Abstract
Over a 4-month period from November 2002 to February 2003, 27 ceftazidime-resistant or cefotaxime-resistant nonrepetitive Enterobacter cloacae isolates were collected from 27 patients hospitalized at HuaShan Hospital, Shanghai, People's Republic of China. The Etest did not detect extended-spectrum beta-lactamases (ESBLs) in those 27 isolates; however, screening by the NCCLS ESBL disk test and confirmatory tests detected ESBLs in 4 of 27 isolates and PCR detected ESBLs in 23 of 27 isolates. The majority of ESBL producers exhibited the same repetitive extragenic palindromic PCR pattern but harbored different ESBL genes. CTX-M-3 was the most prevalent ESBL in our study. Interestingly, 12 clonally related E. cloacae isolates possessed a novel blaVEB-type beta-lactamase, blaVEB-3. BlaVEB-3 was encoded by the chromosome and was located in an integron. Nine of the 12 isolates harbored both the blaVEB-3 and the blaCTX-M-3-like ESBLs. This is the first report of a VEB-1-like ESBL in China and the first report of the simultaneous presence of VEB-1 and CTX-M-3-like ESBLs in an isolate.
Enterobacter cloacae is an important opportunistic pathogen known to cause nosocomial septicemia and urinary tract and respiratory tract infections (9, 25). Reports of multidrug-resistant isolates have increased during the last decade, probably as a result of the extensive use of broad-spectrum antibiotics. In some patient populations, the derepressed production of the AmpC beta-lactamase is a mechanism of beta-lactam resistance in E. cloacae strains (25, 28). However, the occurrence of extended-spectrum beta-lactamases (ESBLs) in members of the family Enterobacteriaceae that possess inducible Bush group 1 chromosomal beta-lactamases is also increasingly reported worldwide (1, 3, 4, 5, 6, 8, 12).
A 4-month survey was carried out to evaluate the prevalence of diverse ESBLs among E. cloacae isolates in HuaShan Hospital, Shanghai, People's Republic of China. This study identified a novel VEB-type ESBL, VEB-3. This is also the first report of VEB-1-like ESBLs from nosocomial isolates in the People's Republic of China. The first reported VEB-type ESBL, VEB-1, was detected in Pseudomonas aeruginosa strains from Southeast Asia (22). Unlike most of the ESBL genes, blaVEB-1 is part of a gene cassette and is located in class 1 integrons of various structures (10, 11, 15, 16).
MATERIALS AND METHODS
Bacterial isolates.
Twenty-seven nonrepetitive ceftazidime-resistant or cefotaxime-resistant E. cloacae clinical isolates were consecutively collected in the bacteriology laboratory of the Center of Laboratory Medicine, Huashan Hospital, Shanghai, from November 2002 to February 2003. During this period, 58 isolates of the species were obtained from patient specimens (22 from sputum, 22 from urine, 6 from blood, and 8 from other specimens). The isolates were identified with the API 20E system (bioMerieux SA, Marcy-1′Etoile, France). Escherichia coli ATCC 25922 was used as a negative control. Klebsiella pneumoniae ATCC 700603 was used as an ESBL-positive control. E. cloacae 029 and E. cloacae 029 M were used as control strains for the inducible production and the hyperproduction of AmpC, respectively. PET28 was used as the VEB-3 expression vector, and E. coli BL 21 were used as the expression host.
Antimicrobial susceptibility testing and screening for ESBLs and the AmpC enzyme.
Antibiotic susceptibilities were initially determined by the NCCLS disk diffusion method, and the results were interpreted according to the guidelines of the NCCLS (17). The MICs of imipenem, cefepime, amikacin, ceftazidime, cefotaxime, cefoperazone-sulbactam, ciprofloxacin, ceftazidime-clavulanic acid, cefoxitin, and cefotaxime-clavulanic acid were determined for 27 E. cloacae isolates, E. coli BL 21 with PET28a, and E. coli BL 21 with the VEB-3-PET28a recombinant plasmid by the Etest method (AB Biodisk, Solna, Sweden). The isolates were investigated for the presence of ESBLs by the NCCLS-recommended screening and confirmatory tests (17).
PCR amplification of ESBLs and sequencing.
A series of primers was designed for the detection of Ambler class A beta-lactamase genes. Tests were performed for the detection of genes encoding TEM (TEM-A and TEM-B), SHV (SHV-A and SHV-B), PER-1 and PER-2 (PER-A and PER-B), VEB-1 (VEB-1A and VEB-1B) (10, 11), and CTX-M (primers M13U and M13L for CTX-M-3, primers M25U and M25L for CTX-M-2, and primers M9U and M9L for CTX-M-9). The primers used for the detection of class 1 integrons (primers 5′-CS and 3′-CS) hybridized to sequences located in the 5′ and 3′ conserved segments. Combinations of primer 5′-CS or 3′-CS and blaVEB-1-specific primers (primers 5′-CS and VEB-INV1 or primers 3′-CS and VEB-INV2) were also used to determine the genetic contents of the class 1 integrons (15, 16).
