Abstract
Primers for herpes simplex virus type 1 (HSV 1)-specific loop-mediated isothermal amplification (LAMP) method amplified HSV-1 DNA, while HSV-2-specific primers amplified only HSV-2 DNA; no LAMP products were produced by reactions performed with other viral DNAs. The sensitivities of the HSV-1- and HSV-2-specific LAMP methods, determined by agarose gel electrophoresis, reached 500 and 1,000 copies/tube, respectively. The turbidity assay, however, determined the sensitivity of the HSV-1- and HSV-2-specific LAMP methods to be 1,000 and 10,000 copies/tube, respectively. After initial validation studies, 18 swab samples (in sterilized water) collected from patients with either gingivostomatitis or vesicular skin eruptions were examined. HSV-1 LAMP products were detected by agarose gel electrophoresis in the 10 samples that also demonstrated viral DNA detection by real-time PCR. Nine of these 10 samples exhibited HSV-1 LAMP products by turbidity assay. Furthermore, both the agarose gel electrophoresis and the turbidity assay directly detected HSV-1 LAMP products in 9 of the 10 swab samples collected in sterilized water. Next, we examined the reliability of HSV type-specific LAMP for the detection of viral DNA in clinical specimens (culture medium) collected from genital lesions. HSV-2 was isolated from all of the samples and visualized by either agarose gel electrophoresis or turbidity assay.
Viral isolation and serological assays are standard methods of herpes simplex virus (HSV) diagnosis. Both viral isolation and serological testing, however, require substantial time to obtain accurate final results. More rapid detection has been achieved by modification of cell culture techniques by centrifugation of inocula on cell monolayers and the use of immunofluorescence techniques (6). Recent studies have suggested that detection of HSV DNA by PCR increases the sensitivity of viral infection detection compared to antigenic detection or cell culture methods (3, 4, 11, 13, 14). While quantitative analysis of viral DNA by real-time PCR may become a valuable tool for bedside monitoring of HSV infection and progression (1, 2, 7, 10, 17, 21, 22), it has not yet become a common procedure in hospital laboratories due to the requirement of specific expensive equipment (a thermal cycler).
Recently, Notomi et al. (18) reported a novel nucleic acid amplification method, termed loop-mediated isothermal amplification (LAMP), which is used to amplify DNA under isothermal conditions with high specificity, efficiency, and speed. The most significant advantage of LAMP is the ability to amplify specific sequences of DNA between 63 and 65°C without thermocycling. Thus, the technique requires only simple and cost-effective equipment amenable to use in hospital laboratories. The LAMP method also exhibits both high specificity and high amplification efficiency. As the LAMP method uses four primers which recognize six distinct target DNA sequences, the specificity is extremely high. This method also exhibits extremely high amplification efficiency, due in part to its isothermal nature; as there is no time lost due to changes in temperature and the reaction can be conducted at the optimal temperature for enzyme function, the inhibition reactions that often occur at later stages of typical PCR amplifications are less likely to occur. Thus, this method could potentially be a valuable tool for the rapid diagnosis of infectious diseases (5, 8, 9, 12, 19, 23) in both commercial and hospital laboratories. In this study, we sought to establish a LAMP-based HSV type-specific DNA amplification method and examine its reliability for the detection of HSV DNA from clinical specimens.
HSV-1 (KOS) DNA and HSV-2 (186) DNA were used as positive controls to determine the appropriate conditions for HSV type-specific LAMP and to establish the baseline sensitivity and specificity levels. HSV-1 (KOS), HSV-2 (186), varicella-zoster virus (VZV) (Oka), human cytomegalovirus (HCMV) (AD-169), human herpesvirus type 6B (HHV-6B) (Z29), and HHV-7 (RK) DNA were used to determine the specificity of HSV type-specific LAMP. Plasmids containing the HSV-1 and HSV-2 target sequences were used to determine the assay sensitivity.
