum, and is essential for the invasion of luminal bacteria into intestinal epithelial cells. In contrast, expression of T3SS2 is induced only after entry into host cells, where it requires conditions that prevail in endosomal/phagosomal compartments.27 T3SS2 mediates the translocation of more than 30 effectors required for intracellular survival.28 A subset of these effectors hijack the host endocytic pathway, and remodel the phagosome into a nondegradative compartment referred to as the Salmonella-containing vacuole (SCV). Temporally, expression of T3SS1 and its effectors largely is down-regulated 1–2 hours after internalization, whereas expression of T3SS2 increases over time, consistent with SCV maturation.29, 30 During infection of epithelial cells, expression of the 2 sets of effectors can overlap and cooperate in the remodeling of the SCV.29
Induction of Autophagy by Salmonella
The active invasion of epithelial cells by Salmonella triggers the autophagic response, at least in part, by insertion of the T3SS1 translocation apparatus into the plasma membrane.31 Pores formed by T3SS1 allow leakage of amino acids from the cell, causing an acute starvation response. Amino acid starvation then acts through a complex signaling cascade to reduce the activity of the kinase mechanistic target of rapamycin 1 (mTORC1), which suppresses autophagy under nonstarvation conditions.31
Upon entry into host cells, S typhimurium can adopt 1 of 2 fates. The majority of bacteria are enclosed in endosomal membranes, which are modified progressively by T3SS2 and its effectors to form mature SCVs. However, a small population manages to escape the vacuole and enter the cytosol, where they can hyper-replicate in the nutrient-rich environment.32, 33 As noted earlier, these cytosolic bacteria are recognized by the host and rapidly become ubiquitylated, recruit adaptor proteins, and are captured efficiently in autophagosomes.32 If the multiplicity of infection is high, these cytosolic bacteria can overwhelm the autophagic machinery and ultimately replicate to the point where they kill the host cell. If the multiplicity of infection is low, the bacteria are cleared by autophagy and the cell survives.
However, even bacteria that remain within vacuoles can be targeted for autophagic degradation. As described earlier, expression of T3SS1 persists for 1–2 hours after internalization and insertion of the translocation pores (translocons) damages the membrane of the SCV just as it does the plasma membrane.34 If this damage is sufficiently severe, it exposes the SCV lumen to cytosolic ubiquitin ligases, including LRSAM1, which directly binds to bacterial surface components.11 In addition to ubiquitylation, damage to the SCV also is detected by a family of cytosolic carbohydrate-binding proteins, the galectins. One of these, galectin-8, recognizes β-galactoside, a sugar present on host glycoproteins lining the luminal surface of the SCV.35 Galectin-8 in turn binds directly and selectively to the adaptor protein NDP52, thereby linking the damaged vacuole to nascent phagophores.35
Brumell and colleagues36 also have shown that diacylglycerol (DAG) accumulates on the surface of SCVs, and that inhibition of DAG synthesis prevents autophagic recognition of the enclosed bacteria. However, how DAG contributes to the autophagic process remains unknown. Together these host responses restrict, but do not completely contain intracellular growth. In agreement with this, mice containing intestinal epithelial cell–specific knockouts of Atg16L1 or Atg5 (components of the LC3 conjugation machinery) are significantly more susceptible to oral infection with S typhimurium.37, 38
Suppression of Autophagy by S typhimurium
Although the autophagic response to S typhimurium described earlier is robust, it also is transient. In cultured epithelial cells or fibroblasts, autophagy peaks within 1 hour of infection, and returns to near baseline within 3 hours.31 Importantly, this timing corresponds with down-regulation of T3SS1 expression and up-regulation of T3SS2. Presumably, down-regulation and degradation of T3SS1 allow restoration of plasma membrane integrity and normalization of amino acid levels. Although it is logical to assume that the induction of T3SS2 and its effectors also contributes to the attenuation of autophagy in epithelial cells, this remains to be tested directly.
