Abstract
Epizootic bovine abortion (EBA) is endemic in California's coastal range and the foothill regions of the Sierra Nevada, where it has been the primary diagnosed cause of abortion in beef cattle for >50 years. Investigation of these losses has defined a specific fetal syndrome characterized by late-term abortion or birth of weak or dead calves. Although the unusual clinical presentation and unique fetal pathology associated with EBA have been recognized since the 1950s, the identity of the etiologic agent is unknown. In this study, suppression-hybridization PCR was used to identify a fragment of the 16S rRNA gene of a previously undescribed bacterium in thymus tissue derived from affected fetuses. Phylogenetic analysis revealed that this pathogen was a deltaproteobacterium closely related to members of the order Myxococcales. A specific PCR was subsequently developed to detect the presence of this bacterium in DNA extracted from fetal thymuses. Using histopathology as the definitive diagnosis for EBA, this PCR demonstrated 100% specificity and 88% sensitivity. The bacterium was also detected in the argasid tick Ornithodoros coriaceus, which is the recognized vector of EBA. These data imply a close association between this novel agent and the etiology of EBA.
Epizootic bovine abortion (EBA), sometimes also called foothill abortion, is endemic in California's coastal range and in the foothill regions of the Sierra Nevada. EBA continues to be the primary diagnosed cause of abortion in beef cattle in California and is estimated to be responsible for the loss of 45,000 to 90,000 beef calves annually. EBA is transmitted to susceptible heifers by the bite of the soft argasid tick, Ornithodoros coriaceus (referred to as the pajaroello tick) (14, 25). The tick lives in tree and brush litter in deer and cattle beds, feeds rapidly (in less than an hour under experimental conditions), and then departs the host. Identification of this vector was facilitated by the demonstration that the geographic distribution of the tick largely parallels that of EBA (14, 25).
EBA is defined as a specific fetal syndrome characterized by late-term abortion or birth of weak or dead calves (8). The unique fetal pathology of EBA has classically served as the only definitive basis of diagnosis (9, 10). EBA-associated lesions are chronic, developing progressively over a period of 3 months or more. Gross lesions include mucosal and thymic hemorrhages, lymphadenopathy, splenomegaly, hepatomegaly, and ascites. Histologic examination of fetal tissues, particularly the lymphoid organs, is required to confirm a diagnosis (9, 10). Thymic lesions unique to EBA include a loss of cortical thymocytes and extensive infiltration of macrophages into the medulla. Widespread inflammatory lesions with a vascular orientation are evident in most tissues. Affected fetuses also have markedly elevated immunoglobulin levels, suggestive of a vigorous fetal immune response.
A number of detailed studies have incriminated a variety of microbes as potential causative agents of EBA, including a member of the chlamydial group (originally called psittacosis-lymphogranuloma-venereum), uncharacterized viruses, Borrelia coriaceae, and an uncharacterized spirochete (3, 11, 12, 15, 16, 17, 18, 19, 20, 27, 28, 31, 32). However, upon more detailed examination, all have been excluded as the probable EBA etiologic agent. The recent development of a challenge system in which inoculation of EBA-diseased fetal thymus into pregnant (90 to 120 days of gestation) heifers could result in consistent and predictable transmission of the EBA agent has facilitated efforts to define the causative organism (29). Concurrent treatment with antibiotics at the time of EBA challenge abolished infection of the fetus and therefore implicated a prokaryotic organism as the likely causative agent (29). Although all bacterial culture attempts have been unsuccessful, this breakthrough provided sufficient direction to develop a molecular approach for the identification of the etiologic agent. Similar approaches have been successfully used to identify microbial pathogens, including those responsible for cat scratch fever, Whipple's disease, human ehrlichiosis, hepatitis C, and hantavirus pulmonary syndrome (5, 21).
MATERIALS AND METHODS
Animals.
Fetal necropsy tissues were obtained from a combination of field-infected and experimentally infected heifers, as previously described (29). The heifers were exposed to the agent of EBA by (i) natural exposure (fetuses were submitted by ranchers), (ii) tick feeding under experimental conditions, or (iii) inoculation with tissue homogenate derived from fetuses previously diagnosed as being EBA positive. Inoculation of heifers with tissue homogenates derived from apparently healthy fetuses was used as a control in these experiments. Fetuses from experimentally exposed heifers were obtained either by surgical removal following dam euthanasia (captive bolt) in the third trimester (100 to 140 days postchallenge) or as dead, weak, or healthy calves following natural parturition.