For direct DNA sequencing, PCR amplifications were performed with whole-cell DNA of 12 E. cloacae clinical isolates that harbored the blaVEB-1 gene by use of primers that allowed amplification of the entire blaVEB-1 gene cassette (primers VEBcas-F and VEBcas-B) (11). The PCR products were purified with a QIAquick PCR purification kit (Qiagen, Hilden, Germany) and sequenced on an ABI PRISM 377 automated sequencer (Applied Biosystems, Foster City, Calif.). The nucleotide and deduced amino acid sequences were analyzed with software available over the Internet (www.ncbi.nlm.nih.gov/).
Cloning experiment and analysis of recombinant plasmids.
For cloning and expression of VEB-3, PCR was first performed with whole-cell DNA of the E. cloacae clinical isolates that harbored the blaVEB-3 gene by use of primers VEBc-R and VEBc-L. The fragment from the PCR product double digested with BamHI and HindIII was ligated into vector PET28a, which was also double digested with BamHI and HindIII. The recombinant plasmid was designated pETVEB3 and was transferred into E. coli BL 21. The E. coli BL 21 isolates with PET28a and pETVEB3 were designated E. coli ET and E. coli EV3, respectively. Plasmid DNA was prepared by using Qiagen columns and was sequenced to confirm that blaVEB-3 was correctly constructed in PET28a.
IEF of β-lactamase.
Crude beta-lactamase extracts were subjected to analytical isoelectric focusing (IEF) on an ampholine polyacrylamide gel (pH 3.5 to 9.5; Pharmacia Biotech). Beta-lactamases were visualized with a 0.2-mg/ml nitrocefin solution (Oxoid Ltd., Basingstoke, England). They were immediately applied to a parallel focused gel and were applied to another gel after the gel was flooded with a 1 mM solution of potassium clavulanate. This allowed the visual distinction between class C beta-lactamases that were insensitive to clavulanate and plasmid-mediated enzymes that were susceptible to clavulanate to be visualized.
Strain typing.
The 27 ceftazidime-resistant or cefotaxime-resistant E. cloacae isolates were typed by repetitive extragenic palindromic PCR (rep-PCR) with primers ERIC2 (13) and REP (7) (Table 1). The PCR products were separated in 1.2% agarose.
TABLE 1.
Sequences of primers used for detection of bla genes or genotyping of strains
| ESBL or region | Primer | Sequence | Position | GenBank accession no. or reference |
|---|---|---|---|---|
| TEM | TEM-A | 5′-ATA AAA TTC TTG AAG AC-3′ | 1-17 | X54604.1 |
| TEM-B | 5′-TTA CCA ATG CTT AAT CA-3′ | 1075-1059 | ||
| SHV | SHV-FOR | 5′-TGGTTATGCGTTATATTCGCC-3′ | 69-89 | X98100.1 |
| SHV-REV | 5′-GCTTAGCGTTGCCAGTGCT-3′ | 936-918 | ||
| PRE-1 | PER-1-FOR | 5′-AATTTGGGCTTAGGGCAGAA-3′ | 278-306 | Z21957 |
| PER-1-REV | 5′-ATGAATGTCATTATAAAAGC-3′ | 1211-1192 | ||
| VEB-1 | VEB-1-FOR | 5′-CGACTTCCATTTCCCGATGC-3′ | 226-245 | AF220758 |
| VEB1-REV | 5′-GGACTCTGCAACAAATACGC-3′ | 868-849 | ||
| CTX-M-3 | M13U | 5′-GGTTAAAAAATCACTGCGTC-3′ | 65-84 | X92506 |
| M13L | 5′-TTGGTGACGATTTTAGCCGC-3′ | 928-909 | ||
| CTX-M-9 | M9U | 5′-ATGGTGACAAAGAGAGTGCA-3′ | 1-20 | AF252621.2 |
| M9L | 5′-CCC TTC GGC GAT GAT TCT C-3′ | 870-852 | ||
| CTX-M-2 | M25U | 5′-ATGATGACTCAGAGCATTCG-3′ | 304-323 | AJ416343.1 |
| M25L | 5′-TGGGTTACGATTTTCGCCGC-3′ | 1169-1150 | ||
| VEBcas | VEBcas-F | 5′-GTTAGCGGTAATTTAACCAGATAG-3′ | 11-34 | AY027870.1 |
| VEBcas-B | 5′-CGGTTTGGGCTATGGGCAG-3′ | 1081-1063 | ||
| Rep-PCR | ERIC2 | 5′-AAGTAAGTGACTGGGGTGAGCG-3′ | 22 | |
| REP 2-1 | 5′-ICG ICT TAT CIG GCC TAC-3′ | 2 | ||
| VEBINV1 | 5′-CAGTTTGAGCATTTGAATACAC-3′ | 16 | ||
| VEBINV2 | 5′-AGC GTA TTT GTT GCA GAG TCC-3′ | |||
| Variable region of integron | 5-CS | 5′-GGC ATC CAA GCA GCA AGC-3′ | 1163-1180 | AF205943 |
| 3-CS | 5′-AAG CAG ACT TGA CCT GAT-3′ | 9231-9214 | ||
| VEB clone | VEBc-R | 5′-CCGGGATCCATGAAAATCGTAAAAAGGATATTATT-3′ | AF220758 | |
| VEBc-L | 5′-CCCAAGCTTTTATTTATTCAAATAGTAATTCCACG-3 |
Transfer of ceftazidime or cefotaxime resistance markers.