To determine the reliability of HSV type-specific LAMP for detection of viral DNA from clinical samples, 18 swab samples (sample numbers 1 to 18) were collected from patients with either gingivostomatitis or vesicular skin eruptions. Swabs were collected from patients at the outpatient clinic of the Fujita Health University hospital and the Central Hospital of the Tokai Medical Institute and placed into 1 ml of sterilized water. Five swab samples (sample numbers 19 to 23) were also collected from patients with genital herpes at Teikyo University Mizonokuchi Hospital outpatient clinic. Swabs were collected from the lesions and placed into culture medium. HSV-2 was isolated from all of these samples. We attempted detection of HSV-1 and HSV-2 DNA from either post-DNA extraction or without DNA extraction by using HSV-1-specific and HSV-2-specific LAMP. The results of HSV type-specific LAMP were compared with results obtained by the previously established technique of HSV type-specific real-time PCR to assess the reliability of the methods for the rapid diagnosis of HSV infection.
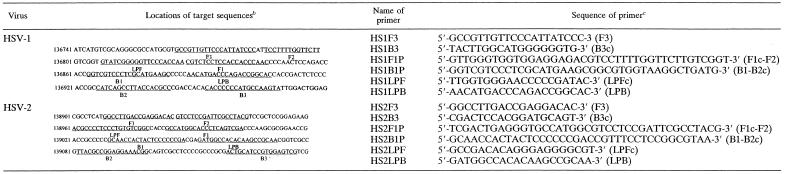
LAMP reactions were conducted as described previously by Notomi et al. (18) and Nagamine et al. (16). The LAMP method requires a set of four primers (B3, F3, BIP, and FIP) that recognize a total of six distinct sequences (B1 to B3 and F1 to F3) within the target DNA. Primers for HSV-1 and HSV-2 LAMP were designed against the HSV-1 glycoprotein G (gG) and HSV-2 gG genes, respectively, by using Primer Explorer V software (FUJITSU, Tokyo, Japan), the locations and sequences of which are shown in Table 1. Primer BIP for the gG genes of HSV-1 (HS1BIP) and HSV-2 (HS2BIP) contained the B1 direct sequence and B2 complementary sequence, each specific for the respective strains. Primer FIP for the gG genes of HSV-1 (HS1FIP) and HSV-2 (HS2FIP) contained the F1 complementary sequence and the F2 direct sequence. Primers B3 (HS1B3 and HS2B3) and F3 (HS1F3 and HS2F3) for the HSV-1 and HSV-2 gG genes were located outside the F2-B2 regions. As additional loop primers increase the amplification efficiency (16), loop primers specific for the HSV-1 gG (HS1LPB and HS1LPF) and HSV-2 gG (HS2LPB and HS2LPF) genes were also synthesized. HS1LPB and HS2LPB contained the LPB sequence, while HS1LPF and HS2LPF contained the LPF complementary sequence. The LAMP reaction was performed by using a Loopamp DNA amplification kit (Eiken Chemical, Tochigi, Japan). Reaction mixtures (25 μl) contained 1.6 μM each FIP and BIP primer, 0.8 μM each outer primer (F3 primer and B3 primer), 0.8 μM each loop primer (LPF primer and LPB primer), 2× reaction mix (12.5 μl), Bst DNA polymerase (1 μl), and 5 μl of each sample. The mixture was incubated at 63°C for 30 min. Next, a TERAMECS LA200 (Teramecs, Kyoto, Japan) was used to measure turbidity after 30 min of LAMP (15). After turbidity measurement, LAMP products were subjected to electrophoresis on 1.5% agarose gels. Gels were visualized under UV light after ethidium bromide staining. To avoid contamination between samples, different rooms were used for DNA extraction, LAMP setup, and gel analysis using filter-containing pipette tips for aerosol protection. As the turbidities of five negative samples were demonstrated to be 0.01 ± 0.02, we defined 0.1 as the cutoff value for discrimination between positive and negative samples.