An Alternative Role for Autophagy in Epithelial Cells
Although xenophagy generally is thought to benefit the host, recent evidence has indicated that the autophagic machinery in epithelia actually can benefit S typhimurium by repairing damage to the SCV caused by T3SS1. By using a high-throughput small interfering RNA screen for host proteins required for SCV biogenesis, Hardt and colleagues34 identified a large number of autophagy-related genes whose depletion inhibited the induction of a T3SS2 reporter (a sensitive readout of SCV maturation). Mechanistically, they found that SCVs in autophagy-deficient cells failed to retain fluid-phase tracers, indicating a loss of membrane integrity. A leaky SCV membrane would result in dissipation of the proton gradient essential for compartment acidification (and therefore T3SS2 induction). Conversely, membrane repair would allow re-acidification of the compartment lumen and restoration of T3SS2-inducing conditions. In agreement with this hypothesis, maturation of SCVs containing T3SS1-deficient bacteria (which do not damage the membrane) did not require autophagy proteins for induction of T3SS2.34 However, how the autophagic machinery actually repairs the SCV membrane remains unknown, and will be an important area for future investigation.
At first glance, it is difficult to reconcile these observations with the large body of evidence that autophagy restricts, rather than enhances, intracellular growth of bacteria. However, it is plausible, at least in the case of S typhimurium, that autophagy benefits the pathogen during the early stages of SCV biogenesis, and benefits the host by clearing the fraction of bacteria that escape the vacuole and hyper-replicate in the cytosol. In most reports in the literature, the quantitative assessment of bacterial replication has not distinguished between vacuolar and nonvacuolar populations. This issue clearly will require clarification as the field moves forward.
The Autophagic Response to Salmonella in Macrophages
Although T3SS1 is essential for penetration and colonization of the intestinal epithelium, it is dispensable for the systemic phase of infection.27 At this stage, bacteria that enter the lamina propria are captured by professional phagocytes (primarily macrophages and neutrophils) that are recruited into the tissue in response to infection. It generally is believed that bacteria that have passed through the epithelium no longer express T3SS1, and in fact T3SS1 is toxic to macrophages, where it induces rapid apoptosis.39 In the absence of T3SS1, a significant fraction of bacteria survives within phagocytes in a T3SS2-dependent manner and uses these cells as vehicles for dissemination through the lymphatic system. Work from our laboratory has shown that even in the absence of T3SS1 the presence of intracellular Salmonella triggers an autophagic response in macrophages.40 Although the mechanisms of induction are not known, this response is transient (as it is in epithelial cells), and its down-regulation corresponds temporally with the expression of T3SS2.
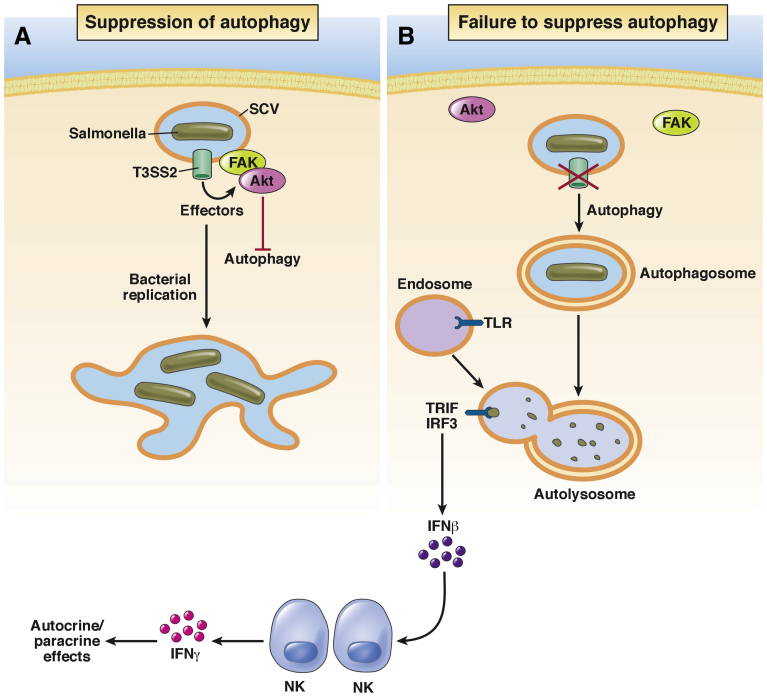
Our laboratory recently identified a T3SS2-dependent signaling pathway that is essential for the observed suppression of the macrophage autophagic response (Figure 1). Focal adhesion kinase (FAK) is a nonreceptor tyrosine kinase best known for its role in adhesion-mediated signaling. In that context, adhesion of cells to the extracellular matrix induces FAK activation and the generation of downstream signals that promote cell proliferation (through the Ras/mitogen-activated protein kinase pathway) and survival (through the phosphatidylinositol-3 kinase/Akt pathway).41, 42, 43, 44 Importantly, Akt is also a central regulator of autophagy, where it controls mTORC1 function; when Akt activity is high, mTORC1 activity is high and autophagy is suppressed. When Akt activity is low, mTORC1 activity also is low, and autophagy is induced.45
Figure 1.