Plasmid DNA was extracted with a Qiagen Plasmid Midi kit. The plasmid extracts were electroporated into E. coli DH5α, and recombinant strains were selected on Luria-Bertani agar plates containing ceftazidime (2 μg/ml) or cefotaxime (2 μg/ml) (20).
Plasmid DNA analysis, Southern blotting, and hybridization.
Plasmid and chromosomal DNAs were extracted as described previously (13, 20) and were separated by agarose gel electrophoresis. Plasmids isolated from E. coli strain NCTC50192 was used as standard size markers. Plasmids and chromosomal DNAs were transferred from the agarose gel to a nylon membrane by the method of Southern (26) and were hybridized with digoxigenin-labeled blaVEB-1 and intI1 gene fragments with the PCR DIG detection system (Roche Diagnostics GmbH).
Nucleotide sequence accession number.
The nucleotide sequence data for the new blaVEB-1 gene variant (blaVEB-3) were submitted to the GenBank nucleotide sequence database and assigned accession number AY536519.
RESULTS
Twenty-seven (46.5%) of the 58 E. cloacae isolates were found to be resistant to ceftazidime or cefotaxime by disk diffusion susceptibility testing. The MICs of β-lactams, amikacin, and ciprofloxacin for the 27 isolates are presented in Table 2. All isolates were resistant to cefoxitin and susceptible to imipenem, 23 of 58 E. cloacae isolates were resistant to cefoperazone-sulbactam. Of the 27 isolates tested, only 0 and 4 isolates were positive for ESBL production by the Etest and the NCCLS-recommended screening and confirmatory tests, respectively, which indicates that ESBL production was detected in this species at a very low frequency by conventional methods.
TABLE 2.
MICs of selected antibiotics for representative E. cloacae clinical isolates
| E. cloacae isolate | Etest MIC (μg/ml)a
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| IPM | FEP | AK | CAZ | CTX | PTc | CIP | TZ-TZL | FOX | CT-CTL | |
| 72 | 0.38 | 3 | 12 | 48 | 32 | 4 | 0.13 | >32/>4 | >256 | >16/>1 |
| 64 | 0.25 | 4 | 8 | 48 | >256 | 2 | 0.25 | >32/>4 | >256 | >16/>1 |
| 5 | 0.25 | >256 | 2 | >256 | >256 | >256 | >32 | >32/>4 | >256 | >16/>1 |
| 57 | 0.25 | 24 | >256 | >256 | >256 | >256 | >32 | >32/>4 | >256 | >16/>1 |
| 136 | 0.25 | 3 | 1.5 | 12 | >256 | 6 | 0.75 | >32/>4 | >256 | >16/>1 |
| 42 | 0.5 | 128 | >256 | >256 | >256 | >256 | >32 | >32/>4 | >256 | >16/>1 |
| 150 | 0.38 | >256 | >256 | >256 | >256 | >256 | >32 | >32/>4 | 48 | >16/>1 |
| 108 | 0.25 | >256 | >256 | >256 | >256 | >256 | >32 | >32/>4 | >256 | >16/>1 |
| 39 | 0.75 | >256 | >256 | >256 | >256 | >256 | >32 | >32/>4 | >256 | >16/>1 |
| 103 | 0.25 | 0.75 | 4 | 96 | 32 | 96 | 0.16 | >32/>4 | >256 | >16/>1 |
| 212 | 0.5 | 2 | >256 | 3 | 6 | 48 | >32 | >32/>4 | >256 | >16/>1 |
| 142 | 0.25 | 3 | 2 | >256 | >256 | >256 | 0.06 | >32/>4 | >256 | >16/>1 |
Abbreviations: IPM, imipenem; FEP, cefepime; AK, amikacin; CAZ, ceftazidime; CTX, cefotaxime; PTc, cefoperazone-sulbactam; CIP, ciprofloxacin; TZ-TZL, ceftazidime-ceftazidime-clavulanic acid; FOX, cefoxitin; CT-CTL, cefotaxime-cefotaxime-clavulanic acid.
