Abstract
Line-scanning hyperspectral imaging (LHSI) is known to have a higher acquisition rate but lower sectioning capability than point-scanning hyperspectral imaging. To further increase the axial imaging contrast of LHSI, structured illumination was integrated into line excitation to remove the off-focus and scattered on-focus fluorescence signals. In an unsectioned leaf, the imaging contrast can be enhanced by 8 times, while in sectioned mouse skin tissues, a 4.5-fold enhancement can be achieved. With a spectral resolution of 1.15 nm, the fluorophores with seriously-overlapped spectra was proved to be separated without cross-talk by applying linear unmixing to the recorded spectral information.
OCIS codes: (170.2520) Fluorescence microscopy, (110.4234) Multispectral and hyperspectral imaging, (180.5810) Scanning microscopy
1. Introduction
Hyperspectral imaging (HSI) has been widely applied in various fields, such as astronomy [1,2], food quality and safety control [3,4], remote sensing [5,6], pharmaceuticals [7–9] and much more, due to its outstanding ability to distinguish different molecular compositions. In recent years, this technique has also gathered a huge amount of attention in biomedical fields [10–18]. With its ability to obtain a hypercube containing two-dimensional spatial information, along with additional spectral information of the target specimen, the combination of imaging and spectroscopy has made such technique emerge as a useful imaging modality for biomedical applications. Hyperspectral imaging can be simply divided into two types of scanning methods, namely the spectral scanning and the spatial scanning methods [19], a category of non-scanning method [20,21]. In the spectral scanning method, the detector collects images containing two-dimensional spatial information within a wavelength band. Devices such as filter wheels, acousto-optical tunable filters and liquid crystal tunable filters are most often used to select the wavelength band. The hypercube is then completed by repeating the process within different wavelength bands throughout the target region. In each and every process, most portion of the excited signals are blocked and wasted, the specimen is required to be excited repeatedly, so that the photodamage issue would become more considerable. On the other hand, the spatial scanning method utilizes a dispersive component to expand the spectral information of the incoming signals in spatial dimension. In each process, full spectral information of the excited region can be obtained without information waste. A complete hypercube can then be filled out by two different approaches, point-scanning and line-scanning. In contrast to both the spectral and spatial scanning methods, non-scanning method can obtain a complete hypercube with a snapshot [20,21]. Without the scanning process, the non-scanning method has its unsurpassable superiority in acquisition speed. However, based on wide-field illumination, the non-scanning method has difficulties in investigating thick samples due to lack of optical sectioning power.
Among these methods, the spatial scanning methods can be easily combined with optical sectioning modalities to obtain high axial contrast within thick samples [22–28]. Based on the point-scanning method, confocal [22,23] and two-photon [24] hyperspectral microscopy has been introduced to increase the axial contrast. Yet the excitation light was to scan point-by-point throughout a 2-dimensional area, which leads to a rather long acquisition time. The acquisition time can be concerning in some cases that desires a shorter one. Unlike the point-scanning method, line-scanning method utilizes a line-shaped excitation light along one dimension and a slit is placed in front of the spectrometer instead of a pinhole. Line-scanning methods have been applied in previous studies of confocal hyperspectral microscopy [25,26] and two-photon hyperspectral microscopy [27,28]. By using a 2D camera to perform the parallel recording, the acquisition speed can be further improved. However, in the case of line-scanning confocal hyperspectral microscopy, the off-focus fluorescence signals are allowed to pass through the slit’s long-side and perform as background noises. At shallow depths, both the point-scanning and line-scanning methods can provide similar axial contrast. As the depth gets deeper, due to the loss of confocality along one dimension, the axial contrast of the line-scanning method becomes much lower than that of the point-scanning method [29]. Although the two-photon excitation has intrinsic optical sectioning power, the background noises can also be contributed by the on-focus fluorescence signals scattered by surrounding tissues. Therefore, in the line-scanning methods, the image’s axial contrast are usually poorer than those of the point-scanning methods, especially in thick specimens.