In an additional experiment, six heifers were challenged with aliquots of a single thymus tissue pool previously demonstrated to be capable of transmitting EBA (29). Three of these heifers were simultaneously treated with penicillin for 1 week and tetracycline for 3 weeks, while the remaining three were left untreated to serve as challenge controls (29). Two of the three healthy calves born from antibiotic-treated dams were euthanized immediately (prior to consumption of colostrum), and tissues were collected for PCR analysis.
Collection of tissues.
Necropsy tissues were collected using a sterile technique. Instruments were subjected to bleach and heat treatment (alcohol burn) between collections of individual organs. Tissues collected for histopathology (formalin) included brain, lung, heart, liver, kidney, thymus, lymph nodes, spleen, adrenal, muscle, and digestive tract samples. Serum was also collected, and total immunoglobulin G was determined using radial immunodiffusion (California Animal Health and Food Safety Laboratory, Davis, Calif.). A diagnosis of EBA was established on the basis of histopathology and elevated serum immunoglobulin G, as previously defined (9, 10, 23). Fetal blood (buffy coat) and small pieces of fresh fetal tissues, including brain, thymus, lymph node (both prescapular and mesenteric), spleen, liver, kidney, lung, heart, adrenal gland, tongue, diaphragm, and thigh muscle, were placed in sterile microcentrifuge tubes on dry ice and then stored at −80°C.
Identification of the etiologic agent by suppression hybridization PCR (shPCR).
Two different fetal bovine thymic pools containing tissue homogenates from two EBA-positive (tester) or three EBA-negative (driver) individual cases were prepared by phenol-chloroform extraction (22). One hundred nanograms of extracted tester DNA was subjected to PCR using 16S rRNA primers (2) with linker adapter sequences added to the 5′ ends: 5′-AGC ACT CTC CAG AGA GTT TGA TCM TGG CTC A-3′ and 5′-CCG ACT ATC CAT CTA CCA GGG TAT CTA ATC C-3′. After two to four cycles (94°C for 40 s, 58°C for 40 s, and 72°C for 120 s), unincorporated reaction components were removed (QIAquick PCR purification kit; QIAGEN, Valencia, Calif.), after which the DNA was added to a 10-fold excess of driver DNA that had been subjected to prior restriction enzyme digestion (BamHI; Gibco BRL, Grand Island, N.Y.). Tester template was amplified by PCR using linker-adapter primers (5′-AGC ACT CTC CAG AGA G-3′ and 5′-CCG ACT ATC CAT CTA C-3′). Fifty-microliter reaction mixtures contained 2 mM MgCl2, 50 mM KCl, 10 mM Tris-HCl (pH 8.0), 0.2 mM (each) deoxynucleoside triphosphate, 50 pmol of each primer, and 2.5 U of Taq DNA polymerase (Promega, Madison, Wis.). The amplification conditions were 35 cycles of 94°C for 40 s, 57°C for 40 s, and 72°C for 120 s. It has been hypothesized that driver-tester hybridization suppresses the amplification of ubiquitous bacterial 16S rRNA sequences, allowing unique bacterial sequences present only in the EBA-positive tissue samples to be amplified. Amplicons identified on 1.2% agarose gels were cloned into pGEM-T Easy (Promega), selected by restriction enzyme digestion, and sequenced by chain termination (ABI Prism).
Sequence analysis.
Phylogenetic analysis was performed using maximum-likelihood and distance methods of the PHYLIP package (version 3.57c; Department of Genetics, University of Washington) on a nucleotide alignment generated using CLUSTAL X (version 1.8) (30). Transition/transversion ratios were calculated using TREE-PUZZLE version 5.0 (24).
Detection of etiologic agent of EBA in fetal tissues, bacterial cultures, and ticks by PCR.