To determine the frequency of ESBL production relatively exactly, we analyzed the beta-lactamases of these isolates by PCR experiments with a series of primers specific for blaTEM, blaSHV, blaCTX-M-3, blaCTX-M-9, blaCTX-M2, blaPER-1, and blaVEB-1. Five of the seven genes were found alone or in various combinations. blaTEM-1-like and blaSHV-1-like genes were found in 20 and 2 of the 27 isolates, respectively. blaCTX-M-3-like, blaCTX-M-9-like, and blaVEB-1-like genes were detected in 13,7, and 12 of the 27 isolates, respectively (Table 3). Determination of the sequences of all the PCR products confirmed the identities of the genes. Interestingly, 12 of 27 isolates harbored a blaVEB-1-like gene, and 9 of them also harbored a blaCTX-M-3-like gene. This is the first report of a blaVEB-1-like gene in China and the first report of the detection of blaVEB-1-like and blaCTX-M-3-like genes in the same isolate.
TABLE 3.
Genotyping of rep-PCR products and beta-lactamase genes of 27 ceftazidime-resistant or cefotaxime-resistant E. cloacae clinical isolates and correlation with phenotypic ESBL tests
| Type | Isolate(s) | Result for bla gene for:
|
Result by NCCLS K-Ba method | ||||
|---|---|---|---|---|---|---|---|
| TEM-1 | CTX-M-3 | CTX-M-9a | VEB-1 | SHV-5a | |||
| A | 5, 25 | − | − | + | − | − | +b |
| B | 64 | − | − | + | − | − | + |
| 12, 150 | + | − | + | − | − | − | |
| 33 | + | + | − | − | + | + | |
| 39, 40, 42, 71 | + | + | − | + | − | − | |
| 236, 57, 108 | + | + | − | + | − | − | |
| 77, 88 | + | + | − | + | − | − | |
| 157, 205, 189 | + | − | − | + | − | − | |
| 17, 224 | + | + | − | − | − | − | |
| C | 72 | + | + | − | − | − | + |
| D | 103 | − | − | − | − | − | − |
| E | 136 | + | − | + | − | − | − |
| 216 | − | − | + | + | − | − | |
| F | 140, 212 | − | − | − | − | − | − |
| G | 142 | + | − | − | − | − | − |
K-B, Kirby-Bauer disk diffusion.
Isolate 5 was negative.
Next, work was focused on the 12 blaVEB-1-like gene-positive isolates. External primers specific for the veb-1 cassette were used for PCR amplification, with the whole-cell DNAs of three randomly selected blaVEB-1-like positive isolates (isolates 39, 40, and 157) used as template DNA, and then the entire blaVEB-like genes were sequenced. External blaVEB-1-specific primers gave 1,066-bp PCR fragments when the DNAs from these three E. cloacae isolates were used as templates and when both strands were sequenced. The deduced amino acid sequences, obtained over the Internet, identified VEB-1-like sequences that shared 99% amino acid identity with VEB-1 (Fig. 1). Compared with the amino acid sequences of VEB-1, VEB-1a, VEB-1b, and VEB-2, amino acid changes in VEB-3 from E. cloacae 39, 40, and 157 occurred at position 56, located in the N-terminal sequence of the mature protein, and positions 18 and 19, located in the putative leader peptide sequence (22).
FIG. 1.
Comparison of the amino acid sequences of the VEB-1, VEB-1a, VEB-1b, VEB-2, and VEB-3 beta-lactamases from P. aeruginosa JES from Thailand, P. aeruginosa KU-1 and KU-2 from Kuwait, P. aeruginosa 16 from Thailand, and E. cloacae 39 from China. Dashes indicate identical amino acids. The arrow indicates the position of the putative cleavage site of the leader peptide. Numbering is according to Ambler.
IEF analysis showed that E. cloacae isolate 39 expresses several beta-lactamases with pIs of 8.8, 7.7, 7.45, and 5.4. The pI 8.8 beta-lactamase corresponds to a CTX-M-3-like enzyme (inhibited by clavulanate), the pI 7.7 beta-lactamase corresponds to AmpC (not inhibited by clavulanate), and the pI 5.4 beta-lactamase corresponds to a TEM-1-like enzyme (inhibited by clavulanate). The β-lactamase with pI of 7.45 was somewhat inhibited by clavulanate, and recombinant bacterium E. coli EV3 expressed only a single beta-lactamase with a pI of 7.45, which shows that a pI of 7.45 corresponds to VEB-3.
