Abstract
Raman spectroscopy (RS) has shown great potential in noninvasive cancer screening. Statistically based algorithms, such as principal component analysis, are commonly employed to provide tissue classification; however, they are difficult to relate to the chemical and morphological basis of the spectroscopic features and underlying disease. As a result, we propose the first Raman biophysical model applied to in vivo skin cancer screening data. We expand upon previous models by utilizing in situ skin constituents as the building blocks, and validate the model using previous clinical screening data collected from a Raman optical fiber probe. We built an 830nm confocal Raman microscope integrated with a confocal laser-scanning microscope. Raman imaging was performed on skin sections spanning various disease states, and multivariate curve resolution (MCR) analysis was used to resolve the Raman spectra of individual in situ skin constituents. The basis spectra of the most relevant skin constituents were combined linearly to fit in vivo human skin spectra. Our results suggest collagen, elastin, keratin, cell nucleus, triolein, ceramide, melanin and water are the most important model components. We make available for download (see supplemental information) a database of Raman spectra for these eight components for others to use as a reference. Our model reveals the biochemical and structural makeup of normal, nonmelanoma and melanoma skin cancers, and precancers and paves the way for future development of this approach to noninvasive skin cancer diagnosis.
OCIS codes: (300.6450) Spectroscopy, Raman; (170.3880) Medical and biological imaging; (170.1790) Confocal microscopy; (170.6510) Spectroscopy, tissue diagnostics; (170.1610) Clinical applications
1. Introduction
As the most common type of malignancy, skin cancer accounts for over 5.4 million cases and 10K deaths per year in the US alone [1]. At present, the clinical diagnosis of skin cancer relies on visual inspection of suspicious lesions followed by biopsy and histopathology. The biopsies are performed in a dermatologist’s office and then sent to the histopathology lab for further examination. A few additional days may be needed to deliver the final results. These biopsy procedures are invasive, inefficient, and inconvenient. More importantly, the process has low diagnostic accuracy (49-81% among dermatologists for melanoma, the most deadly form [2]). The number of pigmented lesions needed to be excised to identify one melanoma ranges from 22 to 59 for experienced versus new general practitioners [3]. Large numbers of biopsies are performed on benign skin, leading to a substantial financial burden to the healthcare system and the patient, alone with considerable patient discomfort. As a result, a critical need exists to develop a noninvasive, accurate, fast, and inexpensive method for early skin cancer screening.
Raman spectroscopy (RS) is a noninvasive optical technique sensitive to the molecular composition of biological tissues. Recent advances in near-infrared lasers, optical filters, fiber optics and CCD cameras have greatly improved its sensitivity in detecting the chemical composition of biological tissues. In the most recent decade, the Raman optical fiber probe has allowed for fast and accurate cancer diagnostics, including ex vivo study of breast [4, 5], prostate [6], lung [7] and skin [8, 9], and in vivo study of breast [10], cervical [11], and skin [2, 12]. Recent clinical studies from our group [12] and others [2] have demonstrated that RS has high diagnostic accuracy in discriminating skin melanoma from nonmelanoma pigmented lesions.
To date, most fiber probe-based approaches have used statistical algorithms to describe the spectral differences of RS data, such as principal component analysis (PCA) [13] and independent component analysis (ICA) [14]. However, the principal or independent components are difficult to relate to the biophysical origin of disease, such as the microstructural organization of proteins and lipids and the functional state of cellular metabolism. These microstructural changes are what pathologists and dermatologists use to make diagnostic decisions and decide on the most appropriate treatment [10].
As a result, we describe for the first time a Raman “biophysical model” of human skin. This inverse model derives the morphological and biochemical composition of skin tissue from its Raman spectrum. The building blocks of our model are Raman active components extracted from skin in situ. Previous biophysical models used model components either measured directly from synthetic/purified chemicals [9, 15–18], or extracted from tissue sections in situ [19–21]. The advantage of using synthetic/purified chemicals as model components is that they can be easily measured without the need for Raman micro-imaging. This concept has been applied successfully to a previous Raman model of excisional skin biopsies [9] and has given us some prior knowledge about skin composition. Other groups used in situ constituents to build biophysical models of other disease processes, such as coronary atherosclerosis [19], breast cancer [20], and brain tumor [21]. Here, we expand upon this approach by developing a biophysical human skin model using in situ skin constituents. In situ constituents better represent the milieu of biological tissues that cannot be recapitulated in a synthetic environment. In human skin many constituents are present in various forms and each has a slightly different Raman spectrum. For instance, both collagen type I and III are abundant in human dermis; however, if both of them are included in the model, it may lead to overfitting and unstable results. In addition, skin constituents synthesized in the lab or from commercial sources are not in their natural states as in the human skin. For instance, the Raman features of a protein may change when it is exposed to organic solvents during synthesis. As a result, a single spectrum of skin constituent extracted from its microenvironment can provide a more general picture of the biophysical origins of skin spanning normal and abnormal disease states.
Furthermore, we applied the model for the first time to in vivo human skin cancer screening data that covers a wide range of normal, nonmelanoma and melanoma skin cancers and precancers [12]. Previous Raman models of human skin were either built for ex vivo tissue specimens [9] or in vivo normal skin [22]; however, Raman biophysical models have not been applied to in vivo skin cancer screening to interpret biophysical changes between pathologies.
In this study, we performed Raman imaging of tissue sections using a custom confocal Raman microscope. Multivariate curve resolution (MCR) analysis [23] was used to resolve in situ skin constituents from Raman images. Our results suggest that eight skin constituents are the most relevant building blocks, illustrating some variances with their corresponding synthetic components. The basis spectra of those skin constituents were then combined linearly to describe in vivo human skin spectra. The fit coefficients provided insight into the biochemical and structural composition of normal, benign and malignant skin tissues. Our model revealed the most important skin constituents representing the spectral features of skin tissues, and provided significant guidance to develop diagnostic algorithms for real-time noninvasive skin cancer diagnosis in future.
2. Materials and methods
2.1 Raman instrumentation
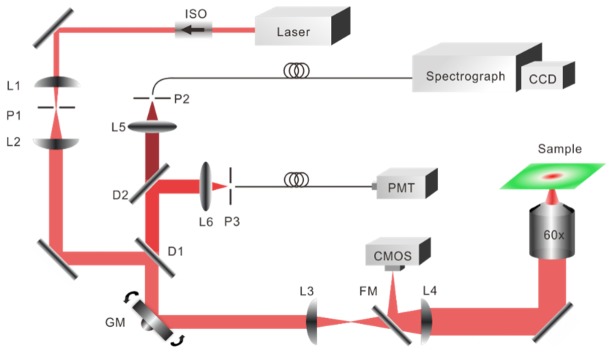
Raman images were collected with a custom built confocal Raman microscope illustrated in Fig. 1. The system was also integrated with a confocal laser-scanning microscope (CLSM) and a bright-field microscope, which provided the morphology image for assisting in locating the region of interest for Raman micro-imaging. The Raman excitation source was an 830nm single mode diode laser (LM830-PLR200, Ondax). The laser beam was reshaped, expanded and delivered to the sample through a microscope objective (Olympus, NA = 1.2, 60x). The galvanometer mirror performed 2D raster scanning on the tissue. The backscattered Raman signal was collected by a spectrograph (f/1.8i, Kaiser) and a deep cooling CCD camera (IDUS, Andor) through an optical fiber (50 μm, NA = 0.22), which also acted as a pinhole. The Rayleigh scattering light was collected by a PMT (C10709, Hamamatsu), and amplified by a current preamplifier (SR570, Stanford Research Systems). A data acquisition board (PCIe-6351, National Instruments) and LabVIEW software (National Instruments) were used to control the system. The power delivered to the sample was approximately 45mW. Lateral resolution was measured from the FWHM (full width at half maximum) of the point spread function using 0.2 and 0.5 μm microbeads. Axial resolution was measured from the FWHM of the intensity profile by translating a mirror towards the objective. The lateral, axial and spectral resolution of the system was around 1 μm, 8 μm and 8 cm−1, respectively.
