Abstract
Israeli Mycobacterium marinum isolates from humans and fish were compared by direct sequencing of the 16S rRNA and hsp65 genes, restriction mapping, and amplified fragment length polymorphism analysis. Significant molecular differences separated all clinical isolates from the piscine isolates, ruling out the local aquaculture industry as the source of human infections.
Mycobacterium marinum is a recognized fish pathogen (1, 11, 17) that can also infect endothermic organisms including humans (20). In human infections, M. marinum gains access through skin abrasions and generally produces superficial and self-limiting lesions involving the cooler parts of the body such as hands, forearms, elbows, and knees (9, 12, 22).
In Israel, several cases of human infections in people tending home aquaria (12) or bathing in natural pools on the shores of the Dead Sea (9) have been reported. In addition, since the first report in 1990 of the occurrence of a systemic mycobacteriosis in the European sea bass Dicentrarchus labrax cultured in the Red Sea (5), the disease has been detected in more than 20 species of marine and freshwater fish (cultured as well as wild) and in a captive sea turtle (6, 25; A. Colorni, M. Ucko, and W. Knibb, Abstr. Proc. Int. Workshop Seabass Seabream Culture Prob. Prospects, Eur. Aquacult. Soc., p. 259-261, 1996).
Elsewhere in the world, rapid development of fish farming and the ornamental fish industry has similarly led to a worldwide increase in the number of reports of M. marinum infections in fish (2, 10), thus intensifying the risk of infections for people who handle fish (7, 15). Since it is reasonable to presume that only certain strains pose a health hazard, the ability to distinguish among them is of great importance.
Molecular characterization of two M. marinum genes, 16S rRNA and hsp65, recently enabled us to detect strain variation in M. marinum fish isolates from various sources (25). In the present study, these two genes were analyzed in Israeli human isolates of M. marinum. A whole-genome fingerprinting technique was also used and tested for its discriminatory potential. The aim was to find out whether more than one strain of M. marinum is involved in human infections in Israel and whether the Israeli fish isolates are identifiable with any of them, thus enabling us to assess the degree of hazard to which fish farmers and consumers are exposed.
The sources of the bacteria are summarized in Table 1. Twenty clinical M. marinum isolates (H1 to H20), stored in the collection of the National Center for Mycobacteria in Tel Aviv, originated from various Israeli hospitals and clinics between 1992 and 1999. Israeli piscine isolates, covering approximately the same period (1990 to 2000), included seven marine isolates from cultured and wild fish and two freshwater isolates from cultured fish. In addition, one marine isolate from a captive hawksbill sea turtle (Eretmochelys imbricata) from the Red Sea was examined. For purposes of comparison, foreign M. marinum isolates from both human and piscine lesions were included. Clinical strain 98100 was isolated in Sardinia, Italy. Three European piscine isolates, two from Greece and one from Denmark (marine), and two piscine isolates from Thailand (freshwater) were also included. Reference strain ATCC 927, isolated from an unspecified marine fish in the United States, was purchased directly from the American Type Culture Collection, Manassas, Va. In addition, a clinical Mycobacterium ulcerans isolate (strain 93147172, from Australia) was used, due to the fact that its phylogeny is thought to be close to that of M. marinum (18, 24).
TABLE 1.
