Abstract
The anti-malarial drug artesunate is a semi-synthetic derivative of artemisinin, the principal active component of a medicinal plant Artemisia annua. It is hypothesized to attenuate allergic asthma via inhibition of multiple signaling pathways. We used a comprehensive approach to elucidate the mechanism of action of artesunate by designing a novel biotinylated dihydroartemisinin (BDHA) to identify cellular protein targets of this anti-inflammatory drug. By adopting an untargeted proteomics approach, we demonstrated that artesunate may exert its protective anti-inflammatory effects via direct interaction with multiple proteins, most importantly with a number of mitochondrial enzymes related to glucose and energy metabolism, along with mRNA and gene expression, ribosomal regulation, stress responses and structural proteins. In addition, the modulatory effects of artesunate on various cellular transcription factors were investigated using a transcription factor array, which revealed that artesunate can simultaneously modulate multiple nuclear transcription factors related to several major pro- and anti-inflammatory signalling cascades in human bronchial epithelial cells. Artesunate significantly enhanced nuclear levels of nuclear factor erythroid-2-related factor 2 (Nrf2), a key promoter of antioxidant mechanisms, which is inhibited by the Kelch-like ECH-associated protein 1 (Keap1). Our results demonstrate that, like other electrophilic Nrf2 regulators, artesunate activates this system via direct molecular interaction/modification of Keap1, freeing Nrf2 for transcriptional activity. Altogether, the molecular interactions and modulation of nuclear transcription factors provide invaluable insights into the broad pharmacological actions of artesunate in inflammatory lung diseases and related inflammatory disorders.
TOC image
Introduction
Artemisinins are a family of herbal-derived anti-malarial agents, known for their anti-parasitic efficacy and well-established safety profile in humans. Artesunate (Arts), a semi-synthetic analogue of artemisinins, is currently the most commonly studied derivative of the family, due to its improved solubility and enhanced pharmacological profile. As discussed in our recent review,1 artemisinins are increasingly being explored for treatment of many non-malarial disease conditions and exhibit broad protective effects in various cancers, inflammatory conditions and pathogenic infections. Among current investigations of artemisinins, concerted efforts are being made to elucidate how members of this family of anti-inflammatory compounds are effective against various inflammatory lung diseases, such as lung cancers, asthma and chronic obstructive pulmonary disease (COPD). In studies of asthma, we first demonstrated that artesunate is effective in preventing development of various hallmarks of the disease, particularly airway inflammation, excessive mucus production and airway hyper-responsiveness.2 In a subsequent comparison with dexamethasone, a potent corticosteroid, artesunate was shown to have comparable protective effects and could differentially modulate pulmonary antioxidants and pro-oxidants, to reduce oxidative stress and related redox lung damage in experimental asthma.3 Follow-up studies further revealed that artesunate could also effectively reverse asthma-related metabolic changes and inhibit disease-related cellular proliferation in human airway smooth muscle cells.4,5 Collectively, our and others research has revealed that artesunate can simultaneously modulate multiple pro- and anti-inflammatory signaling cascades, particularly PI3K/Akt, Syk-PLCγ and Nrf2, which results in broad and potent protective effects in allergic asthma.2,3,5–7 While the safety profile, molecular effects and physiological endpoints of the drug effects in lung diseases have been extensively documented, the mechanistic actions of artesunate on various signaling pathways and its cellular protein binding targets have not been fully elucidated. This is a major limiting factor in the eventual clinical adoption of this anti-inflammatory drug for treating asthma and its extended therapeutic uses in other related inflammatory diseases.
Artesunate can bind to proteins in an iron-dependent and independent manner.8 In malaria, iron catalyzes the activation of endoperoxide bridge of artesunate which leads to the production of free radicals9,10 and reactive aldehydes.11 These reactive molecules can alkylate thiols and amine moieties of albumin8 as well as other proteins.12 However, definitive evidence is lacking to show that this process mediates the activity of artemisinins in non-malarial disease conditions.
In this study, we investigated the mechanistic behavior of artesunate in order to elucidate and explain its broad protective actions in related lung diseases. Applying a hybrid strategy, we combined the artesunate metabolite, dihydroartemisinin (DHA), with biotin to form a single molecular framework that would allow us to identify protein targets of artemisinin using untargeted proteomics in human bronchial epithelial cells (Figure 1). The modulatory effects of artesunate on various cellular transcription factors were investigated using a transcription factor array. The collective results revealed that artesunate can bind to a variety of cellular protein targets, related to glucose metabolism, mRNA and gene expression, ribosomal regulation, stress responses, which further resulted in simultaneous modulation of multiple pro- and anti-inflammatory transcription factor-mediated signaling cascades.
Figure 1.
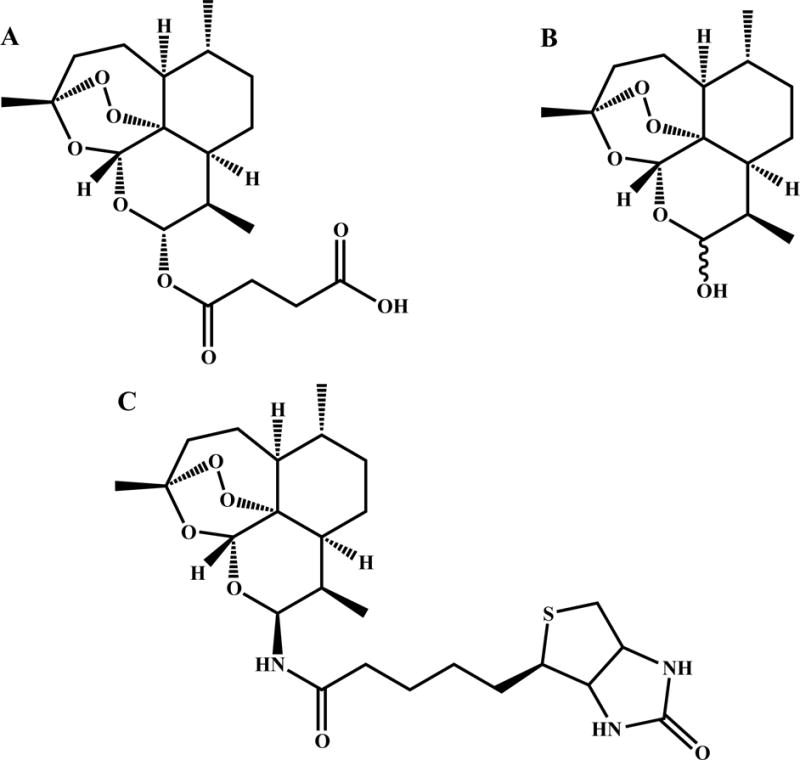
Structure of A. Artesunate, B. dihydroartemisinin (DHA), C. Biotinylated dihydroartemisinin (BDHA).
Materials and methods
Chemicals
Artesunate, dihydroartemisinin (DHA), bromotrimethylsilane (TMBS), sodium azide (NaN3), tetrahydrofuran (THF), triphenyl phosphine (PPh3), 1-hydroxybenzotriazole hydrate, 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide, acetyl chloride, N,N-dimethylformamide (DMF), dimethyl-sulfoxide (DMSO), dichloromethane (CH2Cl2), hydrogen peroxide (H2O2), and Protease cocktail were purchased from Sigma-Aldrich (St. Louis, MO, USA). Imm. Drystrip pH 3–11 (Cat No. 17600377) and buffer (Cat No. 17600440) purchased from GE. RIPA lysis and extraction buffer was purchased from Pierce Biotechnology (Cat. No. 89900). Coomassie stain solution is from Bio-Rad (Cat. No. 161-0436). Nrf2 and corresponding secondary antibodies are from Santa Cruz Biotechnology (Cat. No. sc-722, sc-2030), Keap1 antibody is from Cell Signalling Technology (Product No. 4617S, 8047S), ECL reagent is from GE Healthcare (Cat. No. RPN2232), UltraLink Immobilized Streptavidin beads are from Thermo Scientific (Cat. No. 20349), Centrifuge Columns are from Pierce (Cat. No. 89896), OMIX tips are from Agilent technologies (Part No. A57003100). Unless otherwise noted, all the materials were obtained from commercially available sources and were used without further purification.
Human Bronchial Epithelial Cell Culture
Beas-2B, transformed human bronchial epithelial cells (American Type Culture Collection, Rockville, MD, USA) were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) in a humidified CO2 incubator at 37°C. Beas-2B cells were incubated for 3 hrs with 0.02% DMSO, Arts (30μM) or probes (30μM) dissolved in FBS-free RPMI 1640 medium as previously described.3 Nuclear and cytosolic protein extractions were performed to obtain corresponding protein lysates for subsequent proteomic and transcription factor array experiments.
WST-1 Cellular Proliferation Assay
In brief the WST-1 assay measures colorimetrically the cleavage of tetrazolium salts to formazan by mitochondrial dehydrogenases in metabolically active cells. Fixed numbers of Beas-2B cells (2 × 104 per well) were plated in 96 well cell culture plates in the above-described culture conditions. DMSO, Arts (30μM), BDHA (30μM) and H2O2 (50 mM) were added 24 hrs later. The medium was aspirated 3 hrs later and replaced with complete culture medium to incubate for 21 hrs. The cells were incubated with WST-1 for 1 hr and cell proliferation was measured using a microplate reader at 450 nm.