TABLE 1.
Primers targeting the HSV-1 and HSV-2 gG genesa
Names and sequences of each primer used for HSV type-specific LAMP are shown, as are locations of target sequences within the gG genes of HSV-1 and HSV-2. B2c, sequence complementary to B2; F1c, sequence complementary to F1; LPFc, sequence complementary to LPF; B3c, sequence complementary to B3.
Numbers at left of sequences are nucleotide positions. GenBank accession numbers are as follows: NC001806 (HSV-1 complete genome) and NC001798 (HSV-2 complete genome).
Designations in parentheses correspond to definitions and sequences described in footnote a.
Real-time PCR quantitated the amount of either HSV-1 or HSV-2 DNA in each sample. The genes encoding HSV-1 and HSV-2 glycoprotein G were selected for HSV type-specific real-time PCR. The sequences of the primers and probes used for these experiments were described previously by Pevenstein et al. (20).
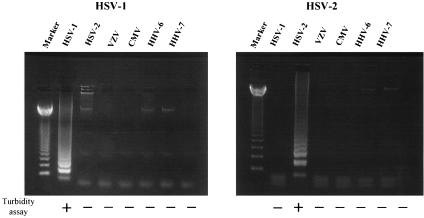
To develop an effective assay for rapid measurement of HSV DNA content, we first evaluated the specificity of our HSV type-specific primers. HSV type-specific LAMP was performed on DNA extracted from HSV-1 (KOS)-, HSV-2 (186)-, VZV (Oka)-, HCMV (AD-169)-, HHV-6B (Z29)-, and HHV-7 (RK)-infected cells. As the LAMP products contained several inverted-repeat structures, positive samples exhibit multiple bands of different sizes upon agarose gel electrophoresis. HSV-1-specific primers amplified only HSV-1 DNA and HSV-2-specific primers amplified only HSV-2 DNA (Fig. 1), and no LAMP products were detected in reactions performed with DNA from other viral infections. We also tested the specificity of the primers by using a turbidity assay. The use of HSV-1-specific primers elevated sample turbidity only in HSV-1 DNA-containing samples. Similar specificity was observed for HSV-2-specific primers (Fig. 1).
FIG. 1.
DNA extracted from HSV-1 (KOS)-, HSV-2 (186)-, VZV (Oka)-, HCMV (AD-169)-, HHV-6 B (Z29)-, and HHV-7 (RK)-infected cells was amplified by using HSV-1- and HSV-2-specific LAMP to determine method specificity. The detection of LAMP products was assessed by agarose gel electrophoresis and turbidity assay using an LA-200. Marker, 123-bp DNA ladder marker.
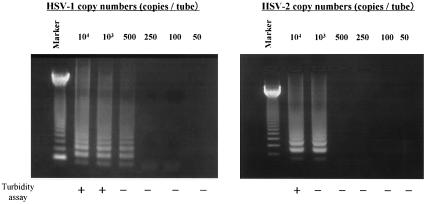
We also determined the sensitivity of this method. Serial dilutions of either pGEMHS1 or pGEMHS2 plasmid containing the target sequences determined the detection limits of HSV type-specific LAMP. The sensitivities of the HSV-1- and HSV-2-specific LAMP determined by agarose gel electrophoresis were 500 and 1,000 copies/tube, respectively (Fig. 2). Detection by the turbidity assay, however, produced sensitivity levels of 1,000 and 10,000 copies/tube for HSV-1- and HSV-2-specific LAMP, respectively.
FIG. 2.
To determine the respective sensitivities of each assay, serial dilutions of pGEMHS1 and pGEMHS2 plasmid DNAs were amplified by the HSV-1- and HSV-2-specific LAMP, respectively. The detection of LAMP products was assessed by agarose gel electrophoresis and turbidity assay using an LA-200. Marker, 123-bp DNA ladder marker.