Suppression of autophagy permits intracellular survival and replication. (A) Effector proteins translocated by T3SS2 recruit FAK and Akt to the surface of the SCV. Akt activation then suppresses autophagy by phosphorylating mTORC1. SCV maturation is allowed to proceed, and bacteria replicate inside mature SCVs. (B) In the absence of T3SS2 or its effectors, or in the absence of FAK, Akt activity remains low, autophagy is not suppressed, and bacteria are killed in autolysosomes. Bacterial products released into the lumen of autolysosomes interact with Toll-like receptors to stimulate IFNβ production. Secreted IFN stimulates production of IFNγ by neighboring natural killer (NK) cells, increasing both cell-autonomous and non–cell-autonomous bacterial killing. IRF, interferon regulatory factor 3.
Surprisingly, we found that both FAK and Akt are recruited to the surface of SCVs in infected macrophages.40 This recruitment requires T3SS2, and corresponds temporally with the activation of both kinases, peaking at 5 hours after infection. Importantly, mTORC1 activity also increases in parallel, as measured by phosphorylation of its substrate, p70S6 kinase. In the absence of FAK (primary macrophages from FAK-deficient mice) both Akt activity and mTORC1 activity remain low even at 5 hours after infection. As a consequence, autophagy is not suppressed and the number of bacteria captured in autophagosomes increases more than 4-fold. Under these conditions bacterial survival is reduced proportionally. Notably, survival can be restored to wild-type levels by knockdown of Atg5, showing that the observed reduction in survival is entirely owing to autophagy.40
To show the physiological relevance of this mechanism, we made use of a mouse model in which FAK is deleted selectively from macrophages.46 Oral infection of these mice with a wild-type Salmonella strain (SL1344) showed a significant defect in virulence; colonization of the ileal lamina propria was reduced by more than 50-fold, systemic dissemination to the liver and spleen was attenuated proportionally, and survival of the host was prolonged.40 Taken together, these observations suggest that active suppression of autophagy is an important virulence mechanism used by Salmonella to infect their animal hosts.
