ABSTRACT
Amyloids and amyloid-based prions are self-perpetuating protein aggregates which can spread by converting a normal protein of the same sequence into a prion form. They are associated with diseases in humans and mammals, and control heritable traits in yeast and other fungi. Some amyloids are implicated in biologically beneficial processes. As prion formation generates reproducible memory of a conformational change, prions can be considered as molecular memory devices. We have demonstrated that in yeast, stress-inducible cytoskeleton-associated protein Lsb2 forms a metastable prion in response to high temperature. This prion promotes conversion of other proteins into prions and can persist in a fraction of cells for a significant number of cell generations after stress, thus maintaining the memory of stress in a population of surviving cells. Acquisition of an amino acid substitution required for Lsb2 to form a prion coincides with acquisition of increased thermotolerance in the evolution of Saccharomyces yeast. Thus the ability to form an Lsb2 prion in response to stress coincides with yeast adaptation to growth at higher temperatures. These findings intimately connect prion formation to the cellular response to environmental stresses.
KEYWORDS: actin, amyloid, heat shock, Lsb1, Lsb2, prion, Sup35, ubiquitin, yeast (Saccharomyces cerevisiae)
INTRODUCTION
This extra-view is related to our recent paper describing a new yeast prion that can carry cellular memories of stress.1 Highly ordered fibrous cross-β protein aggregates (amyloids) and/or oligomers (protofibrils) involved in amyloid generation are associated with a variety of human and animal diseases, including Alzheimer (AD), Parkinson (PD) and Huntington diseases.2,3 Amyloids spread by immobilizing the protein molecules of the same sequence into amyloid fibrils and converting them into an amyloid form. Frequently, amyloids can be transmitted between cells within one organism, and sometimes even between organisms, resulting in protein-based infection.3 The prototype of an infectious amyloid is the prion protein (PrP), associated with transmissible spongiform encephalopathies (TSEs), such as sheep scrapie, mad cow disease, and human Creutzfeldt-Jakob and kuru diseases.4 Recent data show that many amyloid diseases possess at least some prion-like properties. 5-7 Many amyloidoses, including the majority of the cases of AD and PD, are idiopathic and not associated with any DNA mutations.8,9 Although environmental agents have been linked to amyloidoses, e. g. certain pesticides to PD10 and certain metal ions to AD,11 systematic information about environmental factors promoting or inhibiting amyloids formation is lacking, resulting in the absence of clearly defined prophylactic strategies.
Notably, some amyloids play biologically positive functions. Amyloids help in the attachment of bacterial or fungal cells to the substrate and/or to other cells, possess structure-forming properties (e. g. silks), are used for storage (e. g. peptide hormones), and form scaffolds of covalent polymers, such as melanin.12-18 Possibly the most fascinating example of the biological consequences of amyloid-style polymerization is the role of self-perpetuating oligomers of CPEB protein in long-term memory.12-15,19 Indeed, amyloids and prions are capable of fixing and propagating change in protein conformation, making them efficient “memory machines.” How then can a prion-like mechanism be used for maintaining a memory of environmental changes?
A simple tractable model system is needed to investigate the impact of environmental factors on amyloids. Endogenous amyloids of the yeast Saccharomyces cerevisiae (termed “yeast prions”) provide such a system.20 About 10 yeast proteins convert to the amyloid-based prion form in vivo, and many more candidate prion proteins are suspected.21-23 Some yeast prions control detectable phenotypic traits, making them an excellent model system for studying general mechanisms of amyloid formation and propagation. At least 1/3 of wild or industrial isolates of yeast possess traits that are inherited in a prion-like fashion.24 Most yeast prion proteins contain domains (usually Q/N-rich), responsible for prion properties and termed prion domains, or PrDs.21 Notably, yeast cells can also form multimolecular assemblies that are not heritable but are maintained by a mother cell and carry a “memory” of certain events; memory of the deceptive mating encounter depends on the Whi3 protein.25 Prion-like QN-rich domains are involved in the formation of a maternally retained super-assembly in case of Whi3. This indicates that stably inherited yeast prions may represent only a tip of the iceberg, providing extreme examples of phenomena involved in a broader spectrum of cellular memories.
In spite of a heated discussion regarding the biological roles of prions, it is now becoming clear that both pathogenic,16,26,27 and beneficial22,28 prions can be found in yeast and other fungi. Some yeast prions control phenotypic traits that can be easily monitored.20,21 For example, [PSI+] a prion form of the yeast translation termination factor Sup35 (eRF3), causes a partial defect of translation termination due to aggregation of a termination factor. Formation of the prion results in translational readthrough of nonsense-codons (so called nonsense-suppression), allowing prion detection by growth on selective medium and/or by color on complete medium in the specially designed yeast strains.29 The availability of such convenient assays makes yeast a powerful system for studying the mechanisms of prion formation and maintenance.