Fig. 1.
Schematic of confocal Raman and confocal laser-scanning microscope used for skin measurements. The flip mirror and CMOS camera were used for bright-field imaging. ISO, isolator; D1, D2: dichroic mirror; P1 – P3: pinhole; L1 – L6, lens; GM, galvanometer mirror; FM, flip mirror.
2.2 Tissue preparation and Raman micro-imaging
Our study was approved by the Institutional Review Board at The University of Texas at Austin and Seton Medical Center. Fresh frozen human skin tissue samples were acquired from biopsy specimens during routine skin cancer surgery at Austin Dermatologic Surgery Center. After being transferred to the lab, the samples were stored at −80C. They were then thawed to −22C in a cryostat and sliced into 10 μm thin sections. The sections were transferred to magnesium fluoride slides (Edmund Optics) for the Raman measurement, and serial sections were transferred on standard microscope slides for hematoxylin and eosin (H&E) staining. Prior to Raman imaging, the sections were warmed to room temperature and kept moisturized with 0.9% saline solution.
Next, we performed Raman imaging on skin sections. Typical integration time at each pixel location was 2s. Typical step size in both the x and y directions was 1μm, but sometimes to achieve a large of view a step size of 5 μm was used. Imaging area varied from 30 × 30 μm2 to 150 × 150 μm2. We then correlated the Raman image with the histopathology image of the serial stained section. A board certified dermatologist assisted in identifying and confirming the morphology and biochemical components measured. In total, we collected more than 40 Raman images from samples of different disease states, including 24 images from 11 basal cell carcinoma (BCC) patients, 15 images from 5 squamous cell carcinoma (SCC) patients and 4 images from 1 malignant melanoma (MM) patient.
2.3 Data preprocessing and MCR analysis
Raman data preprocessing was performed using MATLAB (R2015b, MathWorks). All spectra underwent wavelength calibration, background subtraction, cosmic ray removal and smoothing. The system spectral response was calibrated using a tungsten halogen lamp (LS-1-CAL, Ocean Optics). The fluorescence background was then removed by modifying a 5th order polynomial fitting routine [24]. The effective spectral range was 800 to 1800 cm−1.
A multivariate curve resolution (MCR) method was employed to resolve individual morphological or biochemical components from the Raman image. This method has been successfully applied to stimulated Raman imaging data by Zhang et al. [25]. The basic concept of MCR is to decompose the raw spectra matrix (unfolded from Raman imaging) into the product of two smaller matrices and by a bilinear model:
| (1) |
corresponds to the matrix of the pure spectra, is the related concentration profiles for each of the components and is the error matrix. As an unsupervised learning method, the number of components contributing to was determined either by prior knowledge or by assessing the results obtained using singular value decomposition (SVD). After initial estimation is given for , the and are optimized iteratively using an alternative least-squares algorithm (ALS) until convergence is reached.
Here, we used a MATLAB based MCR-ALS toolbox [23] to determine and. The initial estimates of and were determined by means of a purest variable detection method [26]. The basic idea is to resolve highly overlapping near-infrared spectra with baseline problems by using the second-derivative spectra [26]. A nonnegative constraint and a 10% of tolerance were added to the ALS optimization. The concentration images for each individual component were reconstructed from , and the corresponding basis spectra were obtained from . We then categorized the basis spectra according to their biochemical or structural origin, such as elastic fibers, collagen fibers and cell nucleus.
We obtained a library of basis spectra from various skin sections spanning normal skin, and various skin disease states. The spectra in the same category were then averaged to create a single basis spectrum to represent that biochemical or structure. Although the basis spectra collected from different patients had minor differences, after averaging spectra from many patients we could ensure that the inter-patient variation was minimized.
2.4 Clinical screening data description
In vivo human skin spectra came from our previous skin cancer screening study [12]. Data were collected from an optical fiber probe [27] integrated in a multimodal spectroscopy system [28] on different sites, such as scalp, nose, earlobe, shoulder and thigh. Lesion types including basal cell carcinoma (BCC, 19 lesions), squamous cell carcinoma (SCC, 38 lesions), actinic keratosis (AK, 14 lesions), benign pigmented lesion (PL, 17 lesions) and malignant melanoma (MM, 12 lesions). BCC and SCC are the most common types of nonmelanoma skin cancers, whereas AK and PL are the most common precancers of SCC and MM, respectively. Raman spectra of adjacent normal skin for each individual lesion were also collected. Although normal skin measurements were not verified by histopathology, they were visually verified to be normal by an experienced dermatologist/physician assistant.
2.5 Model establishment
A sample’s Raman spectrum can be represented as a linear combination of the Raman spectra of the sample’s individual constituents. The signal intensity is then proportional to the chemical concentration [29]. Therefore, if one knows the spectra of the basis tissue constituents a priori, one can determine the concentration of those basis constituents. We used linear least-squares fitting with a nonnegative restraint for model fitting, according to the following equation:
| (2) |
While is the sample’s spectrum (in vivo human skin spectrum). is the spectra matrix of the sample’s individual constituents. is the relative spectral contribution (fit parameter) predicted by the model. is the noise related with the clinical RS system. Next, a combination of forward selection and backward elimination methods was performed to derive the most relevant basis constituents to the spectroscopic model. Finally, after applying the model to all the in vivo human skin data, we could obtain the biochemical and structural makeup of tissues spanning normal and various disease states.
One important factor that may influence the performance of the model is collinearity of the basis spectra. Collinearity is a common issue in linear regression that may lead to an unstable result [30]. The following equation is to calculate the collinearity coefficients between two basis spectra, and:
| (3) |
A value of 0 means the two basis spectra x and y do not have collinearity, and 1 indicates the two vectors are the same. This equation was used for the initial evaluation of the model components.
3. Results
3.1 Extraction of primary skin constituents in situ
The basis spectra were categorized into: (1) cellular components, (2) epidermal extracellular matrix (ECM), (3) dermal ECM, (4) lipids, (5) pigments, and (6) miscellaneous. We will illustrate each category in the following part of this section.