DNAs from bovine fetal tissues, pure bacterial cultures, and ticks were prepared using a QIAmp DNA extraction kit (QIAGEN). Bacterial cultures were obtained from either the American Type Culture Collection or the California Animal Health and Food Safety Laboratory System (Table 1). In the tick studies, ticks were collected from locations where EBA was considered to be endemic, including the eastern slope of the Sierra Nevada mountain range in California and southeast Oregon, using dry ice as an attractant (6, 14, 25). The ticks were halved longitudinally with a disposable razor prior to DNA extraction. The DNA was subjected to PCR using EBA agent-specific primers (5′-CAC GTG GAT AAT TTC CCC T-3′ and 5′-GCA AGG TAT TTG CTT GCA TT-3′) designed to amplify a 373-bp region of the 16S rRNA gene fragment. These primer regions showed the most variation compared with other bacterial sequences in multiple-nucleotide alignments. The buffers and the concentrations of the PCR components used were identical to those described above for shPCR. This PCR for the agent of EBA and the control PCRs used to validate negative results (described below) were amplified using identical cycling conditions: 94°C for 180 s, followed by 35 cycles of 94°C for 40 s, 61°C for 40 s, and 72°C for 40 s. Chain elongation was extended to 10 min during the final cycle, after which the reactions were held at 4°C until the products were analyzed by agarose gel electrophoresis.
TABLE 1.
Analysis of EBA PCR specificity
| Organism | ATCC no.a | PCR resultb
|
|
|---|---|---|---|
| 16S universal primers | EBA primers | ||
| EBA fetal thymus | NA | + | + |
| B. coriaceae | 43381 | + | − |
| Streptococcus agalactiae | 12386 | + | − |
| Staphylococcus aureus | 25923 | + | − |
| Listeria monocytogenes | 35152 | + | − |
| Campylobacter jejuni | 29428 | + | − |
| Salmonella group D1 | NA | + | − |
| Pseudomonas aeruginosa | 27853 | + | − |
| Pasteurella multocida | NA | + | − |
| Mannheimia haemolytica | NA | + | − |
| Moraxella bovis | 10900 | + | − |
| Escherichia coli | NA | + | − |
| Arcanobacterium pyogenes | 19411 | + | − |
| P. cellulosum subsp. fulvum Mishustin | 25523 | + | − |
ATCC, American Type Culture Collection. NA, not applicable (these cultures had no ATCC designation).
+, positive; −, negative.
Since nucleic acid samples extracted from ticks can often exhibit activity inhibitory to PCR, it was vital to establish an internal amplification control to discriminate true- and false-negative PCR results. A 1,660-bp fragment (AF096274) of the 18S rRNA gene of O. coriaceus was amplified, cloned, and sequenced using conserved primers based on 18S rRNA genes of members of the Argasidae tick family: (5′ CTT GTC TCA AAG ATT AAG CC 3′ and 5′ AAG ACC TCA CTA AAT CAT CC 3′). A control PCR was developed using the primers 5′-CAT CAT GCC TTC TAT CCT C-3′ and 5′-GCT CTC AAT CTG TCA ATC C-3′, which amplify a 561-bp fragment of this 18S rRNA gene of O. coriaceus. Parallel control reactions were also performed for bovine tissues using the primers (5′-TCT CTC TGC AGC ACA TTT CCT-3′ and 5′-TCA CCT CGC CGC TGC AC-3′) for exon 2, BoLA (1), to ensure that the DNA extracted was not degraded and did not contain substances inhibitory to PCR.
Nucleotide sequence accession numbers.
The nucleotide sequence data reported in this paper have been submitted to GenBank and have been assigned the following accession numbers: AF503916 (16S rRNA fragment of the EBA agent), AF096274 (18S rRNA fragment of O. coriaceus), and AF016401 (16S rRNA fragment of the novel betaproteobacterium).