HETEROLOGOUS PRION CROSS-SEEDING
Formation of a yeast prion de novo can be induced by a transient overproduction of the prion-forming protein or its PrD.20,30 However, typically this process is efficient only if there is another Q/N-rich protein aggregate in the same cell.31-34 For example, de novo formation of the Sup35 prion, [PSI+] after transient overproduction of Sup35 protein is promoted by the presence of a non-Mendelian element named [PIN+], for [PSI+] inducibility.31,32 This [PIN+] element, initially defined in genetic experiments, was later identified as a prion form of Rnq1 protein and named [RNQ+].32,33,35 Furthermore, overproduction of other aggregating Q/N-rich proteins can substitute for [RNQ+] and confer the [PSI+] inducibility) (Pin+) phenotype to the yeast cells lacking the Rnq1 prion.32-34,36,37 Some of these proteins are known to form prions by themselves, however such evidence is lacking for others.21,38 Increased amyloid formation by one protein in the presence of the amyloid form of another protein has also been reported or suspected in mammalian systems, for instance, for tau and Aβ proteins in case of AD.6,39
The molecular foundations of cross-seeding on prion formation by other prion proteins are still insufficiently studied. Proposed models include: a) direct heterologous cross-seeding of one prion by another, and b) sequestration of folding cofactors (such as chaperones) that promotes misfolding and prion formation.32,33 Evidence exists for both mechanisms, and it is possible that different prions may act in different ways.40
One of QN-rich proteins whose overproduction is shown to promote formation of the [PSI+] prion by excess Sup35 in strains lacking the [RNQ+] prion is the Lsb2 protein, also called Pin332,41. Lsb2 is a stress-inducible short-lived protein that is degraded via the ubiquitin-proteasome system.41 Lsb2 interacts with overproduced Sup35 protein and promotes its accumulation and aggregation at actin patches. Apparently Lsb2 can also promote formation of other QN-rich prions, as [RNQ+] isolates have also been obtained from cultures overproducing Lsb241. Lsb2 is associated with actin cytoskeleton through interaction with the Las17 protein (yeast homolog of WASP)42 via SH3 domains. Mutations in Lsb2 abolishing this interaction also abolish its ability to form cytologically detectable puncta, promote aggregation of Sup35 and formation of the Sup35 prion.41 Our new (previously unpublished) data confirm that excess Lsb2 is not able to promote [PSI+] induction in the las17Δ strain (Fig. 1). Thus, association of Lsb2 with actin cytoskeleton via Las17 is crucial for prion induction by Lsb2. Apparently, Lsb2 confines misfolded Sup35 (and possibly some other misfolded proteins) to the cytoskeleton-associated deposits, thus increasing local concentrations of Sup35 and facilitating prion nucleation. Notably, Lsb2 has a close paralog, Lsb1, which also interacts with Las17 but is not capable of promoting prion formation by Sup35.41,43
Figure 1.
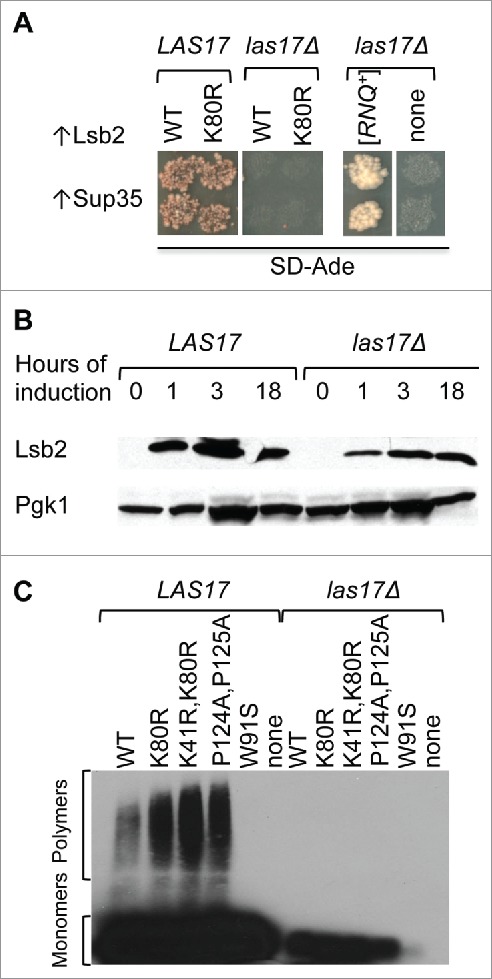
Association with Las17 is required for prion induction by Lsb2. A. Lsb2 cannot induce formation of prion [PSI+] by overexpression of Sup35 in the strain depleted of Las17 (las17Δ). Depletion of Las17 does not affect formation of [PSI+] by overexpression of Sup35 in the presence of prion aggregates of Rnq1 protein ([RNQ+]). [PSI+] formation is manifested by growth on SD-Ade medium. B. Total levels of Lsb2 protein are slightly decreased in Las17 strain as detected by western blotting. Different time of Lsb2 induction from plasmid copper inducible promoter is indicated. C. Wild type Lsb2 and more stable ubiquitination deficient mutants (K80; K80R, K41R; P124A, P125A) do not form detergent resistant amyloid-like aggregates in the strain depleted of Las17 as detected by SDD-AGE. Lsb2 W91S mutant unable to bind Las17 does not form amyloid-like aggregates in the presence of Las17 (WT).
Prion Cross-Seeding by LSB2 is due to Formation of a Metastable Prion, [LSB+]
Despite its ability to cross-seed other proteins into a prion state, earlier studies failed to detect prion formation by Lsb2 itself.38 In our recent paper,1 we have shown that transient overexpression of Lsb2 results in generation of a fraction (3–4%) of the cells that retain ability to induce conversion of Sup35 into [PSI+] after overexpression of Lsb2 is turned off. This prion-inducing phenotype was inherited through an indefinite number of mitotic divisions, however only in a fraction of the progeny, indicating a low mitotic stability. The prion-inducing phenotype was dominant and inherited in a non-Mendelian fashion through mating and meiosis, being retained by a fraction (17%) of ascospores. Semi-denaturing detergent agarose gel electrophoresis, SDD-AGE44 confirmed that cultures that inherited the prion inducing phenotype contain a small but detectable fraction of Lsb2 protein in the form of detergent-resistant polymers, similar to those formed (in a larger amount) in the cells containing overproduced Lsb21. We have designated this heritable aggregated state of Lsb2 protein as a metastable prion, [LSB+]. The low stability of the [LSB+] prion could be due at least in part to the proteolytic instability of Lsb2 protein, resulting in low levels of this protein in non-stressed cells. Indeed, mutants of Lsb2 that are deficient in ubiquitination, form [LSB+] aggregates that are inherited by a larger fraction of cells, compared with the aggregates formed by a wild type protein.1 This agrees with our finding that the efficiency of [PSI+] induction by Lsb2 is increased by a defect in protein ubiquitination.41 Also, in agreement with previous data45 we have shown that the replacement of 8Q stretch by 8N stretch in the Lsb2 protein significantly increases prion-inducing efficiency and decreases average size (as typical of more efficiently propagated prions) of the [LSB+] aggregates. This agrees with the findings that an increase in the proportion of N residues versus Q residues favors more efficient prion propagation by QN-rich proteins.