Nuclear Transcription Factor Profiling Array
Nuclear protein extracts (15 μg) were incubated in a commercial 96-well sixteen transcription factor (TF) activation profiling array (Signosis, Inc, CA) in accordance with manufacturer’s instructions. Briefly, nuclear TF are incubated for 30 min at room temperature with TF binding buffer mix and corresponding TF DNA probes to form TF DNA complexes. Bound TF DNA probe complexes are separated from free probes using an isolation column and eluted TF DNA complexes are hybridized to a hybridization plate overnight at 42°C. Detection of the bounded TF complexes were performed by adding Streptavidin-HRP conjugate (1:500, 95 μL) for 45 min and corresponding substrates (95 μL) for 1 min. Relative nuclear TF levels were measured by luminometry using a luminometer plate-reader.
Statistical Analysis
One-way ANOVA, followed by Dunnett’s test, validated by Bonferroni test, was used to determine significant differences in WST-1 fold changes and nuclear TF protein fold levels between groups, with significant levels at p < 0.05.
Extraction of nuclear and cytosolic proteins
Beas-2B cells were treated with different artemisinin derivatives at a dose of 30 μM for 3hrs, then washed with cold PBS and harvested by centrifugation of the collected cell suspensions into the cold PBS (2000 rpm for 5 minutes at 4 °C; the supernatant was then aspirated and the pellets kept on ice.) Cells were resuspended at 4 °C in cytosolic extraction buffer (10mM HEPES (pH 7.9), 10mM KCl, 0.1mM EDTA, 0.3% NP-40 and 1X protease inhibitor cocktail), allowed to swell on ice for 10 min, and then Vortexed for 10 sec. Samples were centrifuged and the supernatant containing the cytosolic fraction was stored at −80 °C. The pellet was resuspended with cytosolic extraction buffer (without NP-40), centrifuged, and supernatent was removed. The pellet was resuspended in nuclear extraction buffer (20mM HEPES (pH 7.9), 400 mM NaCl, 1mM EDTA, 25% glycerol, and 1X protease inhibitor cocktail) and incubated on ice for 20 min for high salt extraction. Cellular debris was removed by centrifugation (12000 rpm for 2 min at 4 °C), and the supernatant fraction was stored at −80 °C. Total protein contents were determined by Bio-Rad protein estimation kits.
Detection of biotinylated proteins
The total cell lysates were treated with different artemisinin derivatives with RIPA lysis and extraction buffer. Equal amounts of proteins (25 μg) were loaded into the SDS-PAGE gel for electrophoresis. The proteins were transferred into polyvinylidene difluoride membranes (PVDF) and the blot was probed against streptavidin-HRP to detect the biotinylated proteins. The bands were captured using Fluorchem™ 8900 from Alpha Innotech.
Nrf2 Immuno-blot
For measuring levels of Nrf2, equal amounts of cytosolic and nuclear extracts were denatured and electrophoresed on SDS-PAGE and transferred to PVDF membrane. The membranes were blocked in TBS-0.05% Tween 20 with 5% non-fat milk, incubated with primary Nrf2 antibody, and after extensive wash, incubated with a horseradish peroxidase (HRP)-conjugated secondary antibody. Protein bands were visualized using the ECL reagent.
Keap1 interaction with BDHA
To detect the binding of BDHA to Keap1, Beas-2B cells were treated with the biotinylated compound or artesunate for 3 hr. Cells were then washed with cold PBS, harvested by scraping, and lysed with RIPA and extraction buffers containing a protease inhibitor cocktail. Cell lysates containing 1 mg of protein were denatured by addition of 10 μL of 10% SDS in 100 μL and then incubated with 50 μl UltraLink Immobilized Streptavidin beads overnight at 4 °C with constant shaking. Next, beads were washed and the biotinylated protein fraction released from beads by boiling in presence of electrophoresis sample loading buffer, followed by SDS-PAGE and transfer to PVDF membranes. Membranes were incubated with monoclonal Keap1 antibody and, after extensive wash, incubated with a HRP-conjugated detection antibody. Protein bands were visualized with ECL reagent.
Co-immuno-precipitation of Keap1 and BDHA
Interaction between Keap1 and BDHA was further confirmed with a co-immuno-precipitation assay. Briefly, after treatment with BDHA or artesunate, cells were lysed in RIPA buffer and cell extracts containing equal amounts of protein incubated overnight at 4 °C with (600 μg) Keap1 antibody. Immuno-precipitated complexes were captured by incubation with protein A-agarose beads for 2 h, followed by four washes with RIPA buffer, elution in the presence of electrophoresis sample buffer for 5 minutes at 95 °C, SDS-PAGE, and transfer to PVDF membranes. Co-precipitation of BDHA with Keap1 was verified by detection of biotinylated DHA on these membranes using avidin-HRP conjugate (Vector Labs) and ECL reagent.
Pre-processing of proteins for In-gel and pseudo-shotgun proteomics experiments
Prior to processing the samples for In-gel digestion and pseudo-shotgun proteomics, the proteins were denatured with 10 μL of 10% SDS in 100 μL, boiling at 95 °C for 5 min followed by centrifugation for 5 min at 12000 rpm. The clear supernatant transferred into 1.7 mL low-binding eppendorf tubes and precipitated with chloroform/methanol. The pellets were rehydrated with 8M urea (in 50mM ammonium bicarbonate (ABC)). The rehydrated proteins were reduced with 10 mM tris(2-carboxyethyl)phosphine (TECP), alkylated with 20 mM iodoacetamide, then processed once again for chloroform/methanol precipitation. After the precipitation, the pellet was dissolved stepwise in 2% SDS. The dissolved proteins were added into the streptavidin column and processed for pull-down.
Streptavidin affinity chromatography of cell lysates
Immobilized Streptavidin was packed into Pierce Centrifuge Columns. The final column volume was 2 mL. The beads were pre-washed with pull-down buffer (50mM ABC with 0.1% SDS). The pre-processed protein samples were directly added to the columns and pull-down was carried out at room temperature for 1hr. After the pull-down the beads were washed with 5 mL pull-down buffer (50mM ABC, 0.1% SDS), 2 mL 4M urea (in 50mM ABC), and then 5 mL deionized water. Bound proteins were eluted by the addition of 4 column volumes of 0.4% TFA/80% acetonitrile. Elution fractions were combined, neutralized the pH using 100 mM ammonium bicarbonate (pH 8.5), lyophilized and stored at −20 °C. Eluted proteins were processed for pseudo-shotgun proteomics. For In-gel digestion, the beads were directly boiled with 2X loading dye and loaded into the SDS-PAGE gel.
In-gel digestion
After pull-down the biotinylated proteins were subjected to SDS-PAGE analysis followed by Coomassie staining. Briefly, after electrophoresis, the gel was fixed in 50% methanol and 5% acetic acid for 30 min and washed with 50% methanol for 10 min and then with water for 10 min. The gel was incubated in Coomassie stain solution for 90 min at room temperature with rocking. Then gel was washed three times with deionized water for 10 min each. The protein bands were then excised for in-gel digestion. Each band was cut into 1mm3 blocks with a surgical scalpel and kept in a 1.5 mL Eppendorf tube. The gel blocks were washed in deionized water for 15 min and then washed three times in 50 mM ABC/50% CH3CN for 30 min. Gel plugs were dehydrated in 100% CH3CN for 10 min with vortex mixing. The supernatants were removed, and gel plugs were dried in the SpeedVac®. Trypsin (1 μg/50 μL) in 50 mM ABC was added, and gel plugs were allowed to rehydrate for 30 min on ice and allowed to digest overnight at 37 °C. The samples were then centrifuged, and the supernatant was removed. The pellet was resuspended in CH3CN with 1% TFA, Vortexed, and sonicated for 30 min to release hydrophobic peptides. The supernatant was removed and combined with the previous supernatant, lyophilized and stored at −20 °C until ready for MS/MS analysis.
Pseudo-shotgun proteomics
The eluted proteins were digested overnight with trypsin in the ratio of 1:50 at 37 °C. After digestion, trypsin was inactivated by addition of 20% trifluoroacetic acid to a final concentration of 0.5%. Digested proteins were concentrated and desalted with OMIX tips and concentrated in a SpeedVac®.
The desalted peptides were fractionated based on their isoelectric points by using Agilent off-gel fractionator with IPG strips (pH 3–11) according to the manufacturer’s instructions. After fractionation, the total - 24 fractions - were pooled into 12 fractions. All fractions were dried in a SpeedVac® prior to re-suspension in 20 μL of 98% H2O, 2% acetonitrile, and 0.1% formic acid for LC-MS analysis as described below.