RESULTS
The present study employed an shPCR technique in an attempt to identify a bacterial agent present in fetal thymic tissues collected from EBA cases. This approach was used because preliminary cloning experiments using universal bacterial primers yielded sequences identical to those of Pseudomonas sp. and Escherichia coli. Since subsequent PCR showed that these agents were present in both EBA-positive and -negative tissues, their presence was assumed to be the result of postmortem contamination of the samples. The use of shPCR allowed prokaryotic sequences derived from bovine tissues to be amplified in preference to contaminating bacteria. Furthermore, since this technique utilizes gene-specific primers directed toward the commonly sequenced 16S rRNA gene, phylogenetic comparisons with other bacteria were possible. Three separate shPCR experiments were performed using the pooled positive (tester) and negative (driver) material, yielding 17 plasmid clones containing inserts of approximately the correct size (Table 2). From the first experiment, sequences corresponding to a novel betaproteobacterium and Ochrobactrum anthropi were identified. Two specific PCRs were subsequently developed to detect these agents separately in a panel of individual bovine thymus tissues derived from five EBA-positive and four EBA-negative animals. The primer pairs used were 5′-TAA CAG GCC TTT CGG GGT GCT GA-3′ and 5′-CCC CTG TTC AGG AAA GCG ATT TCG-3′ or 5′-CGT ACC TTT TGC TAC GGA ATA ACT CAG-3′ and 5′-ACC ATA CTC AAG ACT TCC AGT ATC AAA G-3′ for the detection of the betaproteobacterium and O. anthropi, respectively. Although these agents were detected in some of the DNA samples, the pattern for neither bacterium paralleled the clinical and histopathological diagnosis of EBA (data not shown).
TABLE 2.
Summary of shPCR experiments used to identify the etiologic agent of EBA
| Agent | No. of clones
|
Identity of clone (GenBank accession no.) | Comments | ||
|---|---|---|---|---|---|
| Expt 1 | Expt 2 | Expt 3 | |||
| 1 | 3 | Novel β-proteobacterium (AF016401) | 89.2% identity to AY219713 | ||
| 2 | 1 | O. anthropi | 100% identity to D12794 | ||
| 3 | 2 | 4 | Novel δ-proteobacterium (AF382826) | Putative agent of EBA | |
| 4 | 1 | Novel bacterium in phylum Bacteroidetes | 81.7% identity to AJ252610a | ||
| 5 | 3 | Novel β-proteobacterium | 98.0% identity to AF125877a | ||
| 6 | 1 | Corynebacterium sp. | 99.2% identity to AJ438048a | ||
| 7 | 1 | Novel β-proteobacterium | 99.0% identity to AB069809a | ||
| 8 | 1 | Novel α-proteobacterium | 95.3% identity to D13947a | ||
Partial sequence of insert.
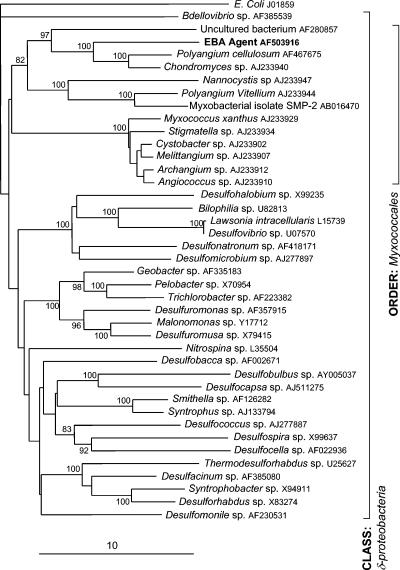
Second and third repetitions of the shPCR experiments were performed. In these experiments, an additional 13 clones were sequenced. Of these plasmid sequences, six were identical, containing a 767-bp fragment of the 16S rRNA gene of a previously unrecognized deltaproteobacterium (AF382826). Partial sequencing of the remaining clones (Table 2) revealed sequences similar to those of a Corynebacterium sp., two additional novel betaproteobacteria, a novel alphaproteobacterium, and a bacterium possibly in the phylum Bacteroidetes. The deltaproteobacterium corresponding to AF382826 was the most frequently recognized agent and the only sequence identified in multiple experiments. A specific PCR was developed (described in Materials and Methods) to detect this deltaproteobacterium in bovine tissues. Initial results using the small panel of nine bovine tissues (see above) showed complete concordance with the EBA status of the animals. In light of these data, this bacterium was chosen as a primary candidate for the putative agent of EBA. This 767-bp sequence was extended to 1,480 bp (AF503916) by PCR using gene-specific and conserved 16S rRNA primers. Nucleotide analysis (http://www.ebi.ac.uk/fasta33/nucleotide.html) demonstrated that this sequence had the closest identity (89.4%) to the 16S rRNA sequence of Polyangium cellulosum (AF467675). Phylogenetic analysis (Fig. 1) showed that this novel bacterium is a member of the Deltaproteobacteria, sharing the closest relationships with members of the order Myxococcales.