In order to determine the biochemical character of VEB-3, the open reading frame of VEB-3 was cloned into vector PET28 and expressed in E. coli BL 21. The MICs of β-lactams for recombinant bacteria E. coli EV3 and E. coli ET (Table 4) showed that VEB-3 has ESBL activity.
TABLE 4.
MICs of beta-lactams for recombinant bacteria E. coli EV3 and E. coli ET
| E. coli strain | Etest MIC (μg/ml)a
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| IPM | FEP | CAZ | CTX | PTc | CAZ-TZL | FOX | CTX-CTL | AM | |
| EV3 | 0.16 | 3 | >256 | 24 | 0.38 | >32/0.25 | 1.5 | 8/0.25 | >256 |
| ET | 0.125 | 0.032 | 0.47 | 0.16 | 0.38 | 0.5/0.094 | 1.5 | 0.5/0.094 | 2 |
Abbreviations: IPM, imipenem; FEP, cefepime; CAZ, ceftazidime; CTX, cefotaxime; PTc, cefoperazone-sulbactam; TZL, ceftazidime-clavulanic acid; FOX, cefoxitin; CTL, cefotaxime-clavulanic acid, AM, ampicillin.
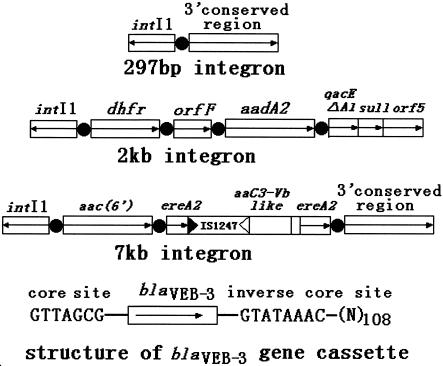
Since the blaVEB-1 gene cassette had been reported to be located on an integron, PCR amplifications were performed to identify its precise location. Isolates were first screened for the presence of class 1, 2, and 3 integrons, as described previously (19). The intI1 gene was detected in all 12 blaVEB-3-positive E. cloacae isolates. Class 2 or 3 integrons were not detected. By using primers 5′-CS and 3′-CS, a 7-kb PCR fragment was obtained from each E. cloacae isolate, an additional 0.3-kb PCR fragment was obtained from E. cloacae 57, and an additional 2-kb PCR fragment was obtained from E. cloacae 42. The sequences of these three integrons were determined, and their structures are shown in Fig. 2, which indicates that they do not harbor the blaVEB-3 gene. Combinations of primers 5′-CS and VEB-INV1 or primers 3′-CS and VEB-INV2 were also used for determination of the genetic contents of the class 1 integrons, but these two amplifications failed. Analysis of the genetic environment of blaVEB-3 revealed key signatures of gene cassettes. The presence of a core site (GTTAGCG) at positions 1 to 7 and the presence of an inverse core site (GTATAAAC) 3′ of blaVEB-3, followed by the remainder of a 59-base element, suggested that blaVEB-3 is encoded on a gene cassette and could thus be part of the variable region of an integron. This suggests that there is another integron in these blaVEB-3-positive E. cloacae isolates. Whether these blaVEB-3-positive E. cloacae isolates possess more plasmids requires further research.
FIG. 2.
Schematic representation of the three integrons found in E. cloacae 39, 42, and 57 and the structure of the blaVEB-3 gene cassette. Gene cassettes are shown as boxes, with arrows indicating the orientation of transcription and black circles indicating the 59-base element. The 5′ conserved segment contains the intI1 gene, which encodes integrase. The ereA2 gene was interrupted by IS1247, which are indicated by filled and empty triangles, respectively.
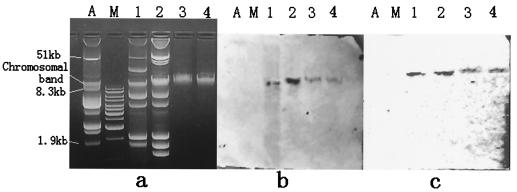
Preparation of plasmid DNA from E. cloacae isolates that harbored the blaVEB-3 gene revealed the presence of six or seven plasmids. Transfer of the ceftazidime resistance marker by electroporation failed with the E. coli DH5α recipient strains. Plasmid DNAs and chromosomal DNAs were separated by agarose gel electrophoresis, transferred from the agarose gel to a nylon membrane by the method of Southern (26), and hybridized with digoxigenin-labeled blaVEB-1 and intI1 gene fragments with the PCR DIG detection system. The results of the Southern experiment are shown in Fig. 3. The hybridization signals for both the blaVEB-3 and the intI1 genes were on the chromosomal DNA, indicating that the blaVEB-3 gene of E. cloacae isolate 39 is located on the chromosome.