LC-MS parameters and protein profiling
An Agilent 6530 quadrupole time-of-flight (QTOF) mass spectrometer equipped with an electrospray ionization (ESI) source was used. All samples were analyzed using an Agilent 1290 series Ultra Performance Liquid Chromatography system (UPLC) (Agilent Technologies, Santa Clara, CA, USA) containing a binary pump, degasser, well-plate auto-sampler with thermostat, and thermostatted column compartment. Mass spectra were acquired in the 3200 Da extended dynamic range mode (2 GHz) using the following settings: ESI capillary voltage, 3800 V; fragmentor, 150 V; nebulizer gas, 30 psig; drying gas, 8 L/min; drying temperature, 380°C. Data were acquired at a rate of 6 MS spectra per second and 3 MS/MS spectra per second in the mass ranges of m/z 100 – 2000 for MS, and 50 – 2500 for MS/MS and stored in profile mode with a maximum of five precursors per cycle. Fragmentation energy was applied at a slope of 3.0 V/100 Da with a 3.0 offset. Mass accuracy was maintained by continually-sprayed internal reference ions, m/z 121.0509 and 922.0098, in positive mode.
Agilent ZORBAX 300SB-C18 RRHD column 2.1 × 100 mm, 1.8μm (Agilent Technologies, Santa Clara, CA) was used for all analyses. LC parameters: autosampler temperature, 4°C; injection volume, 20 μL; column temperature, 40°C; mobile phases were 0.1% formic acid in water (phase A) and 0.1% formic acid in acetonitrile (phase B). The gradient started at 2% B at 400 μL/min for 1 min, increased to 50%B from 1 to 19 min with a flow rate of 250 μL/min, then increased to 95%B from 19 to 23 min with an increased flow rate of 400 μL/min and held up to 27 min at 95%B before decreasing to 2%B at 27.2, ending at 30 min and followed by a 2 minute post run at 2%B.
Data processing
Raw data was extracted and searched using the Spectrum Mill search engine (B.04.00.127, Agilent Technologies, Palo Alto, CA). “Peak picking” was performed within Spectrum Mill with the following parameters: signal-to-noise was set at 25:1, a maximum charge state of 7 is allowed (z = 7), and the program was directed to attempt to “find” a precursor charge state. During searching the following parameters were applied: genome of NCBInr, carbamidomethylation as a fixed modification, trypsin, maximum of 2 missed cleavages, precursor mass tolerance +/−20 ppm, product mass tolerance +/−50 ppm, and maximum ambiguous precursor charge = 3. Data were evaluated and protein identifications were considered significant if the following confidence thresholds were met: protein score > 13, individual peptide scores of at least 10, and Scored Peak Intensity (SPI) of at least 70%. The SPI provides an indication of the percent of the total ion intensity that matches the peptide’s MS/MS spectrum. A reverse (random) database search was simultaneously performed and manual inspection of spectra was used to validate the match of a spectrum with the predicted peptide fragmentation pattern, hence increasing confidence in the identification. Standards were run at the beginning of each day and at the end of a set of analyses for quality control.
Protein expression values (spectrum counts) were calculated in Scaffold software with the imported peptide hits from Spectrum Mill. The threshold for including a protein was a minimum of three distinct peptides with 95% confidence. To compare between samples, spectrum counts for each protein were divided by the sum of spectrum counts in respective samples, resulting in protein expression values as a percent of total.
Bioinformatics
GO and KEGG pathway enrichment (P<0.05) analyses were performed by using the functional annotation tool DAVID.13 A professional software ClueGO, Cytoscape plug-in, was used to facilitate the functional and pathway analysis for the BDHA targets and to create networks and charts.14
Results
Artesunate and probes exhibit no observable inhibition of cellular proliferation in human bronchial epithelial cells
Prior to investigating the mechanistic actions of artesunate and the synthesized probes, we assessed their toxicity in Beas-2B cells using the WST-1 cell proliferation assay. No significant inhibition of cellular proliferation was observed after exposure to Arts and BDHA probes at concentration ranges of 10 μM to 100 μM (Figure 2). Similarly, no observable toxic effects occurred after exposure to DMSO, used to dissolve artesunate and the chemical probes. Notably, our positive control for the assay, 50 mM H2O2, caused over 50% inhibition of cellular proliferation, further validating the functionality of the toxicological assay. A concentration of 30 μM Arts and BDHA was selected for subsequent mechanistic studies, consistent with our previous experiments.2,3,6
Figure 2. Effects of Artesunate and Chemical Probes on Human Bronchial Epithelial Cell Proliferation.
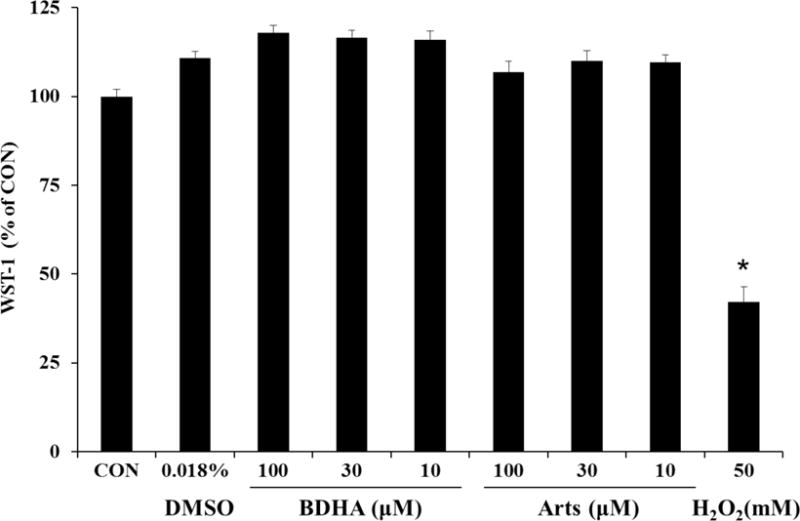
Beas-2B cells were incubated for 3 hrs in the indicated concentrations of DMSO, Arts, BDHA, or H2O2. Cellular proliferation was investigated via metabolic activity with the WST-1 assay. * Indicates a significant difference from CON, p < 0.05, n = 8 per treatment group.
Artesunate and chemical probes could modulate multiple signalling pathways in human bronchial epithelial cells
To further elucidate the mechanistic actions of the artesunate and BDHA probes, we studied the modulatory effects of these drugs on sixteen major nuclear transcription factors using a nuclear transcription factor array. Based on the barchart which shows the detailed effects of both drugs on the levels of the TFs (Figure 3), we noted that BDHA and Arts had similar effects at promoting nuclear levels of transcription factors, forkhead box protein O1 (FOXO1), nuclear factor (erythroid-derived 2)-like 2 (Nrf2), serum response factor (SRF) and signal transducer and activator of transcription 3 (STAT3). While both drugs similarly had no significant effects on activating transcription factor 4 (ATF4), early growth response protein 1 (EGR1), E26-transformation specific (ETS), hypoxia-inducible factor (HIF) and p53, Arts showed inductive effects on activator protein 1 (AP-1), interferon regulatory factors (IRF), nuclear factor of activated T-cells (NFAT) and nuclear factor kappa-light-chain-enhancer of activated B cells (NFkB), while BHA promoted core binding factor/nuclear transcription factor Y (CBF/NFY), heat shock factor (HSF) and Smad. As observed, while the probe BHDA and Arts had minor differences in molecular structures, the addition of biotin might result in some differences on their biological effects on the transcription factors. Collectively, the results demonstrate that artesunate and related artemisinins probe can modulate levels of multiple nuclear transcription factors and the protective effects of artemisinins may result from their involvement in major signalling pathways represented here.
Figure 3. Artesunate and Probes Modulate Nuclear Transcription Factor Protein Levels.
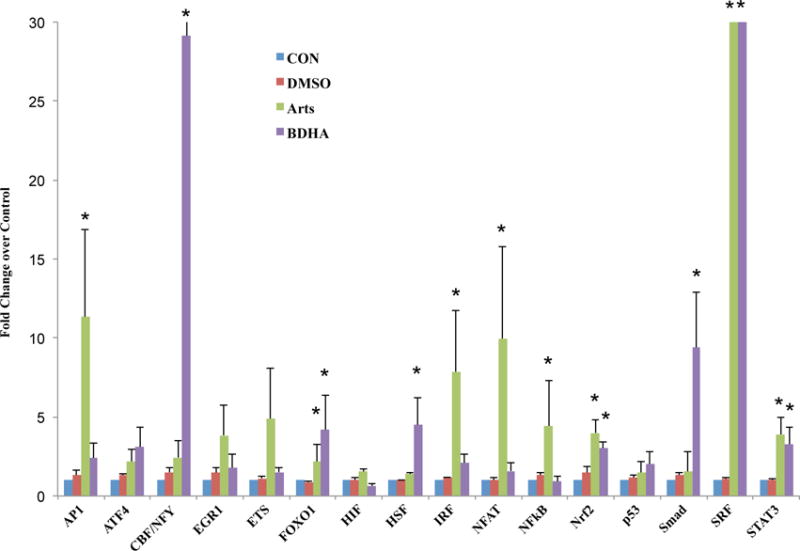
Beas-2B cells were incubated with or without Arts (30μM), artesunate probes (30μM) and 0.018% DMSO (drug vehicle) for 3 hours. Nuclear protein extracts were analysed using a Signosis Transcription Factor Protein Array and fold changes over CON were expressed in a barchart. * indicates a statistical significant difference in Arts/BDHA over CON, p < 0.05, n = 3 per treatment group.
