Summary
Basolateral amygdala (BLA) principle cells are capable of driving and antagonizing behaviors of opposing valence. BLA neurons project to the central amygdala (CeA), which also participates in negative and positive behaviors. However, the CeA has primarily been studied as the site for negative behaviors and the causal role for CeA circuits underlying appetitive behaviors is poorly understood. Here we identified several genetically distinct populations of CeA neurons that mediate appetitive behaviors and dissected the BLA to CeA circuit for appetitive behaviors. Protein phosphatase 1 regulatory subunit 1B+ BLA pyramidal neurons to dopamine receptor 1+ CeA neurons define a pathway for promoting appetitive behaviors, while R-spondin 2+ BLA pyramidal neurons to dopamine receptor 2+ CeA neurons define a pathway for suppressing appetitive behaviors. These data reveal genetically defined neural circuits in the amygdala that promote and suppress appetitive behaviors analogous to the direct and indirect pathway of the basal ganglia.
Keywords: Central amygdala, Basolateral amygdala, Direct and indirect pathway, Appetitive, Reward, Fear, Amygdala circuit, Feeding, Drinking, Freezing
eToc Blurb
Kim & Zhang et al. dissect a genetically-defined circuit for appetitive behaviors from the basolateral amygdala to central amygdala that is genetically analogous to the direct and indirect pathway of the cortex and striatum.
Introduction
The basolateral (BLA) and central amygdala (CeA) are involved in the control of emotional behaviors (Gallagher and Chiba, 1996; Swanson and Petrovich, 1998). The BLA contains two spatially segregated, genetically distinct populations of cortical-like excitatory pyramidal neurons—Protein phosphatase 1 regulatory subunit 1B+ (Ppp1r1b+ also known as DARPP-32) parvocellular neurons and R-spondin 2+ (Rspo2+) magnocellular neurons (Carlsen and Heimer, 1988; Hemmings et al., 1984; Kim et al., 2016; McDonald, 1984; Pitkanen et al., 1997; Swanson and Petrovich, 1998). BLA Ppp1r1b+ neurons elicit appetitive behaviors, inhibit defensive behaviors, and send projections to the lateral (CeL) and medial (CeM) nucleus of the CeA. BLA Rspo2+ neurons elicit defensive behaviors, inhibit appetitive behaviors, and send projections to the capsular (CeC) nucleus of the CeA (Kim et al., 2016). The CeA consists of GABAergic striatal medium spiny-like neurons, and similar to the BLA, is critical for appetitive and defensive behaviors (Gallagher and Chiba, 1996; McDonald, 1991; Swanson and Petrovich, 1998).
The CeA has been mainly studied on the basis of its role in innate and learned fear-related behaviors (Davis, 1992; Duvarci and Pare, 2014; Ehrlich et al., 2009; Goosens and Maren, 2001; Herry and Johansen, 2014; Hitchcock and Davis, 1986; Killcross et al., 1997; LeDoux et al., 1988). Cell-type specific studies have shown evidence for the involvement of several genetically defined CeA neurons in aversive behaviors such as defensive responses and anxiogenesis (Andero et al., 2014; Han et al., 2015; Haubensak et al., 2010; Isosaka et al., 2015; Li et al., 2013; McCall et al., 2015; Pomrenze et al., 2015; Sanford et al., 2017). However, despite early evidence suggesting the involvement of the CeA in appetitive behaviors (Galaverna et al., 1993; Gallagher et al., 1990; Parkinson et al., 2000; Ritter and Hutton, 1995) and more recent activation studies demonstrating a modulatory role of the CeA in appetitive behaviors (Cai et al., 2014; Robinson et al., 2014; Seo et al., 2016), how appetitive behavior integrates into a structural and functional model of amygdala has yet to be established. For instance, a genetically defined population of CeA neurons that are positive mediators of appetitive behavior has not been identified. Given the strong projections from BLA Ppp1r1b+ parvocellular neurons to the CeL and CeM (Kim et al., 2016), we hypothesized that the CeA may contain neurons that are positive mediators of appetitive behavior. Therefore, we first examined the role of genetically distinct CeA populations in both appetitive and defensive behaviors. Furthermore, although the BLA and CeA are both important for appetitive and defensive behaviors, it is not known how BLA Ppp1r1b+ parvocellular and BLA Rspo2+ magnocellular neurons are connected to genetically defined CeA neurons. Therefore, we also examined the connectivity from genetically defined neurons from the BLA to CeA. Lastly, cytoarchitectural studies suggest that the BLA and CeA are structurally similar to the cortex and striatum, respectively (Carlsen and Heimer, 1988; Swanson and Petrovich, 1998). For this reason, we explored the expression pattern of striatal markers in the CeA to examine if there exists, an organizing principle in the BLA to CeA circuit that is common with the cortex and striatum.
Results
Prkcd, Nts, Sst, Tac2 define distinct cellular populations in the CeA
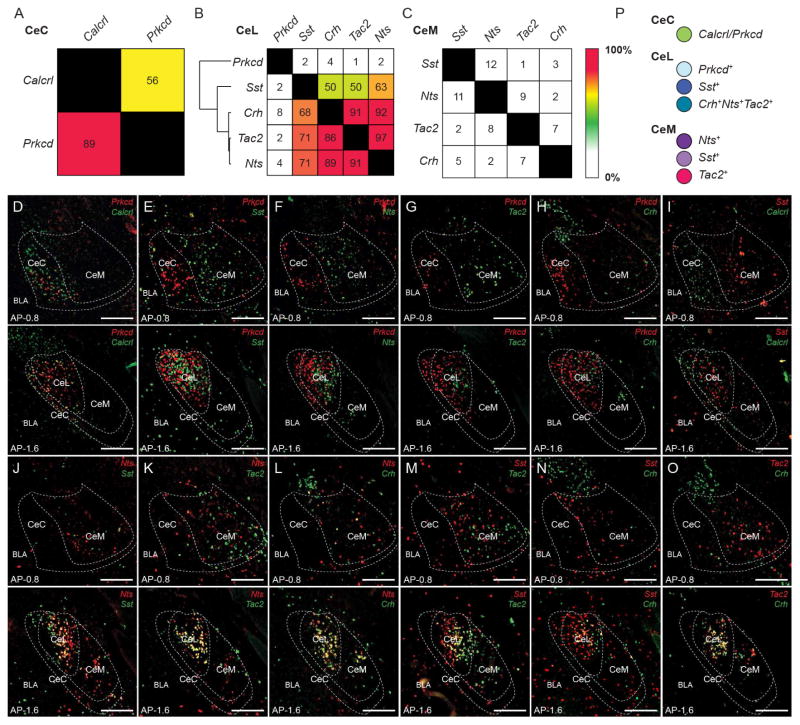
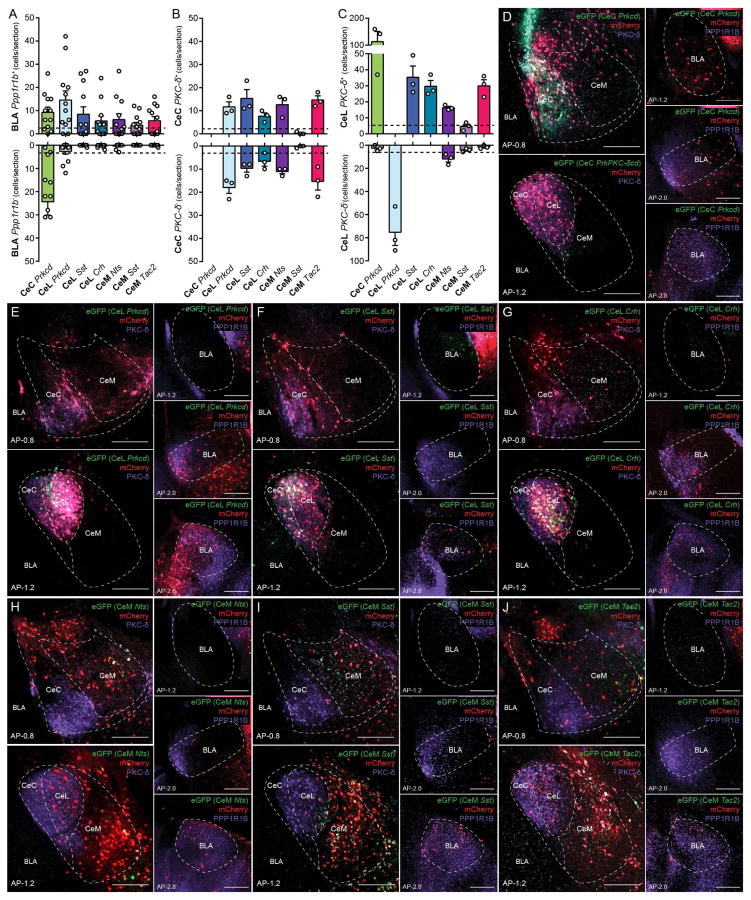
Gene expression studies have shown a wide range of genetic diversity within the CeA (Cornea-Hebert et al., 1999; Moga and Gray, 1985; Skofitsch and Jacobowitz, 1985; Warden and Young, 1988; Zirlinger et al., 2001). To identify genetically distinct populations in the CeA, we performed single molecule in situ hybridization (smFISH) (ACDBio RNAscope) of genes that are expressed in the CeA—calcitonin receptor-like (Calcrl), corticotropin-releasing hormone (Crh), serotonin receptor 2a (Htr2a), neurotensin (Nts), protein kinase C-δ (Prkcd), somatostatin (Sst), and tachykinin 2 (Tac2). It should be noted that for the CeC only the anterior region was quantified due to the ambiguity of the CeC in the posterior CeA (Figure S1B). Calcrl and Prkcd were expressed in the CeC; Crh, Htr2a, Nts, Prkcd, Sst, and Tac2 were expressed in the CeL; Htr2a, Nts, Sst, and Tac2 were expressed in the CeM (Figure S1A and 1SB). Overlap of the expression of Calcrl, Crh, Nts, Prkcd, Sst, and Tac2 was examined in the CeA. In the CeC, Calcrl labeled 89%of Prkcd neurons, while Prkcd labeled 56% of Calcrl+ neurons (Figure 1A and 1D). Calcrl+ neurons were non-overlapping with Sst (which delineates the CeL) in the CeA (Figure 1I), further indicating that Calcrl+ neurons reside in the CeC rather than the CeL. In the CeL, several of the genes had significantly high levels of overlap (>50%) (Figure 1B, 1J–1O). Using the levels of overlap among pairs of genes, hierarchical clustering revealed 3 major clusters. The first containing Prkcd, the second containing, Sst, and third containing Crh, Tac2, and Nts (Figure 1B). In the CeM, Crh, Nts, Sst, and Tac2 were minimally (<15%) overlapping (Figure 1C). Htr2a expression was weak and difficult to quantify with a high degree of confidence in the CeA. However, based upon a few double label smFISH, several Htr2a+ neurons were found to coexpress Crh, Prkcd, Nts, Sst, and Tac2 in the CeL and Nts, Sst, Tac2 in the CeM (Figure S1D). Therefore, Htr2a may be less specific for labelling a distinct population compared to the other genes. Collectively, characterization using these sets of genes revealed as many as 8 or 9 genetically and regionally distinguishable populations of neurons in the CeA.
Figure 1. Identification of genetically distinct populations in the CeA.
(A–C) Quantification of overlap of Prkcd and Calcrl in the CeC (A). Quantification of overlap of Prkcd, Sst, Crh, Tac2, and Nts in the CeL (B). Quantification of overlap of Sst, Nts, Tac2, and Crh in the CeM (C). Values represent percent labelling of overlap of genes in column amongst genes in rows. For example, 56% of CeC Calcrl neurons coexpress Prkcd (A). Values represent percentage of labeling from totaling all cells counted from n = 3 mice. Hierarchical clustering was performed in the CeL using the percent overlap profile of each gene (B).
(D–O) Representative histology of CeA expression of Prkcd and Calcrl (D), Prkcd and Sst (E), Prkcd and Nts (F), Prkcd and Tac2 (G), Prkcd and Crh (H), Sst and Calcrl (I), Nts and Sst (J), Nts and Tac2 (K), Nts and Crh (L), Sst and Tac2 (M), Sst and Crh (N), Tac2 and Crh (O) in the anterior CeA (anterior-posterior (AP) distance from Bregma −0.8 mm) and posterior CeA (AP distance from Bregma −1.6 mm). Scale bar, 250 μm.
(P) 7 major population of neurons that were selected for examination and color selection for subsequent data presentation, CeC Prkcd+ neurons (green), CeL Prkcd+ (light blue), CeL Sst+ (dark blue), CeL Crh+Nts+Tac2+ (teal), CeM Sst+ (light purple), CeM Nts+ (dark purple), and CeM Tac2+ (magenta) neurons.
For this study, we have chosen to study 7 major neuronal populations of the CeA, distinguishable based on gene expression and region—CeC Prkcd+, CeL Prkcd+, CeL Crh+Nts+Tac2+, CeL Sst+, CeM Nts+, CeM Sst+, CeM Tac2+ (Figure 1P). To determined what proportion of neurons CeL Prkcd+, CeL Crh+Nts+Tac2+, and CeL Sst+ neurons constitute in the CeL and what proportion of neurons CeM Nts+, CeM Sst+, and CeM Tac2+ neurons constitute in the CeM, the expression of Nts, Prkcd, Sst, and Tac2 with glutamate decarboxylase 1 (Gad1), a marker of inhibitory neurons, was examined. Prkcd, Sst, and Tac2 collectively labeled 96% of Gad1+ neurons in the CeL, whereas Prkcd+, Sst+, and Tac2+ neurons were 100% Gad1+ (Figure S1C). Nts, Sst, and Tac2 collectively labeled 95% of Gad1+ neurons in the CeM, whereas Nts+, Sst+, and Tac2+ were 100% Gad1+ (Figure S1C). This suggests that CeL Prkcd+, CeL Crh+Nts+Tac2+, and CeL Sst+ neurons constitute the majority of GABAergic neurons in the CeL and that CeM Nts+, CeM Sst+, and CeM Tac2+ neurons constitute the majority of GABAergic neurons in the CeM.