After these initial validation studies, we determined the reliability of this HSV type-specific LAMP as a method of viral DNA detection from clinical specimens. Eighteen swab samples (sample numbers 1 to 18) collected from patients with either gingivostomatitis or vesicular skin eruptions were examined (Table 2). Neither HSV-1 nor HSV-2 LAMP products were detected in samples (sample numbers 1 to 8) from which no HSV DNA could be detected by real-time PCR. In contrast, HSV-1 LAMP products were detected by agarose gel electrophoresis in the 10 HSV-1-positive samples, correlating perfectly with the results of real-time PCR. When the turbidity assay was used, HSV-1 LAMP products were detected in all but 1 (sample number 10) of these 10 positive samples. No HSV-2 DNA could be detected in these samples by either real-time PCR or LAMP. As rapidity and simplicity of the method are critical for commercial and hospital laboratory use, we investigated the requirement for DNA extraction in HSV type-specific LAMP. Either agarose gel electrophoresis or turbidity assay directly detected HSV-1 LAMP product in all 10 swab samples (sample numbers 9 to 18) (sterilized water), with the exception of sample number 10, regardless of the presence or absence of DNA extraction (Table 2). As genital herpes is another important clinical manifestation of herpes infection, we next examined the reliability of HSV type-specific LAMP for the detection of viral DNA in clinical specimens (culture medium) collected from genital lesions (sample numbers 19 to 23) (Table 2). High copy numbers of HSV-2 DNA (ranging between 641 and 443,963 copies/tube) were detected in these samples by HSV-2 type-specific real-time PCR. Both agarose gel electrophoresis and turbidity assay detected HSV-2 LAMP products in all of the samples. To determine the necessity of DNA extraction by this method, we again tried to detect HSV-2 LAMP products in the samples with or without (culture medium) DNA extraction. In contrast, while HSV-2 LAMP products were detected in samples after DNA extraction, no HSV-2 LAMP products were detected in the samples without DNA extraction (Table 2). To determine if the culture medium contained an inhibitor of LAMP, we attempted to detect HSV LAMP products from both sterilized water and culture medium containing plasmid DNA which contained the target sequences. Although both HSV-1 and HSV-2 LAMP products could be detected in sterilized water containing the target sequences, no LAMP products were detected in culture medium containing these DNAs (data not shown).
TABLE 2.
Comparison between HSV type-specific real-time PCR and HSV type-specific LAMP for detection of HSV DNA in swab samples collected from patients with gingivostomatitis or vesicular skin eruptions (samples 1 to 18) and genital HSV infection (samples 19 to 23)
| Sample no. | HSV-1a
|
HSV-2a
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Real-time PCR (copies/tube) | LAMP (DNA extraction) result
|
LAMP (directb) result
|
Real-time PCR (copies/tube) | LAMP (DNA extraction) result
|
LAMP (directb) result
|
|||||
| Agarose gel electrophoresis | Turbidity assay | Agarose gel electrophoresis | Turbidity assay | Agarose gel electrophoresis | Turbidity assay | Agarose gel electrophoresis | Turbidity assay | |||
| 1 | 0 | − | − | − | − | 0 | − | − | ND | ND |
| 2 | 0 | − | − | − | − | 0 | − | − | ND | ND |
| 3 | 0 | − | − | − | − | 0 | − | − | ND | ND |
| 4 | 0 | − | − | − | − | 0 | − | − | ND | ND |
| 5 | 0 | − | − | − | − | 0 | − | − | ND | ND |
| 6 | 0 | − | − | − | − | 0 | − | − | ND | ND |
| 7 | 0 | − | − | − | − | 0 | − | − | ND | ND |
| 8 | 0 | − | − | − | − | 0 | − | − | ND | ND |
| 9 | 6,350 | + | + | + | + | 0 | − | − | ND | ND |
| 10 | 9,200 | + | − | − | − | 0 | − | − | ND | ND |
| 11 | 29,450 | + | + | + | + | 0 | − | − | ND | ND |
| 12 | 129,650 | + | + | + | + | 0 | − | − | ND | ND |
| 13 | 194,050 | + | + | + | + | 0 | − | − | ND | ND |
| 14 | 283,000 | + | + | + | + | 0 | − | − | ND | ND |
| 15 | 669,950 | + | + | + | + | 0 | − | − | ND | ND |
| 16 | 2,629,400 | + | + | + | + | 0 | − | − | ND | ND |
| 17 | 3,769,650 | + | + | + | + | 0 | − | − | ND | ND |
| 18 | 86,136,700 | + | + | + | + | 0 | − | − | ND | ND |
| 19 | 0 | − | − | ND | ND | 641 | + | + | − | − |
| 20 | 0 | − | − | ND | ND | 22,430 | + | + | − | − |
| 21 | 0 | − | − | ND | ND | 89,929 | + | + | − | − |
| 22 | 0 | − | − | ND | ND | 413,117 | + | + | − | − |
| 23 | 0 | − | − | ND | ND | 443,963 | + | + | − | − |
+, positive; −, negative.