Effects of Environmental Stresses on Yeast Prions
Propagation of yeast prions in cell division occurs via repetitive cycles of fibril fragmentation and growth.20,46,47 Prion fragmentation depends strictly on the cellular chaperone disaggregation machinery, composed of the Hsp104, Hsp70 and Hsp40 proteins.20,23,48-52 Certain environmental conditions destabilize prions and cause prion loss, possibly due to alterations in the chaperone balance.53,54 Different prions, and even different variants (“strains”) of a prion formed by one and the same protein may exhibit different responses to stress. Probably the best studied case of prion destabilization by an environmental stress is an effect of a short-term mild heat shock (39–42°C) on some “weak” variants of the Sup35 prion, [PSI+].55 Yeast [PSI+] cells, exponentially growing at 25–30°C, then heat shocked for a short period of time (30–60 min) and shifted back to normal temperature, produce a small fraction of colonies that have lost prion and a large fraction (30–40%) of mosaic colonies, pointing to high frequency of prion loss in cell divisions following stress treatment. If yeast cells are incubated at high temperature for longer periods of time (2–4 hrs), prion destabilization is decreased. Maximal destabilization coincides with the period of maximal imbalance between the Hsp104 chaperone (that is present at very low background levels in exponential cultures and quickly accumulated during heat shock) and the Hsp70-Ssa chaperone (that takes a longer time to achieve a significant fold increase over relatively high background levels). This agrees with data showing that the stoichiometric Hsp104/70/40 complexes promote prion fragmentation and proliferation, while a selective increase in Hsp104 leads to [PSI+] loss.56,57 Analysis of individual cells indicates that prion loss is associated with an asymmetric distribution in post-heat shock cell divisions,43,55,58 that was explained either by asymmetry in distribution of prion aggregates per se20,55 or by asymmetric distribution of the chaperone Hsp10458. These explanations are not mutually exclusive, as Hsp104 is known to be associated with asymmetrically distributed aggregates after stress,59 and excess Hsp104 promotes asymmetric partitioning of the [PSI+] prion units.60
Importantly, Lsb2 protein is induced during the short-term heat shock,41,61 while its paralog Lsb1 is released from the association with ER membrane into a cytosol via heat shock inducible proteolytic processing.43 In the absence of either Lsb1 or Lsb2, heat shock mediated destabilization of [PSI+] is significantly increased.43 Moreover, cells lacking both Lsb proteins do not show an amelioration of prion destabilization during a longer exposure to high temperature. These data indicate that Lsb proteins antagonize the effect of stress on prion aggregates and/or asymmetric distribution of prion/chaperone complexes in cell divisions after stress.
Various environmental stresses were also reported to increase de novo formation of yeast prions, such as [URE3], [RNQ+], [PSI+] and [MOT3+].20,22,23,62-65 However, the molecular basis of the effects of environmental conditions on prion formation remains unknown in most cases. As Lsb2 is a stress-inducible protein, we have investigated if its prion-inducing properties are influenced by stress.
[LSB+] Prion Is Induced by Stress
Indeed, our results (1) show that about 0.1% of colonies produced by yeast cells that were heat shocked at 39°C exhibited a metastable [PSI+]-inducibility phenotype, indicative of the [LSB+] prion. The frequency of such colonies was increased up to 0.6% in the strain containing the 8Qto8N derivative of Lsb2, but they were not detected in the lsb2Δ strain. These data confirm that the [LSB+] prion can be generated (without an artificial protein overproduction) as a result of the physiological increase of Lsb2 levels in response to heat stress.
Thus, Lsb2 serves as a sensor of stress, which can maintain a cellular memory of stress in subsequent generations via converting into a heritable (but metastable) prion form. It should be noted that while Lsb2 was proposed to play a role in endocytosis,66 this role is obviously minor and dispensable. In contrast, our data show that Lsb2 facilitates the assembly of other misfolded proteins at specific cytoskeleton-associated sites.41 This mechanism may have arisen as a protective tool intended to minimize the pathogenic effects of inducing misfolded proteins throughout the cell, and may also help prevent degradation of essential proteins under unfavorable conditions, as proposed previously.63 Indeed, analysis of individual cells shows that heat shock induced cell death is increased in the absence of both Lsb1 and Lsb243. As assembly of other aggregates occurs through interaction with punctate structure formed by aggregated Lsb2, this process is likely to be facilitated by the conversion of Lsb2 into a prion form. In addition, prion aggregates of Lsb2 are likely more proteolytically stable, compared with a non-prion protein, as judged from analogy with other known prions.67-69 Thus, cells bearing the [LSB+] prion likely contain more Lsb2 protein compared with [lsb−] cells, and also contain it in the form that is ready for performing its stress-related functions. The biological consequence of the formation of the metastable [LSB+] prion is a subpopulation of cells, which “remember” the stress and are therefore, may be better prepared for the return of stress conditions. This increases the possibility of survival of the population as a whole in such a case.