Genetically distinct CeA populations that drive appetitive and defensive behaviors
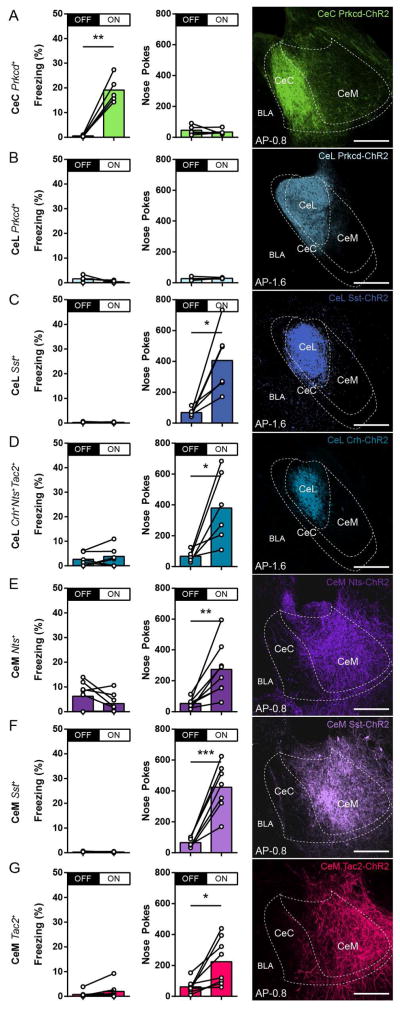
The CeA has been shown to participate in defensive behaviors and appetitive behaviors (Galaverna et al., 1993; Gallagher et al., 1990; Goosens and Maren, 2001; Killcross et al., 1997; LeDoux et al., 1988; Parkinson et al., 2000). Therefore, each of the 7 CeA neuronal populations was subjected to optogenetic stimulation experiments to assess these functions. CeC Prkcd+ and CeL Prkcd+ neurons were targeted using a Cre-dependent channelrhodopsin (ChR2) virus injected into the CeC and CeL, respectively, of the Prkcd-Cre mice; CeL Sst+ and CeM Sst+ using injections into the CeL and CeM, respectively, of the Sst-Cre mice; CeL Crh+Nts+Tac2+ neurons using injections into the CeL of Crh-Cre mice; CeM Nts+ neurons usings injections into the CeM of Nts-Cre mice; CeM Tac2+ neurons using injections into the CeM of the Tac2-Cre mice (Figure S2A). Control mice were Cre− mice that underwent identical surgical procedures in the CeC (Prkcd-Cre− mice), CeL (Sst-Cre− mice), and CeM (Tac2-Cre− mice). In the optogenetic freezing test, mice were placed into a neutral conditioning chamber, where no light stimulation occurred during the 0- to 3-min period (OFF) followed by 20-Hz blue light stimulation during the 3- to 6-min period (ON). Stimulation of CeC Prkcd+ neurons elicited freezing (Figure 2A), measured by an increase in freezing during the ON period compared to the OFF period, while stimulation of CeL Prkcd+, CeL Crh+Nts+Tac2+, CeL Sst+, CeM Nts+, CeM Sst+, and CeM Tac2+ neurons did not elicit freezing (Figure 2B–2G). It should be noted, that stimulation of CeM Tac2+ neurons elicited immobility-like behaviors, but did not reflect stereotypical freezing. Rather, this immobility-like behavior due to activation of CeM Tac2+ neurons coincided with biting behavior in 5 out of 8 mice. Cre− control mice did not demonstrate light-induced freezing (Figure S2B). Following the freezing test, mice were tested for self-stimulation. In the optogenetic self-stimulation test, mice were freely allowed to poke into two nose ports for a single session of 60 min where only one port delivered 20-Hz blue light stimulation. Activation of CeL Sst+, CeL Crh+Nts+Tac2+, CeM Nts+, CeM Sst+, and CeM Tac2+ neurons resulted in self-stimulation, based on an increased in nose pokes in the light stimulated port (ON) compared to the unstimulated port (OFF) (Figure 2C–2G), while CeC Prkcd+ and CeL Prkcd+ neurons did not result in self-stimulation (Figure 2A–2B). Cre− control mice did not demonstrate light induced self-stimulation (Figure S2B). These optogenetic stimulation experiments demonstrate that CeL Sst+, CeL Crh+Nts+Tac2+, CeM Nts+, CeM Sst+, and CeM Tac2+ neurons are capable of driving appetitive behaviors, CeC Prkcd+ neurons are capable of driving defensive behaviors, while CeL Prkcd+ neurons are not capable of driving either appetitive or defensive behaviors.
Figure 2. Genetically distinct CeA neurons drive appetitive and defensive behaviors.
(A–G) Behavioral assessment of percent freezing without (OFF) or with (ON) photostimulation (first column) and total number of nose pokes in unstimulated (OFF) or photostimulated (ON) port in self-stimulation experiments (second column) from optogenetic activation of CeC Prkcd+ (n = 5) (A), CeL Prkcd+ (n = 4) (B), CeL Sst+ (n = 6) (C), CeL Crh+Nts+Tac2+ (n = 6) (D), CeM Nts+ (n = 7) (E), CeM Sst+ (n = 7) (F), and CeM Tac2+ (n = 8) (G) neurons. Representative histology of ChR2 expression in the targeted CeA neurons (third column). Anterior-posterior distribution of ChR2 expression found in Figure S2A. All animals underwent the optogenetic freezing test, followed by the optogenetic self-stimulation test. ChR2 expression was pseudocolored in correspondence with selected color scheme (Figure 1P). Significance for paired t-test, *P< 0.05, **P<0.01, ***P<0.001 (A–G). Anterior-posterior (AP) distance from Bregma (mm), scale bar, 250 μm.
Appetitive and threatening stimuli activate distinct CeA populations
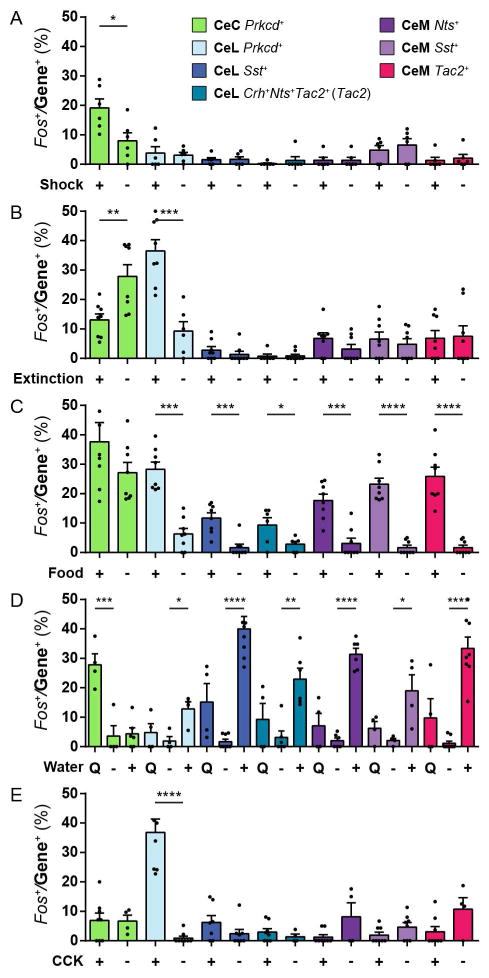
Distinct CeA neurons are capable of eliciting defensive behaviors and appetitive behaviors. Therefore, to assess how these populations are activated by external stimuli, the expression of the activity-dependent gene, Fos (Dubner and Ruda, 1992), was measured in each of the 7 CeA neuronal populations in response to five different conditions associated with defensive or appetitive behaviors. Wild-type mice were exposed to either footshocks or no foosthocks; contextual fear extinction recall or contextual fear recall without fear extinction; ad libitum food or no food in food-deprived mice; ad libitum water, quinine water, or no water in water-deprived mice; injection of cholecystokinin (CCK, an agent that induces satiety) or saline in mice 30 min prior to sacrifice (see Methods). The percentages of Fos labelling within each of the CeA populations were measured. Fos expression was increased in CeC Prkcd+ neurons in response to footshocks compared to corresponding control (Figure 3A) and contextual fear recall compared to contextual fear extinction (Figure 3B). Fos expression was also measured in CeC Calcrl+ neurons in response to footshocks and was found to be significantly increased in CeC Calcrl+ neurons (Figure S3B). This is consistent with the observation that CeC Prkcd labels a subpopulation of CeC Calcrl+ neurons (Figure 1A). Fos expression was increased in CeL Prkcd+ neurons in response to contextual fear extinction recall compared contextual fear recall (Figure 3B). Fos expression was increased in CeL Prkcd+, CeL Sst+, CeL Crh+Nts+Tac2+, CeM Nts+, CeM Sst+, and CeM Tac2+ neurons in response to ad libitum food and ad libitum water compared corresponding controls (Figure 3C–3D). Fos expression was increased in CeL Prkcd+ in response to CCK (Figure 3E). Fos expression was increased in CeC Prkcd+ in response to quinine in water-deprived mice (Figure 3D). These results suggest that CeC Prkcd+ neurons are activated by threatening stimuli and aversive tastes. CeL Prkcd+ neurons are activated by states of suppression of defensive behaviors and stimuli that suppress appetitive behaviors. CeL Sst+, CeL Crh+Nts+Tac2+, CeM Nts+, CeM Sst+, and CeM Tac2+ neurons are activated by stimuli that elicit appetitive behaviors.
Figure 3. Genetically distinct CeA neurons are activated by distinct stimuli.
(A–E) The percent overlap of Fos within CeC Prkcd+, CeL Prkcd+, CeL Sst+, CeL Crh+Nts+Tac2+, CeM Nts+, CeM Sst+, and CeM Tac2+ neurons in response to Shock (+) or No Shock (−) (A); Contextual Extinction Recall (+) or Contextual Fear Recall (−) (B); ad libitum Food (+) or No Food (−) in food-deprived mice (C); Quinine Water (Q), No Water (−) or ad libitum Water (+) in water-deprived mice (D); Cholecystokinin (CCK) (+) or Saline (−) injection (E). CeL Crh+Nts+Tac2+ neurons were measured by quantifying Fos in Tac2+ neurons in the CeL. Significance for unpaired t-test (A,B,C, E) and one-way ANOVA with Bonferroni’s multiple hypothesis correction comparing experimental groups with no water control (D), *P< 0.05, **P< 0.01, ***P<0.001, ****P<0.0001 (A–G). Values are mean ± s.e.m. from 1–2 sections per mouse from up to n = 4 mice, individual values are shown by black dots, some values that are too large and beyond the limit of the y-axis are not shown.
Silencing CeA populations in feeding, drinking, and freezing
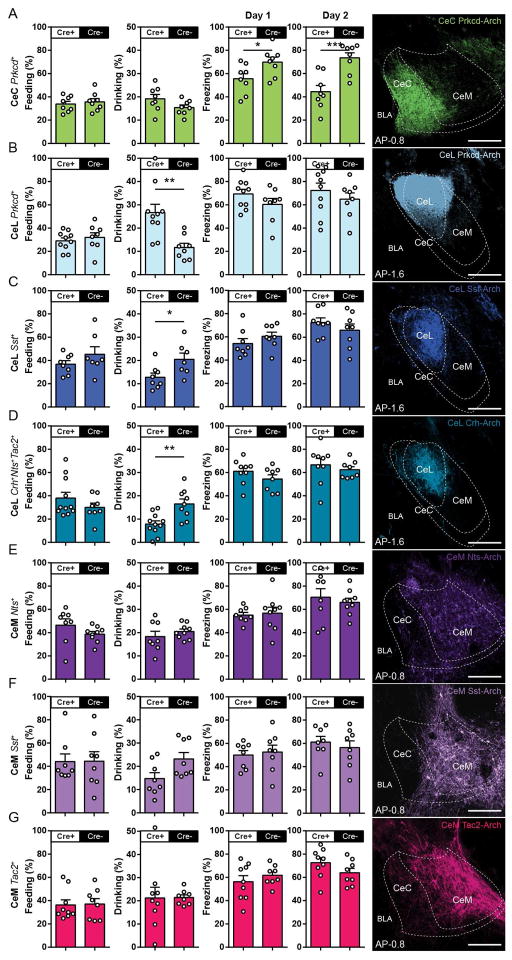
Differential Fos expression in the CeA in response to food, water, and footshock suggests there may be differential involvement of these 7 CeA populations in feeding, drinking, and freezing behaviors. Therefore, to dissociate the contribution of distinct CeA populations in these behaviors, each of the 7 CeA neuronal populations was subjected to a series of optogenetic inhibition experiments (see Methods). For targeting these populations, a Cre-dependent archaerhodopsin (Arch) virus was injected in the same fashion as in the previous optogenetic stimulation experiments, while littermate Cre− mice that underwent identical procedures were used as controls. When presenting ad libitum food in food-deprived mice during a 10-min trial, inhibition of each of the CeA populations—CeC Prkcd+, CeL Prkcd+, CeL Sst+, CeL Crh+Nts+Tac2+, CeM Nts+, CeM Sst+, and CeM Tac2+—did not result in any significant changes in the duration of feeding behavior compared to corresponding controls (Figure 4A–G). Subsequently, when presenting ad libitum water in water-deprived mice during a 5-min trial, inhibition of CeL Prkcd+ neurons resulted in an increase in drinking behavior (Figure 4B), inhibition of CeL Sst+ and CeL Crh+Nts+Tac2+ resulted in a decrease in drinking behavior (Figure 4C and 4D), and inhibition of CeC Prkcd+, CeM Nts+, CeM Sst+, and CeM Tac2+ did not result in changes to drinking compared to corresponding controls (Figure 4A,4E–G). Finally, mice underwent contextual fear conditioning in which CeA neurons were inhibited during the presentation of 3 footshocks in a contextual fear conditioning protocol (Day 1) and were subsequent reexposed to the conditioning chamber 24 hr later with no light inhibition (Day 2). Inhibition of CeC Prkcd+ neurons resulted in a minor, but statistically significant, reduction in freezing on Day 1 and subsequently reduced levels of freezing on Day 2 compared to corresponding controls (Figure 4A). Although there is a trend for increased freezing levels from inhibition of CeL Prkcd+ neurons as previously reported (Haubensak et al., 2010), inhibition of the any of the CeA populations did not affect freezing on Day 1 or 2 compared to corresponding controls (Figure 4B–G). Together, these data suggest that CeL Prkcd+ and CeL Prkcd− (CeL Sst and CeL Crh+Nts+Tac2+) neurons have opposing roles on drinking, CeC Prkcd+ neurons are required for defensive behaviors, while inhibition of any one of these populations does not affect feeding behavior in food-deprived mice.