Swab samples were directly (without DNA extraction) used for LAMP reaction. ND, not done.
HSV-1- and HSV-2-specific LAMP specifically amplified HSV-1 and HSV-2 DNA, respectively, exhibiting no cross-reactivity with other human herpesviruses, including another member of the subfamily Alphaherpesvirinae, VZV (Fig. 1). This specificity was confirmed by agarose gel electrophoresis and turbidity assay. Although the capability to distinguish between HSV-1 and HSV-2 infection is not crucial for correct administration of antiviral drugs, this discrimination is important from an epidemiological or public health standpoint. As a consequence of the experiment used for determination of the assay sensitivity, it was suggested that the turbidity assay is a less sensitive detection method than agarose gel electrophoresis, as previously suggested (23). However, the turbidity assay is more appropriate for bedside monitoring due to its ease and rapidity. Additionally, turbidity measurement of LAMP products allows a reduction in operation time and reduces contamination risks because of the absence of agarose gel electrophoresis.
We also evaluated the reliability of HSV type-specific LAMP in the detection of viral DNA from different clinical specimens. Although HSV-1 LAMP products were detected by agarose gel electrophoresis in 10 of the 18 swab samples (sterilized water) collected from patients with vesicular skin lesions and gingivostomatitis suspected as HSV infection, no HSV-2 LAMP products were detected in these samples. All five swab samples (culture medium) collected from the lesions of patients with genital herpes contained HSV-2 LAMP products. These results corresponded well with those from real-time PCR analysis, suggesting that HSV type-specific LAMP is a reliable method for the detection of viral DNA in clinical samples. Although an HSV-1 LAMP product could not be detected in one HSV-1-positive sample by turbidity assay, this inconsistency is probably due to low copy numbers. As the majority of clinical samples (e.g., skin eruptions, oral ulcers, and genital lesions) contain large quantities of viral DNA, the sensitivity of type-specific LAMP by turbidity assay is likely sufficient for the evaluation of most clinical samples. Moreover, all amplification steps are completed within 30 min with an LA-200, and it is a cheaper piece of equipment than that required for real-time PCR, which are major advantages for hospital laboratory use.