BDHA directly interacts with Beas-2B proteins
To investigate the ability of BDHA to bind proteins in our system, after treatment of Beas-2B cells with BDHA, cell extracts were made and equal amounts of protein were resolved by electrophoresis and analysed by probing with streptavidin-HRP. As shown on Figure 4, biotin was detected in numerous bands in extracts from cells treated with BDHA. The specificity of this detection approach was validated by the virtually undetectable biotin signal in untreated cells, as well as after DMSO and artesunate treatments. These results indicate that BDHA can stably and directly interact with proteins in these epithelial cells.
Figure 4.
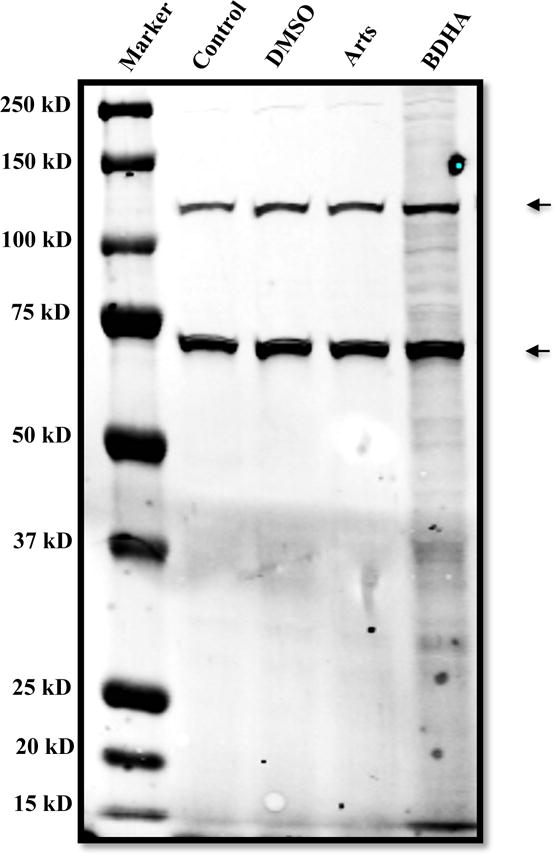
Visualization of BDHA binding to proteins on PVDF membrane. Total proteins (25 μg) from each compound-treated sample were subjected to electrophoresis. The biotinylated proteins were visualized by probing against Streptavidin-HRP. The two strong biotin bands seen in all lanes correspond to endogenously biotinylated proteins, and in this case incidentally verify the consistent protein loading across the four treatments.
Activation of Nrf2 by artemisinin analogues
Transcriptional activity of Nrf2 triggers a vast array of important antioxidant mechanisms. Keap1 is known to bind Nrf2, sequester it in the cytoplasm and repress its activity. Under oxidative and electrophilic stress Nrf2 is released from Keap1, stabilized via reduced proteasomal degradation and accumulates in the nucleus.15 In addition, Keap1 may also promote degradation of Nrf2 in the nucleus via the proteasome. Nrf2 controls constitutive and inducible expression of ARE-driven genes through a dynamic pathway involving nucleocytoplasmic shuttling by Keap1.16 To determine whether artemisinin modulated Nrf2, we analysed the relative amounts of Nrf2 in cytoplasmic and nuclear fractions of cells after treatment with artesunate or its derivatives for 3 hr. As shown in Figure 5, all forms of artesunate promoted significant increases in nuclear, but not cytosolic, Nrf2 levels. The newly designed BDHA derivative was as potent in its ability to induce nuclear Nrf2 accumulation as the parent artesunate compound, confirming BDHA suitability for the artesunate-targeted proteomic studies presented below.
Figure 5.
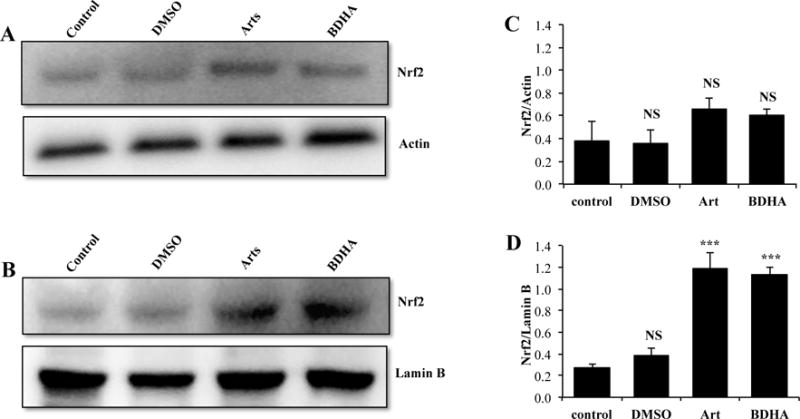
Nuclear up-regulation of Nrf2 by artemesinin and BDHA. Cells were submitted to the indicated treatments for 3 hr, followed by preparation of cytoplasmic and nuclear extracts and western blot analysis of equal amounts of protein (25μg). Representative western blots of Cytoplasm (A) and Nucleus (B) are shown. Graphs display density analysis of bands as a ratio of Nrf2 over actin or lamin B (C and D). Bars represent the mean of 3 experiments plus SEM. NS: P>0.05 vs respective control; ***: P<0.001 vs respective control (ANOVA followed by Tukey-Kramer multiple comparisons test).
Keap1 directly interacts with BDHA
A major challenge in understanding the significance of protein alkylation-induced signalling is to define a relationship between adduction and functional biochemical changes. To our understanding, the artemisinin analogues investigated here should demonstrate chemical properties typical of that exhibited by a number of endogenous electrophiles and electrophilic drug metabolites. Keap1 is reported to undergo adduction with electrophiles at multiple sites in vitro and in vivo,17,18 although how this controls the fate and activity Nrf2 is unclear.
Based on the above result (Figure 5), we investigated the hypothesis that artesunate modulates Nrf2 nuclear levels indirectly via interaction with Keap1. For the first strategy to demonstrate the interaction between Keap1 and artesunate, homogenates from bronchial epithelial cells treated with the parent compound or BDHA were submitted to biotin pull-down, then the captured and the biotin-containing fraction was investigated for the presence of Keap1 by western blotting. Keap1 was found in the fraction obtained from cells treated with BDHA, but not under non-biotinylated-artesunate treatment (Figure 6A). This finding was confirmed by showing that biotin (i.e., BDHA) co-immunoprecipitated with Keap1 in cells treated with BDHA but not with Arts (Figure 6B). These results indicate that Keap1 forms adducts with the biotinylated form of artesunate.
Figure 6.
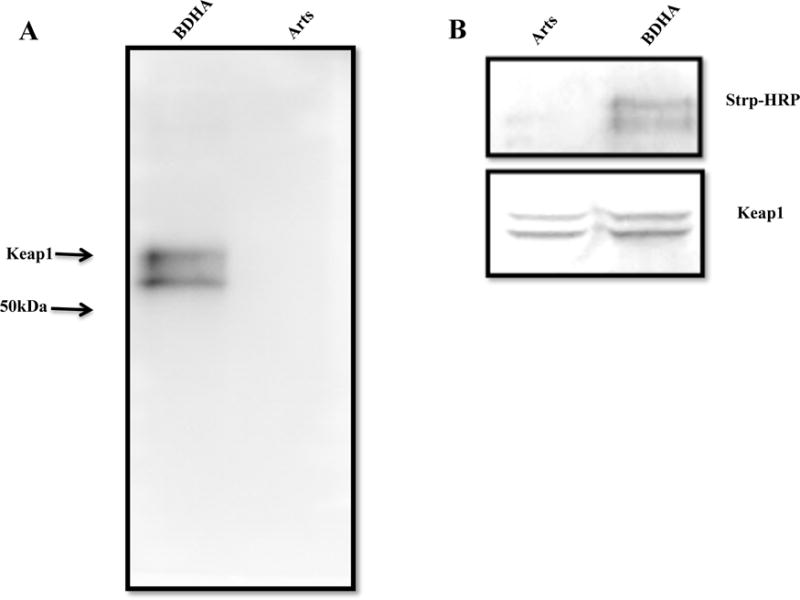
Interaction of BDHA with Keap1: A. The cell lysate of BDHA- and Arts-treated samples were subjected to Streptavidin-biotin pull-down, followed by probing against anti-Keap1 monoclonal antibody. The bands were visualized by using ECL reagent. B. Alternatively, co-immunoprecipitation of Keap1 in cells treated with BDHA and Arts, followed by probing against Streptavidin-HRP (upper panel). The same blot was stripped and probed against Keap1 antibody to detect the co-precipitation efficiency (lower panel).