To regain the confocality and improve the axial contrast, the off-focus signals collected along the slit’s long-side has to be eliminated, and the concept of structured illumination microscopy (SIM) can be applied. SIM was introduced by Neil et al. in 1997 and is a powerful optical sectioning method [30–32]. SIM’s main concept is to project sinusoidal patterns onto specimens, either by using two-beam interference, a spatial light modulator (SLM), or a digital light processing (DLP) projector [33–35]. By utilizing a high NA objective to focus the patterns, the patterns fade away rapidly while defocusing. Having obtained a sequence of fluorescence images under patterned illuminations with different phases, the on-focus information can thus be extracted by
| (1) |
where I1, I2 and I3 are the images with three different phases of 0, 2π/3 and 4π/3, and If is the extracted on-focus image. And by applying the three images to
| (2) |
the constructed image Is can represent the image under non-patterned illumination. However, the quality of the reconstructed image relies strongly on the phase accuracy and intensity stability. Either phase error or intensity fluctuation can lead to residual patterns in the reconstructed image. Mechanically translating the optical elements to change the phases is more time-consuming and has issues of mechanical error, which may cause the intensity fluctuation and phase error, respectively [30]. In contrast, by electrically controlling the optical array elements such as SLM and DLP projector instead [33–35], not only the higher pattern refreshing rate can reduce the intensity fluctuation but almost no phase errors need to be considered.
In this study, the concept of SIM was combined with the line-scanning hyperspectral imaging (LHSI) to make having simultaneous high axial resolution and high acquisition rate possible. A line-shaped excitation light with sequenced patterns was generated by a DLP projector, and a 2-D camera was used to acquire the spatio-spectral fluorescence images of the line-shaped excitation region. A complete hypercube was accomplished by scanning the sample throughout the area of interest. To demonstrate the system’s ability of axial contrast enhancement in specimens with different thickness, sectioned mouse skin and unsectioned Epipremnum aureum leaves were used as specimens. The recorded spectral information, including chlorophyll fluorescence and the fluorescence signals of the mouse skin, was then analyzed by applying linear unmixing to show the system’s power to separate the molecular components clearly even with seriously overlapped spectra.
2. System overview
2.1 System setup
Figure 1(a) shows the system setup, which is based on a traditional confocal line-scanning hyperspectral system [25,26]. A DLP projector (LightCrafter, Texas Instruments; 684*608 pixels) is used to generate a sequence of patterned line-shaped excitation light along the y-direction, with three phases, 0, 2π/3 and 4π/3 (Fig. 1(b)-1(d)). The DLP projector can provide excitation lights with embedded red, green and blue LEDs, each with its spectral peak at 460 nm, 540 nm and 617 nm. In this research, only blue and green excitations are employed. To achieve optical sectioning, the excitation light is focused by a high NA objective (UPLSAPO 20X, NA 0.75, Olympus) onto the specimens. The objective then gathers the backward fluorescence signals. The signals are separated from the excitation light by a dichroic beam splitter (FF509-FDi01 for blue and FF532-FDi01 for green, Semrock) and then focused upon the slit. After the slit, the signals are collimated along the x-direction and hit onto the grating so that the spectra of the signals are dispersed spatially. Imaged by L2, a 2-D y-λ spatio-spectral image can be obtained with a sCMOS camera (ORCA-Flash4.0 V2, Hamamatsu). In front of the sCMOS, a long-pass filter (BLP01-514R-25 for blue and BLP01-532-25 for green, Semrock) is inserted to block the excitation light and background noises. To obtain a complete hypercube, a series of y-λ images is required. Thus, a scanning stage moving the specimen along the x-direction is utilized to accomplish the task. By setting 200 nm each step for the x-direction scanning and a magnification of 33.3X for the y-direction imaging, the corresponding size at the sample plane of each sCMOS pixel can be calculated as 200 nm and 195.2 nm in the x- and y-direction. At every x position, the sCMOS captures the three sequenced images of different phases. And by applying the three images to Eq. (1), an optical sectioning y-λ image can be extracted.
Fig. 1.
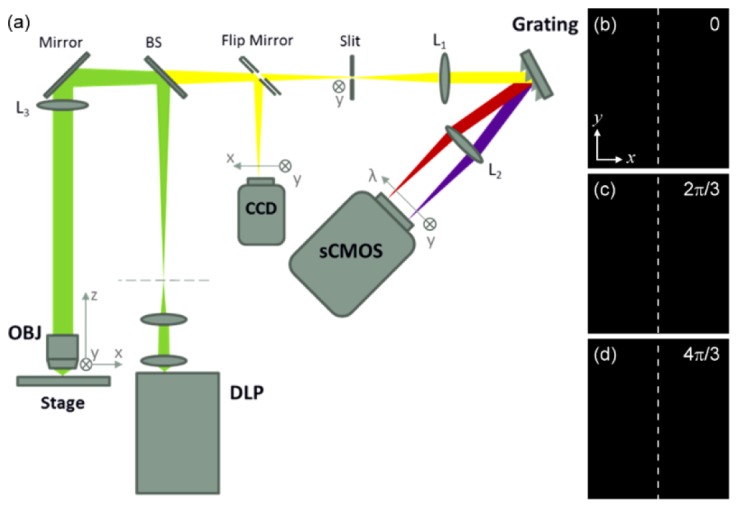
(a) Schematic diagram of the LHSI system. OBJ: objective; BS: beamsplitter; L: lens. (b)-(d) Line-shaped excitation light with three phases of 0, 2π/3 and 4π/3, generated by DLP.