Notably, the protective aggregate assembly mediated by Lsb2 in a prion form may generate self-perpetuating amyloid forms (prions) of other proteins as a by-product, as we observe in case of [PSI+] induction, and possibly, for [RNQ+].41 Indeed, increased de novo formation of the [PSI+] prion has been detected in certain stress conditions, including incubation at high temperature.64 It is possible that induction of the transient [LSB+] prion contributes to this phenomenon. Other prions triggered by [LSB+] could be pathogenic, however the fast degradation of Lsb2 protein after stress (or after adaptation to stress conditions) and the metastable nature of the [LSB+] prion ensure induction of such prions would occur only in a fraction of the population and therefore minimize potential negative effects. On the other hand, stress-dependent induction of prion formation increases overall phenotypic variability and may generate prion-controlled traits that become enhance survival of a subpopulation of cells. Known examples of yeast prion increasing survival in certain stressful conditions include: [MOT3+],22 that increases resistance of yeast cells to ethanol stress, and [MOD+], that increases resistance to fluconazole.70
Evolutionary Acquisition of Prion-Forming Properties by LSB2
The Lsb2 paralog, Lsb1 also directly interacts with Sup35 PrD in the yeast 2-hybrid assay43 and forms puncta that colocalize with the Sup35 aggregates.41,43 However in contrast to Lsb2, overproduction of Lsb1 does not promote prion formation.43 Surprisingly, we have found that the difference in a single amino acid residue N213 (Lsb2) vs. S239 (Lsb1) within the otherwise conserved C-terminal region confers the prion-inducing ability to Lsb2 in comparison to Lsb1 (Fig. 2).1 The N213S substitution completely abolished the ability of overproduced Lsb2 to promote [PSI+] induction (and therefore, its ability to form an [LSB+] prion), while the reciprocal S239N substitution enabled overproduced Lsb1 to become [PSI+] inducer. Remarkably, the N to S substitution disrupts an “amyloid stretch” consensus sequence formed by the C-terminal hexapeptide of Lsb2. Hexapeptides conforming to this consensus were shown to be a characteristic feature of the proteins capable of forming amyloids in vitro.71 Lsb2 and Lsb1 possess 2 additional amyloid stretch hexapeptides, however only the presence of the third, C-proximal amyloid stretch (found only in Lsb2) strongly correlates with the prion-inducing ability (Fig. 2).
Figure 2.
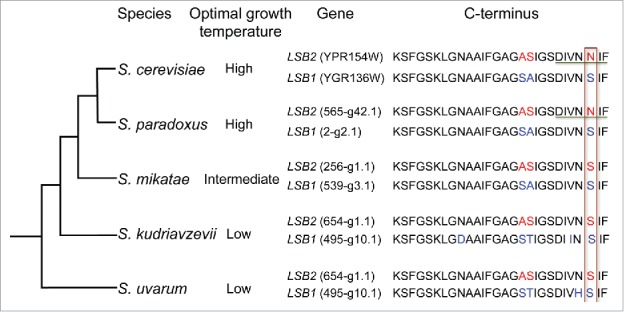
Prion–inducing activity of Lsb2 coincides with yeast adaptation to higher growth temperature. Schematic shows phylogenetic relationships among some members of the Saccharomyces sensu stricto genus. The preferred growth temperature of each species is indicated.74,75 CLUSTALW-formatted multiple sequence alignment of C-terminal of Lsb1/Lsb2 is shown. Difference in amino acids is indicated in red (Lsb2) and blue (Lsb1). Residue essential for prion induction is bordered. Amyloid stretch hexapeptide is underlined.
Notably, only the Saccharomyces sensu stricto clade contains 2 paralogs of Lsb, while other Saccharomyces species possess only Lsb1 (data available at http://www.yeastgenome.org). Moreover, within the S. sensu stricto clade, only the Lsb2 proteins of S. cerevisiae and its sister species, S. paradoxus contain N residue at the position 213, while Lsb2s of more distantly related species, S. mikatae and S. bayanus bear S (Fig. 2). This indicates that the prion-inducing activity of Lsb2 is a relatively recent evolutionary acquisition. It has been reported previously that the prion forming ability can be confined to some yeast proteins by individual amino acid substitutions in these proteins.72 We demonstrate that the substitutions producing a prion-forming potential indeed arise and can be fixed in real phylogenetic lineages. It is worth noting that both S. cerevisiae and S. paradoxus are characterized by increased optimal growth temperatures, compared with other species of the S. sensu stricto clade. Thus, acquisition of the prion-forming ability by the Lsb2 protein coincides with the acquisition of increased thermotolerance in evolution of Saccharomyces yeast.
A switch of the S. cerevisiae / S. paradoxus branch to the increased growth temperatures should have resulted in an increased exposure to the high temperature stress conditions. As described above, prion-forming potential of Lsb2 and prion-mediated stress memory could have become beneficial for the population survival in such conditions. Therefore, it is possible that the acquisition of prion-forming capabilities by Lsb2 protein played an adaptive role in yeast evolution.
CONCLUSIONS AND PERSPECTIVES
Our data uncover a new mechanism of stress memory that is based on the ability of a stress-inducible protein Lsb2 to acquire a metastable prion state in response to stress and to maintain it for the indefinite number of cell divisions but only in a fraction of the clonal population (Fig. 3). The ability of aggregated Lsb2 to promote assembly of other aggregated proteins in the cytoskeleton-associated deposits indicates that the prion-forming potential of Lsb2 may play a protective role during a proteotoxic stress, and that prion-mediated stress memory can produce a subpopulation that is better prepared for the return of stressful conditions. Acquisition of the prion-forming potential by Lsb2 is a relatively recent evolutionary event that coincides with the adaptation of Saccharomyces yeast to higher growth temperatures. Further work will show if such a prion-based stress memory could also be mediated by other proteins. Remarkable similarities between amyloidogenic formations of yeast and higher eukaryotes suggest that prion-based stress memories could be applicable beyond yeast. Understanding general mechanisms of the environmental and physiological modulation of prion aggregation may pave the way for developing the anti-amyloid prophylactic strategies in humans.16,23,73
Figure 3.