Interestingly, when the swabs were collected in sterilized water, HSV LAMP products could be detected directly from the samples without DNA extraction. In contrast, LAMP products could not be detected directly from the culture medium containing viral DNA, regardless of the HSV strain. DNA target sequences, however, became detectable in samples after DNA extraction, suggesting that culture medium contains inhibitors of the LAMP reaction. As the DNA extraction step requires approximately 30 min, omission of DNA extraction could save both time and labor for preparing the samples for LAMP, a major advantage for rapid diagnosis in hospital laboratories. To our knowledge, this is the first report to demonstrate direct amplification of viral DNA from sterilized water containing viral nucleic acids without DNA extraction. We emphasize that the swab should be placed into sterilized water for direct amplification of viral DNA by LAMP. Direct amplification from swab samples in combination with assessment by turbidity assay would accomplish the entire amplification within 30 min. This system would therefore allow large increases in throughput, which is highly relevant for clinical laboratory use. Furthermore, as HSV DNA could be directly detected from swab samples without DNA extraction, the lesions of HSV infection may contain a large quantity of naked viral DNA as well as complete virions. Thus, direct detection of viral DNA from swab samples may be possible by additional DNA amplification methods such as real-time PCR. Further investigation will be necessary to confirm this hypothesis in the future.
Acknowledgments
We thank Eiken Chemical for their contributions to this work. We also thank Akiko Yoshikawa and Maki Sawamura for their technical assistance.
This work was supported in part by a grant-in-aid for the 21st Century COE Program of Medicine of Fujita Health University and the Open Research Center of Fujita Health University from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and also by a grant-in-aid for scientific research from the Ministry of Education, Science, and Culture of Japan.
REFERENCES
- 1.Aldea, C., C. P. Alvarez, L. Folgueira, R. Delgado, and J. R. Otero. 2002. Rapid detection of herpes simplex virus DNA in genital ulcers by real-time PCR using SYBR green I dye as the detection signal. J. Clin. Microbiol. 40:1060-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asano, S., T. Yoshikawa, H. Kimura, Y. Enomoto, M. Ohashi, H. Terasaki, and Y. Nishiyama. 2004. Monitoring of herpesviruses DNA in three cases of acute retinal necrosis by real-time PCR. J. Clin. Virol. 29:206-209. [DOI] [PubMed] [Google Scholar]
- 3.Cone, R. W., A. C. Hobson, J. Palmer, M. Remington, and L. Corey. 1991. Extended duration of herpes simplex virus DNA in genital lesions detected by the polymerase chain reaction. J. Infect. Dis. 164:757-760. [DOI] [PubMed] [Google Scholar]
- 4.Cone, R. W., A. C. Hobson, Z. Brown, R. Ashley, S. Berry, C. Winter, and L. Corey. 1994. Frequent detection of genital herpes simplex virus DNA by polymerase chain reaction among pregnant women. JAMA 272:792-796. [PubMed] [Google Scholar]
- 5.Enosawa, M., S. Kageyama, K. Sawai, K. Watanabe, T. Notomi, S. Onoe, Y. Mori, and Y. Yokomizo. 2003. Use of loop-mediated isothermal amplification of the IS900 sequence for rapid detection of cultured Mycobacterium avium subsp. paratuberculosis. J. Clin. Microbiol. 41:4359-4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Espy, M. J., A. D. Wold, D. J. Jespersen, M. F. Jones, and T. F. Smith. 1991. Comparison of shell vials and conventional tubes seeded with rhabdomyosarcoma and MRC-5 cells for the rapid detection of herpes simplex virus. J. Clin. Microbiol. 29:2701-2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Espy, M. J., J. R. Uhl, P. S. Mitchell, J. N. Thorvilson, K. A. Svien, A. D. Wold, and T. F. Smith. 2000. Diagnosis of herpes simplex virus infections in the clinical laboratory by LightCycler PCR. J. Clin. Microbiol. 38:795-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ihira, M., T. Yoshikawa, Y. Enomoto, S. Akimoto, M. Ohashi, S. Suga, Y. Nishimura, T. Ozaki, Y. Nishiyama, T. Notomi, Y. Ohta, and Y. Asano. 