Figure S6.
Glycans and Positively Selected Residues Do Not Influence VR3/Lysin Complex Formation, Related to Figure 3
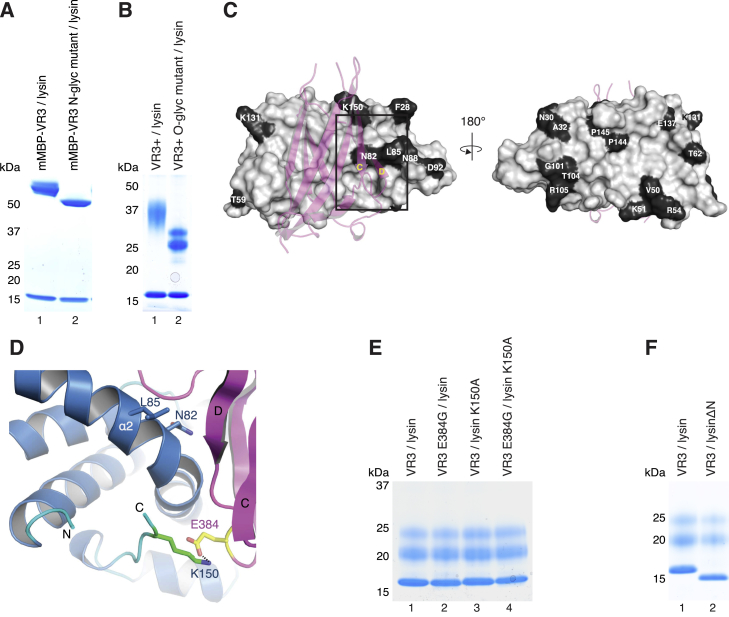
(A and B) VR3 or VR3+ constructs carrying mutations of N-glycosylation sites (N373Q, N417Q, N438Q) or interdomain linker O-glycosylation sites (T456G, T459G, S460G, T467A, T470G, S471G, S472G, S478G, S479G, S485G, S489G) are significantly less glycosylated than counterpart wild-type proteins, but bind lysin equally well.
(C) Surface representation of lysin bound to VR3 (depicted as dark pink cartoon), with positively selected residues highlighted in black. Only N82, L85 and K150 lie at the complex interface.
(D) Close-up of the trigonal VR3/lysin complex, with the three positively selected residues of lysin at the interface with VR3 shown as sticks. Whereas N82 and L85 are conserved among all Californian species of abalone, K150 - which makes an ion pair with VR3 E384 - is variable. Note how the N-terminal region of lysin, which is only defined in the electron density map from F28 onward, is positioned away from interface with VR3.
(E) Disruption of the lysin K150/VR3 E384 ion pair by individual or combined mutation of the interacting residues does not hinder complex formation.
(F) A lysin mutant lacking hypervariable N-terminal residues R19-L29 is pulled down by VR3 as efficiently as wild-type lysin.