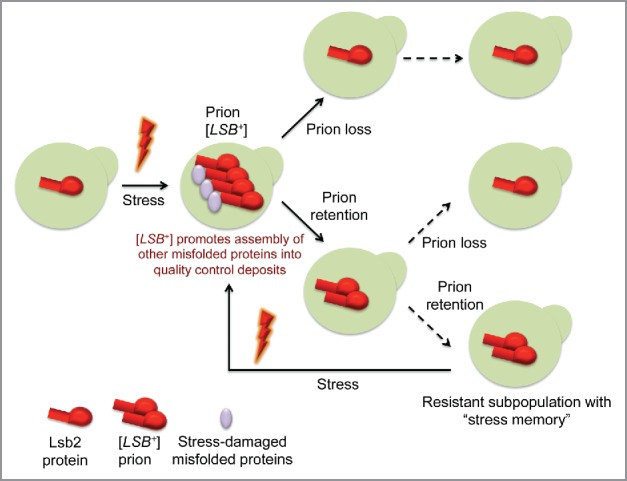
Model “Metastable prion [LSB+] controls stress memory.” Subpopulation of cells with [LSB+] maintains a memory of stress and is better adapted to it. See comments in the text.
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
No potential conflicts of interest were disclosed.
Acknowledgment
We are grateful to John Shanks for assistance with SDD-AGE experiments.
FUNDING
This work supported by grant GM093294 from National Institutes of Health to KDW, and by grants MCB 1516872 from National Science Foundation and 14–50–00069 from Russian Science Foundation to YOC.
REFERENCES
- [1].Chernova TA, Kiktev DA, Romanyuk AV, Shanks JR, Laur O, Ali M, Ghosh A, Kim D, Yang Z, Mang M, et al.. Yeast short-lived actin-associated protein forms a metastable prion in response to thermal stress. Cell Rep 2017; 18(3):751-61; PMID:28099852; https://doi.org/ 10.1016/j.celrep.2016.12.082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Aguzzi A, O'Connor T. Protein aggregation diseases: pathogenicity and therapeutic perspectives. Nat Rev Drug Discov 2010; 9(3):237-48; PMID:20190788; https://doi.org/ 10.1038/nrd3050 [DOI] [PubMed] [Google Scholar]
- [3].Prusiner SB. Biology and genetics of prions causing neurodegeneration. Ann Rev Genet 2013; 47:601-23; PMID:24274755; https://doi.org/ 10.1146/annurev-genet-110711-155524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Haik S, Brandel JP. Infectious prion diseases in humans: cannibalism, iatrogenicity and zoonoses. Infect Genet Evol 2014; 26:303-12; PMID:24956437; https://doi.org/ 10.1016/j.meegid.2014.06.010 [DOI] [PubMed] [Google Scholar]
- [5].Aguzzi A, Rajendran L. The transcellular spread of cytosolic amyloids, prions, and prionoids. Neuron 2009; 64(6):783-90. Epub 2010/January/13; PMID:20064386 [DOI] [PubMed] [Google Scholar]
- [6].Jucker M, Walker LC. Pathogenic protein seeding in Alzheimer disease and other neurodegenerative disorders. Ann Neurol 2011; 70(4):532-40; PMID:22028219; https://doi.org/ 10.1002/ana.22615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Prusiner SB. Cell biology. A unifying role for prions in neurodegenerative diseases. Science 2012; 336(6088):1511-3; PMID:22723400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Savitt JM, Dawson VL, Dawson TM. Diagnosis and treatment of Parkinson disease: molecules to medicine. J Clin Invest 2006; 116(7):1744-54; PMID:16823471; https://doi.org/ 10.1172/JCI29178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science 2002; 297(5580):353-6; PMID:12130773; https://doi.org/ 10.1126/science.1072994 [DOI] [PubMed] [Google Scholar]
- [10].Allen MT, Levy LS. Parkinson's disease and pesticide exposure–a new assessment. Critical Rev Toxicol 2013; 43(6):515-34; PMID:23844699; https://doi.org/ 10.3109/10408444.2013.798719 [DOI] [PubMed] [Google Scholar]
- [11].Duce JA, Bush AI. Biological metals and Alzheimer's disease: implications for therapeutics and diagnostics. Prog Neurobiol 2010; 92(1):1-18; PMID:20444428; https://doi.org/ 10.1016/j.pneurobio.2010.04.003 [DOI] [PubMed] [Google Scholar]
- [12].Fowler DM, Koulov AV, Balch WE, Kelly JW. Functional amyloid–from bacteria to humans. Trends Biochem Sci 2007; 32(5):217-24; PMID:17412596; https://doi.org/ 10.1016/j.tibs.2007.03.003 [DOI] [PubMed] [Google Scholar]
- [13].Maji SK, Perrin MH, Sawaya MR, Jessberger S, Vadodaria K, Rissman RA, Singru PS, Nilsson KP, Simon R, Schubert D, et al.. Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science 2009; 325(5938):328-32. Epub 2009/June/23; PMID:19541956; https://doi.org/ 10.1126/science.1173155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Si K, Lindquist S, Kandel ER. A neuronal isoform of the aplysia CPEB has prion-like properties. Cell 2003; 115(7):879-91; PMID:14697205; https://doi.org/ 10.1016/S0092-8674(03)01020-1 [DOI] [PubMed] [Google Scholar]
- [15].Majumdar A, Cesario WC, White-Grindley E, Jiang H, Ren F, Khan MR, Li L, Choi EM, Kannan K, Guo F, et al.. Critical role of amyloid-like oligomers of Drosophila Orb2 in the persistence of memory. Cell 2012; 148(3):515-29; PMID:22284910; https://doi.org/ 10.1016/j.cell.2012.01.004 [DOI] [PubMed] [Google Scholar]
- [16].Holmes WM, Klaips CL, Serio TR. Defining the limits: Protein aggregation and toxicity in vivo. Crit Rev Biochem Mol Biol 2014; 49(4):294-303; PMID:24766537; https://doi.org/ 10.3109/10409238.2014.914151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Fowler DM, Kelly JW. Functional amyloidogenesis and cytotoxicity-insights into biology and pathology. PLoS Biol 2012; 10(12):e1001459; PMID:23300381; https://doi.org/ 10.1371/journal.pbio.1001459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Di Martino P. Bap: A New Type of Functional Amyloid. Trends Microbiol 2016; 24(9):682-4; PMID:27451288; https://doi.org/ 10.1016/j.tim.2016.07.004 [DOI] [PubMed] [Google Scholar]