3.1.1 Cellular components
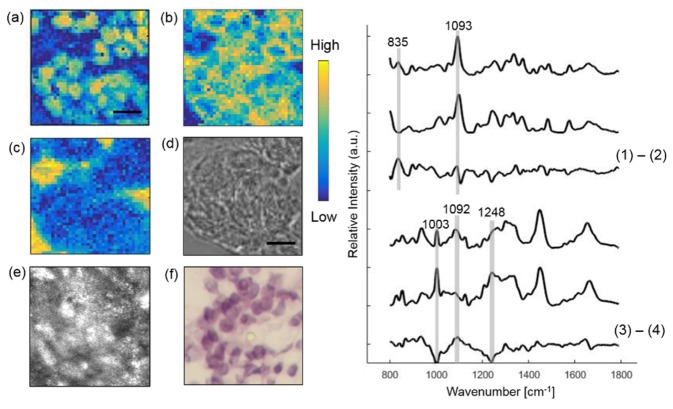
To identify cellular tumor components, Raman micro-imaging was performed within a tumor cluster in a BCC section (Fig. 2). Using MCR analysis, we reconstructed three concentration images (Fig. 2a-c) corresponding to cell nucleus, cell cytoplasm, and the Raman substrate. Those structures correlated well with the bright field, CLSM, and histopathology images (Fig. 2d-f), and their Raman spectra had similar characteristic peaks with the known spectra measured from the pure chemicals (Fig. 2). This approach was used to resolve the other skin constituents in the following sections as well. As seen from the plots on the right, the basis spectra of in situ nucleus and synthetic DNA (Sigma-Aldrich) are similar, which both have the pronounced contribution from phosphodioxy group PO2−1 at 1093 cm−1. However, the difference spectrum shows that in situ nucleus has substantial differences from synthetic DNA. For example, in situ nucleus appears to have a higher contribution from DNA backbone at 835 cm−1 [31]. The spectra of in situ cytoplasm and synthetic actin also have high similarity, but major differences can be found at 1003 cm−1 phenylalanine peak, 1081 and 1092 cm−1 lipid band. Numerous other peaks can also be appreciated in the difference spectra. These differences indicate the morphologically derived basis spectra of nucleus and cytoplasm include features related to other elements found in the cell.
Fig. 2.
Extracting cellular components from a BCC lesion. Raman images of nucleus (a) and cytoplasm (b) and Raman substrate (c) are compared with bright-field image (d), CLSM image (e) and histopathology image (f). The boxes on (e) and (f) mark the location of Raman imaging. The contrast of the CLSM image was provided by the relative difference in refractive index of cells and the surrounding stroma. Plots on the right show Raman spectra of in situ nucleus, synthetic DNA and their difference spectrum. Also in situ cytoplasm, synthetic actin and their difference spectrum. Scale bar: 10 μm.
3.1.2 Epidermal ECM
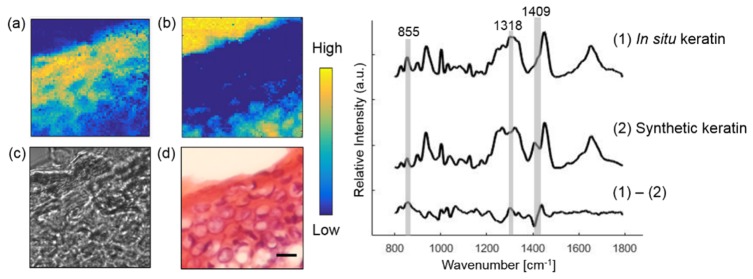
The epidermal layer of skin provides the barrier to water permeation and abrasion resistance. It is produced by continuous cell division of keratinocytes in the basal layer. Ultimately, the keratinocytes cornify and produce the stratum corneum, which is the dead, flattened cells at the outermost layer of the skin [32]. Because keratin is the main chemical component of epidermal ECM, we use in situ keratin to represent epidermal ECM.
Figure 3 illustrates Raman imaging performed on epidermis from a normal skin section. Tissue architecture correlates well with the histopathology image. The concentration images of epidermal ECM and the Raman substrate were reconstructed using MCR analysis. The Raman spectra of in situ and synthetic keratin are similar with substantial differences found at the protein bands at 855, 1318 and 1409 cm−1 [33].
Fig. 3.
Extracting the epidermal component from a normal skin section. Raman images of in situ keratin (a) and Raman substrate (b) are compared with bright-field image (c) and histopathology image (d). Plots on the right show Raman spectra of in situ, synthetic keratin and their difference spectrum. Scale bar:10 μm.
3.1.3 Dermal ECM
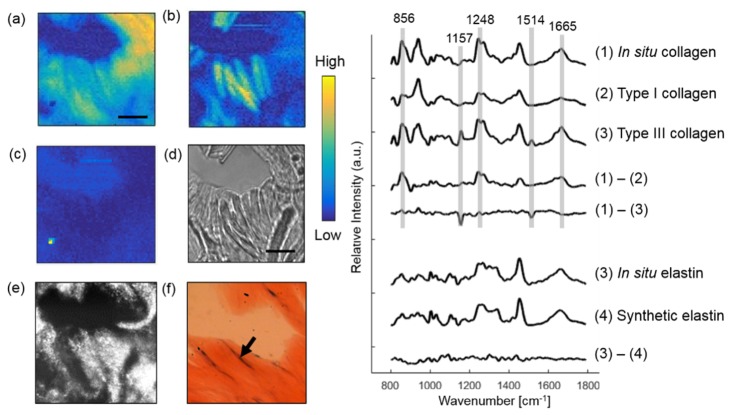
Dermal ECM comprises fibrillar collagens and associated proteins. Collagen fibers account for about 70% of the weight of dry dermis, while elastin maintains skin elasticity through a durable cross-linked array. Large diameter elastin-rich elastic fibers reside in the reticular dermis [34].
Figure 4 illustrates Raman imaging performed on a BCC skin section to extract dermal ECM proteins. The in situ collagen (collagen fiber), in situ elastin (elastic fiber), dye, and Raman substrate were resolved from the image by MCR analysis. The thin blue-gray elastic fibers and the pink collagen fibers can be identified from the histopathology image. The plots on the right compares the Raman spectrum of in situ collagen with synthetic type I and III collagen. Major differences are found at 856, 1248 and 1665 cm−1 protein bands between in situ and type I collagen, and 1157 and 1514 cm−1 between in situ and type III collagen. In situ and synthetic elastin have very similar spectra, which indicates that elastin is the major chemical component of elastic fibers.
Fig. 4.
Extracting dermal components from a BCC skin section. In situ collagen (a) and elastin (b) are resolved from the image. The dye used by the dermatologist to mark the orientation of the tissue was also detected (c). Raman images are compared with the bright-field image (d), CLSM image (e) and histopathology image (f). The box on (e) marks the location of Raman imaging. The arrow in (f) points to a thin blue-gray elastic fiber. Plots on the right displays Raman spectrum of in situ collagen, synthetic collagen and the difference spectrum. Also Raman spectrum of in situ elastin, synthetic elastin, and their difference spectrum. Scale bar: 10 μm.
3.1.4 Lipids
Skin’s epidermal surface is comprised of sebaceous and stratum corneum lipids. Epidermal lipids act like a cement to fill the spaces between the cells. The major constituents of sebaceous lipids are triglycerides (triolein), wax esters and squalene, while the epidermal lipids are a mixture of ceramides, free fatty acids and cholesterol [35, 36]. Ceramide is an important epidermal surface lipid as it composes almost half of the SC lipids [32].
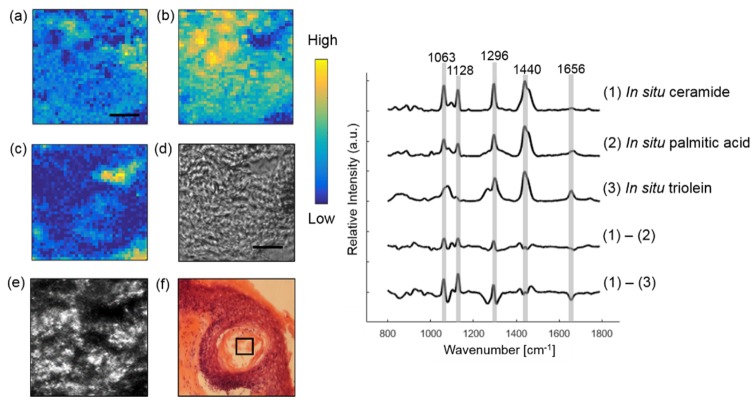
Raman imaging was also performed to derive the basis spectra of lipids. Figure 5 illustrates extracting in situ ceramide and triolein within a hair follicle from a SCC skin section. The synthetic spectra are not shown because they look similar to in situ spectra. Instead, we compare the difference spectra between in situ lipids. Although in situ ceramide and palmitic acid look similar, they have different spectral intensity in C-C stretching mode at 1063 and 1128 cm−1, CH2 twisting mode at 1296 cm−1, CH2 bending mode at 1440 cm−1 and C = C stretching mode at 1656 cm−1. Larger variance was observed in those bands between in situ ceramide and triolein. Triolein is not only abundant in skin lipid, but also in subcutaneous fat [20]. As triolein has a very strong Raman scattering cross-section, it contributes greatly to Raman spectrum of human skin.