Figure 4. Inhibition of genetically defined CeA neurons during feeding, drinking, and defensive behaviors.
(A–G) Behavioral assessment of percent feeding during food presentation in food-deprived mice (first column); percent drinking during water presentation in water-deprived mice (second column); percent freezing during presentation of footshocks on Day 1 (third column) and contextual recall without optogenetic inhibition on Day 2 (fourth column) from optogenetic inhibition of CeC Prkcd+ (n = 8, 8) (A), CeL Prkcd+ (n = 10, 8) (B), CeL Sst+ (n = 8, 8) (C), CeL Crh+Nts+Tac2+ (n = 8, 8) (D), CeM Nts+ (n = 9, 8) (E), CeM Sst+ (n = 8, 8) (F), and CeM Tac2+ (n = 9, 8) (G) neurons. All animals underwent the feeding test, followed by the drinking test, followed by contextual fear conditioning. Representative histology of eArch 3.0 expression and optic fiber placement in the targeted CeA neurons (fifth column). eArch 3.0 expression was pseudocolored in correspondence with selected color scheme (Figure 1P). Significance for unpaired t-test, *P< 0.05, **P<0.01, ***P<0.001, sample size (n = experimental,control) (A–G). Anterior-posterior (AP) distance from Bregma (mm), scale bar, 250 μm.
Inhibition of CeM Nts+, CeM Sst+, or CeM Tac2+ neurons did not affect feeding, drinking or freezing behaviors. Therefore, the effects of collectively inhibiting all three CeM populations were assessed. CeM Nts+, CeM Sst+, and CeM Tac2+ neurons collectively constitute almost all CeM Drd1+ neurons (Figure S7E). Thus, a Cre-dependent Arch virus was injected into the CeM of Drd1-Cre mice and underwent the same procedures as the previous inhibition experiments (Figure 4). Inhibition of CeM Drd1+ neurons resulted in reduction of feeding behavior in food-deprived mice, reduction of drinking in water-deprived mice, and showed no differences in freezing in response to footshocks compared to corresponding controls (Figure S4). Therefore, CeM Drd1+ neurons are critical for both feeding and drinking behavior and suggest that CeM Nts+, CeM Sst+, and CeM Tac2+ neurons may collectively function to mediate feeding and drinking.
CeA mediators of appetitive behavior project to the midbrain
The CeA is one of the output structures of the amygdala and is known to project to several brain regions including the periaqueductal gray (PAG) and it has been widely hypothesized that PAG-projecting CeA neurons mediate freezing (Duvarci and Pare, 2014; Ehrlich et al., 2009; Herry and Johansen, 2014; Penzo et al., 2014). Therefore, the relationship between PAG-projecting CeA neurons and genetically distinct CeA populations was examined. The retrograde tracer, cholera toxin subunit B (CTB), was injected into the PAG. This resulted in CTB+ neurons in the CeL and CeM that were Sst+, Tac2+, and Nts+ (Figure S5A–C). In contrast, CTB retrograde tracing from the PAG did not label Prkcd+ neurons in the CeL (Figure S5A–C) which is consistent with previous reports (Penzo et al., 2014). Using ChR2 mice (Figure 2), anterograde tracing fibers were found in the PAG of CeL-Sst-ChR2, CeL-Crh-ChR2. CeM-Nts-ChR2, CeM-Sst-ChR2, and CeM-Tac2-ChR2 mice, but not CeC-Prkcd-ChR2 and CeL-Prkcd-ChR2 mice (Figure S5D). These data suggest that CeL Sst+, CeL Crh+Nts+Tac2+, CeM Nts+, CeM Sst+, and CeM Tac2+ neurons project to the PAG, while CeC Prkcd+ and CeL Prkcd+ neurons do not project the PAG. These findings suggest that, independent of whether or not appetitive functions are necessarily mediated by transmission from the CeA to PAG, PAG projections may not be a unique structural feature of CeA neurons that mediate defensive function.
BLA Ppp1r1b+ neurons form monosynaptic, while BLA Rspo2+ neurons form disynaptic connections to CeA mediators of appetitive behavior
Genetically defined BLA pyramidal neurons that are capable of driving defensive and appetitive behaviors send projections to the CeA (Kim et al., 2016). Therefore, the anatomical relationship between the BLA and CeA was examined using cell-type specific monosynaptic rabies tracing in the 7 CeA neuronal populations (Kohara et al., 2014; Wickersham et al., 2007). To target these populations, AAV helper virus, AAV1-synP-FLEX-sTpEpB(Kohara et al., 2014), (construct containing Cre-dependent TVA, Rabies G-protein, eGFP) was injected in the same fashion as in the optogenetic stimulation experiments, incubated for 3 weeks prior to injection of the G-deleted rabies mCherry virus(Kohara et al., 2014), then sacrificed 1 week later. Tissues were labeled using antibodies against PPP1R1B to determine the BLA cell-type—Ppp1r1b+ or Ppp1r1b− (as a measure of Rspo2+ neurons because Rspo2+ and Ppp1r1b+ constitute virtually all BLA excitatory neurons (Kim et al., 2016)) (Figure 5D–J). Monosynaptic tracing from CeL Prkcd, CeL Sst+, CeL Crh+Nts+Tac2+, CeM Nts+, CeM Sst+, and CeM Tac2+ neurons resulted in retrograde labeled neurons in the BLA that are predominantly PPP1R1B+ (Figure 5A). In contrast, monosynaptic tracing from CeC Prkcd+ neurons resulted in retrograde labeled neurons in the BLA that were PPP1R1B− but also PPP1R1B+ (Figure 5A). These results suggest that BLA Rspo2+ neurons directly project to CeC Prkcd+ neurons, while BLA Ppp1r1b+ neurons directly projects to all CeA neurons that were examined. It should be noted that the connectivity from BLA Ppp1r1b+ neurons to CeC neurons was not observed in our previous study (Kim et al., 2016). This apparent contradiction may be explained by the fact that we previously targeted the dorsal portion of the CeC for retrograde labelling, whereas CeC Prkcd+ neurons reside more ventrally in the CeC (Figure S1B).
Figure 5. Monosynaptic retrograde tracing from genetically defined CeA neurons.
(A) Quantification of rabies-mediated retrograde labeled PPP1R1B+ and PPP1R1B − neurons in the BLA from genetically defined CeA neurons. Individual points represent the number of retrograde labeled neurons per section in the BLA from n = 8 sections per mouse from n = 3 mice, bars represent mean ± s.e.m.. Arbitrary threshold of 2.5 neurons per section was used to construct a connectivity model (Figure 8A).
(B) Quantification of rabies-mediated retrograde labeled PKC-δ+ and PKC-δ − neurons in the CeC from genetically defined CeA neurons. Individual points represent the number of retrograde labeled neurons per single section in the CeC from n = 3 sections from 3 mice, bars represent mean ± s.e.m.. Arbitrary threshold of 2.5 neurons per section was used to construct a connectivity model (Figure 8A).
(C) Quantification of rabies-mediated retrograde labeled PKC-δ + and PKC-δ − neurons in the CeL from genetically defined CeA neurons. Individual points represent the number of retrograde labeled neurons per single section in the CeL from n = 3 sections from 3 mice, bars represent mean ± s.e.m.. Arbitrary threshold of 5 neurons per section was used to construct a connectivity model (Figure 8A).
(D–J) Representative histology of rabies-mediated retrograde tracing from CeC Prkcd+ neurons (D), CeL Prkcd+ neurons (E), CeL Sst+ neurons (F), CeL Crh+Nts+Tac2+ neurons (G), CeM Nts+ neurons (H), CeM Sst+ neurons (I), CeM Tac2+ neurons (J) in the CeA and BLA. Starter cells are labeled with eGFP (green) and mCherry (red) and mouse line of starter cells are noted (italicized), Rabies virus is labeled with mCherry (red). Prkcd protein (purple) was labeled in the CeA. Ppp1r1b protein (purple) was labeled in BLA. The anterior-posterior (AP) distance from Bregma (mm), scale bar, 250 μm.
Monosynaptic tracing experiments were further analyzed using an antibody against PKC-δ for determining the retrograde labeled CeA cell-type (Prkcd+ or Prckd−). Monosynaptic tracing from CeL Prkcd, CeL Sst+, CeL Crh+Nts+Tac2+, CeM Nts+ and CeM Tac2+ neurons resulted in several (>5 neurons/section) retrograde labeled neurons in the CeC that were PKC-δ+ and PKC-δ− (Figure 5B). Monosynaptic tracing from CeC Prkcd, CeL Sst+, CeL Crh+Nts+Tac2+, CeM Nts+ and CeM Tac2+ neurons resulted in several (>5 neurons/section) retrograde labeled neurons in the CeL that were PKC-δ+ neurons (Figure 5C). Monosynaptic tracing from CeM Nts+ neurons resulted in several (>5 neurons/section) retrograde labeled neurons in the CeL that were PKC-δ− neurons (Figure 5C). Although no leaky viral expression was found (Figure S5E), due to possibilities of differential tropism across mouse lines, nonspecific targeting (Figure S5F), and unmeasurable leakiness, there may be more or less connectivity than demonstrated by these rabies experiments. Nevertheless, using an arbitrary threshold for retrograde labelling (>2.5 neurons/section in the BLA and >5 neurons/section in the CeA), a model of the BLA to CeA connectivity was generated (Figure 8A). Overall, these results demonstrate monosynaptic connections from BLA Ppp1r1b+ neurons to the CeA neurons that mediate appetitive behaviors and a monosynaptic connection from BLA Rspo2+ neurons primarily to CeC neurons that inhibit several of the CeA neurons that are capable of eliciting appetitive behavior.
Figure 8. Summary of anatomical and genetic results.
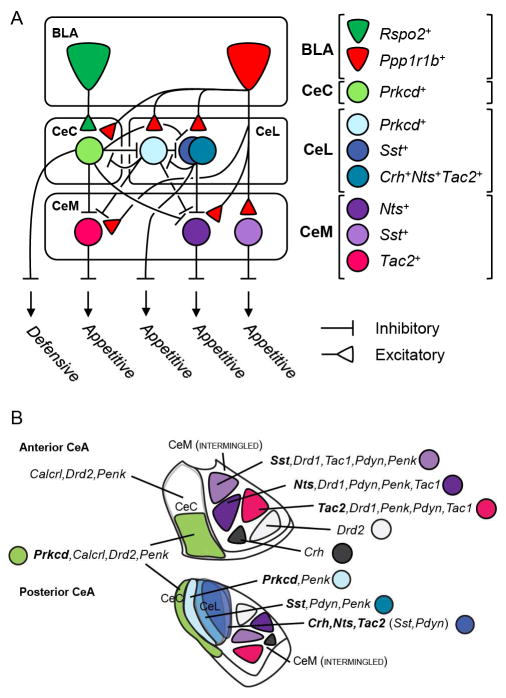
(A) Structural and functional model of cell-type specific BLA to CeA connectivity derived from monosynaptic rabies tracing experiments (Figure 2 and 5). BLA Rspo2+ neurons mainly innervate CeC Prkcd neurons, which in turn innervate several CeA neurons that mediate appetitive behaviors. BLA Ppp1r1b+ neurons innervate all CeA neurons in the model, CeA neurons that mediate appetitive behaviors as well as CeA Prkcd+ neurons, which in turn innervate CeA neurons that mediate appetitive behaviors.
(B) Graphical summary of genetically distinct CeA populations and their spatial distribution within the CeA. CeM neurons (Figure 1, 7), though not portrayed intermingled, are intermingled. CeL neurons are overall intermingled but also have a slight spatial segregation as depicted in the cartoon.