A Non–Cell-Autonomous Role for Autophagy in the Control of Salmonella Infection
In a follow-up study, we discovered that the ability of S typhimurium to evade autophagic killing has broader consequences for the innate immune response. Measurement of cytokine production in wild-type vs FAK-deficient macrophages showed that although induction of canonical proinflammatory cytokines (tumor necrosis factor-α, interleukin 6, interleukin 12) was unaffected, FAK-deficient cells produced significantly higher levels of the type I interferon (IFN)β in response to infection.47 This increase required the Toll-like receptor (TLR)3 and TLR4, and the signaling adaptor TRIF, which interacts with both receptors in endosomal compartments,48 suggesting that autophagy increases the association of both TLR3 and TLR4 with bacterial products. This presumably occurs within autolysosomes, although this remains to be determined directly. In agreement with the earlier-described hypothesis, a similar increase in IFNβ production was observed in wild-type macrophages treated with the mTORC1 inhibitor rapamycin, but only if they were infected.47 Thus, xenophagy, but not autophagy alone, leads to increased IFNβ production. Consistent with these in vitro observations, FAK-deficient macrophages isolated from orally infected mice also showed increased expression of IFNβ.47
An important consequence of IFNβ production by infected macrophages is the ability of this cytokine to induce the expression of the more potently antimicrobial IFNγ in bystander cells. Although IFNγ was barely detectable in the lamina propria of infected wild-type mice, it was up-regulated significantly in FAK-deficient mice, where it was produced predominantly by local natural killer cells.47 IFNγ is an important element of antibacterial, antiviral, and antitumor responses, and also functions as a link between innate and adaptive immunity.49 The relationship between IFNβ and IFNγ and their importance in the control of Salmonella infection was shown by antibody blockade experiments. We found that blockade of the IFNα/β-receptor IFNAR1 inhibited downstream production of IFNγ and led to a striking 100-fold increase in colonization of the lamina propria after oral infection. Direct blockade of IFNγ with a function-blocking antibody resulted in a quantitatively similar increase in colonization. In contrast, induction of IFNβ expression in wild-type mice by injection of the TLR3 ligand poly(I:C) before infection was protective, decreasing colonization by nearly 100-fold.47
Although IFNγ potently enhances the intrinsic microbicidal activity of many cell types (including macrophages), it also stimulates the production of CXC chemokines that promote the recruitment of other leukocytes including neutrophils into infected tissue.50 In this way, local production of IFNγ stimulates both cell-autonomous and non–cell-autonomous clearance of invading bacteria.
Summary and Perspectives
Similar to other bacterial pathogens, S typhimurium shows remarkable adaptations to intracellular life, exemplified by the mechanisms that have evolved to avoid autophagy. Despite our growing understanding of these mechanisms, many unresolved questions remain to be explored. For instance, how does the autophagic machinery facilitate repair of the damage to SCVs mediated by T3SS1, and how does this relate to virulence in vivo? How do intravacuolar bacteria trigger an autophagic response in macrophages in the absence of T3SS1? Does T3SS2 also damage the SCV? Although T3SS2 does insert a translocation pore into the limiting membrane of the SCV, we have not observed either ubiquitin or adaptors on SCVs in infected macrophages, suggesting that T3SS2 does not inflict the same kind of damage as T3SS1.40 Which of the more than 30 effector proteins translocated by T3SS2 mediate recruitment/activation of FAK at the surface of the SCV, and how is this achieved? Does a related mechanism operate in epithelial cells? In an era of increasing antibiotic resistance, understanding at the molecular level how bacterial pathogens manipulate their hosts will provide new potential targets for therapeutic intervention.
Footnotes