FIG. 3.
(a) Plasmid DNA profiles of clinical isolates E. cloacae 39 and 57; (b) Southern blots hybridized with an intI1-specific probe; (c) Southern blots hybridized with a blaVEB-1-like specific probe. Lanes A, V517 plasmid size marker; lanes M, 1-kb DNA marker; lanes 1, plasmid DNA profiles of E. cloacae 39; lanes 2, plasmid DNA profiles of E. cloacae 57; lanes 3, chromosomal DNA of E. cloacae 39; lanes 4, chromosomal DNA of E. cloacae 57.
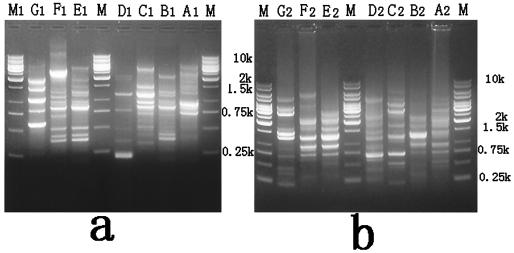
The relationship between the 27 ceftazidime-resistant or cefotaxime-resistant isolates was studied by rep-PCR by using, independently, primer ERIC2 (entrobacterial repetitive intergenic consensus sequences) and primer REP (repetitive extragenic palindomic elements). Amplification with both primer ERIC2 and primer REP gave seven distinct profiles for the strains (Fig. 4). The majority of the E. cloacae isolates that possessed ESBL genes exhibited the same pattern (type B). The exceptions were E. cloacae 5 and 25 (blaCTX-M-9a), which belonged to type A; E. cloacae 72 (blaCTX-M-3 and blaTEM-1), which belonged to type C; and E. cloacae 136 (blaCTX-M-9a and blaTEM-1), which belonged to type E. Although the majority of the E. cloacae isolates that possessed ESBLs genes exhibited the same rep-PCR pattern, they harbored different ESBL genes: E. cloacae 39, 40, 42, 57, 71, 77, 88, 108, and 236 possessed both the blaCTX-M-3 and the blaVEB-1 genes; and E. cloacae 157, 205, and 189 possessed only the blaVEB-1 gene (Table 3).
FIG. 4.
(a) rep-PCR profiles of E. cloacae clinical isolates with primer ERIC2; (b) rep-PCR profiles of E. cloacae clinical isolates with primer REP. Lanes M, DNA molecular size markers; lanes A to G, rep-PCR profiles of seven distinct types of E. cloacae clinical isolates, respectively. Subscript numerals: 1, ERIC2 type; 2, REP PCR type.
DISCUSSION
Plasmid-mediated ESBLs are most frequently found in K. pneumoniae and E. coli, with TEM-1- and SHV-1-derived ESBLs being the most common types found in these species (18). ESBLs have rarely been reported among E. cloacae isolates. This may be because E. cloacae is a species in which the usual mechanism of beta-lactam resistance is overexpression of the chromosomal AmpC beta-lactamase (25, 28), a trait which makes the detection of ESBLs by clavulanic acid-based methods difficult (27). The spread of ESBL-producing E. cloacae isolates may be enhanced by underdetection and underreporting. Many attempts have been made to detect these organisms (4, 6, 8, 24, 27). Although the presence of ESBLs in E. cloacae has been documented previously, the reported numbers of ESBL producers are generally low (4, 6, 8, 24, 27). The results of this study showed a high frequency (23 of 58 E. cloacae isolates) of ESBL producers among the E. cloacae isolates in HuaShan Hospital. This frequency is similar to that detected by Tzelepi et al. (27) (42.9%) but higher than that detected by Canton and Champs (2) (0.4%). The ESBL most frequently encountered in this study was a CTX-M-3-like beta-lactamase, which is widely disseminated among clinical strains of K. pneumoniae and E. coli in China (14). Unexpectedly, a novel VEB-1-like beta-lactamase (VEB-3) was detected in 12 E. cloacae isolates, with 9 of them also possessing the blaCTX-M-3-like ESBL. This is the first report of a blaVEB-1-like beta-lactamase in China and the first report of the simultaneous detection of blaVEB-1-like and blaCTX-M-3-like enzymes in E. cloacae. These blaVEB-3 gene-positive E. cloacae isolates came from four different departments in HuaShan Hospital, but all isolates had the same rep-PCR pattern, suggesting that blaVEB-3-containing E. cloacae isolates have spread in HuaShan Hospital and that the dissemination was caused by clonally related E. cloacae isolates. During dissemination the strains appear to have acquired different ESBL genes. In addition, two isolates were found to harbor the blaSHV-5a gene.