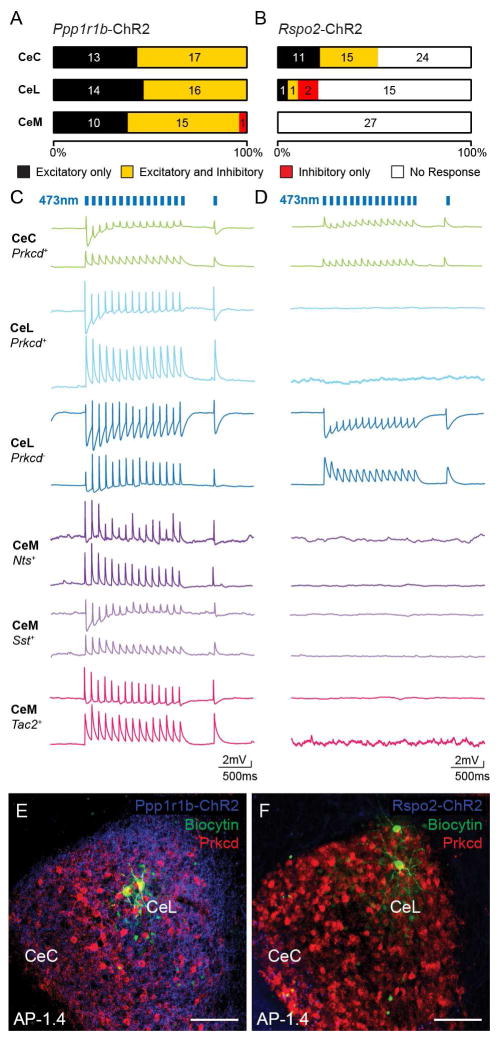
The BLA to CeA connectivity was further assessed using slice patch clamp recordings in conjunction with cell-type specific optogenetic stimulation of the BLA. Cre-dependent ChR2 virus was injected into the BLA of Rspo2-Cre and Cartpt-Cre mice for targeting BLA Rspo2+ (Rspo2-ChR2) and BLA Ppp1r1b+ (Ppp1r1b-ChR2) neurons, respectively (Kim et al., 2016). Patch clamped CeA neurons were recorded in response to ChR2 stimulation and were genetically identified using biocytin filling followed by immunohistochemistry against PKC-δ in the CeL or cytosolic harvesting followed by quantitative polymerase chain reaction (qPCR) in the CeC and CeM. The electrical properties were also measured of these neurons (Figure S6A–D). Blue-light stimulation in Ppp1r1b-ChR2 slices resulted in monosynaptic excitation, determined by latency (Figure S6F–S6I), in 100% of neurons in the CeC, 100% of neurons in the CeL, and 97% of neurons in the CeM. Among these neurons, disynaptic inhibition was also observed following monosynaptic excitation in 57% of neurons in the CeC, 53% of neurons in the CeL, and 60% of neurons in the CeM (Figure 6A). Connections with only disynaptic inhibition were observed in 3% of neurons in the CeM. Based on genetic marker-based confirmation (Figure S6E and S6J), monosynaptic excitatory connections were observed in CeC Prkcd+, CeL Prkcd+, CeM Nts+, CeM Sst+, and CeM Tac2+ neurons (Figure 6C and S6J). Blue light stimulation in Rspo2-ChR2 slices resulted in monosynaptic excitation in 52% of neurons in the CeC, 11% of neurons in the CeL, and 0% of neurons in the CeM. Among these neurons, disynaptic inhibition following monosynaptic excitation was observed in 58% of neurons in the CeC and 50% of neurons in the CeL. Connections with only disynaptic inhibition were observed in 11% of CeL neurons (Figure 6B). Based on genetic marker-based confirmation (Figure S6E and S6K), monosynaptic excitatory connections were observed in CeC Prkcd+ neurons as well as a CeL Prkcd− neuron (Figure 6D and S6K). These results show that BLA Rspo2+ neurons mainly innervate CeC Prkcd+ neurons, while making minimal connections to the CeL and CeM. In contrast, BLA Ppp1r1b+ neurons innervate CeC Prkcd+, CeL Prkcd+, CeM Nts+, CeM Sst+, and CeM Tac2+ neurons. In addition, 100% connectivity from BLA Ppp1r1b+ neurons to CeL neurons suggests that BLA Ppp1r1b+ neurons also innervate CeL Sst+ and CeL Crh+Nts+Tac2+. Although the identity of the neurons that mediates the polysynaptic inhibitory responses cannot be identified from these experiments, these functional experiments confirm the results of the rabies tracing experiments (Figure 5) and connectivity model of the BLA to CeA connectivity (Figure 8A).
Figure 6. BLA Ppp1r1rb + and Rspo2+ neurons make monosynaptic excitatory and disynaptic inhibitory connections to distinct CeA neurons.
(A–B) The proportion and number of CeC, CeL, and CeM neurons that receive only excitatory (black), both excitatory and inhibitory (yellow), only inhibitory (red), or no response (white) from blue light stimulation of BLA Ppp1r1b-ChR2 fibers (A) or BLA Rspo2-ChR2 fibers (B). Numbers inside bars represent total number of neurons for each case.
(C–D) Example voltage clamped traces of IHC or qPCR-confirmed CeC Prkcd+, CeL Prkcd+, CeL Prkcd−, CeM Nts+, CeM Sst+, and CeM Tac2+ neurons in response to blue light stimulation of BLA Ppp1r1b-ChR2 fibers (C) or BLA Rspo2-ChR2 fibers (D). Bottom traces represent responses at ~−70 mV and top traces represent responses at ~−50 mV. Counts of genetically confirmed neurons found in Figure S6J and S6K.
(E–F) Representative histology of IHC confirmation of CeL Prkcd+ neurons in BLA Ppp1r1b-ChR2 slices (E) and CeL Prkcd− neurons in BLA Rspo2-ChR2 slices (F). The anterior-posterior (AP) distance from Bregma (mm), scale bar, 100 μm.
The appetitive and aversive BLA to CeA projections are analogous to cortical projections to striatial direct- and indirect-pathway neurons
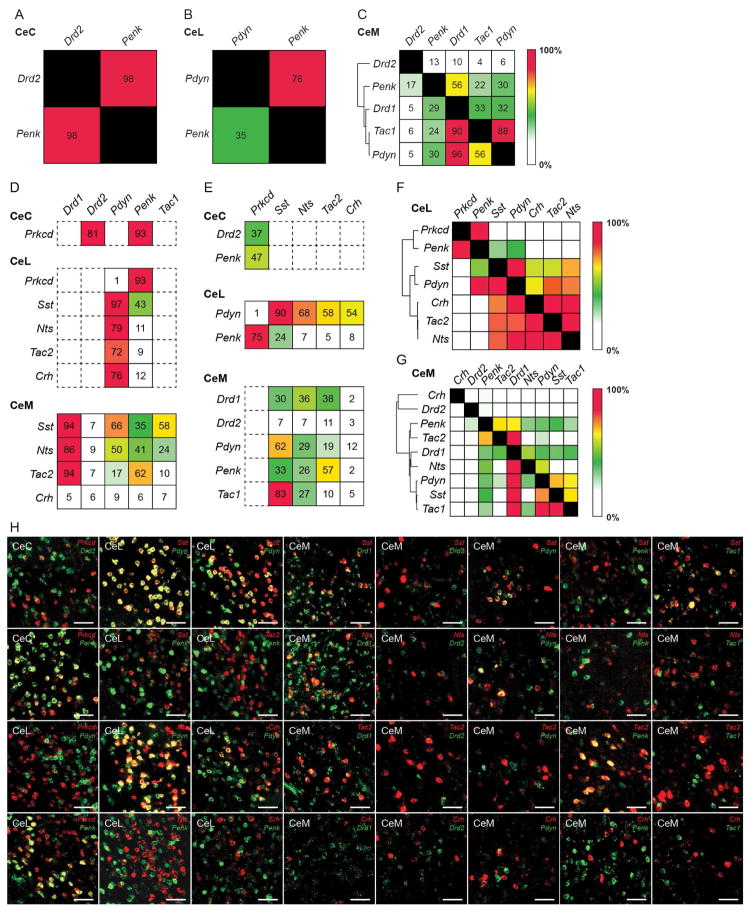
The BLA and CeA are cytoarchitecturally similar to the cortex and striatum, respectively (Carlsen and Heimer, 1988; Swanson and Petrovich, 1998). In the cortex, the direct pathway promotes movement and is characterized by intratelencephalic-type (IT-type) cortical neurons innervating dopamine receptor 1+ (Drd1+), dynorphin+ (Pdyn+), and substance P+ (Tac1+) striatonigral medium spiny neurons. The indirect pathway inhibits movement and is characterized by pyramidal tract-type (PT-type) neurons innervating dopamine receptor 2+ (Drd2+) and enkephalin+ (Penk+) striatopallidal medium spiny neurons (Gerfen et al., 1990; Reiner et al., 2010; Shepherd, 2013; Smith et al., 1998). Therefore, the expression of Drd1, Drd2, Pdyn, Penk, and Tac1 was examined in the CeA to examine how the BLA to CeA circuit is organized compared to the direct and indirect pathway of the cortex and striatum. Drd2 and Penk were expressed in the CeC. Pdyn and Penk were expressed in the CeL. Drd1, Drd2, Pdyn, Penk, and Tac1 were expressed in the CeM (Figure S7A and S7B). Overlap of the expression of these striatal markers was examined in the CeA (Figure 7A–B, S7C). In the CeC, Penk and Drd2 were highly (>90%) overlapping (Figure 7A). In the CeL, Penk labeled 78% of Pdyn neurons, while Pdyn labeled 35% of Penk neurons (Figure 7B,). In the CeM, hierarchical clustering using overlaps of the genes showed 2 major clusters. The first was Drd2, which was minimally overlapping (<15%) with the other markers. The second contained, Drd1, Pdyn, and Penk, which all moderately overlap with one another (~30–60%) except for Pdyn neurons, with which most or all (>90%) coexpressed Drd1 (Figure 7C). Using these sets of striatal markers, 7 or 8 genetically and regionally distinct populations can be identified. It should be noted that Drd2 expression in the CeM is questionable. Though Drd2 is expressed within the bounds of the CeM, its sparseness and expression pattern may reflect expression in the ventromedial extent of the CeC rather than the CeM. Further assessment using CTB retrograde tracing from the PAG resulted in no detectable CTB+ neurons that were Drd2+ (data not shown). Thus, like CeC neurons, Drd2+ neurons likely do not project to the PAG and therefore these CeM Drd2+ neurons may reflect CeC Drd2+ neurons.
Figure 7. BLA to CeA pathway for appetitive behavior is genetically analogous to corticostriatal circuits.
(A–C) Expression of striatal genetic markers in the CeA. Quantification of overlap of Drd2 and Penk in the CeC (A). Quantification of overlap of Pdyn and Penk in the CeL (B). Quantification of overlap of Drd2, Penk, Drd1, Tac1, and Pdyn in the CeM (C). Values represent percent labelling of overlap of genes in column amongst genes in rows. For example, 35% of CeL Penk labeled neurons coexpress Pdyn (B). Values represent percentage of labeling from totaling all cells counted from n = 3 mice. Hierarchical clustering was performed in the CeM using the percent overlap profile of each gene (C).
(D–E) Quantification of overlap of striatal markers—Drd1, Drd2, Pdyn, Penk and Tac1—amongst Prkcd+ neurons in the CeC; Prkcd+, Sst+, Nts+, Tac2+, and Crh+ neurons in the CeL; Sst+, Nts+, Tac2+, and Crh+ neurons in the CeM (D). Quantification of overlap of Prkcd, Sst, Nts, Tac2, Crh amongst Drd1+ and Penk+ neurons in the CeC; Pdyn+ and Penk+ neurons in the CeL; Drd1+, Drd2+, Pdyn+, Penk+ and Tac1+ neurons in the CeM (E). Values represent percentage of labeling from totaling all cells counted from n = 3 mice.
(F–G) Overlap matrix of genes expressed in the CeL (F) and CeM (G). Values represent percentage of labeling from totaling all cells counted from n = 3 mice and include the percentage of labeling values found in Figure 1. Hierarchical clustering was performed from using the overlap profile of each gene.
(H) Representative histology of CeA expression of CeA genetic markers (Prkcd, Sst, Nts, Tac2, Crh) with striatal markers (Drd1, Drd2, Pdyn, Penk Tac1). Scale bar, 50 μm.
The relationship between striatal markers and the CeA markers that were behaviorally and functionally characterized was examined using smFISH (Figure 7D–H). In the CeC, Drd2 and Penk were coexpressed in the majority (>80%) of Prkcd+ neurons (Figure 7D), while Prkcd was coexpressed in a subset (~40%) of Drd2 and Penk neurons (Figure 7E). In the CeL, Penk labeled the vast majority (>90%) of Prkcd+ neurons and a subpopulation of Sst+ neurons. Pdyn labeled virtually all (97%) of Sst+ neurons and the majority (70–80%) of Nts+, Tac2+, and Crh+ neurons (Figure 7D). In the CeL, heirarchical clustering using overlaps between all the genes revealed 3 major clusters, the first containing Penk and Prkcd, the second containing Pdyn and Sst, and the third containing Crh, Nts, and Tac2 (Figure 7F). In the CeM, Drd1 was coexpressed in the majority (>85%) of Nts+, Sst+, and Tac2+ neurons. Pdyn and Penk were expressed in a subpopulation of Nts+, Sst+, and Tac2+ neurons, with slightly more expression of Pdyn in Sst+ neurons and more expression of Penk in Tac2+ neurons (Figure 7D). In the CeM, heirarchical clustering using overlaps between all the genes revealed 2 majors clusters, the first containing Drd2, the second containing Drd1, which can be further clustered into 3 groups—Penk and Tac2; Drd1 and Nts; Pdyn, Sst and Tac1 (Figure 7G). Results of gene expression in the CeA were summarized in a model (Figure 8B). These results show that CeC Prkcd+ neurons express the striatal markers for the corticostriatal indirect pathway, Drd2+ and Penk+, while CeM Nts+, CeM Sst+, and CeM Tac2+ neurons mainly express the striatal markers for the corticostriatal direct pathway, Drd1, Pdyn and Tac1 (Gerfen et al., 1990; Smith et al., 1998).
Discussion
Here, we identified a set of CeA neurons that are positive mediators for appetitive behaviors—CeL Sst+, CeL Crh+Nts+Tac2+, CeM Nts+, CeM Sst+, and CeM Tac2+. These mediators of appetitive behavior receive monosynaptic input from BLA Ppp1r1b+ neurons, while several of them also receive disynaptic input from BLA Rspo2+ neurons. Given that BLA Ppp1r1b+ neurons are capable of driving appetitive behaviors and BLA Rspo2+ neurons are capable of suppressing appetitive behaviors (Kim et al., 2016), these two pathways from the BLA to CeA delineate an opposing circuit in the amygdala for the control of appetitive behaviors.