- [19].Fioriti L, Myers C, Huang YY, Li X, Stephan JS, Trifilieff P, Colnaghi L, Kosmidis S, Drisaldi B, Pavlopoulos E, et al.. The persistence of hippocampal-based memory requires protein synthesis mediated by the prion-like protein CPEB3. Neuron 2015; 86(6):1433-48; PMID:26074003; https://doi.org/ 10.1016/j.neuron.2015.05.021 [DOI] [PubMed] [Google Scholar]
- [20].Liebman SW, Chernoff YO. Prions in yeast. Genetics 2012; 191(4):1041-72; PMID:22879407; https://doi.org/ 10.1534/genetics.111.137760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Tuite MF. The natural history of yeast prions. Adv Appl Microbiol 2013; 84:85-137; PMID:23763759 [DOI] [PubMed] [Google Scholar]
- [22].Holmes DL, Lancaster AK, Lindquist S, Halfmann R. Heritable remodeling of yeast multicellularity by an environmentally responsive prion. Cell 2013; 153(1):153-65; PMID:23540696; https://doi.org/ 10.1016/j.cell.2013.02.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Chernova TA, Wilkinson KD, Chernoff YO. Physiological and environmental control of yeast prions. FEMS Microbiol Rev 2014; 38(2):326-44; PMID:24236638; https://doi.org/ 10.1111/1574-6976.12053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Halfmann R, Jarosz DF, Jones SK, Chang A, Lancaster AK, Lindquist S. Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature 2012; 482(7385):363-8; PMID:22337056; https://doi.org/ 10.1038/nature10875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Caudron F, Barral Y. A super-assembly of Whi3 encodes memory of deceptive encounters by single cells during yeast courtship. Cell 2013; 155(6):1244-57; PMID:24315096; https://doi.org/ 10.1016/j.cell.2013.10.046 [DOI] [PubMed] [Google Scholar]
- [26].McGlinchey RP, Kryndushkin D, Wickner RB. Suicidal [PSI+] is a lethal yeast prion. Proc Natl Acad Sci U S A 2011; 108(13):5337-41; PMID:21402947; https://doi.org/ 10.1073/pnas.1102762108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wickner RB, Edskes HK, Bateman D, Kelly AC, Gorkovskiy A. The yeast prions [PSI+] and [URE3] are molecular degenerative diseases. Prion 2011; 5(4):258-62; PMID:22052353; https://doi.org/ 10.4161/pri.17748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Saupe SJ. The [Het-s] prion of Podospora anserina and its role in heterokaryon incompatibility. Semin Cell Dev Biol 2011; 22(5):460-8; PMID:21334447; https://doi.org/ 10.1016/j.semcdb.2011.02.019 [DOI] [PubMed] [Google Scholar]
- [29].Serio TR, Cashikar AG, Moslehi JJ, Kowal AS, Lindquist SL. Yeast prion [psi +] and its determinant, Sup35p. Methods Enzymol 1999; 309:649-73. Epub 1999/October/03; PMID:10507053 [DOI] [PubMed] [Google Scholar]
- [30].Chernoff YO, Derkach IL, Inge-Vechtomov SG. Multicopy SUP35 gene induces de-novo appearance of psi-like factors in the yeast Saccharomyces cerevisiae. Curr Genet 1993; 24(3):268-70. Epub 1993/September/01; PMID:8221937; https://doi.org/ 10.1007/BF00351802 [DOI] [PubMed] [Google Scholar]
- [31].Derkatch IL, Bradley ME, Zhou P, Chernoff YO, Liebman SW. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics 1997; 147(2):507-19. Epub 1997/October/23; PMID:9335589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: the story of [PIN(+)]. Cell 2001; 106(2):171-82. Epub 2001/August/21; PMID:11511345; https://doi.org/ 10.1016/S0092-8674(01)00427-5 [DOI] [PubMed] [Google Scholar]
- [33].Osherovich LZ, Weissman JS. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI(+)] prion. Cell 2001; 106(2):183-94. Epub 2001/August/21; PMID:11511346; https://doi.org/ 10.1016/S0092-8674(01)00440-8 [DOI] [PubMed] [Google Scholar]
- [34].Derkatch IL, Uptain SM, Outeiro TF, Krishnan R, Lindquist SL, Liebman SW. Effects of Q/N-rich, polyQ, and non-polyQ amyloids on the de novo formation of the [PSI+] prion in yeast and aggregation of Sup35 in vitro. Proc Natl Acad Sci U S A 2004; 101(35):12934-9. Epub 2004/August/25; PMID:15326312; https://doi.org/ 10.1073/pnas.0404968101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sondheimer N, Lindquist S. Rnq1: an epigenetic modifier of protein function in yeast. Mol Cell 2000; 5(1):163-72. Epub 2000/March/11; PMID:10678178; https://doi.org/ 10.1016/S1097-2765(00)80412-8 [DOI] [PubMed] [Google Scholar]