Fig. 5.
Extracting lipids within a hair follicle from a SCC skin section. In situ ceramide (a), triolein (b) and Raman substrate (c) are resolved from the image. Raman images are compared with the bright-field image (d), CLSM image (e) and histopathology image (f). Some lipids in (f) were lost during the staining processing. The box on (e) and (f) marks the location of Raman imaging. Difference spectrum between in situ ceramide and palmitic acid and difference spectrum between in situ ceramide and triolein are also shown. Scale bar: 10 μm.
3.1.5 Pigments
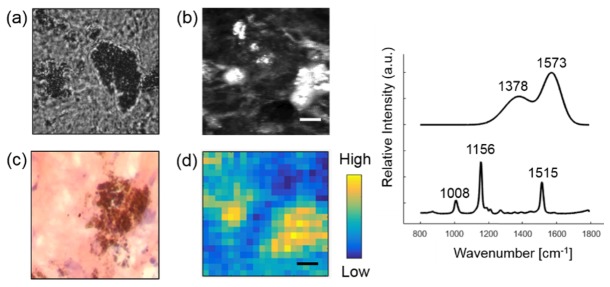
Skin pigments include melanin and beta carotene. Melanin is produced by melanocytes in the basal layer of the epidermis. In Fig. 6, we identified melanin from a MM skin section. As expected, melanin provides strong contrast in CLSM image [37]. We lowered the laser excitation to 20mW to reduce tissue burning caused by strong absorption of melanin. As this led to a worsening in the SNR, we further smoothed the melanin spectrum by fitting it to Gaussian functions [38]. The two broad peaks located at 1378 cm−1 and 1573 cm−1 were consistent with the spectrum of in vivo cutaneous melanin [38]. Beta carotene is a plant-derived carotenoid. It was extracted from skin sections adjacent to fatty tissue. The characteristic peaks of beta carotene at 1008, 1156 and 1515 cm−1 are consistent with a previous study [39].
Fig. 6.
Extracting melanin from a MM skin section. Raman image of melanin (d) are compared with bright field image (a) CLSM image (b) and histopathology image (c). Basis spectra of melanin and beta carotene are shown on the right. Scale bar: 50 μm.
3.1.6 Miscellaneous
In Raman imaging, water came from the saline used to keep the skin section moist. We found water plays an important role in fitting the broad Raman band at 1645 cm−1. Hemoglobin and calcium hydroxyapatite (CaH) were only detected in one skin section but were included in our library. Morphologies such as hair follicle (HF) and keratin pearl (KP) were also obtained. KP was extracted from SCC lesions with acceleration of keratinization. Figure 7 shows the basis spectra of these constituents. Although the spectra of HF and KP are similar, the difference spectrum suggested that the former contained cellular information (DNA backbone at 835 cm−1 and phosphodioxy group PO2−1 at 1093 cm−1). Finally, we included a spectrum of fiber background generated from the Raman optical fiber probe. This component is used to fit the broad peak between 1000 – 1100 cm−1 in the in vivo data.
Fig. 7.
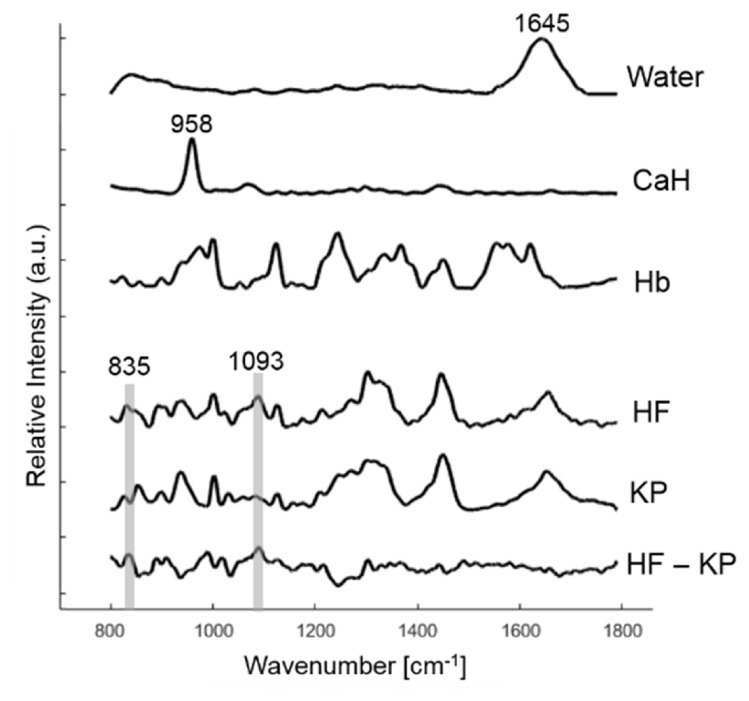
Basis Raman spectra of water, calcium hydroxyapatite (CaH), hemoglobin (Hb), hair follicle (HF) and keratin pearl (KP) collected in situ are displayed. The difference spectrum between HF and KP is also shown.
3.2 Biophysical modeling results
A total of fifteen candidate model components were derived. The basis spectra were peak normalized with a minimum value of 0 and maximum value of 1. Their collinearity coefficients are displayed in Table 1. Beta carotene and calcium hydroxyapatite are not shown because they have low collinearity (< 0.50) with other components. Several components have high collinearity, such as in situ keratin in epidermis, keratin pearl (KP), and hair follicle (HF), likely because keratin dominates their chemical composition. As a result, we selected only one model component to represent keratin. In addition, we observed high collinearity between cell cytoplasm (Cyt) and other protein-rich components (elastin (Ela), keratin, KP, and HF). These components share common features of many functional groups, such as C-C stretching around 939 cm−1, Amide III around 1270 cm−1, CH modes around 1454 cm−1 and amide I around 1660 cm−1. Considering that (1) cell cytoplasm has a much smaller Raman scattering cross-section and less quantity than keratin, and (2) the spectrum of keratin may contain some cell features due to their close proximity, we finally excluded cytoplasm from our model. In addition, Raman spectrum of palmitic acid (PA) has a high degree of overlap with triolein (0.94) and ceramide (0.96), so PA was also excluded from the model. In total, we arrive at eight primary Raman active components: collagen, elastin, triolein, cell nucleus, keratin, ceramide, melanin and water (Fig. 8). The peak positions of main Raman bands are displayed in Table 2.