The CeA has an integral role in appetitive behaviors. The PAG has been shown to be a key site for executing defensive behaviors (Bandler and Depaulis, 1988; Kim et al., 1993) and several amygdala models hypothesized that the PAG-projecting CeA neurons mediate defensive behaviors (Duvarci and Pare, 2014; Fanselow, 1991; Herry and Johansen, 2014; Tovote et al., 2016). Although Cre transgenics and targeted virus injections would not necessarily give absolute selectivity when it come to the functional study of neurons, none of the CeA neurons that were identified in this study that project to the PAG (Figure S5A–D)—CeL Sst+, CeL Crh+Nts+Tac2+, CeM Nts+, CeM Sst+, and CeM Tac2+ neurons—elicit or are required for defensive behaviors or respond (using the expression of Fos) to stimuli that elicit defensive behaviors (Figure 2–4). Rather, these PAG-projecting populations elicit appetitive behaviors (though not necessarily through their projections to the PAG) and were activated by stimuli that elicit or are associated with appetitive behaviors (Figure 2,3). Furthermore, although the global action of several of the genes (Crh, Nts, Pdyn, Sst, Tac2, Tac1) that were found to be expressed in these PAG-projecting CeA neurons have been traditionally thought to be involved in negative behaviors and affective states, several studies that have examined the roles of these genes in the CeA and/or PAG have shown an opposing effect of these neuropeptides on negative behaviors or a positive role in appetitive behaviors, which is consist with our findings (Bilkei-Gorzo et al., 2012; Cui et al., 2004; Helmchen et al., 1995; Laszlo et al., 2010; Merali et al., 1998; Rosen et al., 2004). Collectively, the populations that were examined in this study constitute almost all neurons (>90%) in the CeL and CeM, reinforcing the idea that the CeA participates in appetitive behaviors. Moreover, these data suggest that the main route for conveying defensive information from the BLA to PAG may involve an alternative circuit that does not involve the CeL or CeM. Nevertheless, we do not disregard the role of the CeA in defensive behavior, as CeC Prkcd+ neurons participate in defensive behaviors (Figure 2A,4A). In addition, neither do we preclude the role of PAG-projecting CeA neurons in defensive or conditioned defensive behaviors; nor the possibility of an unidentified neuronal population or a subpopulation of one of these populations for regulating defensive behaviors. However, in light of our findings and previous conflicting reports on the role of the CeA in defensive behavior (Cai et al., 2014; Koo et al., 2004; LeDoux et al., 1988; Li et al., 2013), further studies will be required to resolve how, if at all, defensive behaviors are positively mediated by the pathway from the CeA to PAG. Nevertheless, the involvement of several distinct CeA projection neurons as positive mediators of appetitive behavior validates the integral role of the CeA in reward-related function (Badrinarayan et al., 2012; Gallagher and Chiba, 1996; Gallagher et al., 1990; Knapska et al., 2006; Parkinson et al., 2000; Ron and Barak, 2016).
CeC Prkcd+ and CeL Prkcd+ neurons as regulators of defensive and appetitive behaviors
Studies on Prkcd+ neurons in the CeA have suggested that there may be functional diversity among Prkcd+ CeA neurons. An early study on Prkcd+ neurons suggested a role in inhibition of defensive behavior (Haubensak et al., 2010). More recent studies have shown that CeA Prkcd+ neurons inhibit feeding behavior (Cai et al., 2014) and that a subpopulation of Prkcd+ neurons, CeA Calcrl,+ neurons, elicit and are required for defensive behavior (Han et al., 2015). Here, based on the gene expression patterning of Calcrl and Prkcd, Calcrl+ neurons define neurons in the CeC (Figure S1B) (D’Hanis et al., 2007) rather than what was previously reported as the CeL in the study that described CeA Calcrl+ neurons in defensive behaviors (Han et al., 2015). Prkcd+ neurons reside in the CeL as well as define a subpopulation of Calcrl+ neurons in CeC (Figure 1A) (Han et al., 2015). Considering this structural distinction, experiments between these two types of Prkcd+ CeA neurons yielded behavioral, functional, and connectivity dissociations. CeC Prkcd+ neurons elicit and are required for defensive behaviors and are activated by stimuli that drive defensive behaviors (Figure 2A and 3A). This is consistent with the reported role of CeA Calcrl,+ neurons in defensive behaviors (Han et al., 2015). In contrast, CeL Prkcd+ neurons do not drive defensive behaviors, but are activated by contextual fear extinction (Figure 2B and 3B). This is consistent with the initially hypothesized role of inhibition of defensive behaviors (Haubensak et al., 2010). Based on findings from retrograde rabies experiments, CeC Prkcd+ and CeL Prkcd+ are reciprocally connected (Figure 8A). Thus, Prkcd+ neurons in the CeA represent two distinct populations that have opposing functions on defensive behavior and we speculate that CeL Prkcd+ neurons and CeC Calcrl+ neurons, rather than CeL Prkcd+ and CeL Sst+ neurons (Duvarci and Pare, 2014; Herry and Johansen, 2014; Li et al., 2013), may represent the electrophysiological opposing units for fear-related responses in the CeA (Ciocchi et al., 2010; Haubensak et al., 2010).
With regard to appetitive behaviors, CeC Prkcd+ and CeL Prkcd+ neurons both directly inhibit mediators of appetitive behavior in the CeL and CeM (Figure 8A). Both Prkcd+ populations are connected in such a way that they can support an inhibitory role on appetitive behaviors, but may be functionally distinct based on differences in Fos activation profile and anatomical inputs. CeC Prkcd+ neurons are activated by threatening stimuli and aversive tastes (Figure 3A and 3D) and receive input from neurons that respond to aversive stimuli, BLA Rspo2+ neurons (Figure 8A) and calcitonin-related polypeptide, alpha (Calca) expressing neurons of the lateral parabrachial nucleus (by consideration that CeC Prkcd+ neurons are a subpopulation of CeC Calcrl+ neurons) (Carter et al., 2013; Han et al., 2015; Kim et al., 2016). In contrast, CeL Prkcd+ neurons are activated by states of satiety (Cai et al., 2014) (Figure 3E) and receive input from BLA Ppp1r1b+ neurons (Figure 8A), which respond to reward-related stimuli and Calca+ neurons of the lateral parabrachial nucleus, which in addition to responding to threat, also respond to states of satiety (Campos et al., 2016; Kim et al., 2016). Therefore, furthering previous findings and proposals on the role of CeA Prkcd+ neurons (Cai et al., 2014), these results suggest that the role of CeC Prkcd+ (and by extension CeC Calcrl+) neurons is to signal the inhibition of appetitive behaviors mainly in response to aversive stimuli such as threat and aversive tastes, while the role of CeL Prkcd+ neurons is to signal the inhibition of appetitive behaviors in response to positive states such as satiety.
CeL Sst+ and CeL Crh+Nts+Tac2+ neurons as positive mediators for drinking
CeL Sst+ and CeL Crh+Nts+Tac2+ neurons are Prkcd− neurons of the CeL. CeL Prkcd− neurons have been hypothesized to be positive mediators of fear-related behaviors (Duvarci and Pare, 2014; Herry and Johansen, 2014). A previous study has demonstrated that CeL Sst+ neurons elicit and are required for defensive behaviors (Li et al., 2013). Contrary to these early findings, a more recent study suggested that activation of CeL Prkcd− neurons promotes feeding (Cai et al., 2014), while another study demonstrated that activation of CeL Prkcd− neurons suppresses defensive behaviors evoked by innate threatening odors (Isosaka et al., 2015). Here, we were unable to find evidence for CeL Sst+ or CeL Crh+Nts+Tac2+ neurons as mediators of defensive behavior (Figure 2–4). We speculate the main differences in our results from the original work on CeL Sst+ neurons in defensive behaviors is likely attributed to our use of lower volume of virus, 100 nL of ~1012 vs 300 to 800 nL of ~1012 viral particles, resulting in more specific targeting of the CeL (Li et al., 2013). Although a disinhibitory pathway that supports defensive behaviors from BLA Rspo2+ neurons (positive mediators of defensive behaviors) to CeL Prkcd− neurons is anatomically identifiable (BLA Rspo2+ to CeC Prkcd+ to CeL Prkcd+ to CeL Prkcd−), a disynaptic inhibitory connection is also identifiable from BLA Rspo2+ neurons, through CeC Prkcd+ neurons to CeL Prkcd− neurons, which does not support defensive behaviors (Figure 8A). Moreover, CeL Prkcd− neurons receive direct innervation from BLA Ppp1r1b+ neurons (Figure 8A), which suppress defensive behaviors and elicit appetitive behaviors (Kim et al., 2016). Thus, the anatomical and functional connectivity of Prkcd− neurons in relation to BLA neurons does not fully support a positive role in defensive behavior.
With regard to appetitive behaviors, CeL Sst+ and CeL Crh+Nts+Tac2+ neurons are capable of eliciting appetitive behaviors, are critical for drinking, and are strongly activated by water in addition to food (Figure 2C,2D,3D,4C,4D). Moreover, inhibition of CeL Prkcd+ neurons, which are reciprocally connected to CeL Prkcd− neurons, results in the enhancement of drinking behavior (Figure 4B) and a previous study demonstrated that activation of CeL Prkcd+ neurons suppresses drinking behavior (Cai et al., 2014). Together, these results indicate that CeL Sst+ and CeL Crh+Nts+Tac2+ neurons are positive mediators of appetitive behavior and mainly participate in drinking-related function. These results bring insight to previous studies that have implicated the CeL in drinking and alcohol-related behaviors (Dalmasso et al., 2015; Hwang et al., 2004; Kissler et al., 2014; Nie et al., 2004; Pereira-Derderian et al., 2010). With regard to the function of the two types of Prkcd− neurons, examination of CeL Sst+ and CeL Crh+Nts+Tac2+ neurons revealed no functional, behavioral, or connectivity differences. CeL Sst+ and CeL Crh+Nts+Tac2+ are highly overlapping populations; ~50% of CeL Sst+ are CeL Crh+Nts+Tac2+ and ~70% of CeL Crh+Nts+Tac2+ are CeL Sst+ (Figure 1B). Therefore, it was not unexpected that a structural or functional dissociation was not found.
CeM Nts+, Sst+, and Tac2+ neurons as positive mediators for appetitive behaviors
Genetically distinct CeM populations—Nts+, Sst+, Tac2+—were found to participate in appetitive behaviors. Though gene expression of striatal markers in the CeM suggests there may be alternative ways to divide CeM populations, these markers define 3 major well segregated Drd1+ neurons (Figure 1C, 7D). Behaviorally, these populations elicit appetitive behaviors and collective silencing of these neurons results in reduced feeding and drinking (Figure S4). This is in contrast to CeL mediators of appetitive behavior, which appear to have a specific role in appetitive drinking behavior. Therefore, CeM neurons may have a more general role in appetitive behaviors. Alternatively, but not exclusively, the mediators of appetitive behavior in the CeL and CeM may function together to execute and regulate distinct behavioral programs for different types of appetitive behaviors and reward-related states. Although we did not assess the nuances of the different aspects of appetitive behaviors and reward-related phenotypes (Berridge and Robinson, 2003) or evaluate more long-term effects of silencing these neurons, further studies will be required to further dissociate the role of these distinct populations in appetitive behaviors.
Direct and indirect pathways of the amygdala for appetitive behavior
The BLA to CeA circuit that promotes and suppresses appetitive behaviors is analogous to the cortex and striatum of the direct and indirect pathway of the basal ganglia that promotes and suppresses movement (Smith et al., 1998). CeM mediators of appetitive behavior express Drd1, Pdyn, and Tac1 (Figure 7D). Although Penk is expressed in a subset of CeM mediators of appetitive behavior, Drd2 is only minimally expressed (Figure 7D). CeM mediators of appetitive behavior directly receive monosynaptic input from excitatory Ppp1r1b+ parvocellular neurons of the BLA (Figure 8A), which are also capable of promoting appetitive behaviors (Kim et al., 2016). Hence, the pathway for promoting appetitive behaviors from the BLA to CeA is genetically and structurally analogous to cortex and striatum of the direct pathway, which involves the direct innervation of Drd1+, Pdyn+, and Tac1+ striatonigral neurons from excitatory IT-type neurons of the cortex (Gerfen et al., 1990; Reiner et al., 2010; Shepherd, 2013; Smith et al., 1998). The BLA to CeA pathway that supports the suppression of appetitive behavior emanates from BLA Rspo2+ neurons and also may involve a subset of BLA Ppp1r1b+ neurons in consideration of the connection from BLA Ppp1r1b+ to CeA Prkcd+ neurons (Figure 8A). Although BLA Rspo2+ neurons are capable of driving defensive behaviors, they are also capable of suppressing appetitive behaviors (Kim et al., 2016) and form disynaptic inhibitory connections to mediators of appetitive behavior in the CeM and CeL (Figure 8A). The pathway from BLA Rspo2+ neurons to CeA mediators of appetitive behavior involves an intermediate step in the CeC that express Drd2 and Penk, but does not express Drd1, Pdyn, or Tac1 (Figure 7D). Hence, the pathway for suppressing appetitive behaviors, from excitatory Rspo2+ magnocellular neurons of the BLA to the CeA, is functionally and genetically analogous to the cortex and striatum of the indirect pathway, which involves the direct innervation of Drd2+ and Penk+ striatopallidal neurons from PT-type neurons of the cortex (Gerfen et al., 1990; Reiner et al., 2010; Shepherd, 2013; Smith et al., 1998). Interestingly, the IT-type cortical neurons (direct pathway) are smaller in soma size than PT-types cortical neurons (indirect pathway) (Reiner et al., 2003), while BLA Ppp1r1b+ parvocellular neurons are smaller in soma size as BLA Rpso2+ magnocellular neurons (Kim et al., 2016). This suggests that the two types of BLA neurons may also be morphologically analogous to the two types of corticostriatal neurons. Although an analogy can be described between the BLA and CeA with the cortex and striatum, the output structures and circuitry of the CeA were not fully examined in this study. Thus, future studies will be required to examine if the output of the CeA share any organizing principle with the output circuitry of the striatum of the direct and indirect pathway.
Overall, the dissection of the connectivity from the BLA to CeA and examination of the expression of striatal genetic markers in the CeA revealed a corticostrial-like direct and indirect pathway between the BLA and CeA for the promotion and suppression of appetitive behaviors. The duplication and then specialization of structure and function is a common paradigm found in development and evolution. In the brain, this type of phenomenon is apparent in the stereotypic laminar organization of neurons across the cortex and between the glomeruli of the olfactory bulb (Ramón y Cajal, 1995). The similarities in architecture, genetics, and connectivity between the BLA/CeA compared to the cortex/striatum suggest that such an organizing principle of duplication and specialization may have occurred to give rise to what may be considered a corticostriatal neural circuit motif in the amygdala for appetitive behaviors.