FIG. 1.
Consensus phylogram of 2,000 neighbor-joining trees generated for a 1,360-bp fragment of the 16S rRNA gene of the putative agent causing EBA compared with sequences from 39 representative genera (7) of the class Deltaproteobacteria. GenBank accession numbers are shown. Escherichia coli, a member of the class Gammaproteobacteria, was included as an outgroup in this analysis. Significant (>70%) bootstrap confidence values for tree nodes are shown. The scale represents percent nucleotide substitutions per site.
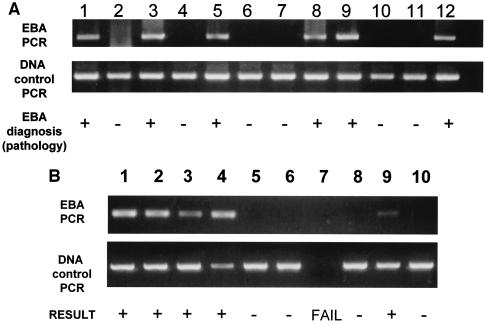
PCR was used to detect the presence of the putative EBA agent in DNA extracted from 68 fetal thymuses (Fig. 2A and Table 3). This PCR analysis was performed blind using coded DNA samples and demonstrated 100% specificity and 88% sensitivity using histopathology as the definitive diagnosis. A combination of direct sequencing (373 bp from 25 EBA-positive fetuses representing 16 field cases, four experimental tick transmissions, and five experimental tissue transmissions, the last using thymus homogenate derived from an EBA-positive fetus) and Southern blotting of these amplicons was used to confirm PCR specificity. Furthermore, the PCR failed to amplify DNA extracted from pure cultures of a selection of bacterial pathogens typically encountered in veterinary medicine or even P. cellulosum, a close phylogenetic relative of the agent of EBA (Table 1).
FIG. 2.
Representative 1.2% agarose gels showing the identification of the putative EBA agent in DNA extracted from (A) diseased bovine fetal thymuses and (B) selected free-ranging O. coriaceus isolates collected in California and Oregon. The DNA in panel B, lane 7, was unsuitable for analysis, as no PCR product was observed using primers for the tick 18S rRNA gene.
TABLE 3.
Analysis of EBA PCR sensitivity and specificity
| Heifer exposure | n | Diagnosisa | No. PCR+/no. PCR− |
|---|---|---|---|
| Diagnostic submission | 31 | 18 EBA | 15/3 |
| 12 non-EBA | 0/12 | ||
| 1 inconclusive | 0/1 | ||
| Experimental tick bite | 5 | 3 EBA | 2/1 |
| 1 negative | 0/1 | ||
| 1 inconclusive | 0/1 | ||
| Inoculation with EBA necropsy tissue | 25 | 21 EBA | 19/2 |
| 4 negative | 0/4 | ||
| Inoculation with negative control necropsy tissue | 4 | 2 normal | 0/2 |
| 2 inconclusive | 0/2 | ||
| No exposure | 3 | 3 normal | 0/3 |
All animals were subjected to standard gross, histopathologic, and microbiologic examinations. Non-EBA diagnoses included bovine viral diarrhea, neosporosis, Haemophilus somnus, and unknown etiologies.
Three heifers experimentally challenged with thymus tissue and simultaneously treated with antibiotics demonstrated the EBA agent to be prokaryotic (29). These three heifers gave birth to healthy calves; two were euthanized immediately (prior to consumption of colostrum) for PCR analysis; all tissues were PCR negative. In contrast, the three challenge control fetuses whose dams did not receive antibiotics all presented with classic EBA-associated lesions, and their tissues were PCR positive.