Table 1. Collinearity measurement for candidate model componentsa.
| Col | Ela | Ker | KP | HF | Cyt | Nuc | Trio | Cer | PA | Mel | Hb | Water | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (a) | Col | 1.00 | |||||||||||||
| Ela | 0.89 | 1.00 | |||||||||||||
| Ker | 0.88 | 0.93 | 1.00 | ||||||||||||
| KP | 0.86 | 0.94 | 0.97 | 1.00 | |||||||||||
| HF | 0.77 | 0.90 | 0.94 | 0.95 | 1.00 | ||||||||||
|
| |||||||||||||||
| (b) | Cyt | 0.86 | 0.93 | 0.95 | 0.97 | 0.95 | 1.00 | ||||||||
| Nuc | 0.64 | 0.76 | 0.70 | 0.75 | 0.78 | 0.77 | 1.00 | ||||||||
|
| |||||||||||||||
| (c) | Trio | 0.70 | 0.78 | 0.82 | 0.82 | 0.84 | 0.86 | 0.59 | 1.00 | ||||||
| Cer | 0.56 | 0.62 | 0.66 | 0.66 | 0.70 | 0.72 | 0.44 | 0.85 | 1.00 | ||||||
| PA | 0.65 | 0.75 | 0.79 | 0.79 | 0.82 | 0.83 | 0.51 | 0.94 | 0.96 | 1.00 | |||||
|
| |||||||||||||||
| (d) | Mel | 0.47 | 0.58 | 0.56 | 0.61 | 0.56 | 0.57 | 0.48 | 0.39 | 0.33 | 0.42 | 1.00 | |||
|
| |||||||||||||||
| (e) | Hb | 0.64 | 0.73 | 0.68 | 0.72 | 0.67 | 0.69 | 0.61 | 0.41 | 0.34 | 0.44 | 0.78 | 1.00 | ||
| Water | 0.56 | 0.58 | 0.58 | 0.65 | 0.57 | 0.62 | 0.48 | 0.33 | 0.20 | 0.29 | 0.55 | 0.60 | 1.00 | ||
Components are sorted according to their major composition: (a) protein, (b) cell, (c) lipid, (d) pigment, (e) miscellaneous. Components include collagen (Col), elastin (Ela), keratin (Ker), keratin peal (KP), hair follicle (HF), cell cytoplasm (Cyt), cell nucleus (Nuc), triolein (Trio), ceramide (Cer), palmitic acid (PA), melanin (Mel), hemoglobin (Hb) and water.
Fig. 8.
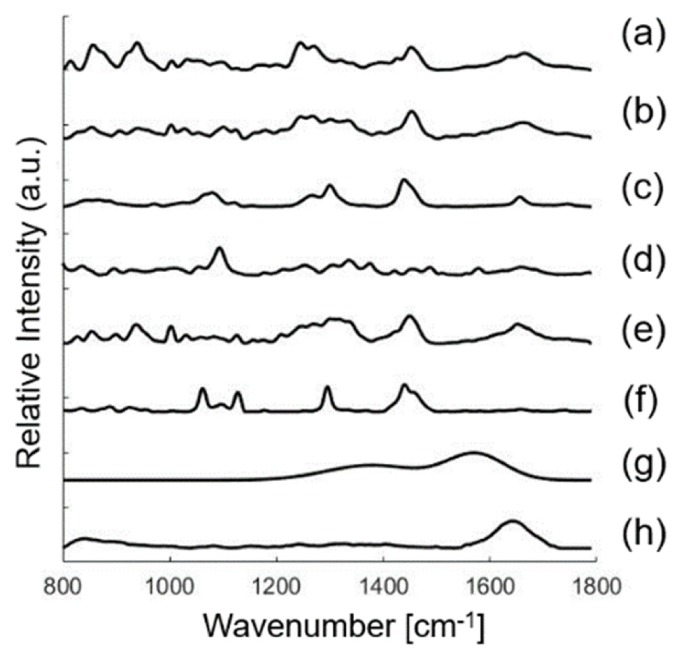
Basis spectra used in the biophysical model of skin. Model components include collagen (a), elastin (b), triolein (c), nucleus (d), keratin (e), ceramide (f), melanin (g), water (h). See Data File 1 (106.8KB, CSV) for underlying values.
Table 2. Peak positions of main Raman bands in the Raman active components (Refs. 2, 9, 19, 31, 33, 38).
| Raman peaks [cm−1] |
Band assignments | Components |
|---|---|---|
| 835 | DNA backbone: O-P-O/ tyrosine | nucleus |
| 855 | CCH bending (aromatic) of protein | elastin, keratin |
| 856 | C-C vibration of the collagen backbone | collagen |
| 937 | C-C stretching of proline and valine and protein backbone | keratin |
| 940 | C-C stretching of protein backbone | collagen, elastin |
| 1003 | C-C vibration of phenyl ring | collagen, elastin, keratin |
| 1063 | C-C asymmetric skeletal stretching of lipids (trans-conformation); | ceramide |
| 1080 | C-C skeletal stretching | triolein |
| 1093 | O-P-O symmetric stretching vibration of the DNA backbone | nucleus |
| 1128 | C-C symmetric skeletal stretching | ceramide |
| 1248 | Amide III (β-sheet and random coil conformations) | collagen, elastin |
| 1254 | β sheet/ thymine/ cytosine (DNA base/ DNA & RNA base) | nucleus |
| 1269 | Amide III (α-helix conformation), C-N stretching, N-H in-plane bending | collagen, elastin, keratin |
| 1301 | C-H modes (CH2 twisting and wagging) of lipids; CH2/CH3 bands | triolein |
| 1336 | desmosine/isodesmosine | elastin |
| 1337 | adenine, guanine (DNA & RNA base) | nucleus |
| 1378 | linear stretching of the C-C bonds within the rings | melanin |
| 1440 | CH2/CH3 bands | triolein, ceramide |
| 1450 | C-H bending of proteins | keratin |
| 1454 | C-H stretching, C-H asymmetric deformation | collagen, elastin |
| 1573 | in-plane stretching of the aromatic rings | melanin |
| 1645 | O-H bending mode of liquid water | water |
| 1653 | C-O stretching model of amide I | keratin |
| 1656 | C-C lipids | triolein |
| 1665 | C-O amide I vibration | collagen, elastin |
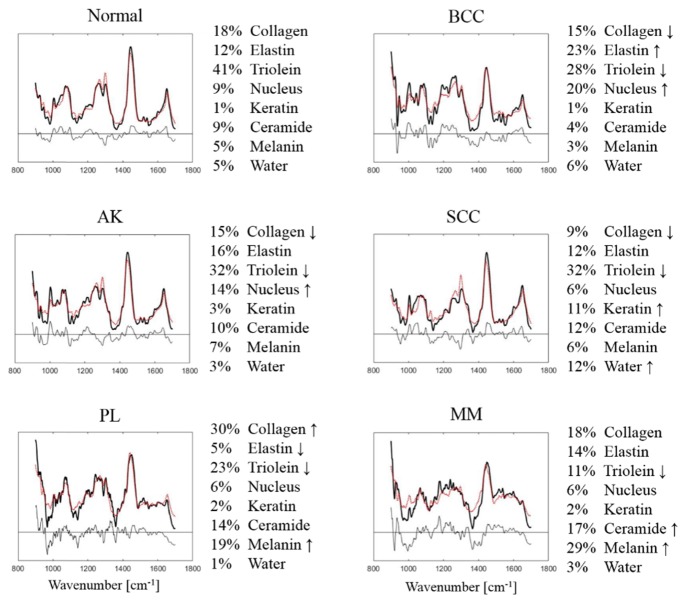
In an effort to validate that these eight components captured the primary skin constituents as measured on in vivo human skin cancers, we fit this linear component model to the clinical data set. We determined the relative contribution from the eight model components of each of the pathology groups. Figure 9 shows the fitting result of the mean Raman spectra. Considering the order of magnitude of the residuals with respect to the bulk tissue spectra, most of the spectroscopic features are well represented. The fit coefficients across each model component are normalized to sum to 1.