Identification of BDHA-binding proteins by in-gel digestion and pseudo-shotgun proteomics
To identify the target proteins of artesunate, we applied both in-gel and pseudo-shotgun proteomic methods. Beas-2B cells were treated with BDHA, while a control group was treated only with artesunate, after which cells were lysed, pelleted and pre-processed as described in Materials and Methods. The released proteins were separated by 1D SDS-PAGE and processed for Coomassie blue-staining. The 12 most intensely stained spots were located on the gel and processed for in-gel digestion (Figure S3), LC-MS/MS identification, and analysis by QTOF. Proteins were identified by Spectrum Mill and scored by Scaffold. A complete list of BDHA-interacting proteins is presented in Supporting Information (Table S1).
To further validate the detected proteins by In-gel digestion, pseudo-shotgun proteomics was performed. The BDHA-adducted proteins were digested with trypsin, and tryptic peptides were fractionated based on their isoelectric point by Agilent off-gel electrophoresis. The initial 24 fractions were pooled into 12 fractions, analysed by LC-MS/MS, and proteins were identified by Spectrum Mill and Scaffold. The number of proteins identified by this method was larger than by the typical in-gel digestion method (Table S1). As a negative control, we similarly processed protein extracts from nonbiotinylated-artesunate-treated cells. Proteins that were pulled down in this negative control were later subtracted during the analysis of BDHA samples. The proteins identified from in-gel digestion are in close agreement with pseudo-shotgun proteomics analysis. All the proteins are listed in Table S1.
Discussion
Asthma is a major non-communicable respiratory disease that affects over 300 million people worldwide, and uncontrolled asthma has been reported to contribute to over 250 thousand deaths annually.19 There is a clear and unmet need to accelerate the discovery and development of effective anti-inflammatory therapeutics for improved control of this chronic inflammatory airway disease. Apart from being the mainstream therapeutic drug to treat malaria, artesunate has potent protective effects in many inflammatory conditions, particularly allergic asthma,2–4 anaphylaxis,6 rheumatoid arthritis20–22 and many others. While the pharmacological and therapeutic endpoints of artesunate in allergic asthma and various inflammatory conditions have been relatively well-established, comprehensive mechanistic actions of artesunate have not been fully elucidated. Our previous studies,2,3,6 along with others,23–25 have shown that artesunate mediates anti-inflammatory effects via modulation of the PI3K/Akt, Syk-PLCγ, NFkB and Nrf2 signalling cascades. We postulated in a recent review,1 that the anti-inflammatory actions of artesunate are likely to be mediated via the simultaneous modulation of multiple inflammatory cascades, resulting in an effective and broad-based protective effect, comparable to potent corticosteroids.26 In the present work, we identified a large number of the molecular targets with which artesunate interacts directly, providing a foundation for further elucidation of its molecular mode(s) of action.
In this study we synthesized a biotinylated analogue of DHA and applied an untargeted proteomics approach to discover molecular binding targets of artesunate in human bronchial epithelial cells. Our results demonstrated that artesunate binds to multiple proteins related to glucose metabolism, mRNA and protein synthesis, ribosomal regulation, stress response, structural components, and several others (Figure 8 and Table S1). To elucidate the comprehensive modulatory effects of artesunate on various signalling cascades, we investigated its effects using a nuclear transcription factor array. The results supported our hypothesis that artesunate mediates its anti-inflammatory actions via simultaneous modulation of multiple nuclear transcription factors, notably FOXO1, AP-1, IRF, NFAT, NFkB, Nrf2, SRF and STAT3. These findings are consistent with our previous studies which similarly described the molecular actions of artesunate on the Nrf2 and NFkB signalling cascades.2,3 The biotinylated analogue shared a similar profile of activation, but further promoted activation of nuclear factors CBF/NFY, HSF and Smad.
Figure 8.
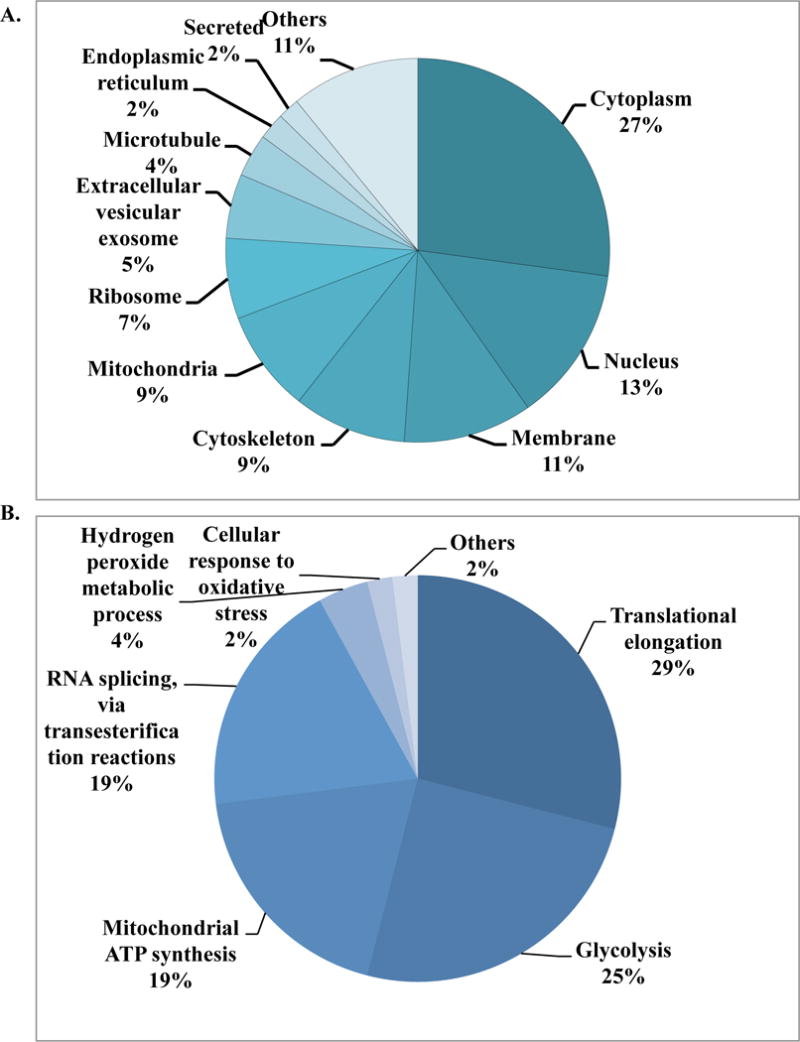
Overview of location and function of proteins captured by BDHA in Beas 2B cells. (A) Cellular distribution of captured proteins. (B) Classification of identified proteins based on relevant functional process (Gene Ontology terms).
Figures 9 and S2 summarize a collection of proteins we identified as capable of direct molecular interactions with artesunate, transcription factors activated under treatment with this compound, and gene expression modulation and phenotypic outcomes of exposure to artesunate.2–6 These drug-protein molecular interactions illustrate the broad impact of artesunate on a variety of cellular functions. For example, a major protective mechanism promoted by artesunate is the activation of the transcription factor Nrf2, a master regulator of endogenous antioxidant defences, whose activity decreases oxidative stress and related redox signalling via the up-regulation of multiple antioxidants and suppression of pro-oxidants in respiratory diseases.25,27 FOXO1 is a corresponding downstream transcription factor related to Nrf2,28 which may transduce pro-inflammatory gene transcription. Consistent with our previous report,3 activation of nuclear transcription factors, particularly Nrf2 and FOXO1, ameliorated oxidative stress and related lung damage caused by suppressed expression of genes encoding pulmonary pro-oxidants, such as inducible nitric oxide synthase (iNOS), NADPH oxidases (NOX1-4), as well as enhancement of endogenous antioxidants, particularly superoxide dismutases (SODs) and catalase.
Figure 9. Proposed Mechanism of Action of Artesunate in Lung Diseases.
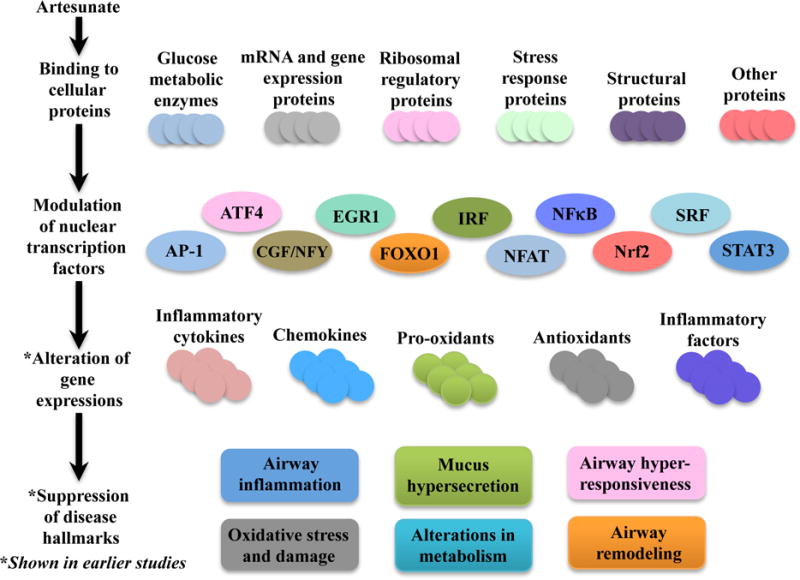
As supported by our data, artesunate is able to bind to various proteins related to glucose metabolism, mRNA and gene expressions, ribosomal regulatory proteins, stress responses proteins, structural proteins and others. Artesunate also led to modulation of multiple nuclear transcription factors, related to major inflammatory signalling cascades. This in turn leads to the alteration of various pro-inflammatory and anti-inflammatory gene expressions, which results in the suppression of the respective hallmarks of asthma.