Multiple tissues from a subset of the experimentally infected fetuses (n = 7) were used to further define the tissue distribution of the EBA agent (Table 4). Thymuses from all seven fetuses were PCR positive. Six of seven lymph nodes (both prescapular and mesenteric) and six of the seven spleens were also positive. All other tissues tested, including kidney, liver, brain, lung, heart, adrenal gland, tongue, diaphragm, thigh muscle, and blood (buffy coat), were variably positive (Table 4). These preliminary results suggest that the agent is preferentially associated with lymphoid tissues.
TABLE 4.
PCR-based tissue distribution of EBA agent in diseased fetuses
| Tissue | Presence of agent in fetusa:
|
||||||
|---|---|---|---|---|---|---|---|
| 1c | 2c | 3c | 4c | 5c | 6c | 7b | |
| Thymus | + | + | + | + | + | + | + |
| Spleen | + | − | + | + | + | + | + |
| Liver | + | − | + | − | − | − | + |
| PSLNd | + | − | + | + | + | + | + |
| MesLNe | + | − | + | + | + | + | + |
| Kidney | + | − | + | − | + | + | + |
| Brain | + | − | + | − | NAf | + | + |
| Lung | + | − | − | − | + | − | + |
| Heart | + | − | + | − | − | + | + |
| Adrenal gland | + | − | − | − | − | + | + |
| Tongue | + | − | + | − | − | + | + |
| Diaphragm | + | + | + | − | + | − | + |
| Caudal thigh muscle | + | + | + | − | − | + | − |
| Buffy coat | + | − | + | − | − | + | + |
+, present; −, absent.
Fetus infected via tick feeding.
Three different thymuses were used for the challenge inoculum for fetuses 1; 2 to 4; and 5 and 6.
Prescapular lymph node.
Mesenteric lymph node.
NA, tissue unavailable for analysis.
In addition to identifying this novel agent in diseased fetal bovine tissues, PCR has also demonstrated the presence of the deltaproteobacterium in the tick vector, O. coriaceus (Fig. 2B). Preliminary data demonstrated the EBA agent to be present in 4 of 11 nymphs tested and 2 of 8 female ticks tested; The nucleic acid sequences were the same (five were directly sequenced to confirm the identity of the PCR product) as that identified in EBA fetal bovine tissues.
DISCUSSION
This study demonstrates a strong association between the presence of a novel deltaproteobacterium and the development of EBA. The agent was detected in thymic tissues from affected fetuses and in the argasid tick vector of EBA. In the absence of a cultivable agent, it is proposed that these molecular data are sufficient evidence to classify this bacterium as the etiologic agent of EBA. The majority of guidelines that have been suggested for sequence-based determination and incriminating of a presumptive microorganism in disease (5) have been satisfied. However, one of the correlates proposed by Fredericks and Relman (5) that was not fulfilled was that “the nature of the microorganism inferred from the available sequence should be consistent with the known biological characteristics of that group of organisms.” This was certainly not true for the bacterium described in this study. The phylogenetic location of the putative EBA etiologic agent was surprising, since none of the closest relatives have been demonstrated to be mammalian pathogens. Furthermore, although the closest relatives are members of the proposed suborder “Sorangineae” (26), phylogenetic analysis suggested that the bacterium may be distinct from all previously characterized Myxococcales. The novel bacterium described in this paper represents only the second member of the delta subdivision of the class Proteobacteria to be a mammalian pathogen (the first was Lawsonia intracellularis [4, 13], the causative agent of porcine proliferative enteropathy).
The PCR developed in this study proved to be a useful tool, allowing the detection of the novel bacterium in DNA extracted from bovine tissues and ticks. The inability to identify the agent in all ticks would be consistent with the low-level infection achieved by experimental transmission of EBA to cattle by tick bite. Previous studies (14, 25, 29) suggest that feeding 100 to 500 ticks on susceptible pregnant heifers will result in ∼50% disease transmission.
In summary, this work demonstrates a close relationship between a novel bacterium and EBA. Efforts are under way to define additional gene sequences of this unique pathogen, to visualize the agent in affected tissues, and to visually link the rRNA sequence to the bacterium using in situ hybridization techniques on diseased fetal necropsy tissue. In addition, with the development of a diagnostic PCR, efforts can be initiated to better define the ecology of EBA, including the identification of additional reservoirs of the bacterium and defining the association between the EBA agent and the tick vector.