Conflicts of interest The author discloses no conflicts.
Funding Work in the author’s laboratory was supported by National Institutes of Health grant RO1DK58536.
References
- 1.Yang Z., Klionsky D.J. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mizushima N., Yoshimori T., Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 3.Yang Z., Klionsky D.J. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang J., Brumell J.H. Bacteria-autophagy interplay: a battle for survival. Nat Rev Microbiol. 2014;12:101–114. doi: 10.1038/nrmicro3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sorbara M.T., Girardin S.E. Emerging themes in bacterial autophagy. Curr Opin Microbiol. 2015;23:163–170. doi: 10.1016/j.mib.2014.11.020. [DOI] [PubMed] [Google Scholar]
- 6.Kohler L.J., Roy C.R. Autophagic targeting and avoidance in intracellular bacterial infections. Curr Opin Microbiol. 2016;35:36–41. doi: 10.1016/j.mib.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Axe E.L., Walker S.A., Manifava M., Chandra P., Roderick H.L., Habermann A., Griffiths G., Ktistakis N.T. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Slobodkin M.R., Elazar Z. The Atg8 family: multifunctional ubiquitin-like key regulators of autophagy. Essays Biochem. 2013;55:51–64. doi: 10.1042/bse0550051. [DOI] [PubMed] [Google Scholar]
- 9.Randow F., Youle R.J. Self and nonself: how autophagy targets mitochondria and bacteria. Cell Host Microbe. 2014;15:403–411. doi: 10.1016/j.chom.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fiskin E., Bionda T., Dikic I., Behrends C. Global analysis of host and bacterial ubiquitinome in response to Salmonella typhimurium infection. Mol Cell. 2016;62:967–981. doi: 10.1016/j.molcel.2016.04.015. [DOI] [PubMed] [Google Scholar]
- 11.Huett A., Heath R.J., Begun J., Sassi S.O., Baxt L.A., Vyas J.M., Goldberg M.B., Xavier R.J. The LRR and RING domain protein LRSAM1 is an E3 ligase crucial for ubiquitin-dependent autophagy of intracellular Salmonella typhimurium. Cell Host Microbe. 2012;12:778–790. doi: 10.1016/j.chom.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tumbarello D.A., Manna P.T., Allen M., Bycroft M., Arden S.D., Kendrick-Jones J., Buss F. The autophagy receptor TAX1BP1 and the molecular motor myosin VI are required for clearance of salmonella typhimurium by autophagy. PLoS Pathog. 2015;11:e1005174. doi: 10.1371/journal.ppat.1005174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng Y.T., Shahnazari S., Brech A., Lamark T., Johansen T., Brumell J.H. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J Immunol. 2009;183:5909–5916. doi: 10.4049/jimmunol.0900441. [DOI] [PubMed] [Google Scholar]
- 14.Thurston T.L.M., Ryzhakov G., Bloor S., von Muhlinen N., Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol. 2009;10:1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- 15.Mostowy S., Sancho-Shimizu V., Hamon M.A., Simeone R., Brosch R., Johansen T., Cossart P. p62 and NDP52 proteins target intracytosolic Shigella and Listeria to different autophagy pathways. J Biol Chem. 2011;286:26987–26995. doi: 10.1074/jbc.M111.223610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verlhac P., Grégoire I.P., Azocar O., Petkova D.S., Baguet J., Viret C., Faure M. Autophagy receptor NDP52 regulates pathogen-containing autophagosome maturation. Cell Host Microbe. 2015;17:515–525. doi: 10.1016/j.chom.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 17.Dong N., Zhu Y., Lu Q., Hu L., Zheng Y., Shao F. Structurally distinct bacterial TBC-like GAPs link Arf GTPase to Rab1 inactivation to counteract host defenses. Cell. 2012;150:1029–1041. doi: 10.1016/j.cell.2012.06.050. [DOI] [PubMed] [Google Scholar]
- 18.Ogawa M., Yoshimori T., Suzuki T., Sagara H., Mizushima N., Sasakawa C. Escape of intracellular Shigella from autophagy. Science. 2005;307:727–731. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- 19.Yoshikawa Y., Ogawa M., Hain T., Yoshida M., Fukumatsu M., Kim M., Mimuro H., Nakagawa I., Yanagawa T., Ishii T. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat Cell Biol. 2009;11:1233–1240. doi: 10.1038/ncb1967. [DOI] [PubMed] [Google Scholar]