With a maximum frame rate of 100 fps provided by the sCMOS camera, only 30 ms is required for acquiring three images with different phases to reconstruct a y-λ image of a scanning line. In contrast to the traditional line-scanning method, 3 times longer total acquisition time is required to perform the structured illumination. To obtain a complete hypercube with 100 μm in the x-direction, 500 scanning lines and a total time of 15 s is at least required. Due to the unlimited scanning range in the x-direction, the total time for a complete hypercube can change with the imaging area. On the other hand, the exposure time may need to extend due to lower fluorescence excitation efficiency and the total imaging time will become longer as well. Since the DLP projector possesses the flexibility to project different patterns, the width of the line-shaped excitation light can be adjusted according to each specimen’s characteristics. For specimens with lower excitation efficiency, widening the line-shaped excitation light in the x-direction can help to increase the fluorescence intensity, yet decrease the spatial resolution in the x-direction. With these tradeoffs, it is important to utilize the proper width for the excitation light in order to achieve the best resolution under sufficient signal-to-noise ratio (SNR).
2.2 System calibration
To calibrate the spectral system, a Hg(Ne) lamp and a diode laser were utilized. Within the spectral range of measurement, the Hg(Ne) lamp consists of 435.84-nm, 546.07-nm, 576.96-nm, and 579.07-nm emission lines (Fig. 2(a)) and the diode laser has a wavelength of 650 nm. Figure 2(b) shows the sCMOS pixel numbers corresponding to each emission wavelength. With linear fitting applied, the relation between wavelengths and the sCMOS pixel numbers can be obtained. To further determine the spectral resolution of the system, Gaussian fitting was applied to the emission lines’ intensity profiles of the Hg(Ne) lamp, as shown in Figs. 2(c) and 2(d). With the nature of narrow bandwidth, the full-width-at-half-maximum (FWHM) of the emission lines can represent the system’s spectral resolution. In Fig. 2(c), the FWHM of the emission line 546.07 nm was measured to be 1.21 nm, while in Fig. 2(d), the FWHM of two emission lines at 576.96 nm and 579.07 nm were 1.12 nm and 1.13 nm. Due to the FWHM being averaged around 1.15 nm, the two emission lines with a separation of 2.1 nm can be well distinguished.
Fig. 2.
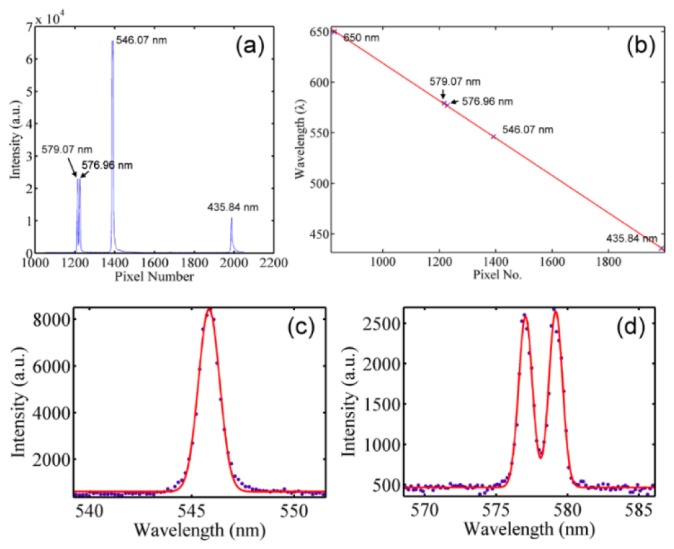
(a) The spectrum of the Hg(Ne) lamp versus the pixel number of the sCMOS; (b) the conversion relation between the pixel number and wavelength obtained by linear fitting the data points 435.84 nm, 546.07 nm, 576.96 nm, and 579.07 nm of the Hg(Ne) lamp and 650nm of a diode laser. (c),(d) The spectra of the Hg(Ne) emission lines at (c) 546.07 nm and (d) 576.96 nm and 579.07 nm. Red lines are the Gaussian fitting curves.