BlaVEB-1 and blaVEB-1-like gene cassettes have previously been documented to be located on an integron and encoded by the plasmid or the chromosome (10, 11, 21, 23). The results of our study show that the chromosome encodes the blaVEB-3 gene. Although we did not acquire the sequence of the whole variable region that includes the blaVEB-3 gene, the genetic environment of the blaVEB-3 gene indicates that the blaVEB-3 gene is located in an undetected integron, that the structure of this integron is different from those of all previously reported blaVEB-containing integrons, and that the integron location may account for the spread of the blaVEB-3 gene in E. coli and K. pneumoniae during the period of this study.
CTX-M-3 mainly confers resistance to cefotaxime, and VEB-1 and SHV-5a confer resistance to both ceftazidime and cefotaxime (22). In our study the E. cloacae isolates that possessed VEB-3 alone or VEB-3 and a CTX-M-3-like enzyme simultaneously were resistant to both ceftazidime and cefotaxime. The results of this study show that ESBLs in E. cloacae have become of increasing concern for therapy for clinical infections and should not be ignored.
Acknowledgments
We are grateful to Kenneth S. Thomson, Creighton University, Omaha, Nebr., for comments and suggestions, and we thank Ellen Smith Moland for performing IEF with E. cloacae isolate 39.
This work was supported by the fund of the 211 project grant “Functional Genomics of Important Pathogenic Microorganisms.”
REFERENCES
- 1.Aripin, C., C. Coze, A. M. Rogues, J. P. Gachie, C. Bebear, and C. Quentin. 1996. Epidemiological study of an outbreak due to multidrug-resistant Enterobacter aerogenes in a medical intensive care unit. J. Clin. Microbiol. 34:2163-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Canton, R., A. Oliver, T. M. Coque, C. Varela Mdel, J. C. Perez-Diaz, and F. Baquero. 2002. Epidemiology of extended-spectrum beta-lactamase-producing Enterobacter isolates in a Spanish hospital during a 12-year period. J. Clin. Microbiol. 40:1237-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chanal, C., D. Sirot, J. P. Romaszko, L. Bret, and J. Sirot. 1996. Survey of extended-spectrum beta-lactamase among Enterobacteriaceae. J. Antimicrob. Chemother. 38:127-132. [DOI] [PubMed] [Google Scholar]
- 4.Coudron, P. E., E. S. Moland, and C. C. Sanders. 1997. Occurrence and detection of extended-spectrum beta-lactamase in members of the family Enterobacteriaceae at a veteran's medical center: seek and you may find. J. Clin. Microbiol. 35:2593-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coudron, P. E., E. S. Moland, and K. S. Thomson. 2000. Occurrence and detection of AmpC beta-lactamase among Escherichia coli, Klebsiella pneumoniae, and Proteus mirabilis isolates at a veterans medical center. J. Clin. Microbiol. 38:1791-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Champs, C., D. Sirot, C. Chanal, M. C. Poupart, M. P. Dumas, and J. Sirot. 1991. Concomitant dissemination of three extended-spectrum beta-lactamases among different Enterobacteriaceae isolated in a French hospital. J. Antimicrob. Chemother. 27:441-457. [DOI] [PubMed] [Google Scholar]
- 7.de la Puente-Redondo, V. A., N. G. del Blanco, C. B. Gutiérrez-Martín, F. J. García-Peña, and E. F. Rodriguez-Ferri. 2000. Comparison of different PCR approaches for typing of Francisella tularensis strains. J. Clin. Microbiol. 38:1016-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emery, C. L., and L. A. Weymouth. 1997. Detection and clinical significance of extended-spectrum beta-lactamase in a tertiary-care medical unit. J. Clin. Microbiol. 35:2061-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fridkin, S. K., and R. P. Gaynes. 1999. Antimicrobial resistance in intensive care units. Clin. Chest Med. 20:303-316. [DOI] [PubMed] [Google Scholar]
- 10.Girlich, D., T. Nass, A. Leelaporn, L. Poirel, M. Fennewald, and P. Nordmann. 2002. Nosocomial spread of the integron-located veb-1-like cassette encoding an extended-spectrum beta-lactamase in Pseudomonas aeruginosa in Thailand. Clin. Infect. Dis. 34:603-611. [DOI] [PubMed] [Google Scholar]