CONTACT FOR REAGENT AND RESOURCE SHARING
Materials, datasets, and protocols are available upon request to the corresponding author, Susumu Tonegawa (tonegawa@mit.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
6- to 12-week-old male mice were used for all experiments, except 5- to 6-week-old male mice were used for slice patch clamp experiments. All Cre transgenic mice were bred using a heterozygous male with females of C57BL/6 background. Crh-Cre, Drd1a-Cre, Rspo2-Cre, Sst-Cre, and Tac2-Cre mice had a C57BL/6 background. Cartpt-Cre, Nts-Cre, and Prkcd-Cre mice were crossed to a C57BL/6 background for at least 2 generations from their original backgrounds from JAX laboratories or GENSAT. Cre-expressing mice were genetic knock-in mice or has been previously been validated for genetic specificity (Haubensak et al., 2010; Kim et al., 2016). BLA Rspo2+ neurons were targeted using Rspo2-Cre mice. BLA Ppp1r1b+ neurons were targeted using Cartpt-Cre mice (Kim et al., 2016). CeC Prkcd+ and CeL Prkcd+ neurons were targeted using Prkcd-Cre mice. CeL Sst+ and CeM Sst+ neurons were targeted using Sst-Cre mice. CeL Crh+Nts+Tac2+ neurons were targeted using Crh-Cre mice. CeM Nts+ neurons were targeted using Nts-cre mice. CeM Tac2+ neurons were targeted using Tac2-Cre mice. CeM Drd1+ neurons were targeted using Drd1a-Cre mice. Mice undergoing all behavioral tests (optogenetic experiments, Fos experiments) were single housed for 1 week prior to experiments and kept on a 12 hr light, 12 hr dark light cycle. Control mice for behavioral experiments underwent identical procedures as experimental mice, but were Cre− mice of the same sex from the same litters. Mice that underwent slice patch clamp experiments or rabies tracing experiments were grouped housed. All mice were maintained and cared in accordance with protocols approved by the Massachusetts Institute of Technology (MIT) Committee on Animal Care (CAC) and guidelines by the National Institutes of Health (NIH).
METHOD DETAILS
Single Molecule Fluorescent In Situ Hybridization
For examination of gene expression and Fos experiments, tissue samples underwent single molecule fluorescent in situ hybridization (smFISH). Isoflurane anesthetized mice were decapitated, brain harvested and flash frozen on aluminum foil on dry ice. Brains were stored at −80 °C. Prior to sectioning, brains were equilibrated to −16 °C in a cryostat for 30 min. Brains were cryostat sectioned coronally at 20 μm and thaw-mounted onto Superfrost Plus slides (25x75 mm, Fisherbrand). Sections from a single brain would be serially thaw-mounted onto 10 slides through the CeA, anterior-posterior distance from Bregma (−0.6 mm to −1.8 mm). Slides were air dried for 60 to 90 min prior to storage at −80 °C. smFISH for all genes examined—Calcrl (ACDBio Cat#452281), Crh (ACDBio Cat#316091), Drd1 (ACDBio Cat#406491), Drd2 (ACDBio Cat#406501), Fos (ACDBio Cat#421981), Gad1 (ACDBio Cat#400951), Htr2a (ACDBio Cat#401291), Nts (ACDBio Cat#420441), Pdyn (ACDBio Cat#318771), Penk (ACDBio Cat#318761), Prkcd (ACDBio Cat#441791), Sst (ACDBio Cat#404631), Tac1 (ACDBio Cat#410351), Tac2 (ACDBio Cat#446391)—was performed using RNAscope Fluorescent Multipex Kit (Advanced Cell Diagnostics) as previously described (Kim et al., 2016). Slides were counterstained for the nuclear marker DAPI using ProLong Diamond Antifade mounting medium with DAPI (ThermosFisher).
Immunohistochemistry
For visualizing Prkcd, Ppp1r1b, Rabies helper virus expression (eGFP), ChR2-eYFP and ArchT-eYFP expression, tissue samples underwent immunohistochemistry (IHC). Mice were euthanized by avertin overdose and underwent a standard perfusion protocol using 1X phosphate-buffered saline (PBS), followed by 4% paraformaldehyde. Mouse brains removed and postfixed overnight at room temperature. Brains were vibratome sectioned at 50 μm and collected through the CeA, anterior-posterior distance from Bregma (−0.6 mm to −1.8 mm). IHC was performed as previously described (Kim et al., 2016) with primary antibodies anti-rabbit PKC-δ (1:1000, Abcam Cat#ab18212), anti-rabbit PPP1R1B (1:1000, Abcam Cat# ab40801), chicken anti-GFP (Invitrogen Cat#A10262) and secondary antibodies Alexa Fluor 647-conjugated Goat anti-Rabbit (1:1000, Invitrogen Cat#A21244), Alexa Fluor 488-conjugated Goat anti-chicken (1:1000, Invitrogen Cat#A11039). In the slice patch clamp experiments, brain slices were fixed for 4 hr at 4 °C prior to undergoing the IHC protocol (Kim et al., 2016), biocytin was labelled using CF555 Strepatvidin (1:100, Biotium Cat#29038) during the secondary antibody incubation step.
Cell Counting
Images of smFISH and retrograde rabies tracing experiments were taken using a standard fluorescent microscope (Zeiss) under a 10X objective. Colors represented in micrographs are false colors and do not necessarily reflect native colors. Images were exported and counted manually using cell counting software (ImageJ). Percent labelling found in Figure 1 and 7 were quantified by counting 1 to 3 sections equal number per mouse, from n = 3 mice and sum totaling all single and double labelled neurons to yield a raw percentage value. The bounds of what considered the CeC, CeL, and CeM were used as shown in cartoons in Figure S1B and S7B. Although CeC in the posterior portion of the CeA (anterior-posterior Bregma −1.6 mm) is depicted in Figures and defined by atlas boundaries lying laterally to the CeL, the posterior CeC was not quantified in any quantification of CeC neurons as the boundary between the CeC and CeL in the posterior CeA is ambiguous.
Stereotaxic Surgeries
Mice underwent standard stereotactic procedures under isoflurane anesthesia. Vectors were injected using a mineral oil filled glass micropipette attached to a 1 μL microsyringe. For optogenetic experiments of the CeA, 100nL of AAV9-Ef1a-DIO-ChR2 or AAV9-Ef1a-DIO-eArch 3.0 was bilaterally injected into the CeC (distance from Bregma, AP-0.8 mm, ML±2.9 mm, DV-5.0 mm), CeL (AP-1.4 mm, ML±2.9 mm, DV-4.7 mm), or CeM (AP-0.8 mm, ML±2.8 mm, DV-5.0 mm) of the different CeA Cre mice. Both ChR2 and Arch viruses were diluted in 1X phosphate buffer (PBS, pH 7.2), one part stock virus, 3 parts PBS to give a final concentration of ~1.0x1012 GC. Subsequent to injections, 5.0 mm Mono fiberoptic cannulas (Doric Lens) were implanted above the site of injection of the CeC (DV-4.4 mm), CeL (DV-4.2 mm), or CeM (DV-4.4 mm). Once positioned above the CeA, the mono fiberoptic cannula was cemented using dental cement (Teets cold cure; A-M Systems) to the skull, which contained 2 screws that were posterior and medial to the injection site. Once the dental cement cured, a protective cap surrounding the implant, made using a 1.5mL black Eppendorf tube, was fixed onto the implant using dental cement. Post-operation, mice received an injection of slow release buprenorphine (1mg/kg). Mice spent 1 week for recovery and then were handled by investigator 2–3 days prior to behavioral experiments. For slice patch clamp experiments, 200nL of AAV9-Ef1a-DIO-ChR2 was into the BLA of Rspo2-Cre (AP-1.3 mm, ML±3.3 mm, DV-4.85 mm) and Cartpt-Cre mice (AP-4.8 mm, ML±0.5 mm, DV-3.0 mm) and incubated for 1 week prior to sacrifice for the slice patch clamp experiments. For CTB retrograde tracing, Alexa Fluor 647-conjugated cholera toxin subunit B (1 μg/μL, Thermofischer Cat#C34778) was unilaterally injected into the PAG (300 nL, AP-1.0 mm, ML+2.9 mm, DV-4.5 mm). 1 week later, mice were sacrificed and brains underwent smFISH.
Optogenetic Activation Experiments
ChR2 virus-injected mice and Cre− control mice underwent an optogenetic freezing test followed by an optogenetic self-stimulation test. These optogenetic activation experiments were performed on cohorts of 6 to 12 mice and took place during first half of the dark cycle. The optogenetic freezing test involved exposing mice to conditioning chamber (Med Associates) for 6 min. During the 0–3-min period, mice did not receive any optogenetic stimulation. During the 3–6-min period, mice received continuous optogenetic stimulation, 10–15-mW 20-Hz 473-nm light stimulation. 1 day later, mice were food deprived for 24 hr prior to the start of the optogenetic self-stimulation test. Videos were captured using Video Freeze software (Med Associates). The optogenetic self-stimulation test took place in an operant conditioning chamber (Med Associates) equipped with a two nose ports. Prior to the trial, one of the two nose ports was randomly assigned to deliver optogenetic stimulation, 5-s duration 10–15-mW 20-Hz 473-nm light stimulation, upon nose poke (ON port), while the other one did not deliver optogenetic stimulation (OFF). Each of the nose points contained a single food pellet to initiate the mouse into the port. Mice were then placed into the operant chamber for 60 min. For the optogenetic freezing test, freezing was scored as the duration of freezing as percentage of total time of the trail on day 2 and from the onset of the first shock to the end of the experiment on day 1. Freezing was scored manually and blind to the condition of the mouse using behavioral scoring software (Solomon Coder). For the optogenetic self-stimulation test, total numbers of pokes on the OFF and ON port were automatically counted and obtained through MED-PC (Med Associates) software. Mice where lack of or non-specific ChR2 expression or improper optic fiber placement occurred were removed from behavioral analysis. 9 out of 52 animals were removed from analysis blind to behavioral results (Figure 2).
Fos Activation Experiments
For all Fos staining, 6 hr prior to sacrifice, food and water were removed from the home cages of C57BL/6 wild-type mice in order to reduce any unintended activation in the Fos experiments. Mice were exposed to a fear conditioning chamber (Med Associates) for 500 s in which 3 footshocks (.75mA) were delivered at the 198 s, 278 s, 358 s time points, while control mice underwent the same procedure, but did not receive any footshocks. Mice were returned to their home cages and 30 min later sacrificed. Mice undergoing fear extinction underwent the same 3 shock fear conditioning protocol, then 24 hr later, were exposed to the fear conditioning chamber without any footshocks 3 times, for 15 min. Mice spent 1 hr in between these 3 15-min extinction sessions. 24 hr later, mice were exposed to the fear conditioning chamber for 5-min, returned to their home cages, and sacrificed 30 min later. Control mice underwent similar procedures, but did not undergo the 3 15-min extinction sessions. Hence, control mice can be considered mice undergoing 48 hr contextual fear recall, rather than contextual fear extinction recall. 24 hr food-deprived mice were transported into an experimental room on a cart and then exposed to ad libitum food, in their home cages. After 30 min from the end of the first feeding bout, mice were sacrificed. Control mice underwent identical procedures but did not receive food. This involved carting the mice to the experimental room and opening the lid of their home cages. 24 hr water-deprived mice that were exposed to water in an analogous way food was presented to food-deprived mice. Quinine water exposed mice were given quinine water (.01% quinine, Sigma) rather than water, only mice that displayed aversion to quinine water (having drinking bouts that were less than 2–3 s) were sacrificed. Control mice underwent identical procedures but did not receive water. Mice were sacrificed 30 min after the exposure to the stimulus. Mice were given CCK injections (5 μg/kg of CCK dissolved in .9% sodium chloride saline solution, Tocris), while control mice received saline injections intraperitoneally within the first hour of the dark cycle. Mice were sacrificed 30 min after the injections. Fos expression in CeC Prkcd+ and CeL Prkcd+ neurons was examined using Prkcd (ACDBio, Cat#441791). Fos expression in CeL Sst+ and CeM Sst+ neurons was examined using Sst (ACDBio, Cat#404631). Fos expression in CeL Crh+Nts+Tac2+ and CeM Tac2+ neurons was examined using Tac2 (ACDBio, Cat#446391). Fos expression in CeM Nts+ neurons was examined using Nts (ACDBio, Cat#420441).
Optogenetic Inhibition Experiments
Arch virus-injected mice underwent a feeding test, followed by a drinking test, then followed by a contextual fear conditioning test. For the feeding test, mice were food-deprived for 24 hr prior to the start of the experiment, which took place during the first half of the dark cycle. For the feeding test, mice were exposed to ad libitum food in their home cages while receiving constant 10–15-mW 532-nm light inhibition for 20 min. After the feeding test, mice returned to their normal diet until the start of the light cycle (approximately 12 hr later) in which mice were water-deprived. Approximately 18 hr later, in the second half of the dark cycle, mice underwent the drinking test. In the drinking test, mice were exposed to ad libitum water for 5 min in a chamber identical to their home cages but without bedding. During the entirety of the 5 min drinking test, mice received constant 10–15-mW 532-nm light inhibition. After the test, mice were returned to their normal diet. The next day, mice underwent contextual fear conditioning. On day 1 of contextual fear conditioning, mice were placed into a fear conditioning chamber (Med Associates) for 500 s and received footshocks (.75mA) during the 198 s, 278 s, 358 s time points. Simultaneously with the onset of the footshock, a constant pulse of 532-nm light (10–15 mW) was delivered through the optical cannulas for duration of 30 s. On day 2, mice were returned to the fear conditioning chamber for 180 s, where no shock or laser was delivered. Feeding, drinking, and freezing were scored manually and blind to the condition of the mouse using behavioral scoring software (Solomon Coder). Behaviors were scored as a percentage, calculated by the duration of the time spend performing the behavior as a proportion of the behavioral trail time. Feeding was scored based on the mouse orally engaging food pellets (for instance, either actively chewing food or biting off small pieces of the larger food pellet) and drinking was scored based on the mouse orally contacting the water spout. Mice where lack of or non-specific Arch expression or improper optic fiber placement occurred were removed from behavioral analysis. 5 out of 137 animals were removed from analysis in a blind manner, prior to the scoring of behaviors (Figure 4 and Supplemental Figure 4).
Retrograde Rabies Tracing
For monosynaptic retrograde rabies experiments, 100nL of rabies helper virus, AAV1-synP-FLEX-sTpEpB, was injected into the CeC, CeL, or CeM or CeA of Cre-expressing mice. 3 weeks later, 100nL EnvA G-protein deleted rabies virus, SAD G-mCherry, was injected into the same location. 1 week later, mice were sacrificed and brains underwent IHC using antibodies against PKC-δ (Abcam, Cat#ab182126) or PPP1R1B (Abcam, Cat#ab40801) and visualized using Alexa Fluor 647-conjugated secondary antibody (Invitrogen, Cat#A21244). Micrographs (Figure 5) of rabies experiment were adjusted so that all immunofluorescent cells signals are able to be visualized.