AP-1, NFAT, NFkB and STAT3 have been consistently highlighted as major pro-inflammatory transcription factors that promote airway inflammation and other associated pathophysiological phenotypes of asthma.26,29–33 The transcription factor ATF4 is also regulated by β2-adrenoreceptor agonists and related to the promotion of AHR in asthma.34 EGR1 has similarly been linked to the pathogenesis of allergic asthma, where it regulates gene transcription in many pro-inflammatory and allergic responses, such as immunoglobulin E (IgE) and TNF production, and also promotes AHR in allergic asthma.35,36 IRF polymorphisms have been reported to confer genetic susceptibility to atopic asthma as well.37 SRF is another transcription factor associated with airway remodelling and promotion of smooth muscle gene transcription and differentiation.38,39 Correspondingly observed in this study, the modulatory effect of artesunate on these nuclear transcription factors may be responsible for its protective actions against various hallmarks of asthma.2 Furthermore, the artesunate-induced signalling alterations are also likely to contribute to the broad suppression of inflammatory and allergic cytokines and chemokines, such as IL-4, IL-5, IL-13, IL-17, IL-12 (p40), MCP-1, G-CSF, eotaxin, ICAM-1, V-CAM-1 and E-selectin, which further results in the suppression of inflammatory cell recruitment and airway hyper-responsiveness.2,3
It has been suggested that the Nrf2-Keap1 interaction acts as a sensor of oxidative stress, and that Keap1 serves as an electrophile-dependent sensor trigger for the activation of Nrf2-regulated genes.15 Electrophile-mediated modification of multiple Keap1 thiols leads to dissociation of the Nrf2-Keap1 complex, and hence Nrf2 transcriptional activity.18 To test the hypothesis that artesunate and its related derivatives modulate Keap1 inhibitory functions due to the electrophilic nature of these chemicals, we used pull-down and co-immunoprecipitation assays whose results strongly support the binding of BDHA to Keap1 (Figure 6 A & B). The literature notes that the degree of affinity of Keap1 for electrophiles is residue-dependent.17 Based on in vitro reactions, some reports show that artesunate and artemisinin derivatives undergo Fe2+-mediated decomposition, thus acquiring the ability to form a covalent bond with the cysteine residue of glutathione.40 These species are presumed to arise via entrapment of the C-4 radicals in rather intricate reactions involving ferrous-cysteine complexes.41,42 However, it seems speculative to assume that this chemistry applies to the interaction of artesunate with Keap1, a protein with 25 cysteine residues, some of which are highly reactive.43 More likely, an iron-independent process leads to activation of the peroxide bridge that promotes the binding of BDHA to Keap1. There is evidence that once the peroxide bridge of artesunate is broken, it reacts with cysteine residues of Keap1, as these highly reactive cysteines in the central linker domain are required for cytoplasmic sequestration of Nrf2.44 The specific nature of the interaction here observed, however, remains to be investigated.
To investigate the interaction of BDHA with proteins, two different techniques were used to identify artemisinin-related protein targets, namely in-gel digestion and pseudo-shotgun (PSG) proteomics (see Supporting information). Proteins listed in Table S1 were detected via both methods, with the criteria of a minimum of 3 peptides and total spectral count of 3 in Scaffold with some relaxed criteria for a few proteins. We see generally good agreement between the proteins identified by both methods except for some of the membrane proteins, which could be explained, for example, by experimental protocol steps leading to loss of the membrane fraction.
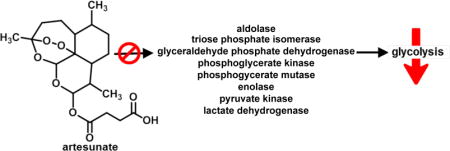
The identification of a number of molecular chaperone proteins, such as heat shock protein (HSP) beta-1, was not surprising since these multifunctional proteins are involved in the transport of a large array of molecules. Based on the cellular abundance of structural proteins, nearly 10% of artesunate binding partners were cytoskeleton components such as actin, tubulin, profilin-1, filamins and others. Half of all identified artesunate binding targets were distributed among nucleus, mitochondria, and cytoplasm, with over 25% found in the latter (Figure 8A). Most importantly, approximately 50% of artesunate-interacting proteins are involved in glycolysis (a cytoplasmic process) and mitochondrial ATP generation (Figure 8B and S1), which strongly suggests that artesunate significantly impacts cellular energy metabolism. This is in agreement with our previous work on asthma, demonstrating that artemisinins affect not only pulmonary, but also systemic metabolism.4,45,46 Notably in this study, artesunate was found to interact directly with eight of the metabolic enzymes involved in glycolysis, namely aldolase, triosephosphate isomerase, glyceraldehyde 3-phosphate dehydrogenases (GAPDH), phosphoglycerate kinase, phosohoglycerate mutase, enolase, pyruvate kinase, and L-lactate dehydrogenase as shown in Figure 7. The interaction of artesunate with these glucose and energy-related metabolic enzymes may be responsible for the broad therapeutic effects of artesunate against localised and systemic asthma-induced metabolic effects that were identified in our recent report.45 Moreover, we speculate that the protective, anti-inflammatory property of artesunate could result from a rerouting of the metabolic pathways due to artesunate’s interaction with components of the glycolysis cascade. In support of this hypothesis, it is known that GAPDH is inactivated in oxidizing circumstances (such as the environment produced by severe inflammation), leading to a metabolic shift from glycolysis to the pentose phosphate pathway, with consequent generation of NADPH.47,48 In addition, the inhibitory property of these molecular interactions in several cellular components has been previously demonstrated. Artemisinin compounds had broader effects than other antimalarials on metabolism, protein synthesis and nucleic acid synthesis in malarial parasites.49 Therefore, along with the modulation of transcriptional factors discussed above, the direct interaction of artesunate with proteins involved in metabolism and synthesis may be key features conferring the modulatory effects of artemisinins in our model.
Figure 7.
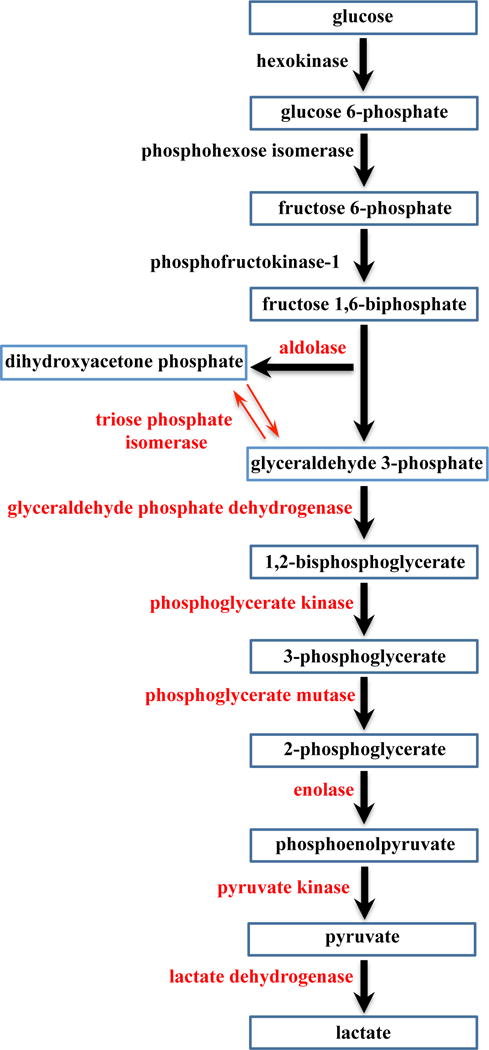
BDHA targeted proteins in the glycolysis pathway. Eight proteins were identified and are highlighted in red.
Considering that we identified several protein targets that interact directly with artesunate, it is noteworthy that many reports indicate that the nature and specificity of these drugs, as well as their subcellular localization, depend mostly on factors such as lipophilicity, ability to interact with thiols and nucleophilic amino groups, thus exhibiting mostly nonspecific molecular targets.8,12,50–52 For example, depending on the experimental model, some drug derivatives concentrate in the endoplasmic reticulum while others localize in the mitochondrion.51 It seems a consensus, based on the structure-function relationship of this class of compounds,52 that artemisinins do not bind specific targets. Instead, the modification of protein function by artesunate, as might to be the case with Keap1, probably involves protein thiol residues and alkylation reactions.43 Regarding our results, it is plausible that the abundance of artesunate targets belonging to mitochondrial ATP synthesis could be explained by a preferential mitochondrial localization of our newly synthesized BDHA, as well as a higher rate of activation of the compound in heme-rich environment within those organelles.