Mycobacterium isolates, their hosts, and their origins
| Code | Isolate | Host | Origina | GenBank accession no. to which identity was found for:
|
|
|---|---|---|---|---|---|
| 16S rRNA | hsp65 | ||||
| Clinical isolates | |||||
| H1-H20b | M. marinum | Human | Israel | AF251565 | AF456474 |
| 98100 | M. marinum | Human | Italy | AF251565 | AF456474 |
| 93147172 | M. ulcerans | Human | Australia | X88926 | AF456475 |
| Fish isolatesc | |||||
| DL240490 | M. marinum Eilaticum | European sea bass (Dicentrarcitus labrax) | Israel, marine (RS), cultured | AF456238 | AF456468 |
| DL150991 | M. marinum Eilaticum | European sea bass (D. labrax) | Israel, marine (MS), cultured | AF456238 | AF456468 |
| DL180892d | M. marinum Eilaticum | European sea bass (D. labrax) | Israel, brackish water, cultured | ||
| CF030494 | M. marinum Eilaticum | Butterflyfish (Chaetodon fasciatus) | Israel, marine (RS), captive | AF456238 | AF456468 |
| SA200695d | M. marinum Eilaticum | Gilthead sea bream (Sparus aurata) | Israel, marine (RS), cultured | ||
| Hyb270995e | M. marinum Eilaticum | Hybrid red sea bream (Pagrus major [female] × S. aurata [male]) | Israel, marine (RS), cultured | AF456238 | |
| SR030597 | M. marinum Eilaticum | Rabbitfish (Siganus rivulatus) | Israel, marine (RS), wild | AF456238 | AF456468 |
| E1100398e | M. marinum Eilaticum | Hawksbill sea turtle (Eretmochelys imbricata) | Israel, marine (RS), captive | AF456238 | |
| DL045 | M. marinum hellenicum | European sea bass (D. labrax) | Greece, marine (MS), cultured | AF456241 | AF456471 |
| DL049 | M. sp. strain Graecum | European sea bass (D. labrax) | Greece, marine (MS), cultured | AF456242 | AF456472 |
| DL/DK | M. marinum | European seabass (D. labrax) | Denmark, marine, cultured | AF456240 | AF456470 |
| CC240299 | M. marinum Cyprinum | Koi (Cyprinus carpio) | Israel, freshwater, cultured | AF456239 | AF456469 |
| BB170200 | M. marinum Cyprinum | Silver perch (Bidyanus bidyanus) | Israel, freshwater, cultured | AF456239 | AF456469 |
| S4 | M. marinum | Snakehead (Channa striata) | Thailand, freshwater | AF251565 | AF456474 |
| S267 | M. marinum | Snakehead (C. striata) | Thailand, freshwater | AF251565 | AF456474 |
| ATCC 927 | M. marinum | Unspecified fish | United States, marine, captive | AF456240 | AF456470 |
RS, Red Sea; MS, Mediterranean Sea.
These 20 isolates originated from various Israeli hospitals (between 1992 and 1999) and were stored in the collection of the National Center for Mycobacteria in Tel Aviv.
With the exception of isolate E1100398, from a sea turtle. All of the Israeli fish isolates were from Israel Oceanographic and Limnological Research Ltd., National Center for Mariculture, Eilat, Israel. The first two letters of the code indicate the initials of the host's genus and species; the digits indicate the date of isolation (two digits each for the day, month, and year).
Not included in the study of 16S rRNA and hsp65 gene sequences.
Not included in the hsp65 sequence study.
DNAs from all human isolates were extracted, and the 16S rRNA (1,522-bp) and hsp65 (1,648-bp) genes were amplified and sequenced as described by Ucko et al. (25) for fish isolates (Table 1).
For the restriction map, a 614-bp PCR product from the 16S rRNA gene was obtained by using primers pA (8) and 266 (3), followed by digestion with the MslI restriction enzyme (New England Biolabs, Beverly, Mass.). Restriction fragments from three representative Israeli clinical isolates (H1 to H3), two piscine isolates representative of the Israeli freshwater and marine environments (M. marinum Cyprinum CC240299 [GenBank accession no. AF456239] and M. marinum Eilaticum DL240490 [GenBank accession no. AF456238]), and two M. marinum isolates of foreign origin (ATCC 927, from an unspecified fish [GenBank accession no. AF456240], and S4 from the snakehead Channa striata, identical to a strain with GenBank accession no. AF251565) were separated on 3.5% NuSieve low-melting-point agarose (BioWhittaker Molecular Applications, Rockland, Maine). A 100-bp molecular ruler (Bio-Rad, Hercules, Calif.) was used as a DNA size marker.