Optogenetic Slice Electrophysiology
Mice were anesthetized by isoflurane and their brains dissected. Using a vibratome (VT1000S, Leica), we prepared 300-μm-thick coronal slices containing the basolateral and central amygdala in oxygenated cutting solution at ~4 °C. Slices were then incubated at ~23 °C in oxygenated artificial cerebrospinal fluid (ACSF) for 45 min to a 1 hr. The cutting solution contained 3 mM KCl, 0.5 mM CaCl2, 10 mM MgCl2, 25 mM NaHCO3, 1.2 mM NaHPO4, 10 mM D-glucose, 230 mM sucrose, saturated with 95% O/5% CO (pH 7.3, osmolarity 340 mOsm). The ACSF contained 124 mM NaCl, 3 mM KCl, 2 mM CaCl2, 1.3 mM MgSO4, 25 mM NaHCO3, 1.2 mM NaHPO4, 10 mM D-glucose, saturated with 95% O/5% CO (pH 7.3, osmolarity 300 mOsm). Slices were transferred to a submerged experimental chamber and perfused with oxygenated 36 °C ACSF at a rate of 3 ml min−1.
Whole-cell recordings in current-clamp mode were performed using a patch clamp setup with an infrared differential interference contrast microscope (BX51, Olympus) with a water immersion 40× objective (N.A. 0.8), four automatic micro-manipulators (Luigs & Neumann) and a CCD camera (Orca R2, Hamamatsu). Borosilicate glass pipettes were fabricated (P97, Sutter Instrument) with a resistances of 8–10 MΩ and filled with the following intracellular solution: 110 mM potassium gluconate, 10 mM KCl, 10 mM HEPES, 4 mM ATP, 0.3 mM GTP, 10 mM phosphocreatine and 0.5% biocytin (pH 7.25, osmolarity 290 mOsm). Biocytin was excluded from the intracellular solution if the cytosol of the patched neuron was aspirated for PCR. Access resistance was monitored throughout the duration of the experiment and data acquisition was suspended whenever the resting membrane potential was depolarized above −50 mV or the access resistance was beyond 20 MΩ. Recordings were amplified using up to two dual-channel amplifiers (Multiclamp 700B, Molecular Devices), filtered at 2 kHz, digitized (20 kHz) and acquired using custom software running on Igor Pro (Wavemetrics). Software and code are available upon request. Gabazine was obtained from Tocris.
Optogenetic stimulation was achieved through a 460-nm LED light source (XLED1, Lumen Dynamics) driven by TTL input with a delay onset of 25 μs (subtracted offline). Light power on the sample was 33 mW mm-2, and only the maximum power was employed. Slices were stimulated by a train of 15 light pulses at 10 Hz repeated 20 times every 6 s. EPSPs, IPSPs and action potentials were measured at resting membrane potential of the patched cell.
The intrinsic electrophysiological properties were measured in current clamp mode with the cell held at −70 mV. Input resistance was estimated by linear fit of the I–V relationship (injection of 10–12 current steps of 1-s duration). Action potential threshold was tested with a current ramp injection. Synaptic connections were verified by taking an average of 10 to 20 individual trials from each patched neuron held at −55mV and −75mV. EPSP amplitude was measured from the average maximum peak response by subtracting a baseline obtained 5 ms before light pulse starts. EPSP onset was measured from the beginning of the light pulse to the starting point of the response. The first differential of the response wave was calculated and the first point of maximum change in voltage (Y-axis) was detected. The corresponding point in time (X-axis) was noted. The difference between the two time points was determined to be the onset latency of the response. If a patched cell had only EPSPs at both −50 and −70mV holding potentials, the cell was determined to be excitatory only, putatively receiving only a direct fiber excitation. If a patched cell showed a combination of E and IPSPs at −50mV and EPSPs at −70mV, the cell was deemed to be excitatory and inhibitory, putatively receiving both direct glutamatergic excitation and the local interneuron generated feedforward inhibition. And finally, if the cell showed no EPSPs in response to light stimulation at both holding potentials, it was classified as inhibitory only. To compute the probability of connection (n successes/n tests) we used only slices with reliable ChR2 expression characterized at least by one responsive postsynaptic cell (principal cell or interneuron).
CeL neurons were filled with biocytin, slices were recovered and fixed in 4% paraformaldehyde for verification of genetic identify using IHC against Prkcd. Analysis for CeL Prkcd− neurons in addition ot CeL Prkcd+ neurons was taken into consideration.
CeC and CeM neurons were harvested for subsequent qPCR analysis. In order to harvest the RNA of recorded neurons, a negative pressure of 250mbar was applied for 5 min followed by a stronger negative pressure of 500mbar also for 5 min. Successful suction of the cytosolic contents was visually confirmed by observing the patched cell shrink on the microscope screen. The negative pressure was maintained until the glass pipette was quickly withdrawn. It was then carefully lowered into a 0.2ml PCR tube until its tip was felt against the bottom of the tube. Finally, gentle positive pressure was applied to expel the contents on the pipette into a qPCR buffer solution (see below).
Single Cell qPCR Genetic Confirmation
Cytosolic harvests was quickly transferred to .2mL PCR tube fill with 10 μL RNase free water, 2 μL oligo(dT), 1 μL dNTP, 1 μL RNaseOUT provided by the SuperScript III CellsDirect cDNA Synthesis Kit (ThermoFisher). Samples were placed on a 70 °C heat block for 5 min, and then chilled on ice. For first strand synthesis, 8 μL of RT mix was added to the sample (6 μL 5x RT Buffer, 1 μL .1M DTT, 1 μL Superscript III RT) and incubated on a 50 °C heat block for 50 min. Next, the reverse transcriptase was inactivated by 10 min incubation on an 85 °C heat block. Samples were stored in −20 °C until quantitative polymerase chain reaction (qPCR).
qPCR was performed using the Taqman Gene Expression Assays (Thermofisher) for Prkcd in CeC neurons or for Nts, Sst, and Tac2 in CeM neurons. Each qPCR reaction consisted of 25 μL 2X TaqMan Gene Expression Master Mix, 7 μL of cDNA template, 17.5 μL of RNase free water, and 2.5 μL of the 20X TaqMan Gene Expression Assay of Prkcd (Cat#Mm00440891_m1), Nts (Cat#Mm00481140_m1), Sst (Cat#Mm00436671_m1), or Tac2 (Cat#Mm01160362_m1). qPCR reaction was performed in an Applied Biosystems 7500 Real-Time PCR System using the Fluorescein (FAM) channel with the standard qPCR reaction protocol for 60 cycles. Any positive amplification signal (ΔRn) within the 60 reaction cycles was considered to be positive confirmation for such gene. In contrast to IHC confirmation of CeL neurons, negative results from CeC and CeM qPCR confirmation was not considered because of the high levels of false negatives in qPCR amplification.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data are represented as mean ± s.e.m.. All histograms display individual points, which represent the values and total number of individual samples. What the individual samples represent is indicated within the figure legends for all experiments. Student’s t-test, paired or unpaired wherever appropriate, was performed on all comparisons with exceptions being the one-way ANOVA with Bonferroni’s correction between the comparison between water and quinine with no water groups (Figure 3D) and one-way ANOVA with Bonferroni’s correction for multiple comparisons (Figure S6A and S6C). 95% confidence interval was used to determine significance. Significance was displayed as *P< 0.05, **P<0.01, ***P<0.001, ****P>0.0001, not significant values were not denoted. Statistical tests were performed using GraphPad Prism 6.0. Hierarchical clustering was performed by generating a matrix containing the percent overlap profile of each gene in a given CeA subdivision, then calculating the distance using the pairwise distance function (pdist), dendrograms were made using the linkage function (linkage) on Matlab8.3. The diagonals on the percent labelling matrices corresponded to identical gene pairs. Therefore, for the cluster, the diagonals of the matrices were denoted as 100%.
Supplementary Material
Highlights.
Several genetically distinct CeA neurons mediate appetitive behaviors
BLA Ppp1r1b+ neurons project to CeA neurons that mediate appetitive behaviors
BLA Rspo2+ neurons project to CeA neurons that suppress appetitive behaviors
BLA to CeA pathways are analogous to corticostriatal direct and indirect pathways
Acknowledgments
We thank Akiko Wagatsuma for designing the operant behavioral chamber. We thank Ian Wickersham for providing the reagents for retrograde rabies tracing. We thank Christopher J. MacDonald and Teru Okuyama for reviewing the manuscript. This work is supported in part by NIH Pre-Doctoral Training Grant T32GM007287 (to J.K.) and by the RIKEN Brain Science Institute, the Howard Hughes Medical Institute, and the JPB Foundation (to S.T.). The authors declare no conflict of interest.
Footnotes
Author Contributions:
Conceptualization, J.K. and S.T.; Methodology, J.K. and X.Z.; Investigation, J.K., X.Z., and S.M.; Writing – Original Draft, J.K.; Writing – Review & Editing, J.K., S.M., X,Z, and S.T.; Resources, J.K., X.Z., S.M., and S.A.L.; Visualization, J.K.; Supervision, J.K. and S.T.; Funding Acquisition, S.T.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andero R, Dias BG, Ressler KJ. A role for Tac2, NkB, and Nk3 receptor in normal and dysregulated fear memory consolidation. Neuron. 2014;83:444–454. doi: 10.1016/j.neuron.2014.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badrinarayan A, Prater KE, Orsini CA. The role of the central amygdala in selecting circuits and responses. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32:8431–8433. doi: 10.1523/JNEUROSCI.1820-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandler R, Depaulis A. Elicitation of intraspecific defence reactions in the rat from midbrain periaqueductal grey by microinjection of kainic acid, without neurotoxic effects. Neuroscience letters. 1988;88:291–296. doi: 10.1016/0304-3940(88)90226-1. [DOI] [PubMed] [Google Scholar]
- Berridge KC, Robinson TE. Parsing reward. Trends in neurosciences. 2003;26:507–513. doi: 10.1016/S0166-2236(03)00233-9. [DOI] [PubMed] [Google Scholar]
- Bilkei-Gorzo A, Erk S, Schurmann B, Mauer D, Michel K, Boecker H, Scheef L, Walter H, Zimmer A. Dynorphins regulate fear memory: from mice to men. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32:9335–9343. doi: 10.1523/JNEUROSCI.1034-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H, Haubensak W, Anthony TE, Anderson DJ. Central amygdala PKC-delta(+) neurons mediate the influence of multiple anorexigenic signals. Nature neuroscience. 2014;17:1240–1248. doi: 10.1038/nn.3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos CA, Bowen AJ, Schwartz MW, Palmiter RD. Parabrachial CGRP Neurons Control Meal Termination. Cell metabolism. 2016;23:811–820. doi: 10.1016/j.cmet.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsen J, Heimer L. The Basolateral Amygdaloid Complex as a Cortical-Like Structure. Brain Res. 1988;441:377–380. doi: 10.1016/0006-8993(88)91418-7. [DOI] [PubMed] [Google Scholar]
- Carter ME, Soden ME, Zweifel LS, Palmiter RD. Genetic identification of a neural circuit that suppresses appetite. Nature. 2013;503:111. doi: 10.1038/nature12596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciocchi S, Herry C, Grenier F, Wolff SBE, Letzkus JJ, Vlachos I, Ehrlich I, Sprengel R, Deisseroth K, Stadler MB, et al. Encoding of conditioned fear in central amygdala inhibitory circuits. Nature. 2010;468:277–U239. doi: 10.1038/nature09559. [DOI] [PubMed] [Google Scholar]
- Cornea-Hebert V, Riad M, Wu C, Singh SK, Descarries L. Cellular and subcellular distribution of the serotonin 5-HT2A receptor in the central nervous system of adult rat. Journal of Comparative Neurology. 1999;409:187–209. doi: 10.1002/(sici)1096-9861(19990628)409:2<187::aid-cne2>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Cui XY, Lundeberg T, Yu LC. Role of corticotropin-releasing factor and its receptor in nociceptive modulation in the central nucleus of amygdala in rats. Brain Res. 2004;995:23–28. doi: 10.1016/j.brainres.2003.09.050. [DOI] [PubMed] [Google Scholar]
- D’Hanis W, Linke R, Yilmazer-Hanke DM. Topography of thalamic and parabrachial calcitonin gene-related peptide (CGRP) immunoreactive neurons projecting to subnuclei of the amygdala and extended amygdala. The Journal of comparative neurology. 2007;505:268–291. doi: 10.1002/cne.21495. [DOI] [PubMed] [Google Scholar]
- Dalmasso C, Antunes-Rodrigues J, Vivas L, De Luca LA. Mapping brain Fos immunoreactivity in response to water deprivation and partial rehydration: Influence of sodium intake. Physiology & behavior. 2015;151:494–501. doi: 10.1016/j.physbeh.2015.08.020. [DOI] [PubMed] [Google Scholar]
- Davis M. The role of the amygdala in fear and anxiety. Annual review of neuroscience. 1992;15:353–375. doi: 10.1146/annurev.ne.15.030192.002033. [DOI] [PubMed] [Google Scholar]
- Dubner R, Ruda MA. Activity-dependent neuronal plasticity following tissue injury and inflammation. Trends in neurosciences. 1992;15:96–103. doi: 10.1016/0166-2236(92)90019-5. [DOI] [PubMed] [Google Scholar]
- Duvarci S, Pare D. Amygdala microcircuits controlling learned fear. Neuron. 2014;82:966–980. doi: 10.1016/j.neuron.2014.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich I, Humeau Y, Grenier F, Ciocchi S, Herry C, Luthi A. Amygdala inhibitory circuits and the control of fear memory. Neuron. 2009;62:757–771. doi: 10.1016/j.neuron.2009.05.026. [DOI] [PubMed] [Google Scholar]