- 20.Choy A., Dancourt J., Mugo B., O’Connor T.J., Isberg R.R., Melia T.J., Roy C.R. The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science. 2012;338:1072–1076. doi: 10.1126/science.1227026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lapaquette P., Bringer M.-A., Darfeuille-Michaud A. Defects in autophagy favour adherent-invasive Escherichia coli persistence within macrophages leading to increased pro-inflammatory response. Cell Microbiol. 2012;14:791–807. doi: 10.1111/j.1462-5822.2012.01768.x. [DOI] [PubMed] [Google Scholar]
- 22.Romagnoli A., Etna M.P., Giacomini E., Pardini M., Remoli M.E., Corazzari M., Falasca L., Goletti D., Gafa V., Simeone R. ESX-1 dependent impairment of autophagic flux by Mycobacterium tuberculosis in human dendritic cells. Autophagy. 2012;8:1357–1370. doi: 10.4161/auto.20881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Al-Younes H.M., Brinkmann V., Meyer T.F. Interaction of Chlamydia trachomatis serovar L2 with the host autophagic pathway. Infect Immun. 2004;72:4751–4762. doi: 10.1128/IAI.72.8.4751-4762.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yasir M., Pachikara N.D., Bao X., Pan Z., Fan H. Regulation of chlamydial infection by host autophagy and vacuolar ATPase-bearing organelles. Infect Immun. 2011;79:4019–4028. doi: 10.1128/IAI.05308-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pujol C., Klein K.A., Romanov G.A., Palmer L.E., Cirota C., Zhao Z., Bliska J.B. Yersinia pestis can reside in autophagosomes and avoid xenophagy in murine macrophages by preventing vacuole acidification. Infect Immun. 2009;77:2251–2261. doi: 10.1128/IAI.00068-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hansen-Wester I., Hensel M. Salmonella pathogenicity islands encoding type III secretion systems. Microbes Infect. 2001;3:549–559. doi: 10.1016/s1286-4579(01)01411-3. [DOI] [PubMed] [Google Scholar]
- 27.Figueira R., Holden D.W. Functions of the Salmonella pathogenicity island 2 (SPI-2) type III secretion system effectors. Microbiology. 2012;158:1147–1161. doi: 10.1099/mic.0.058115-0. [DOI] [PubMed] [Google Scholar]
- 28.Figueira R., Watson K.G., Holden D.W., Helaine S. Identification of salmonella pathogenicity island-2 type III secretion system effectors involved in intramacrophage replication of S. enterica serovar typhimurium: implications for rational vaccine design. MBio. 2013;4:e00065. doi: 10.1128/mBio.00065-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hautefort I., Thompson A., Eriksson-Ygberg S., Parker M.L., Lucchini S., Danino V., Bongaerts R.J.M., Ahmad N., Rhen M., Hinton J.C.D. During infection of epithelial cells Salmonella enterica serovar Typhimurium undergoes a time-dependent transcriptional adaptation that results in simultaneous expression of three type 3 secretion systems. Cell Microbiol. 2008;10:958–984. doi: 10.1111/j.1462-5822.2007.01099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kröger C., Colgan A., Srikumar S., Händler K., Sivasankaran S.K., Hammarlöf D.L., Canals R., Grissom J.E., Conway T., Hokamp K. An infection-relevant transcriptomic compendium for Salmonella enterica Serovar typhimurium. Cell Host Microbe. 2013;14:683–695. doi: 10.1016/j.chom.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 31.Tattoli I., Sorbara M.T., Vuckovic D., Ling A., Soares F., Carneiro L.A.M., Yang C., Emili A., Philpott D.J., Girardin S.E. Amino acid starvation induced by invasive bacterial pathogens triggers an innate host defense program. Cell Host Microbe. 2012;11:563–575. doi: 10.1016/j.chom.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 32.Birmingham C.L., Brumell J.H. Autophagy recognizes intracellular Salmonella enterica serovar Typhimurium in damaged vacuoles. Autophagy. 2006;2:156–158. doi: 10.4161/auto.2825. [DOI] [PubMed] [Google Scholar]