In summary, using a novel chemical probe that allowed for global analysis of artesunate targets, we demonstrate that artesunate may exert its effects on pulmonary epithelial cells by directly interacting with multiple vital proteins, and also by modulating gene expression via transcription factors. While we observed that artesunate interacts with structural as well as other proteins whose cellular functions have not been well described, new targeted studies are required to elucidate the downstream biological effects of these molecular interactions in each particular experimental model. Along with our previous reports, the present findings bring to light an array of prospective molecular targets essential to advance the understanding of the mechanism of action of artemisinins in allergic asthma and possibly other inflammatory disorders.
Supplementary Material
Table S1: List of identified proteins by In-gel and Pseudo-shotgun proteomics methods.
Acknowledgments
We thank Laura J. Trudel for assistance with cell cultures
Funding sources
Wanxing Eugene Ho is a recipient of the Singapore-MIT Alliance Graduate Fellowship. This research was jointly supported by the Singapore-MIT Alliance Graduate Fellowship and SPH Centre for Environmental and Occupational Health Research. Thanks also to the MIT Center for Environmental Health Sciences, NIEHS Grant #P30-ES002109.
Abbreviations
- Arts
Artesunate
- BDHA
Biotinylated dihydroartesunate
- DHA
Dihydroartemisinin
- LC-MS
Liquid Chromatography Mass Spectroscopy
- Keap1
Kelch-like ECH-associated protein 1
- Nrf2
nuclear factor erythroid-2-related factor 2.
Footnotes
Supporting Information Available
Synthesis and characterization of chemical probes.
ClueGO analysis of BDHA-binding proteins.
Coomassie-stained SDS-PAGE gels.
Additional procedures, tables, and figures. This material is available free of charge via the Internet at http://pubs.acs,org
References
- 1.Ho WE, Peh HY, Chan TK, Wong WSF. Artemisinins: pharmacological actions beyond anti-malarial. Pharmacol Ther. 2014;142:126–139. doi: 10.1016/j.pharmthera.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 2.Cheng C, Ho WE, Goh FY, Guan SP, Kong LR, Lai WQ, Leung BP, Wong WSF. Anti-malarial drug artesunate attenuates experimental allergic asthma via inhibition of the phosphoinositide 3-kinase/Akt pathway. PLoS One. 2011;6:e20932. doi: 10.1371/journal.pone.0020932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ho WE, Cheng C, Peh HY, Xu F, Tannenbaum SR, Ong CN, Wong WSF. Anti-malarial drug artesunate ameliorates oxidative lung damage in experimental allergic asthma. Free Radic Biol Med. 2012;53:498–507. doi: 10.1016/j.freeradbiomed.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 4.Ho WE, Xu YJ, Xu F, Cheng C, Peh HY, Huang SM, Tannenbaum SR, Ong CN, Wong WSF. Anti-malarial drug artesunate restores metabolic changes in experimental allergic asthma. Metabolomics. 2015;11:380–390. [Google Scholar]
- 5.Tan SSL, Ong B, Cheng C, Ho WE, Tam JKC, Stewart AG, Harris T, Wong WSF, Tran T. The antimalarial drug artesunate inhibits primary human cultured airway smooth muscle cell proliferation. Am J Respir Cell Mol Biol. 2014;50:451–458. doi: 10.1165/rcmb.2013-0273OC. [DOI] [PubMed] [Google Scholar]
- 6.Cheng C, Ng DSW, Chan TK, Guan SP, Ho WE, Koh AHM, Bian JS, Lau HYA, Wong WSF. Anti-allergic action of anti-malarial drug artesunate in experimental mast cell-mediated anaphylactic models. Allergy. 2013;68:195–203. doi: 10.1111/all.12077. [DOI] [PubMed] [Google Scholar]
- 7.Ng DSW, Liao W, Tan WSD, Chan TK, Loh XY, Wong WSF. Anti-malarial drug artesunate protects against cigarette smoke-induced lung injury in mice. Phytomedicine. 2014;21:1638–1644. doi: 10.1016/j.phymed.2014.07.018. [DOI] [PubMed] [Google Scholar]
- 8.Yang YZ, Asawamahasakda W, Meshnick SR. Alkylation of human albumin by the antimalarial artemisinin. Biochem Pharmacol. 1993;46:336–339. doi: 10.1016/0006-2952(93)90425-v. [DOI] [PubMed] [Google Scholar]
- 9.O’Neill PM, Posner GH. A medicinal chemistry perspective on artemisinin and related endoperoxides. J Med Chem. 2004;47:2945–2964. doi: 10.1021/jm030571c. [DOI] [PubMed] [Google Scholar]
- 10.Haynes RK, Ho WY, Chan HW, Fugmann B, Stetter J, Croft SL, Vivas L, Peters W, Robinson BL. Highly antimalaria-active artemisinin derivatives: biological activity does not correlate with chemical reactivity. Angew Chem Int Ed Engl. 2004;43:1381–1385. doi: 10.1002/anie.200352343. [DOI] [PubMed] [Google Scholar]
- 11.Posner GH, Oh CH. Regiospecifically oxygen-18 labeled 1,2,4-trioxane: a simple chemical model system to probe the mechanism(s) for the antimalarial activity of artemisinin (qinghaosu) J Am Chem Soc. 1992;114:8328–8329. [Google Scholar]
- 12.Ying-Zi Y, Little B, Meshnick SR. Alkylation of proteins by artemisinin. Biochem Pharmacol. 1994;48:569–573. doi: 10.1016/0006-2952(94)90287-9. [DOI] [PubMed] [Google Scholar]
- 13.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 14.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z, Galon J. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Itoh K, Wakabayashi N, Katoh Y, Ishii T, O’Connor T, Yamamoto M. Keap1 regulates both cytoplasmic-nuclear shuttling and degradation of Nrf2 in response to electrophiles. Genes Cells. 2003;8:379–391. doi: 10.1046/j.1365-2443.2003.00640.x. [DOI] [PubMed] [Google Scholar]
- 16.Nguyen T, Sherratt PJ, Nioi P, Yang CS, Pickett CB. Nrf2 controls constitutive and inducible expression of ARE-driven genes through a dynamic pathway involving nucleocytoplasmic shuttling by Keap1. J Biol Chem. 2005;280:32485–32492. doi: 10.1074/jbc.M503074200. [DOI] [PubMed] [Google Scholar]
- 17.Hong F, Sekhar KR, Freeman ML, Liebler DC. Specific patterns of electrophile adduction trigger Keap1 ubiquitination and Nrf2 activation. J Biol Chem. 2005;280:31768–31775. doi: 10.1074/jbc.M503346200. [DOI] [PubMed] [Google Scholar]
- 18.Eggler AL, Liu G, Pezzuto JM, van Breemen RB, Mesecar AD. Modifying specific cysteines of the electrophile-sensing human Keap1 protein is insufficient to disrupt binding to the Nrf2 domain Neh2. Proc Natl Acad Sci U S A. 2005;102:10070–10075. doi: 10.1073/pnas.0502402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masoli M, Fabian D, Holt S, Beasley R. The global burden of asthma: executive summary of the GINA Dissemination Committee report. Allergy. 2004;59:469–478. doi: 10.1111/j.1398-9995.2004.00526.x. [DOI] [PubMed] [Google Scholar]
- 20.Li Y, Wang S, Wang Y, Zhou C, Chen G, Shen W, Li C, Lin W, Lin S, Huang H, Liu P, Shen X. Inhibitory effect of the antimalarial agent artesunate on collagen-induced arthritis in rats through nuclear factor kappa B and mitogen-activated protein kinase signaling pathway. Transl Res. 2013;161:89–98. doi: 10.1016/j.trsl.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 21.Mirshafiey A, Saadat F, Attar M, Di Paola R, Sedaghat R, Cuzzocrea S. Design of a new line in treatment of experimental rheumatoid arthritis by artesunate. Immunopharmacol Immunotoxicol. 2006;28:397–410. doi: 10.1080/08923970600927447. [DOI] [PubMed] [Google Scholar]
- 22.He Y, Fan J, Lin H, Yang X, Ye Y, Liang L, Zhan Z, Dong X, Sun L, Xu H. The anti-malaria agent artesunate inhibits expression of vascular endothelial growth factor and hypoxia-inducible factor-1α in human rheumatoid arthritis fibroblast-like synoviocyte. Rheumatol Int. 2011;31:53–60. doi: 10.1007/s00296-009-1218-7. [DOI] [PubMed] [Google Scholar]
- 23.Xu H, He Y, Yang X, Liang L, Zhan Z, Ye Y, Lian F, Sun L. Antimalarial agent artesunate inhibits TNF-alpha-induced production of proinflammatory cytokines via inhibition of NF-kappaB and PI3 kinase/Akt signal pathway in human rheumatoid arthritis fibroblast-like synoviocytes. Rheumatology (Oxford) 2007;46:920–926. doi: 10.1093/rheumatology/kem014. [DOI] [PubMed] [Google Scholar]