2004. Rapid diagnosis of human herpesvirus 6 infection by a novel DNA amplification method, loop-mediated isothermal amplification. J. Clin. Microbiol. 42:140-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iwamoto, T., T. Sonobe, and K. Hayashi. 2003. Loop-mediated isothermal amplification for direct detection of Mycobacterium tuberculosis complex, M. avium, and M. intracellulare in sputum samples. J. Clin. Microbiol. 41:2616-2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kessler, H. H., G. Muhlbauer, B. Rinner, E. Stelzl, A. Berger, H. W. Dorr, B. Santner, E. Marth, and H. Rabenau. 2000. Detection of herpes simplex virus DNA by real-time PCR. J. Clin. Microbiol. 38:2638-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kimberlin, D. W., F. D. Lakeman, A. M. Arvin, C. G. Prober, L. Corey, D. A. Powell, S. K. Burchett, R. F. Jacobs, S. E. Starr, R. J. Whitley, et al. 1996. Application of the polymerase chain reaction to the diagnosis and management of neonatal herpes simplex virus disease. J. Infect. Dis. 174:1162-1167. [DOI] [PubMed] [Google Scholar]
- 12.Kuboki, N., N. Inoue, T. Sakurai, F. Di Cello, D. J. Grab, H. Suzuki, C. Sugimoto, and I. Igarashi. 2003. Loop-mediated isothermal amplification for detection of African trypanosomes. J. Clin. Microbiol. 41:5517-5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lakeman, F. D., R. J. Whitley, et al. 1995. Diagnosis of herpes simplex encephalitis: application of polymerase chain reaction to cerebrospinal fluid from brain-biopsied patients and correlation with disease. J. Infect. Dis. 171:857-863. [DOI] [PubMed] [Google Scholar]
- 14.Mitchell, P. S., M. J. Espy, T. F. Smith, D. R. Toal, P. N. Rys, E. F. Berbari, D. R. Osmon, and D. H. Persing. 1997. Laboratory diagnosis of central nervous system infections with herpes simplex virus by PCR performed with cerebrospinal fluid specimens. J. Clin. Microbiol. 35:2873-2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mori, Y., K. Nagamine, N. Tomita, and T. Notomi. 2002. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 289:150-154. [DOI] [PubMed] [Google Scholar]
- 16.Nagamine, K., T. Hase, and T. Notomi. 2002. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes 16:223-229. [DOI] [PubMed] [Google Scholar]
- 17.Ndjoyi-Mbiguino, A., F. Ozouaki, J. Legoff, F. X. Mbopi-Keou, A. Si-Mohamed, I. N. Onas, E. Avoune, and L. Belec. 2003. Comparison of washing and swabbing procedures for collecting genital fluids to assess cervicovaginal shedding of herpes simplex virus type 2 DNA. J. Clin. Microbiol. 41:2662-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Notomi, T., H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino, and T. Hase. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parida, M., G. Posadas, S. Inoue, F. Hasebe, and K. Morita. 2004. Real-time reverse transcription loop-mediated isothermal amplification for rapid detection of West Nile virus. J. Clin. Microbiol. 42:257-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pevenstein, S. R., R. K. Williams, D. McChesney, E. K. Mont, J. E. Smialek, and S. E. Straus. 1999. Quantitation of latent varicella-zoster virus and herpes simplex virus genomes in human trigeminal ganglia. J. Virol. 73:10514-10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ryncarz, A. J., J. Goddard, A. Wald, M. L. Huang, B. Roizman, and L. Corey. 1999. Development of a high-throughput quantitative assay for detecting herpes simplex virus DNA in clinical samples. J. Clin. Microbiol. 37:1941-1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Doornum, G. J., J. Guldemeester, A. D. Osterhaus, and H. G. Niesters. 2003. Diagnosing herpesvirus infections by real-time amplification and rapid culture. J. Clin. Microbiol. 41:576-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshikawa, T., M. Ihira, S. Akimoto, C. Usui, F. Miyake, S. Suga, Y. Enomoto, R. Suzuki, Y. Nishiyama, and Y. Asano. 2004. Detection of human herpesvirus 7 DNA by loop-mediated isothermal amplification. J. Clin. Microbiol. 42:1348-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]