Acknowledgments
This study was supported in part by funds provided by a USDA NRI-Competitive Grant (2002-35204-12365); the Center for Food Animal Health, School of Veterinary Medicine, University of California, Davis; the California Cattlemen's Association; and USDA Western Regional Research Project W-112.
We acknowledge the assistance of Mike Oliver for tick collection and Richard Gniewek and Svetlana Khaiboullina for assistance in identifying the agent in ticks and bovine tissues, respectively.
REFERENCES
- 1.Aldridge, B. M., S. M. McGuirk, R. J. Clark, L. A. Knapp, D. I. Watkins, and D. P. Lunn. 1998. Denaturing gradient gel electrophoresis: a rapid method for differentiating BoLA-DRB3 alleles. Anim. Genet. 29:389-394. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanchard-Channell, M., and J. L. Stott. 1991. Characterization of Borrelia coriaceae antigens with monoclonal antibodies. Infect. Immun. 59:2790-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dale, C. J., E. K. Moses, C. C. Ong, C. J. Morrow, M. B. Reed, D. Hasse, and R. A. Strugnell. 1998. Identification and sequencing of the groE operon and flanking genes of Lawsonia intracellularis: use in phylogeny. Microbiology 144:2073-2084. [DOI] [PubMed] [Google Scholar]
- 5.Fredericks, D. N., and D. A. Relman. 1996. Sequence-based identification of microbial pathogens: a reconsideration of Koch's postulates. Clin. Microbiol. Rev. 9:18-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia, R. 1962. Carbon dioxide as an attractant for certain ticks (Acarina: Argasidae and Ixodidae). Ann. Entomol. Soc. Am. 13:605-606. [Google Scholar]
- 7.Garrity, G. M., M. Winters, and D. B. Searles. 2001. Taxonomic outline of the prokaryotic genera, p. 16-17. In Bergey's manual of systematic bacteriology, 2nd ed. Springer-Verlag, New York, N.Y. [Online.]
- 8.Howarth, J. A., J. E. Moulton, and L. M. Frazier. 1956. Epizootic bovine abortion characterized by foetal hepatopathy. J. Am. Vet. Med. Assoc. 128:441-449. [PubMed] [Google Scholar]
- 9.Kennedy, P. C., H. J. Olander, and J. A. Howarth. 1960. Pathology of epizootic bovine abortion. Cornell Vet. 50:417-429. [PubMed] [Google Scholar]
- 10.Kennedy, P. C., A. P. Casaro, P. B. Kimsey, R. H. BonDurant, R. B. Bushnell, and G. M. Mitchell. 1983. Epizootic bovine abortion: histogenesis of the fetal lesions. Am. J. Vet. Res. 44:1040-1048. [PubMed] [Google Scholar]
- 11.Kimsey, P. B., P. C. Kennedy, R. B. Bushnell, A. P. Casaro, R. H. BonDurant, M. N. Oliver, and J. W. Kendrick. 1983. Studies on the pathogenesis of epizootic bovine abortion. Am. J. Vet. Res. 44:1266-1271. [PubMed] [Google Scholar]
- 12.Lane, R. S., W. Burgdorfer, S. F. Hayes, and A. G. Barbour. 1985. Isolation of a spirochete from the soft tick, Ornithodoros coriaceus, possible agent of epizootic bovine abortion. Science 230:85-87. [DOI] [PubMed] [Google Scholar]
- 13.Lawson, G. H. K., S. McOrist, S. Jasni, and R. A. Mackie. 1993. Intracellular bacteria of porcine proliferative enteropathy: cultivation and maintenance in vitro. J. Clin. Microbiol. 31:1136-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loomis, E. C., E. T. Schmidtmann, and M. N. Oliver. 1974. A summary review on the distribution of Ornithodoros coriaceus Koch in California (Acarina: Argasidae). Calif. Vector Views 21:57-62. [Google Scholar]
- 15.McKercher, D. G., E. M. Wada, E. A. Robinson, and J. A. Howarth. 1966. Epizootiologic and immunologic studies of epizootic bovine abortion. Cornell Vet. 56:433-450. [PubMed] [Google Scholar]