- 24.Lee IS, Ryu DK, Lim J, Cho S, Kang BY, Choi HJ. Artesunate activates Nrf2 pathway-driven anti-inflammatory potential through ERK signaling in microglial BV2 cells. Neurosci Lett. 2012;509:17–21. doi: 10.1016/j.neulet.2011.12.034. [DOI] [PubMed] [Google Scholar]
- 25.Kim J, Cha YN, Surh YJ. A protective role of nuclear factor-erythroid 2-related factor-2 (Nrf2) in inflammatory disorders. Mutat Res. 2010;690:12–23. doi: 10.1016/j.mrfmmm.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 26.Jacques E, Semlali A, Boulet LP, Chakir J. AP-1 overexpression impairs corticosteroid inhibition of collagen production by fibroblasts isolated from asthmatic subjects. Am J Physiol Lung Cell Mol Physiol. 2010;299:L281–7. doi: 10.1152/ajplung.00360.2009. [DOI] [PubMed] [Google Scholar]
- 27.Rangasamy T, Guo J, Mitzner WA, Roman J, Singh A, Fryer AD, Yamamoto M, Kensler TW, Tuder RM, Georas SN, Biswal S. Disruption of Nrf2 enhances susceptibility to severe airway inflammation and asthma in mice. J Exp Med. 2005;202:47–59. doi: 10.1084/jem.20050538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang J, Yue S, Ke B, Zhu J, Shen X, Zhai Y, Yamamoto M, Busuttil RW, Kupiec-Weglinski JW. Nuclear factor erythroid 2-related factor 2 regulates toll-like receptor 4 innate responses in mouse liver ischemia-reperfusion injury through Akt-forkhead box protein O1 signaling network. Transplantation. 2014;98:721–728. doi: 10.1097/TP.0000000000000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Desmet C, Gosset P, Henry E, Garzé V, Faisca P, Vos N, Jaspar F, Mélotte D, Lambrecht B, Desmecht D, Pajak B, Moser M, Lekeux P, Bureau F. Treatment of experimental asthma by decoy-mediated local inhibition of activator protein-1. Am J Respir Crit Care Med. 2005;172:671–678. doi: 10.1164/rccm.200410-1431OC. [DOI] [PubMed] [Google Scholar]
- 30.Simeone-Penney MC, Severgnini M, Tu P, Homer RJ, Mariani TJ, Cohn L, Simon AR. Airway epithelial STAT3 is required for allergic inflammation in a murine model of asthma. J Immunol. 2007;178:6191–6199. doi: 10.4049/jimmunol.178.10.6191. [DOI] [PubMed] [Google Scholar]
- 31.Aggarwal BB, Takada Y, Shishodia S, Gutierrez AM, Oommen OV, Ichikawa H, Baba Y, Kumar A. Nuclear transcription factor NF-kappa B: role in biology and medicine. Indian J Exp Biol. 2004;42:341–353. [PubMed] [Google Scholar]
- 32.Mitsuta K, Matsuse H, Fukushima C, Kawano T, Tomari S, Obase Y, Goto S, Urata Y, Shimoda T, Kondo T, Kohno S. Production of TNF-α by Peripheral Blood Mononuclear Cells through Activation of Nuclear Factor κB by Specific Allergen Stimulation in Patients with Atopic Asthma. Allergy Asthma Proc. 2003;24:19–26. [PubMed] [Google Scholar]
- 33.Monticelli S, Solymar DC, Rao A. Role of NFAT proteins in IL13 gene transcription in mast cells. J Biol Chem. 2004;279:36210–36218. doi: 10.1074/jbc.M406354200. [DOI] [PubMed] [Google Scholar]
- 34.Ladwa SR, Dilly SJ, Clark AJ, Marsh A, Taylor PC. Rapid identification of a putative interaction between beta2-adrenoreceptor agonists and ATF4 using a chemical genomics approach. ChemMedChem. 2008;3:742–744. doi: 10.1002/cmdc.200700317. [DOI] [PubMed] [Google Scholar]
- 35.Li B, Power MR, Lin TJ. De novo synthesis of early growth response factor-1 is required for the full responsiveness of mast cells to produce TNF and IL-13 by IgE and antigen stimulation. Blood. 2006;107:2814–2820. doi: 10.1182/blood-2005-09-3610. [DOI] [PubMed] [Google Scholar]
- 36.Cho SJ, Kang MJ, Homer RJ, Kang HR, Zhang X, Lee PJ, Elias JA, Lee CG. Role of early growth response-1 (Egr-1) in interleukin-13-induced inflammation and remodeling. J Biol Chem. 2006;281:8161–8168. doi: 10.1074/jbc.M506770200. [DOI] [PubMed] [Google Scholar]
- 37.Nakao F, Ihara K, Kusuhara K, Sasaki Y, Kinukawa N, Takabayashi A, Nishima S, Hara T. Association of IFN-gamma and IFN regulatory factor 1 polymorphisms with childhood atopic asthma. J Allergy Clin Immunol. 2001;107:499–504. doi: 10.1067/mai.2001.113051. [DOI] [PubMed] [Google Scholar]
- 38.Liu HW, Halayko AJ, Fernandes DJ, Harmon GS, McCauley JA, Kocieniewski P, McConville J, Fu Y, Forsythe SM, Kogut P, Bellam S, Dowell M, Churchill J, Lesso H, Kassiri K, Mitchell RW, Hershenson MB, Camoretti-Mercado B, Solway J. The RhoA/Rho kinase pathway regulates nuclear localization of serum response factor. Am J Respir Cell Mol Biol. 2003;29:39–47. doi: 10.1165/rcmb.2002-0206OC. [DOI] [PubMed] [Google Scholar]
- 39.Davies DE, Wicks J, Powell RM, Puddicombe SM, Holgate ST. Airway remodeling in asthma: new insights. J Allergy Clin Immunol. 2003;111:215–225. doi: 10.1067/mai.2003.128. [DOI] [PubMed] [Google Scholar]
- 40.Wang DY, Wu YL. A possible antimalarial action mode of qinghaosu (artemisinin) series compounds. Alkylation of reduced glutathione by C-centered primary radicals produced from antimalarial compound qinghaosu and 12-(2,4-dimethoxyphenyl)-12-deoxoqinghaosu. Chem Commun. 2000:2193–2194. [Google Scholar]
- 41.Wu Y, Yue Z, Wu Y. Interaction of Qinghaosu (Artemisinin) with Cysteine Sulfhydryl Mediated by Traces of Non-Heme Iron. Angew Chem Int Ed Engl. 1999;38:2580–2582. doi: 10.1002/(sici)1521-3773(19990903)38:17<2580::aid-anie2580>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 42.Wu WM, Chen YL, Zhai Z, Xiao SH, Wu YL. Study on the mechanism of action of artemether against schistosomes: the identification of cysteine adducts of both carbon-centred free radicals derived from artemether. Bioorg Med Chem Lett. 2003;13:1645–1647. doi: 10.1016/s0960-894x(03)00293-2. [DOI] [PubMed] [Google Scholar]
- 43.Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, Yamamoto M, Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci U S A. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang DD, Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol. 2003;23:8137–8151. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ho WE, Xu YJ, Xu F, Cheng C, Peh HY, Tannenbaum SR, Wong WSF, Ong CN. Metabolomics reveals altered metabolic pathways in experimental asthma. Am J Respir Cell Mol Biol. 2013;48:204–211. doi: 10.1165/rcmb.2012-0246OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ho WE, Xu YJ, Cheng C, Peh HY, Tannenbaum SR, Wong WSF, Ong CN. Metabolomics Reveals Inflammatory-Linked Pulmonary Metabolic Alterations in a Murine Model of House Dust Mite-Induced Allergic Asthma. J Proteome Res. 2014;13:3771–3782. doi: 10.1021/pr5003615. [DOI] [PubMed] [Google Scholar]
- 47.McGettrick AF, O’Neill LAJ. How metabolism generates signals during innate immunity and inflammation. J Biol Chem. 2013;288:22893–22898. doi: 10.1074/jbc.R113.486464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grant CM. Metabolic reconfiguration is a regulated response to oxidative stress. J Biol. 2008;7:1. doi: 10.1186/jbiol63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ter Kuile F, White N, Holloway P, Pasvol G, Krishna S. Plasmodium falciparum: In Vitro Studies of the Pharmacodynamic Properties of Drugs Used for the Treatment of Severe Malaria. Exp Parasitol. 1993;76:85–95. doi: 10.1006/expr.1993.1010. [DOI] [PubMed] [Google Scholar]
- 50.Wang J, Huang L, Li J, Fan Q, Long Y, Li Y, Zhou B. Artemisinin directly targets malarial mitochondria through its specific mitochondrial activation. PLoS One. 2010;5:e9582. doi: 10.1371/journal.pone.0009582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Y, Lok CN, Ko BCB, Shum TYT, Wong MK, Che CM. Subcellular localization of a fluorescent artemisinin derivative to endoplasmic reticulum. Org Lett. 2010;12:1420–1423. doi: 10.1021/ol902890j. [DOI] [PubMed] [Google Scholar]
- 52.O’Neill PM, Barton VE, Ward SA. The molecular mechanism of action of artemisinin-the debate continues. Molecules. 2010;15:1705–1721. doi: 10.3390/molecules15031705. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: List of identified proteins by In-gel and Pseudo-shotgun proteomics methods.