- 33.Knodler L.A. Salmonella enterica: living a double life in epithelial cells. Curr Opin Microbiol. 2015;23:23–31. doi: 10.1016/j.mib.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 34.Kreibich S., Emmenlauer M., Fredlund J., Rämö P., Münz C., Dehio C., Enninga J., Hardt W.-D. Autophagy proteins promote repair of endosomal membranes damaged by the Salmonella type three secretion system 1. Cell Host Microbe. 2015;18:527–537. doi: 10.1016/j.chom.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 35.Thurston T.L.M., Wandel M.P., von Muhlinen N., Foeglein A., Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. 2012;482:414–418. doi: 10.1038/nature10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shahnazari S., Yen W.-L., Birmingham C.L., Shiu J., Namolovan A., Zheng Y.T., Nakayama K., Klionsky D.J., Brumell J.H. A diacylglycerol-dependent signaling pathway contributes to regulation of antibacterial autophagy. Cell Host Microbe. 2010;8:137–146. doi: 10.1016/j.chom.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Conway K.L., Kuballa P., Song J.H., Patel K.K., Castoreno A.B., Yilmaz O.H., Jijon H.B., Zhang M., Aldrich L.N., Villablanca E.J. Atg16l1 is required for autophagy in intestinal epithelial cells and protection of mice from Salmonella infection. Gastroenterology. 2013;145:1347–1357. doi: 10.1053/j.gastro.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benjamin J.L., Sumpter R., Levine B., Hooper L.V. Intestinal epithelial autophagy is essential for host defense against invasive bacteria. Cell Host Microbe. 2013;13:723–734. doi: 10.1016/j.chom.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hueffer K., Galán J.E. Salmonella-induced macrophage death: multiple mechanisms, different outcomes. Cell Microbiol. 2004;6:1019–1025. doi: 10.1111/j.1462-5822.2004.00451.x. [DOI] [PubMed] [Google Scholar]
- 40.Owen K.A., Meyer C.B., Bouton A.H., Casanova J.E. Activation of focal adhesion kinase by Salmonella suppresses autophagy via an Akt/mTOR signaling pathway and promotes bacterial survival in macrophages. PLoS Pathog. 2014;10:e1004159. doi: 10.1371/journal.ppat.1004159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pylayeva Y., Gillen K.M., Gerald W., Beggs H.E., Reichardt L.F., Giancotti F.G. Ras- and PI3K-dependent breast tumorigenesis in mice and humans requires focal adhesion kinase signaling. J Clin Invest. 2009;119:252–266. doi: 10.1172/JCI37160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang D., Khoe M., Befekadu M., Chung S., Takata Y., Ilic D., Bryer-Ash M. Focal adhesion kinase mediates cell survival via NF-kappaB and ERK signaling pathways. Am J Physiol Cell Physiol. 2007;292:C1339–C1352. doi: 10.1152/ajpcell.00144.2006. [DOI] [PubMed] [Google Scholar]
- 43.Sonoda Y., Matsumoto Y., Funakoshi M., Yamamoto D., Hanks S.K., Kasahara T. Anti-apoptotic role of focal adhesion kinase (FAK). Induction of inhibitor-of-apoptosis proteins and apoptosis suppression by the overexpression of FAK in a human leukemic cell line, HL-60. J Biol Chem. 2000;275:16309–16315. doi: 10.1074/jbc.275.21.16309. [DOI] [PubMed] [Google Scholar]
- 44.Zhao J., Guan J.-L. Signal transduction by focal adhesion kinase in cancer. Cancer Metastasis Rev. 2009;28:35–49. doi: 10.1007/s10555-008-9165-4. [DOI] [PubMed] [Google Scholar]
- 45.Manning B.D., Cantley L.C. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Owen K.A., Pixley F.J., Thomas K.S., Vicente-Manzanares M., Ray B.J., Horwitz A.F., Parsons J.T., Beggs H.E., Stanley E.R., Bouton A.H. Regulation of lamellipodial persistence, adhesion turnover, and motility in macrophages by focal adhesion kinase. J Cell Biol. 2007;179:1275–1287. doi: 10.1083/jcb.200708093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Owen K.A., Anderson C.J., Casanova J.E. Salmonella suppresses the TRIF-dependent type I interferon response in macrophages. MBio. 2016;7:e02051–e02115. doi: 10.1128/mBio.02051-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fitzgerald K.A., Rowe D.C., Barnes B.J., Caffrey D.R., Visintin A., Latz E., Monks B., Pitha P.M., Golenbock D.T. LPS-TLR4 signaling to IRF-3/7 and NF-kappaB involves the toll adapters TRAM and TRIF. J Exp Med. 2003;198:1043–1055. doi: 10.1084/jem.20031023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin F., Young H.A. Interferons: success in anti-viral immunotherapy. Cytokine Growth Factor Rev. 2014;25:369–376. doi: 10.1016/j.cytogfr.2014.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van Raemdonck K., Van den Steen P.E., Liekens S., Van Damme J., Struyf S. CXCR3 ligands in disease and therapy. Cytokine Growth Factor Rev. 2015;26:311–327. doi: 10.1016/j.cytogfr.2014.11.009. [DOI] [PubMed] [Google Scholar]