- [36].Meriin AB, Zhang X, He X, Newnam GP, Chernoff YO, Sherman MY. Huntington toxicity in yeast model depends on polyglutamine aggregation mediated by a prion-like protein Rnq1. J Cell Biol 2002; 157(6):997-1004; PMID:12058016; https://doi.org/ 10.1083/jcb.200112104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Derkatch IL, Liebman SW. Prion-prion interactions. Prion 2007; 1(3):161-9. Epub 2007/July/01; PMID:19164893; https://doi.org/ 10.4161/pri.1.3.4837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 2009; 137(1):146-58. Epub 2009/April/07; PMID:19345193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Walker LC, LeVine H 3rd. Corruption and spread of pathogenic proteins in neurodegenerative diseases. J Biol Chem 2012; 287(40):33109-15; PMID:22879600; https://doi.org/ 10.1074/jbc.R112.399378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Yang Z, Stone DE, Liebman SW. Prion-promoted phosphorylation of heterologous amyloid is coupled with ubiquitin-proteasome system inhibition and toxicity. Mol Microbiol 2014; 93(5):1043-56; PMID:25039275; https://doi.org/ 10.1111/mmi.12716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Chernova TA, Romanyuk AV, Karpova TS, Shanks JR, Ali M, Moffatt N, Howie RL, O'Dell A, McNally JG, Liebman SW, et al.. Prion induction by the short-lived, stress-induced protein Lsb2 is regulated by ubiquitination and association with the actin cytoskeleton. Mol Cell 2011; 43(2):242-52; PMID:21777813; https://doi.org/ 10.1016/j.molcel.2011.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Tyler JJ, Allwood EG, Ayscough KR. WASP family proteins, more than Arp2/3 activators. Biochem Soc Trans 2016; 44(5):1339-45; PMID:27911716; https://doi.org/ 10.1042/BST20160176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ali M, Chernova TA, Newnam GP, Yin L, Shanks J, Karpova TS, Lee A, Laur O, Subramanian S, Kim D, et al.. Stress-dependent proteolytic processing of the actin assembly protein Lsb1 modulates a yeast prion. J Biol Chem 2014; 289(40):27625-39; PMID:25143386; https://doi.org/ 10.1074/jbc.M114.582429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Bagriantsev SN, Kushnirov VV, Liebman SW. Analysis of amyloid aggregates using agarose gel electrophoresis. Methods Enzymol 2006; 412:33-48. Epub 2006/October/19; PMID:17046650 [DOI] [PubMed] [Google Scholar]
- [45].Halfmann R, Alberti S, Krishnan R, Lyle N, O'Donnell CW, King OD, Berger B, Pappu RV, Lindquist S. Opposing effects of glutamine and asparagine govern prion formation by intrinsically disordered proteins. Mol Cell 2011; 43(1):72-84; PMID:21726811; https://doi.org/ 10.1016/j.molcel.2011.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Kushnirov VV, Ter-Avanesyan MD. Structure and replication of yeast prions. Cell 1998; 94(1):13-6; PMID:9674422; https://doi.org/ 10.1016/S0092-8674(00)81216-7 [DOI] [PubMed] [Google Scholar]
- [47].Tanaka M, Collins SR, Toyama BH, Weissman JS. The physical basis of how prion conformations determine strain phenotypes. Nature 2006; 442(7102):585-9; PMID:16810177; https://doi.org/ 10.1038/nature04922 [DOI] [PubMed] [Google Scholar]
- [48].Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science 1995; 268(5212):880-4. Epub 1995/May/12; PMID:7754373; https://doi.org/ 10.1126/science.7754373 [DOI] [PubMed] [Google Scholar]
- [49].Newnam GP, Wegrzyn RD, Lindquist SL, Chernoff YO. Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol Cell Biol 1999; 19(2):1325-33. Epub 1999/January/16; PMID:9891066; https://doi.org/ 10.1128/MCB.19.2.1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Allen KD, Wegrzyn RD, Chernova TA, Muller S, Newnam GP, Winslett PA, Wittich KB, Wilkinson KD, Chernoff YO. Hsp70 chaperones as modulators of prion life cycle: novel effects of Ssa and Ssb on the Saccharomyces cerevisiae prion [PSI+]. Genetics 2005; 169(3):1227-42. Epub 2004/November/17; PMID:15545639; https://doi.org/ 10.1534/genetics.104.037168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kummer E, Oguchi Y, Seyffer F, Bukau B, Mogk A. Mechanism of Hsp104/ClpB inhibition by prion curing Guanidinium hydrochloride. FEBS Lett 2013; 587(6):810-7; PMID:23416293; https://doi.org/ 10.1016/j.febslet.2013.02.011 [DOI] [PubMed] [Google Scholar]
- [52].Chernova TA, Wilkinson KD, Chernoff YO. Prions, Chaperones, and Proteostasis in Yeast. Cold Spring Harb Perspect Biol 2017; 9(2):pii: a023663; PMID:27815300; https://doi.org/ 10.1101/cshperspect.a023663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Tuite MF, Mundy CR, Cox BS. Agents that cause a high frequency of genetic change from [psi+] to [psi-] in Saccharomyces cerevisiae. Genetics 1981; 98(4):691-711; PMID:7037537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Cox BS, Tuite MF, McLaughlin CS. The psi factor of yeast: a problem in inheritance. Yeast 1988; 4(3):159-78; PMID:3059716; https://doi.org/ 10.1002/yea.320040302 [DOI] [PubMed] [Google Scholar]
- [55].Newnam GP, Birchmore JL, Chernoff YO. Destabilization and recovery of a yeast prion after mild heat shock. J Mol Biol 2011; 408(3):432-48; PMID:21392508; https://doi.org/ 10.1016/j.jmb.2011.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Winkler J, Tyedmers J, Bukau B, Mogk A. Chaperone networks in protein disaggregation and prion propagation. J Struct Biol 2012; 179(2):152-60; PMID:22580344; https://doi.org/ 10.1016/j.jsb.2012.05.002 [DOI] [PubMed] [Google Scholar]