Amplified fragment length polymorphism (AFLP) analysis was carried out on both human and piscine isolates as described by Vos et al. (26) with the modifications of Kvitt et al. (14). Briefly, DNA was digested with 20 U of EcoRI and 5 U of MseI (New England Biolabs), and preamplification was performed with two AFLP adapters: M1 (MseI-adapter) (5′ GACGATGAGTCCTGAG 3′) and E1 (EcoRI-adapter) (5′CTCGTAGACTGCGTACC3′). The AFLP reaction employed two oligonucleotide primers: M1, corresponding to the MseI ends, and a radioactively labeled EcoRI primer (5′ GACTGCGTACCAATTN 3′), corresponding to the EcoRI ends. The EcoRI primers had one selective nucleotide extension (N stands for G, A, T, or C). In each reaction one selective EcoRI primer was employed. Four primer combinations were used to detect strain variation. Four microliters of each sample was loaded onto 6% denaturing polyacrylamide gels (8 M urea) and subjected to electrophoresis for 1.5 h at 2,000 W. Polymorphism was determined by the presence (scored as 1) or absence (scored as 0) of DNA fragments. The banding patterns of the four primer sets were combined, and the pairwise distances between isolates were calculated by using Nei and Li's method (16) and the PAUP 4 (beta 7) program. A dendrogram was constructed by UPGMA (unweighted-pair group method using average linkages) analysis (21). Bootstrap proportions were used to assess the tree robustness with 1,000 bootstrap replications. Mycobacterium sp. strain Graecum DL049 was used as an outgroup, because this strain has been demonstrated to be a new Mycobacterium species (25).
The 20 Israeli clinical isolates and the clinical isolate from Italy showed identical sequences for both the 16S rRNA and hsp65 genes. These sequences were found to be identical to those of two freshwater-fish isolates from C. striata from Thailand (GenBank accession no. AF251565 and AF456474, respectively). Conversely, when compared with those of the Israeli marine and freshwater fish isolates, both genes shared 99.6% sequence homology.
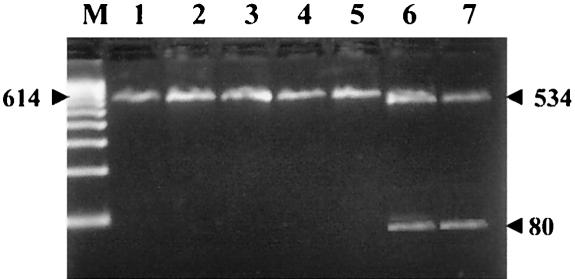
A restriction map was drawn based on the sequence information obtained from the 16S rRNA gene (Fig. 1). MslI digestion of the 614-bp PCR product from the 16S rRNA gene resulted in cleavage products of 80 and 534 bp. Identical patterns for the three representative Israeli clinical isolates, the reference strain ATCC 927, and isolate S4 from C. striata from Thailand were revealed, thus distinguishing the Israeli clinical isolates from the Israeli fish isolates.
FIG. 1.
Enzyme restriction mapping. A 614-bp product from the 16S rRNA gene was cleaved with the restriction enzyme MslI. Lanes: M, 100-bp molecular size marker; 1 to 3, Israeli M. marinum clinical isolates; 4, M. marinum ATCC 927; 5, M. marinum isolate S4, from a snakehead (C. striata); 6, M. marinum Cyprinum CC240299 (GenBank accession no. AF456239); 7, M. marinum Eilaticum DL240490 (GenBank accession no. AF456238).