Fig. 9.
Model fitting results for in vivo human skin spectra categorized as Normal, BCC, SCC, AK, PL and MM. Mean Raman tissue spectra (solid lines), model fits (dotted lines), residuals are plotted on the same scale. Fit coefficients in percentage are listed on the right. The arrow indicates the most characteristic changes for each lesion type.
The results illustrate key biophysical changes of skin with different tissue types. For instance, normal skin has an average of 41% triolein, 30% of dermal ECM (collagen and elastin), and minor contribution from nucleus, ceramide, melanin and water. However, the contribution of triolein drops significantly from normal skin (41%) to nonmelanoma skin cancer/precancer (BCC, SCC, AK) (28%, 32%, 32%) and to MM, PL (11%, 23%). As expected, melanin content is much higher in MM (29%) and PL (19%) compared to other tissue types (3 – 7%). In addition, keratin concentration is higher in SCC (11%) as compared to other tissue types (1 – 3%).
4. Discussion
In this study, we establish a Raman “biophysical model”, an inverse model for determining biophysical skin components using in vivo Raman spectroscopy. We built a confocal Raman microscope to identify eight of the most relevant skin constituents contributing to the spectral differences among different skin malignancies.
Our model components were found to be consistent with previous studies. Some were commonly used in skin and non-skin models. For instance, collagen and triolein are known to be important contributors to the RS signal of breast, gastric, and artery tissues [17, 19, 20]. We demonstrated these two components also played an important role for fitting in vivo skin data. Other components were more specific to skin. For instance, Caspers et al. used ceramide to model epidermal lipid in human stratum corneum layer [40]. Silveira et al. included elastin to model skin dermal protein [9]. Keratin was important for in vivo skin to consider the impact of epidermis [22, 40] but not necessary for excisional skin fragments because the measurement was on the dermis side [9]. Melanin was important only when pigmented lesions were considered, so it was used to model melanoma skin tissue [9].
However, our model is different from previous biophysical Raman skin models in the following two aspects. First, we used skin constituents in their microenvironment as the basis spectra. Our results showed that it was possible to use a single morphologically derived basis spectrum rather than synthetic/purified chemicals. As demonstrated in the Results section, in situ skin constituents had substantial differences from their corresponding synthetic chemicals, even if their major chemical components were the same. Since in situ constituents are extracted from skin in their natural state, without any further processing, they can better represent the skin microenvironment that cannot be recapitulated in a synthetic environment. Second, our model was validated by a previous in vivo clinical screening study [12] acquired by a Raman optical fiber probe [27]. Currently, the only biophysical Raman skin cancer model was based on excised fragments of BCC and melanoma skin tissues [9]. We expanded upon this research to apply our model to in vivo skin spectra study and covered a wider range of nonmelanoma and melanoma skin cancers and precancers.
While we found a total of 15 measurable Raman components in skin, we found the most consistent model outcomes were achieved when minimizing this number to only eight components. Our approach was to select only one Raman constituent to represent those in situ components that were chemically similar and with high collinearity. Similarly, Stone et al. demonstrated that including both amino acids and the proteins containing them in a linear model skewed the fit coefficients [16]. Our experience is that minimizing the number of protein components resulted in the most consistent fit coefficients.
The biophysical changes of skin derived by our model follow known morphological and biochemical changes in skin malignancy. We observed that there is less triolein in skin cancer/pre-cancer lesions relative to the amount of triolein in normal skin. Triolein is a major form of triglyceride in human skin, which presents as subcutaneous fat and epidermal surface lipid. The apparent decrease in triolein as cancer progresses could be due to: (1) the reduction of subcutaneous fat sampled by the probe, caused by the thickening epidermis during lesion formation; and/or (2) the reduction of membrane lipid synthesis induced by UV damage [34]. Because subcutaneous fat exists in a substantial amount and has large Raman scattering cross section [41] we believe (1) is the major reason. The thickening of epidermis originates from the progression of malignancy [42].
While both melanoma and nonmelanoma skin cancers were included in this study, the direct comparison between BCC or SCC and MM is much less clinically relevant [43]. Thus, we compared nonmelanoma (BCC, SCC, AK) versus normal skin and MM versus PL separately. We observed that the amount of collagen was substantially lower in nonmelanoma skin cancer lesions as compared to normal skin. This could be explained by the breakdown of collagen in dermis due to the role metalloproteinases (MMP) play in degrading collagen and prohibiting procollagen biosynthesis [35]. The thickening of the epidermis also leads to reduced collagen signal collected by the probe. Furthermore, we observed the amount of elastin was higher in BCC lesions as compared to normal skin, potentially resulting from the existence of solar elastosis. Elastosis is characterized by the accumulation of disorganized elastic fibers in the dermis and commonly found in photoaging skin [36]. Finally, we found keratin was substantially higher in SCC compared to the other groups, which suggests massive keratinization disorders during SCC tumor progression [37].
By visual inspection, the mean spectra of MM and PL appear very different than the mean spectra of other pathologies. The spectral flattening between 1500 and 1700 cm−1 is caused by increased melanin and pigmentation, indicating RS is sensitive to pigment-related variations. However, discriminating MM from PL remains the most challenging discrimination in skin cancer screening, resulting in high negative biopsy ratios clinically. In our study, we observed melanin content in MM is substantially higher as compared to PL, indicating massive melanocyte proliferation. The significantly lower level of triolein in MM than PL could be explained by both the reasons given above and by the strong absorption of melanin, which further reduced the signal sampled from subcutaneous fat. In addition, collagen is substantially lower in MM than PL. This suggests that tumor formation is closely related to the changes in its stroma microenvironment in favor of its proliferation and eventual metastasis [37, 38]. Our model demonstrated that collagen, triolein, and melanin are the most important cancer identifiers for MM. Future work will explore the diagnostic potential of these biophysical parameters in discriminating skin cancers.
We observed a higher fitting residual in MM than the other tissue types. The basic assumption of our linear fitting model is that the scattering properties of tissue do not significantly distort the Raman spectrum [29], but this assumption may not hold for melanin due to its strong absorption and scattering. Intrinsic Raman spectroscopy may help correct this distortion by relating the observed and intrinsic Raman spectra through diffuse reflectance using light transport model [44]. We will also explore nonlinear fitting models to improve the fitting, such as partial least-squares (PLS) and support vector machine (SVM). Other factors also contribute to the residuals in general. One factor is that the basis spectra and bulk tissue spectra were acquired from two independent Raman systems, which were composed of different detectors, lenses, beam splitters, etc. Spectral response calibration was used to match the spectral response of the two systems, but it could not completely eliminate the differences in the spectra measured by the two systems. Another factor is the signal generated by probe components, such as the fiber background, epoxy and sapphire [27].
In general, we did not find site-specific constituents that are not covered by the current model, but the concentration of the 8 components may vary due to location. For example, when the measurement was taken on the scalp surround with dark hairs we would detect melanin signal. Future work will examine how sensitive our model is in picking up such information.