- 11.Girlich, D., L. Poirel, A. Leelaporn, A. Karin, C. Tribuddharat, M. Fennewald, and P. Nordmann. 2001. Molecular epidemiology of the integron-located VEB-1 extended-spectrum beta-lactamase in nosocomial enterobacterial isolates in Bangkok, Thailand. J. Clin. Microbiol. 39:175-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hibbert-Rodgers, L. C. F., J. Heritage, D. M. Gascoyne-Binzi, P. M. Hawkey, N. Todd, I. J. Lewis, and C. Bailey. 1995. Molecular epidemiology of ceftazidime resistant Enterobacteriaceae from patients on a pediatric oncology ward. J. Antimicrob. Chemother. 36:65-82. [DOI] [PubMed] [Google Scholar]
- 13.Hulton, C. S., C. F. Higgins, and P. M. Sharp. 1991. ERIC sequences: a novel family of repetitive elements in the genomes of Escherichia coli, Salmonella typhimurium and other enterobacteria. Mol. Microbiol. 5:825-834. [DOI] [PubMed] [Google Scholar]
- 14.Munday, C. J., J. Xiong, C. Li, D. Shen, and P. M. Hawkey. 2004. Dissemination of CTX-M type beta-lactamases in. Enterobacteriaceae isolates in the People's Republic of China. Int. J. Antimicrob. Agents 23:175-180. [DOI] [PubMed] [Google Scholar]
- 15.Naas, T., F. Benaoudia, S. Massuard, and P. Nordmann. 2000. Integron-located VEB-1 extended-spectrum beta-lactamase gene in a Proteus mirabilis clinical isolate from Vietnam. J. Antimicrob. Chemother. 46:703-711. [DOI] [PubMed] [Google Scholar]
- 16.Naas, T., L. Poirel, A. Karim, and P. Nordmann. 1999. Molecular characterization of In 50, a class 1 integron encoding the gene for the extended-spectrum beta-lactamase VEB-1 in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 176:411-419. [DOI] [PubMed] [Google Scholar]
- 17.National Committee for Clinical Laboratory Standards. 2002. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 18.Philippon, A., G. Arlet, and P. H. Lagrange. 1994. Origin and impact of plasmid-mediated extended spectrum beta-lactamase. Eur. J. Clin. Microbiol. Infect. Dis. 13(Suppl. 1):S17-S29. [DOI] [PubMed] [Google Scholar]
- 19.Ploy, M.-C., F. Denis, P. Courvalin, and T. Lambert. 2000. Molecular characterization of integrons in Acinetobacter baumannii: description of a hybrid class 2 integron. Antimicrob. Agents Chemother. 44:2684-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poirel, L., M. Guibert, S. Bellais, T. Naas, and P. Nordmann. 1999. Integron- and carbenicillinase-mediated reduced susceptibility to amoxicillin-clavulanic acid in isolates of multidrug-resistant Salmonella enterica serotype Typhimurium DT104 from French patients. Antimicrob. Agents Chemother. 43:1098-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poirel, L., O. Meuteau, N. Agoli, C. Cattoen, and P. Nordmann. 2003. Outbreak of extended-spectrum beta-lactamase VEB-1-producing isolates of Acinetobacter baumannii in a French hospital. J. Clin. Microbiol. 41:3542-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poirel, L., T. Naas, M. Guibert, E. B. Chaibi, R. Labia, and P. Nordmann. 1999. Molecular and biochemical characterization of VEB-1, a novel class A extended-spectrum beta-lactamase encoded by an Escherichia coli integron gene. Antimicrob. Agents Chemother. 43:573-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poirel, L., V. O. Rotimi, E. M. Mokaddas, A. Karim, and P. Nordmann. 2001. VEB-1-like extended-spectrum beta-lactamase in Pseudomonas aeruginosa, Kuwait. Emerg. Infect. Dis. 7:468-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanders, C. C., A. L. Barry, J. A. Washington, C. Shubert, E. S. Moland, M. M. Traczewski, C. Knapp, and R. Mulder. 1996. Detection of extended-spectrum beta-lactamase-producing members of the family Enterobacteriaceae with the Vitek ESBL test. J. Clin. Microbiol. 34:2997-3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanders, W. E., and C. C. Sanders. 1997. Enterobacter spp.: pathogens poised to flourish at the turn of the century. Clin. Microbiol. Rev. 10:220-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Southern, E. M. 1975. Detection of specific sequence among DNA fragments separated by agarose gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 27.Tzelepi, E., P. Giakkoupi, D. Sofianou, V. Loukova, A. Kemeroglou, and A. Tsakris. 2000. Detection of Extended-spectrum beta-lactamase in clinical isolates of Enterobacter cloacae and Enterobacter aerogenes. J. Clin. Microbiol. 38:542-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tzelepi, E., L. S. Tzouvelekis, A. C. Vatopoulos, A. F. Mentis, A. Tsakris, and N. J. Leagakis. 1992. High prevalence of stably derepressed class-I beta-lactamase expression in multi resistant clinical isolates of Enterobacter cloacae from Greek hospitals. J. Med. Microbiol. 37:91-95. [DOI] [PubMed] [Google Scholar]