- Fanselow MS. The Midbrain Periaqueductal Gray as a Coordinator of Action in Response to Fear and Anxiety. Nato Adv Sci I a-Lif. 1991;213:151–173. [Google Scholar]
- Galaverna OG, Seeley RJ, Berridge KC, Grill HJ, Epstein AN, Schulkin J. Lesions of the central nucleus of the amygdala. I: Effects on taste reactivity, taste aversion learning and sodium appetite. Behavioural brain research. 1993;59:11–17. doi: 10.1016/0166-4328(93)90146-h. [DOI] [PubMed] [Google Scholar]
- Gallagher M, Chiba AA. The amygdala and emotion. Current opinion in neurobiology. 1996;6:221–227. doi: 10.1016/s0959-4388(96)80076-6. [DOI] [PubMed] [Google Scholar]
- Gallagher M, Graham PW, Holland PC. The amygdala central nucleus and appetitive Pavlovian conditioning: lesions impair one class of conditioned behavior. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1990;10:1906–1911. doi: 10.1523/JNEUROSCI.10-06-01906.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerfen CR, Engber TM, Mahan LC, Susel Z, Chase TN, Monsma FJ, Jr, Sibley DR. D1 and D2 dopamine receptor-regulated gene expression of striatonigral and striatopallidal neurons. Science. 1990;250:1429–1432. doi: 10.1126/science.2147780. [DOI] [PubMed] [Google Scholar]
- Goosens KA, Maren S. Contextual and auditory fear conditioning are mediated by the lateral, basal, and central amygdaloid nuclei in rats. Learning & memory. 2001;8:148–155. doi: 10.1101/lm.37601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Soleiman MT, Soden ME, Zweifel LS, Palmiter RD. Elucidating an Affective Pain Circuit that Creates a Threat Memory. Cell. 2015;162:363–374. doi: 10.1016/j.cell.2015.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haubensak W, Kunwar PS, Cai H, Ciocchi S, Wall NR, Ponnusamy R, Biag J, Dong HW, Deisseroth K, Callaway EM, et al. Genetic dissection of an amygdala microcircuit that gates conditioned fear. Nature. 2010;468:270–276. doi: 10.1038/nature09553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmchen C, Fu QG, Sandkuhler J. Inhibition of Spinal Nociceptive Neurons by Microinjections of Somatostatin into the Nucleus Raphe Magnus and the Midbrain Periaqueductal Gray of the Anesthetized Cat. Neuroscience letters. 1995;187:137–141. doi: 10.1016/0304-3940(95)11345-2. [DOI] [PubMed] [Google Scholar]
- Hemmings HC, Jr, Greengard P, Tung HY, Cohen P. DARPP-32, a dopamine-regulated neuronal phosphoprotein, is a potent inhibitor of protein phosphatase-1. Nature. 1984;310:503–505. doi: 10.1038/310503a0. [DOI] [PubMed] [Google Scholar]
- Herry C, Johansen JP. Encoding of fear learning and memory in distributed neuronal circuits. Nature neuroscience. 2014;17:1644–1654. doi: 10.1038/nn.3869. [DOI] [PubMed] [Google Scholar]
- Hitchcock J, Davis M. Lesions of the Amygdala, but Not of the Cerebellum or Red Nucleus, Block Conditioned Fear as Measured with the Potentiated Startle Paradigm. Behavioral neuroscience. 1986;100:11. doi: 10.1037//0735-7044.100.1.11. [DOI] [PubMed] [Google Scholar]
- Hwang BH, Stewart R, Zhang JK, Lumeng L, Li TK. Corticotropin-releasing factor gene expression is down-regulated in the central nucleus of the amygdala of alcohol-preferring rats which exhibit high anxiety: a comparison between rat lines selectively bred for high and low alcohol preference. Brain Res. 2004;1026:143–150. doi: 10.1016/j.brainres.2004.08.042. [DOI] [PubMed] [Google Scholar]
- Isosaka T, Matsuo T, Yamaguchi T, Funabiki K, Nakanishi S, Kobayakawa R, Kobayakawa K. Htr2a-Expressing Cells in the Central Amygdala Control the Hierarchy between Innate and Learned Fear. Cell. 2015;163:1153–1164. doi: 10.1016/j.cell.2015.10.047. [DOI] [PubMed] [Google Scholar]
- Killcross S, Robbins TW, Everitt BJ. Different types of fear-conditioned behaviour mediated by separate nuclei within amygdala. Nature. 1997;388:377–380. doi: 10.1038/41097. [DOI] [PubMed] [Google Scholar]
- Kim J, Pignatelli M, Xu S, Itohara S, Tonegawa S. Antagonistic negative and positive neurons of the basolateral amygdala. Nature neuroscience. 2016;19:1636–1646. doi: 10.1038/nn.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JJ, Rison RA, Fanselow MS. Effects of amygdala, hippocampus, and periaqueductal gray lesions on short- and long-term contextual fear. Behavioral neuroscience. 1993;107:1093–1098. doi: 10.1037//0735-7044.107.6.1093. [DOI] [PubMed] [Google Scholar]
- Kissler JL, Sirohi S, Reis DJ, Jansen HT, Quock RM, Smith DG, Walker BM. The one-two punch of alcoholism: role of central amygdala dynorphins/kappa-opioid receptors. Biological psychiatry. 2014;75:774–782. doi: 10.1016/j.biopsych.2013.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapska E, Walasek G, Nikolaev E, Neuhausser-Wespy F, Lipp HP, Kaczmarek L, Werka T. Differential involvement of the central amygdala in appetitive versus aversive learning. Learning & memory. 2006;13:192–200. doi: 10.1101/lm.54706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohara K, Pignatelli M, Rivest AJ, Jung HY, Kitamura T, Suh J, Frank D, Kajikawa K, Mise N, Obata Y, et al. Cell type-specific genetic and optogenetic tools reveal hippocampal CA2 circuits. Nature neuroscience. 2014;17:269–279. doi: 10.1038/nn.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo JW, Han JS, Kim JJ. Selective neurotoxic lesions of basolateral and central nuclei of the amygdala produce differential effects on fear conditioning. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2004;24:7654–7662. doi: 10.1523/JNEUROSCI.1644-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laszlo K, Toth K, Kertes E, Peczely L, Lenard L. The role of neurotensin in positive reinforcement in the rat central nucleus of amygdala. Behavioural brain research. 2010;208:430–435. doi: 10.1016/j.bbr.2009.12.022. [DOI] [PubMed] [Google Scholar]
- LeDoux JE, Iwata J, Cicchetti P, Reis DJ. Different projections of the central amygdaloid nucleus mediate autonomic and behavioral correlates of conditioned fear. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1988;8:2517–2529. doi: 10.1523/JNEUROSCI.08-07-02517.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HH, Penzo MA, Taniguchi H, Kopec CD, Huang ZJ, Li B. Experience-dependent modification of a central amygdala fear circuit. Nature neuroscience. 2013;16:332–339. doi: 10.1038/nn.3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall JG, Al-Hasani R, Siuda ER, Hong DY, Norris AJ, Ford CP, Bruchas MR. CRH Engagement of the Locus Coeruleus Noradrenergic System Mediates Stress-Induced Anxiety. Neuron. 2015;87:605–620. doi: 10.1016/j.neuron.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald AJ. Neuronal organization of the lateral and basolateral amygdaloid nuclei in the rat. The Journal of comparative neurology. 1984;222:589–606. doi: 10.1002/cne.902220410. [DOI] [PubMed] [Google Scholar]
- McDonald AJ. Topographical organization of amygdaloid projections to the caudatoputamen, nucleus accumbens, and related striatal-like areas of the rat brain. Neuroscience. 1991;44:15–33. doi: 10.1016/0306-4522(91)90248-m. [DOI] [PubMed] [Google Scholar]
- Merali Z, McIntosh J, Kent P, Michaud D, Anisman H. Aversive and appetitive events evoke the release of corticotropin-releasing hormone and bombesin-like peptides at the central nucleus of the amygdala. Journal of Neuroscience. 1998;18:4758–4766. doi: 10.1523/JNEUROSCI.18-12-04758.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moga MM, Gray TS. Evidence for corticotropin-releasing factor, neurotensin, and somatostatin in the neural pathway from the central nucleus of the amygdala to the parabrachial nucleus. The Journal of comparative neurology. 1985;241:275–284. doi: 10.1002/cne.902410304. [DOI] [PubMed] [Google Scholar]
- Nie Z, Schweitzer P, Roberts AJ, Madamba SG, Moore SD, Siggins GR. Ethanol augments GABAergic transmission in the central amygdala via CRF1 receptors. Science. 2004;303:1512–1514. doi: 10.1126/science.1092550. [DOI] [PubMed] [Google Scholar]
- Parkinson JA, Robbins TW, Everitt BJ. Dissociable roles of the central and basolateral amygdala in appetitive emotional learning. The European journal of neuroscience. 2000;12:405–413. doi: 10.1046/j.1460-9568.2000.00960.x. [DOI] [PubMed] [Google Scholar]
- Penzo MA, Robert V, Li B. Fear conditioning potentiates synaptic transmission onto long-range projection neurons in the lateral subdivision of central amygdala. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2014;34:2432–2437. doi: 10.1523/JNEUROSCI.4166-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira-Derderian DTB, Vendramini RC, Menani JV, De Luca LA. Water deprivation-induced sodium appetite and differential expression of encephalic c-Fos immunoreactivity in the spontaneously hypertensive rat. Am J Physiol-Reg I. 2010;298:R1298–R1309. doi: 10.1152/ajpregu.00359.2009. [DOI] [PubMed] [Google Scholar]
- Pitkanen A, Savander V, LeDoux JE. Organization of intra-amygdaloid circuitries in the rat: an emerging framework for understanding functions of the amygdala. Trends in neurosciences. 1997;20:517–523. doi: 10.1016/s0166-2236(97)01125-9. [DOI] [PubMed] [Google Scholar]
- Pomrenze MB, Millan EZ, Hopf FW, Keiflin R, Maiya R, Blasio A, Dadgar J, Kharazia V, De Guglielmo G, Crawford E, et al. A Transgenic Rat for Investigating the Anatomy and Function of Corticotrophin Releasing Factor Circuits. Frontiers in neuroscience. 2015;9:487. doi: 10.3389/fnins.2015.00487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramón y Cajal S. Histology of the nervous system of man and vertebrates. New York: Oxford University Press; 1995. [Google Scholar]
- Reiner A, Hart NM, Lei W, Deng Y. Corticostriatal projection neurons - dichotomous types and dichotomous functions. Frontiers in neuroanatomy. 2010;4:142. doi: 10.3389/fnana.2010.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner A, Jiao Y, Del Mar N, Laverghetta AV, Lei WL. Differential morphology of pyramidal tract-type and intratelencephalically projecting-type corticostriatal neurons and their intrastriatal terminals in rats. The Journal of comparative neurology. 2003;457:420–440. doi: 10.1002/cne.10541. [DOI] [PubMed] [Google Scholar]
- Ritter S, Hutton B. Mercaptoacetate-Induced Feeding Is Impaired by Central Nucleus of the Amygdala Lesions. Physiology & behavior. 1995;58:1215–1220. doi: 10.1016/0031-9384(95)02050-0. [DOI] [PubMed] [Google Scholar]
- Robinson MJF, Warlow SM, Berridge KC. Optogenetic Excitation of Central Amygdala Amplifies and Narrows Incentive Motivation to Pursue One Reward Above Another. Journal of Neuroscience. 2014;34:16567–16580. doi: 10.1523/JNEUROSCI.2013-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Barak S. Molecular mechanisms underlying alcohol-drinking behaviours. Nature reviews Neuroscience. 2016;17:576–591. doi: 10.1038/nrn.2016.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen A, Zhang YX, Lund I, Lundeberg T, Yu LC. Substance P microinjected into the periaqueductal gray matter induces antinociception and is released following morphine administration. Brain Res. 2004;1001:87–94. doi: 10.1016/j.brainres.2003.11.060. [DOI] [PubMed] [Google Scholar]
- Sanford CA, Soden ME, Baird MA, Miller SM, Schulkin J, Palmiter RD, Clark M, Zweifel LS. A Central Amygdala CRF Circuit Facilitates Learning about Weak Threats. Neuron. 2017;93:164–178. doi: 10.1016/j.neuron.2016.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo DO, Funderburk SC, Bhatti DL, Motard LE, Newbold D, Girven KS, McCall JG, Krashes M, Sparta DR, Bruchas MR. A GABAergic Projection from the Centromedial Nuclei of the Amygdala to Ventromedial Prefrontal Cortex Modulates Reward Behavior. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2016;36:10831–10842. doi: 10.1523/JNEUROSCI.1164-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd GM. Corticostriatal connectivity and its role in disease. Nature reviews Neuroscience. 2013;14:278–291. doi: 10.1038/nrn3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skofitsch G, Jacobowitz DM. Calcitonin gene-related peptide: detailed immunohistochemical distribution in the central nervous system. Peptides. 1985;6:721–745. doi: 10.1016/0196-9781(85)90178-0. [DOI] [PubMed] [Google Scholar]
- Smith Y, Bevan MD, Shink E, Bolam JP. Microcircuitry of the direct and indirect pathways of the basal ganglia. Neuroscience. 1998;86:353–387. doi: 10.1016/s0306-4522(98)00004-9. [DOI] [PubMed] [Google Scholar]
- Swanson LW, Petrovich GD. What is the amygdala? Trends in neurosciences. 1998;21:323–331. doi: 10.1016/s0166-2236(98)01265-x. [DOI] [PubMed] [Google Scholar]
- Tovote P, Esposito MS, Botta P, Chaudun F, Fadok JP, Markovic M, Wolff SB, Ramakrishnan C, Fenno L, Deisseroth K, et al. Midbrain circuits for defensive behaviour. Nature. 2016;534:206–212. doi: 10.1038/nature17996. [DOI] [PubMed] [Google Scholar]
- Warden MK, Young WS., 3rd Distribution of cells containing mRNAs encoding substance P and neurokinin B in the rat central nervous system. The Journal of comparative neurology. 1988;272:90–113. doi: 10.1002/cne.902720107. [DOI] [PubMed] [Google Scholar]
- Wickersham IR, Lyon DC, Barnard RJ, Mori T, Finke S, Conzelmann KK, Young JA, Callaway EM. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron. 2007;53:639–647. doi: 10.1016/j.neuron.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirlinger M, Kreiman G, Anderson DJ. Amygdala-enriched genes identified by microarray technology are restricted to specific amygdaloid subnuclei. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5270–5275. doi: 10.1073/pnas.091094698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.