In this study, we proposed the first Raman biophysical model that used in situ Raman active components as the building blocks, and applied to in vivo skin cancer screening data. Our results indicate that eight basis spectra derived from collagen, elastin, triolein, cell nucleus, keratin, ceramide, melanin, and water are the most relevant to describe the spectral features of human skin RS data. We make available for download (see supplemental data file) a database of Raman spectra for these eight components for others to use as a reference. Our future work will evaluate the performance of this model in discriminating skin cancer pathologies within the context of ongoing clinical studies of Raman spectroscopy for skin cancer screening in our group. We environ our model being used with the Raman probe for analyzing individual lesions pointed out by patents or providers. We think it would be reasonable to scan the top ten concerning lesions on any patients without affecting the current patient flow in a physician’s office.
Acknowledgements
We would like to thank Seton Medical Center and Austin Dermatological Surgical Center for collaborating with us. We appreciate the time and help of all the staff, physician assistants, and doctors. We would like to thank Sandra Esparza and Leandra Turner for preparing samples, Prof. Aaron Baker for providing histology facility, and Greg Lyness and Esther Maier for H&E staining. This work was conducted at the Biophotonics Laboratory at The University of Texas at Austin.
Funding
Cancer Prevention & Research Institute of Texas (CPRIT) RP130702.
References and links
- 1.Siegel R. L., Miller K. D., Jemal A., “Cancer statistics, 2016,” Cancer J. Clin. 66(1), 7–30 (2016). 10.3322/caac.21332 [DOI] [PubMed] [Google Scholar]
- 2.Lui H., Zhao J., McLean D., Zeng H., “Real-time Raman spectroscopy for in vivo skin cancer diagnosis,” Cancer Res. 72(10), 2491–2500 (2012). 10.1158/0008-5472.CAN-11-4061 [DOI] [PubMed] [Google Scholar]
- 3.English D. R., Del Mar C., Burton R. C., “Factors influencing the number needed to excise: excision rates of pigmented lesions by general practitioners,” Med. J. Aust. 180(1), 16–19 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Barman I., Dingari N. C., Saha A., McGee S., Galindo L. H., Liu W., Plecha D., Klein N., Dasari R. R., Fitzmaurice M., “Application of Raman spectroscopy to identify microcalcifications and underlying breast lesions at stereotactic core needle biopsy,” Cancer Res. 73(11), 3206–3215 (2013). 10.1158/0008-5472.CAN-12-2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sathyavathi R., Saha A., Soares J. S., Spegazzini N., McGee S., Rao Dasari R., Fitzmaurice M., Barman I., “Raman spectroscopic sensing of carbonate intercalation in breast microcalcifications at stereotactic biopsy,” Sci. Rep. 5(1), 9907 (2015). 10.1038/srep09907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crow P., Molckovsky A., Stone N., Uff J., Wilson B., WongKeeSong L.-M., “Assessment of fiberoptic near-infrared raman spectroscopy for diagnosis of bladder and prostate cancer,” Urology 65(6), 1126–1130 (2005). 10.1016/j.urology.2004.12.058 [DOI] [PubMed] [Google Scholar]
- 7.Magee N. D., Villaumie J. S., Marple E. T., Ennis M., Elborn J. S., McGarvey J. J., “Ex vivo diagnosis of lung cancer using a Raman miniprobe,” J. Phys. Chem. B 113(23), 8137–8141 (2009). 10.1021/jp900379w [DOI] [PubMed] [Google Scholar]
- 8.Bodanese B., Silveira F. L., Zângaro R. A., Pacheco M. T. T., Pasqualucci C. A., Silveira L., Jr., “Discrimination of basal cell carcinoma and melanoma from normal skin biopsies in vitro through Raman spectroscopy and principal component analysis,” Photomed. Laser Surg. 30(7), 381–387 (2012). 10.1089/pho.2011.3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silveira L., Jr, Silveira F. L., Bodanese B., Zângaro R. A., Pacheco M. T. T., “Discriminating model for diagnosis of basal cell carcinoma and melanoma in vitro based on the Raman spectra of selected biochemicals,” J. Biomed. Opt. 17(7), 077003 (2012). 10.1117/1.JBO.17.7.077003 [DOI] [PubMed] [Google Scholar]
- 10.Haka A. S., Volynskaya Z., Gardecki J. A., Nazemi J., Lyons J., Hicks D., Fitzmaurice M., Dasari R. R., Crowe J. P., Feld M. S., “In vivo margin assessment during partial mastectomy breast surgery using raman spectroscopy,” Cancer Res. 66(6), 3317–3322 (2006). 10.1158/0008-5472.CAN-05-2815 [DOI] [PubMed] [Google Scholar]
- 11.Mahadevan-Jansen A., Mitchell M. F., Ramanujam N., Utzinger U., Richards-Kortum R., “Development of a fiber optic probe to measure NIR Raman spectra of cervical tissue in vivo,” Photochem. Photobiol. 68(3), 427–431 (1998). 10.1111/j.1751-1097.1998.tb09703.x [DOI] [PubMed] [Google Scholar]
- 12.Lim L., Nichols B., Migden M. R., Rajaram N., Reichenberg J. S., Markey M. K., Ross M. I., Tunnell J. W., “Clinical study of noninvasive in vivo melanoma and nonmelanoma skin cancers using multimodal spectral diagnosis,” J. Biomed. Opt. 19(11), 117003 (2014). 10.1117/1.JBO.19.11.117003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahadevan-Jansen A., Mitchell M. F., Ramanujam N., Malpica A., Thomsen S., Utzinger U., Richards-Kortum R., “Near-Infrared Raman Spectroscopy for In Vitro Detection of Cervical Precancers,” Photochem. Photobiol. 68(1), 123–132 (1998). 10.1111/j.1751-1097.1998.tb03262.x [DOI] [PubMed] [Google Scholar]
- 14.Chen J., Wang X. Z., “A new approach to near-infrared spectral data analysis using independent component analysis,” J. Chem. Inf. Comput. Sci. 41(4), 992–1001 (2001). 10.1021/ci0004053 [DOI] [PubMed] [Google Scholar]
- 15.Shafer-Peltier K. E., Haka A. S., Motz J. T., Fitzmaurice M., Dasari R. R., Feld M. S., “Model-based biological Raman spectral imaging,” J. Cell. Biochem. Suppl. 87(S39), 125–137 (2002). 10.1002/jcb.10418 [DOI] [PubMed] [Google Scholar]
- 16.Stone N., Hart Prieto M. C., Crow P., Uff J., Ritchie A. W., “The use of Raman spectroscopy to provide an estimation of the gross biochemistry associated with urological pathologies,” Anal. Bioanal. Chem. 387(5), 1657–1668 (2007). 10.1007/s00216-006-0937-9 [DOI] [PubMed] [Google Scholar]
- 17.Bergholt M. S., Zheng W., Lin K., Ho K. Y., Teh M., Yeoh K. G., So J. B. Y., Huang Z., “Characterizing variability in in vivo Raman spectra of different anatomical locations in the upper gastrointestinal tract toward cancer detection,” J Biomed Opt 16, 037003 (2011). 10.1117/1.3556723 [DOI] [PubMed] [Google Scholar]