- [57].Winkler J, Tyedmers J, Bukau B, Mogk A. Hsp70 targets Hsp100 chaperones to substrates for protein disaggregation and prion fragmentation. J Cell Biol 2012; 198(3):387-404; PMID:22869599; https://doi.org/ 10.1083/jcb.201201074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Klaips CL, Hochstrasser ML, Langlois CR, Serio TR. Correction: Spatial quality control bypasses cell-based limitations on proteostasis to promote prion curing. eLife 2015; 4:e06494; PMID:25626954; https://doi.org/ 10.7554/eLife.06494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Liu B, Larsson L, Franssens V, Hao X, Hill SM, Andersson V, Hoglund D, Song J, Yang X, Oling D, et al.. Segregation of protein aggregates involves actin and the polarity machinery. Cell 2011; 147(5):959-61; PMID:22118450; https://doi.org/ 10.1016/j.cell.2011.11.018 [DOI] [PubMed] [Google Scholar]
- [60].Ness F, Cox BS, Wongwigkarn J, Naeimi WR, Tuite MF. Over-expression of the molecular chaperone Hsp104 in Saccharomyces cerevisiae results in the malpartition of [PSI+ ] propagons. Mol Microbiol 2017; 104(1):125-43; PMID:28073182; https://doi.org/ 10.1111/mmi.13617 [DOI] [PubMed] [Google Scholar]
- [61].Fang NN, Ng AH, Measday V, Mayor T. Hul5 HECT ubiquitin ligase plays a major role in the ubiquitylation and turnover of cytosolic misfolded proteins. Nat Cell Biol 2011; 13(11):1344-52; PMID:21983566; https://doi.org/ 10.1038/ncb2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Derkatch IL, Bradley ME, Masse SV, Zadorsky SP, Polozkov GV, Inge-Vechtomov SG, Liebman SW. Dependence and independence of [PSI(+)] and [PIN(+)]: a two-prion system in yeast? EMBO J 2000; 19(9):1942-52. Epub 2000/May/03; PMID:10790361; https://doi.org/ 10.1093/emboj/19.9.1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Chernoff YO. Stress and prions: lessons from the yeast model. FEBS Lett 2007; 581(19):3695-701. Epub 2007/May/19; PMID:17509571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Tyedmers J, Madariaga ML, Lindquist S. Prion switching in response to environmental stress. PLoS Biol 2008; 6(11):e294. Epub 2008/December/11; PMID:19067491; https://doi.org/ 10.1371/journal.pbio.0060294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Li L, Kowal AS. Environmental regulation of prions in yeast. PLoS Pathog 2012; 8(11):e1002973; PMID:23166488; https://doi.org/ 10.1371/journal.ppat.1002973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Spiess M, de Craene JO, Michelot A, Rinaldi B, Huber A, Drubin DG, Winsor B, Friant S. Lsb1 is a negative regulator of las17 dependent actin polymerization involved in endocytosis. PloS One 2013; 8(4):e61147; PMID:23577202; https://doi.org/ 10.1371/journal.pone.0061147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Masison DC, Wickner RB. Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p in prion-containing cells. Science 1995; 270(5233):93-5. Epub 1995/October/06; PMID:7569955; https://doi.org/ 10.1126/science.270.5233.93 [DOI] [PubMed] [Google Scholar]
- [68].Paushkin SV, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J 1996; 15(12):3127-34. Epub 1996/June/17; PMID:8670813 [PMC free article] [PubMed] [Google Scholar]
- [69].Colby DW, Prusiner SB. Prions. Cold Spring Harb Perspect Biol 2011; 3(1):a006833; PMID:21421910; https://doi.org/ 10.1101/cshperspect.a006833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Suzuki G, Shimazu N, Tanaka M. A yeast prion, Mod5, promotes acquired drug resistance and cell survival under environmental stress. Science 2012; 336(6079):355-9; PMID:22517861; https://doi.org/ 10.1126/science.1219491 [DOI] [PubMed] [Google Scholar]
- [71].Pastor MT, Esteras-Chopo A, Serrano L. Hacking the code of amyloid formation: the amyloid stretch hypothesis. Prion 2007; 1(1):9-14; PMID:19164912; https://doi.org/ 10.4161/pri.1.1.4100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Paul KR, Hendrich CG, Waechter A, Harman MR, Ross ED. Generating new prions by targeted mutation or segment duplication. Proc Natl Acad Sci U S A 2015; 112(28):8584-9; PMID:26100899; https://doi.org/ 10.1073/pnas.1501072112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Sattlegger E, Chernova TA, Gogoi NM, Pillai IV, Chernoff YO, Munn AL. Yeast studies reveal moonlighting functions of the ancient actin cytoskeleton. IUBMB Life 2014; 66(8):538-45; PMID:25138357; https://doi.org/ 10.1002/iub.1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Sampaio JP, Goncalves P. Natural populations of Saccharomyces kudriavzevii in Portugal are associated with oak bark and are sympatric with S. cerevisiae and S. paradoxus. Appl Environ Microbiol 2008; 74(7):2144-52; PMID:18281431; https://doi.org/ 10.1128/AEM.02396-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Salvado Z, Arroyo-Lopez FN, Guillamon JM, Salazar G, Querol A, Barrio E. Temperature adaptation markedly determines evolution within the genus Saccharomyces. Appl Environ Microbiol 2011; 77(7):2292-302; PMID:21317255; https://doi.org/ 10.1128/AEM.01861-10 [DOI] [PMC free article] [PubMed] [Google Scholar]