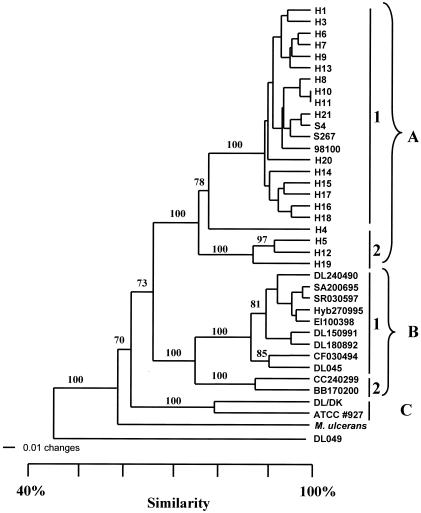
The total number of AFLP bands per primer set ranged from 73 to 130. By summing the bands of the four primer combinations, 380 distinct bands were revealed. Pairwise distances between taxa were calculated by using all 380 bands, and a dendrogram was constructed (Fig. 2). Three distinct main groups, A to C, could be distinguished at a cutoff value of approximately 60% similarity. Group A (n = 23) includes all the Israeli M. marinum clinical isolates, two freshwater fish isolates from Thailand, and the clinical isolate from Italy. At about 80% similarity, group A could be further divided into two subgroups, A1 (n = 19) and A2 (n = 4). Isolates within subgroup A1 shared 92 to 100% similarity, while those in subgroup A2 shared 88% similarity. Group B (n = 11) was distinguished at 68% similarity from group A and includes all the Israeli M. marinum fish isolates from both the marine and freshwater environments. At approximately 80% similarity, group B could be further divided into two subgroups: B1 (n = 9), which includes all the marine fish isolates, the sea turtle isolate, and isolate DL045 from sea bass from Greece, all sharing 93% similarity, and B2 (n = 2), which includes the two Israeli freshwater fish isolates, sharing 91% similarity.
FIG. 2.
Distance tree derived from UPGMA cluster analysis of the AFLP patterns of all strains used in this study. Numbers on the branches indicate bootstrap proportions (1,000 replicates); only bootstrap values above 70% are displayed on the tree. Mycobacterium sp. strain DL049 was used as an outgroup.
Group C (n = 2) formed at a cutoff value of 63% similarity with group A and includes the reference strain ATCC 927 and an isolate from sea bass from Denmark, sharing 84% similarity. M. ulcerans also exhibited 60% similarity with the other groups, whereas the M. sp. strain Graecum isolate (DL049) fell outside these clusters at 46.5% similarity.
Direct sequencing of the two genes 16S rRNA and hsp65 confirmed the identity of all of the 20 Israeli clinical isolates that were isolated during the years 1992 to 1999, characterizing them as M. marinum. Also, these sequences were found to be identical to those from the freshwater species C. striata from Thailand. The more practical and rapid restriction map that previously allowed the distinction of Israeli from foreign fish isolates (25) allowed us, in the present study, to distinguish the Israeli clinical isolates from the Israeli fish isolates of both marine and freshwater environments.
The AFLP whole-genome technique revealed further polymorphism by distinguishing between isolates that shared 100% homology within the 16S rRNA and the hsp65 gene sequences. AFLP band patterns produced several subclusters in each of the three main groups. Also, with this method the resolution within highly homologous isolates was sharper: the Israeli clinical isolates, the clinical isolate from Italy, and the two fish isolates from Thailand could be separated at a similarity as low as 68% from the Israeli fish isolates. M. marinum and M. ulcerans share an apparently close phylogenetic relationship. In fact it was speculated that M. ulcerans might have evolved phylogenetically from the progenitor M. marinum by acquiring foreign DNA from the environment (23), although there is no evidence that M. ulcerans can infect fish. In any case, while sharing >99.8% sequence similarity in the 16S rRNA gene (18, 24, 25), these two species were distinguished by AFLP at 60% similarity. Our results are in agreement with those of Chemlal et al. (4).
M. marinum infections in Thailand are reported as being frequent among fishermen and fishmongers (13). In addition, two of the most economically important fish species cultured in Thailand, the snakehead C. striata and the Siamese fighting fish Betta splendens, have been repeatedly associated with M. marinum (19). The close molecular similarity between the Israeli clinical isolates and those from Thailand suggests that clinical cases in Israel may have derived from the ornamental fish trade associated with home aquaria. This and the lack of records of human infections by our local strains (M. marinum Eilaticum and M. marinum Cyprinum) make it safe to assume that the local aquaculture industry is not the source of human infections in this country, while strengthening the hypothesis that only certain strains of M. marinum have zoonotic potential.