- 16.McKercher, D. G., E. A. Robinson, E. M. Wada, J. K. Saito, and C. D. Franti. 1969. Vaccination of cattle against epizootic bovine abortion. Cornell Vet. 59:210-226. [PubMed] [Google Scholar]
- 17.McKercher, D. G., G. L. Crenshaw, E. M. Wada, C. M. Mauris, and C. D. Franti. 1973. Vaccination against epizootic bovine (chlamydial) abortion. J. Am. Vet. Med. Assoc. 163:889-893. [Google Scholar]
- 18.McKercher, D. G., G. L. Crenshaw, J. H. Theis, E. M. Wada, and C. M. Mau. 1973. Experimentally induced immunity to chlamydial abortion of cattle. J. Infect. Dis. 128:231-234. [DOI] [PubMed] [Google Scholar]
- 19.Osebold, J. W., R. Spezialetti, M. B. Jennings, R. F. Pritchett, and R. B. Bushnell. 1986. Congenital spirochetosis in calves: association with epizootic bovine abortion. J. Am. Vet. Med. Assoc. 188:371-376. [PubMed] [Google Scholar]
- 20.Osebold, J. W., B. I. Osburn, R. Spezialetti, R. B. Bushnell, and J. L. Stott. 1987. Histopathologic changes in bovine fetuses after repeated reintroduction of a spirochete-like agent into pregnant heifers: association with epizootic bovine abortion. Am. J. Vet. Res. 48:627-633. [PubMed] [Google Scholar]
- 21.Relman, D. A. 1998. Detection and identification of previously unrecognized microbial pathogens. Emerg. Infect. Dis. 4:382-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed., p. 6.23-6.27. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Sawyer, M., B. I. Osburn, H. D. Knight, and J. W. Kendrick. 1973. A quantitative serologic assay for diagnosing congenital infections of cattle. Am. J. Vet. Res. 34:1281-1284. [PubMed] [Google Scholar]
- 24.Schmidt, H. A., K. Strimmer, M. Vingron, and A. von Haeseler. 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18:502-504. [DOI] [PubMed] [Google Scholar]
- 25.Schmidtmann, E. T., R. B. Bushnell, E. C. Loomis, M. N. Oliver, and J. H. Theis. 1976. Experimental and epizootiologic evidence associating Ornithodoros coriaceus Koch (Acari: Argasidae) with the exposure of cattle to epizootic bovine abortion in California. J. Med. Entomol. 13:292-299. [DOI] [PubMed] [Google Scholar]
- 26.Sproer, C., H. Reichenbach, and E. Stackebrandt. 1999. The phylogenetic position of Serratia, Buttiauxella and some other genera of the family Enterobacteriaceae. Int. J. Syst. Bacteriol. 49:1255-1262. [DOI] [PubMed] [Google Scholar]
- 27.Storz, J., D. G. McKercher, J. A. Howarth, and O. C. Straub. 1960. Epizootic bovine abortion. J. Am. Vet. Med. Assoc. 137:509-514. [Google Scholar]
- 28.Storz, J., and D. G. McKercher. 1962. Etiological studies on epizootic bovine abortion. Zentbl. Veterinarmed. 9:520-541. [Google Scholar]
- 29.Stott, J. L., M. T. Blanchard, M. Anderson, J. Maas, R. L. Walker, P. C. Kennedy, B. B. Norman, R. H. BonDurant, M. N. Oliver, D. Hanks, and M. R. Hall. 2002. Experimental transmission of epizootic bovine abortion (foothill abortion). Vet. Microbiol. 88:161-173. [DOI] [PubMed] [Google Scholar]
- 30.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wada, E. M., D. G. McKercher, G. Castrucci, and J. H. Theis. 1976. Preliminary characterization and pathogenicity studies of a virus isolated from ticks (Ornithodoros coriaceus) and from tick-exposed cattle. Am. J. Vet. Res. 37:1201-1206. [PubMed] [Google Scholar]
- 32.Zingg, B. C., and R. B. LeFebvre. 1994. Polymerase chain reaction for detection of Borrelia coriaceae, putative agent of epizootic bovine abortion. Am. J. Vet. Res. 55:1509-1515. [PubMed] [Google Scholar]