- 18.Jianhua Zhao H. L., David I., Lean M., Zeng H., “Real time Raman spectroscopy for noninvasive in vivo skin analysis and diagnosis,” in New Developments in Biomedical Engineering (Intech, 2010). [Google Scholar]
- 19.Buschman H. P., Deinum G., Motz J. T., Fitzmaurice M., Kramer J. R., van der Laarse A., Bruschke A. V., Feld M. S., “Raman microspectroscopy of human coronary atherosclerosis: biochemical assessment of cellular and extracellular morphologic structures in situ,” Cardiovasc. Pathol. 10(2), 69–82 (2001). 10.1016/S1054-8807(01)00064-3 [DOI] [PubMed] [Google Scholar]
- 20.Shafer-Peltier K. E., Haka A. S., Fitzmaurice M., Crowe J., Myles J., Dasari R. R., Feld M. S., “Raman microspectroscopic model of human breast tissue: implications for breast cancer diagnosis in vivo,” J. Raman Spectrosc. 33(7), 552–563 (2002). 10.1002/jrs.877 [DOI] [Google Scholar]
- 21.Bergner N., Medyukhina A., Geiger K. D., Kirsch M., Schackert G., Krafft C., Popp J., “Hyperspectral unmixing of Raman micro-images for assessment of morphological and chemical parameters in non-dried brain tumor specimens,” Anal. Bioanal. Chem. 405(27), 8719–8728 (2013). 10.1007/s00216-013-7257-7 [DOI] [PubMed] [Google Scholar]
- 22.Zhao J., Lui H., McLean D. I., Zeng H., “Integrated real-time Raman system for clinical in vivo skin analysis,” Skin Res. Technol. 14(4), 484–492 (2008). 10.1111/j.1600-0846.2008.00321.x [DOI] [PubMed] [Google Scholar]
- 23.Jaumot J., de Juan A., Tauler R., “MCR-ALS GUI 2.0: new features and applications,” Chemom. Intell. Lab. Syst. 140, 1–12 (2015). 10.1016/j.chemolab.2014.10.003 [DOI] [Google Scholar]
- 24.Lieber C. A., Mahadevan-Jansen A., “Automated method for subtraction of fluorescence from biological Raman spectra,” Appl. Spectrosc. 57(11), 1363–1367 (2003). 10.1366/000370203322554518 [DOI] [PubMed] [Google Scholar]
- 25.Zhang D., Wang P., Slipchenko M. N., Ben-Amotz D., Weiner A. M., Cheng J.-X., “Quantitative vibrational imaging by hyperspectral stimulated Raman scattering microscopy and multivariate curve resolution analysis,” Anal. Chem. 85(1), 98–106 (2013). 10.1021/ac3019119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Windig W., Stephenson D., “Self-modeling mixture analysis of second-derivative near-infrared spectral data using the SIMPLISMA approach,” Anal. Chem. 64(22), 2735–2742 (1992). 10.1021/ac00046a015 [DOI] [Google Scholar]
- 27.Motz J. T., Hunter M., Galindo L. H., Gardecki J. A., Kramer J. R., Dasari R. R., Feld M. S., “Optical fiber probe for biomedical Raman spectroscopy,” Appl. Opt. 43(3), 542–554 (2004). 10.1364/AO.43.000542 [DOI] [PubMed] [Google Scholar]
- 28.Sharma M., Marple E., Reichenberg J., Tunnell J. W., “Design and characterization of a novel multimodal fiber-optic probe and spectroscopy system for skin cancer applications,” Rev. Sci. Instrum. 85(8), 083101 (2014). 10.1063/1.4890199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manoharan R., Baraga J. J., Feld M. S., Rava R. P., “Quantitative Histochemical Analysis of Human Artery Using Raman Spectroscopy,” J. Photochem. Photobiol. B 16(2), 211–233 (1992). 10.1016/1011-1344(92)80009-K [DOI] [PubMed] [Google Scholar]
- 30.Wold S., Ruhe A., Wold H., Dunn I., WJ, “The collinearity problem in linear regression. The partial least squares (PLS) approach to generalized inverses,” SIAM J. Sci. Statist. Comput. 5(3), 735–743 (1984). 10.1137/0905052 [DOI] [Google Scholar]
- 31.Jess P. R. T., Garcés-Chávez V., Smith D., Mazilu M., Paterson L., Riches A., Herrington C. S., Sibbett W., Dholakia K., “Dual beam fibre trap for Raman micro-spectroscopy of single cells,” Opt. Express 14(12), 5779–5791 (2006). 10.1364/OE.14.005779 [DOI] [PubMed] [Google Scholar]
- 32.Downing D. T., “Lipid and protein structures in the permeability barrier of mammalian epidermis,” J. Lipid Res. 33(3), 301–313 (1992). [PubMed] [Google Scholar]
- 33.Movasaghi Z., Rehman S., Rehman I. U., “Raman spectroscopy of biological tissues,” Appl. Spectrosc. Rev. 42(5), 493–541 (2007). 10.1080/05704920701551530 [DOI] [Google Scholar]
- 34.Olczyk P., Mencner Ł., Komosinska-Vassev K., “The role of the extracellular matrix components in cutaneous wound healing,” BioMed Res. Int. 2014, 747584 (2014). 10.1155/2014/747584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pappas A., “Epidermal surface lipids,” Dermatoendocrinol 1(2), 72–76 (2009). 10.4161/derm.1.2.7811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caspers P. J., Lucassen G. W., Puppels G. J., “Combined in vivo confocal Raman spectroscopy and confocal microscopy of human skin,” Biophys. J. 85(1), 572–580 (2003). 10.1016/S0006-3495(03)74501-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rajadhyaksha M., Grossman M., Esterowitz D., Webb R. H., Anderson R. R., “In vivo confocal scanning laser microscopy of human skin: melanin provides strong contrast,” J. Invest. Dermatol. 104(6), 946–952 (1995). 10.1111/1523-1747.ep12606215 [DOI] [PubMed] [Google Scholar]
- 38.Huang Z., Lui H., Chen X. K., Alajlan A., McLean D. I., Zeng H., “Raman spectroscopy of in vivo cutaneous melanin,” J. Biomed. Opt. 9(6), 1198–1205 (2004). 10.1117/1.1805553 [DOI] [PubMed] [Google Scholar]
- 39.Vítek P., Osterrothová K., Jehlička J., “Beta-carotene—a possible biomarker in the Martian evaporitic environment: Raman micro-spectroscopic study,” Planet. Space Sci. 57(4), 454–459 (2009). 10.1016/j.pss.2008.06.001 [DOI] [Google Scholar]
- 40.Caspers P. J., Lucassen G. W., Carter E. A., Bruining H. A., Puppels G. J., “In vivo confocal Raman microspectroscopy of the skin: noninvasive determination of molecular concentration profiles,” J. Invest. Dermatol. 116(3), 434–442 (2001). 10.1046/j.1523-1747.2001.01258.x [DOI] [PubMed] [Google Scholar]
- 41.Haka A. S., Shafer-Peltier K. E., Fitzmaurice M., Crowe J., Dasari R. R., Feld M. S., “Diagnosing breast cancer by using Raman spectroscopy,” Proc. Natl. Acad. Sci. U.S.A. 102(35), 12371–12376 (2005). 10.1073/pnas.0501390102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weedon D., “Tumors of the epidermis,” in Skin Pathology (Elsevier, 2002), pp. 753–802. [Google Scholar]
- 43.Lieber C. A., Majumder S. K., Ellis D. L., Billheimer D. D., Mahadevan-Jansen A., “In vivo nonmelanoma skin cancer diagnosis using Raman microspectroscopy,” Lasers Surg. Med. 40(7), 461–467 (2008). 10.1002/lsm.20653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shih W.-C., Bechtel K. L., Feld M. S., “Intrinsic Raman spectroscopy for quantitative biological spectroscopy part I: theory and simulations,” Opt. Express 16(17), 12726–12736 (2008). 10.1364/OE.16.012726 [DOI] [PMC free article] [PubMed] [Google Scholar]