Acknowledgments
We are grateful to A. Lavy, Mycobacterium Reference Laboratory, and A. Felix Public Health Laboratory, Ministry of Health, Tel Aviv, Israel, for providing isolates H1 to H20; L. A. Sechi, Dipartimento di Scienze Biomediche, Sezione di Microbiologia Sperimentale e Clinica, Università di Sassari, for providing isolate 98100; A. Sievers, Mycobacterium Reference Laboratory, Fairfield Hospital, Fairfield, Victoria, Australia, for providing isolate 93147172; H. Nousias, Oceanos, Fish Health Center, Preveza, Greece, for providing isolates 045 and 049; I. Dalsgaard, Danish Institute for Fisheries and Marine Research, Fish Disease Laboratory, Frederiksberg, Denmark, for providing isolate DL/DK; and A. Adams, Institute of Aquaculture, University of Stirling, Stirling, Scotland, for providing isolates S4 and S267.
This research was carried out at the Green-Keiser Fish Health Center, IOLR-NCM, Eilat, Israel, and was funded by Yad haNadiv (grant 524.12).
REFERENCES
- 1.Austin, B., and D. A. Austin. 1999. Mycobacterium spp., p. 52-54. In L. Laird (ed.), Bacterial fish pathogens: disease of farmed and wild fish, 3rd ed. Springer, London, England.
- 2.Belas, R., P. Faloon, and A. Hannaford. 1995. Potential applications of molecular biology to the study of fish mycobacteriosis. Annu. Rev. Fish Dis. 5:133-173. [Google Scholar]
- 3.Böddinghaus, B., T. Rogall, T. Flohr, H. Blöcker, and E. C. Böttger. 1990. Detection and identification of mycobacteria by amplification of rRNA. J. Clin. Microbiol. 28:1751-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chemlal, K., G. Huys, P.-A. Fonteyne, V. Vincent, A. G. Lopez, L. Rigouts, J. Swings, W. M. Meyers, and F. Portaels. 2001. Evaluation of PCR-restriction profile analysis and IS2404 restriction fragment length polymorphism and amplified fragment length polymorphism fingerprinting for identification and typing of Mycobacterium ulcerans and M. marinum. J. Clin. Microbiol. 39:3272-3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colorni, A. 1992. A systemic mycobacteriosis in the European sea bass Dicentrarchus labrax cultured in Eilat (Red Sea). Isr. J. Aquacult. Bamidgeh 44:75-81. [Google Scholar]
- 6.Diamant, A., A. Banet, M. Ucko, A. Colorni, W. Knibb, and H. Kvitt. 2000. Mycobacteriosis in wild rabbitfish Siganus rivulatus associated with cage farming in the Gulf of Eilat, Red Sea. Dis. Aquat. Org. 39:211-219. [DOI] [PubMed] [Google Scholar]
- 7.Durborow, R. M. 1999. Health and safety concerns in fisheries and aquaculture. Occup. Med. 14:373-406. [PubMed] [Google Scholar]
- 8.Edwards, U., T. Rogall, H. Blöcker, M. Emde, and E. C. Böttger. 1989. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17:7843-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Even-Paz, Z., H. Haas, T. Sacks, and E. Rosenmann. 1976. Mycobacterium marinum skin infections mimicking cutaneous leishmaniasis. Br. J. Dermatol. 94:435-442. [DOI] [PubMed] [Google Scholar]
- 10.Falkinham, J. O. 1996. Epidemiology of infection by nontuberculous mycobacteria. Clin. Microbiol. Rev. 9:177-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frerichs, G. N. 1993. Acid-fast fish pathogens, p. 217-233. In V. Inglis, R. J. Roberts, and N. R. Bromage (ed.), Bacterial diseases of fish. Blackwell Scientific Publications Ltd., Oxford, England.
- 12.Huminer, D., S. D. Pitlik, C. Block, L. Kaufman, S. Amit, and J. B. Rosenfeld. 1986. Aquarium-borne Mycobacterium marinum skin infection. Arch. Dermatol. 122:698-703. [PubMed] [Google Scholar]
- 13.Kullavanijaya, P., S. Sirimachan, and P. Bhuddavudhikrai. 1993. Mycobacterium marinum cutaneous infections acquired from occupations and hobbies. Int. J. Dermatol. 32:504-507. [DOI] [PubMed] [Google Scholar]
- 14.Kvitt, H., M. Ucko, A. Colorni, C. Batargias, A. Zlotkin, and W. Knibb. 2002. Photobacterium damselae ssp. piscicida: detection by direct amplification of 16S rRNA gene sequences and genotypic variation as determined by amplified fragment length polymorphism (AFLP). Dis. Aquat. Org. 48:187-195. [DOI] [PubMed] [Google Scholar]
- 15.Lehane, L., and G. T. Rawlin. 2000. Topically acquired bacterial zoonoses from fish: a review. Med. J. Aust. 173:256-259. [DOI] [PubMed] [Google Scholar]
- 16.Nei, M., and W.-H. Li. 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 76:5269-5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noga, E. J. 1996. Fish disease diagnosis and treatment. Mosby, St. Louis, Mo.
- 18.Portaels, F., P. A. Fonteyne, H. de Beenhouwer, P. de Rijk, A. Guédénon, J. Hayman, and W. M. Meyers. 1996. Variability in the 3′ end of the 16S rRNA sequence of the species Mycobacterium ulcerans is related to geographic origins of isolates. J. Clin. Microbiol. 34:962-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Puttinaowarat, S., K. D. Thompson, A. Kolk, and A. Adams. 2002. Identification of Mycobacterium spp. isolates from snakehead, Channa striata (Fowler), and Siamese fighting fish, Betta splendens (Regan), using polymerase chain reaction-reverse cross blot hybridization (PCR-RCBH). J. Fish Dis. 25:235-243. [Google Scholar]
- 20.Smith, D. T., and H. P. Willett. 1980. Other mycobacterium species, p. 674-698. In W. K. Joklik, H. P. Willet, and D. B. Amos (ed.), Zinsser microbiology, 17th ed. Appleton-Century-Crofts, New York, N.Y.
- 21.Sneath, P. H. A., and R. R. Sokal. 1973. Numerical taxonomy: the principles and practice of numerical classification, 2nd ed. W. H. Freeman, San Francisco, Calif.
- 22.Steitz, A., A. Feddersen, C. Freytag, S. Daniello, R. E. Schopf, W. O. Böcher, S. Bhakdi, and M. Husmann. 1997. Rapid identification of Mycobacterium marinum by comparative 16S-rRNA-gene analysis in five cases of progredient cutaneous infections. Eur. J. Dermatol. 7:295-299. [Google Scholar]
- 23.Stinear, T. P., G. A. Jenkin, P. D. R. Johnson, and J. K. Davies. 2000. Comparative genetic analysis of Mycobacterium ulcerans and Mycobacterium marinum reveals evidence of recent divergence. J. Bacteriol. 182:6322-6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tønjum, T., D. B. Welty, E. Jantzen, and P. L. Small. 1998. Differentiation of Mycobacterium ulcerans, M. marinum, and M. haemophilum: mapping of their relationships to M. tuberculosis by fatty acid profile analysis, DNA-DNA hybridization, and 16S rRNA gene sequence analysis. J. Clin. Microbiol. 36:918-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ucko, M., A. Colorni, H. Kvitt, A. Diamant, A. Zlotkin, and W. R. Knibb. 2002. Strain variation in Mycobacterium marinum fish isolates. Appl. Environ. Microbiol. 68:5281-5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vos, P., R. Hogers, M. Bleeker, M. Reijans, T. van de Lee, M. Hornes, A. Freijters, J. Pot, J. Peleman, M. Kuiper, and M. Zabeau. 1995. AFLP: a new concept for DNA fingerprinting. Nucleic Acids Res. 21:4407-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]