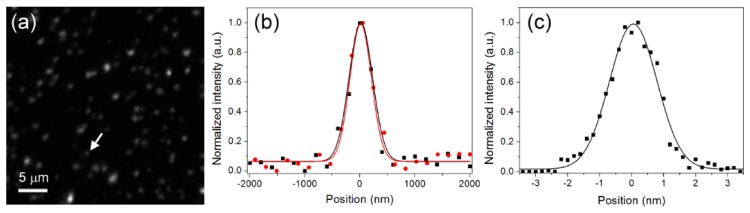
2.3 Spatial resolution
To analyze the spatial resolution of the system, fluorescent beads (FP-0256-2, Spherotech) with an average diameter of 250 nm and emission peak wavelength at 570 nm were chosen as the specimen. In this measurement, green excitation was used and the calculated theoretical spatial resolution was 463.6 nm. Figure 3(a) shows the reconstructed 2D optical sectioning image of the fluorescence beads illuminated by a line-shaped green excitation light with its pattern period of 2.9 μm. Figures 3(b) is the x- and y-direction intensity profiles and Fig. 3(c) is the z-direction intensity profile of the particle shown in Fig. 3(a), indicated with an arrow. Fitting the profiles with Gaussian functions, the FWHM of the particle in x- and y-directions were measured as 498 nm and 500 nm, which can be treated as the lateral resolution of the system. The FWHM of the particle in the z-direction was measured as 1.75 μm, which can be treated as the axial resolution. By applying the Gaussian fitting to a total of 10 particles, the average diameter in x-, y- and z-direction are 523.6 nm, 534.1 nm and 1.79 μm, respectively. The results are larger than the theoretical spatial resolution, which may be caused by the fluorescence beads’ diameter not being smaller enough in contrast to the system resolution or the uncorrected imaging aberration. The reason why the resolution in the x-direction is better than that in the y-direction is due to different image formation mechanisms along these two directions [28]. Since in the x-direction the image is formed by point-by-point recording, the excitation point spread function (PSF) is involved in the image formation. The diffraction limit is 0.61λex/NA, where λex is the excitation wavelength. In the y-direction, the image is effectively formed by wide-field image projection, the PSF of the imaging system instead is involved in the image formation. The diffraction limit is 0.61λem/NA, where λem is the emission wavelength. Due to λex being shorter than λem, the x-direction spatial resolution should be better than the y-direction one.
Fig. 3.
(a) Optical sectioning image of the fluorescence beads; (b) the x-direction (red circle and line) and y-direction (black square and line) intensity profiles of the particle indicated by the arrow in (a); (c) the z-direction intensity profile of the same particle in (b).
3. Sample preparation
3.1 Unsectioned Epipremnum aureum leaf
The leaves were obtained from a potted Epipremnum aureum. The leaves were removed from the plant and cut into 1 cm × 1 cm pieces for imaging, without physical sectioning, fixing, or staining processes. The hypercube were all acquired within 3 hrs after the leaves were taken off the plant.
3.2 Sectioned mouse skin with multiple staining
Sectioned mouse skin specimens were prepared by Dr. Chih-Chiang Chen’s lab in Department of Dermatology, Taipei Veterans General Hospital. Tissues were collected and fixed in 4% paraformaldehyde in phosphate-buffered saline. The fixed tissues were embedded in paraffin and sectioned to have 5-6 μm thickness. Based on the green excitation, Alexa 532 and Alexa 555 were selected to stain the protein antigens, K14 and CD34, respectively, each indicating the basal cells and follicle stem cells.
4. Experimental results
4.1 Unsectioned Epipremnum aureum leaf
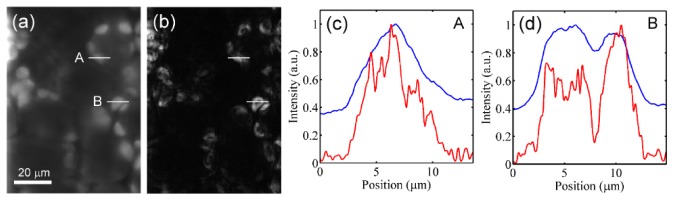
To evaluate the system’s ability to enhance the axial imaging contrast, an unsectioned Epipremnum aureum leaf was used as specimen. Blue excitation light was applied to excite the chlorophyll fluorescence in this case and the pattern period of the excitation line was set to 11.6 μm. Scanning the specimen 72 μm along the x-direction with an exposure time of 300 ms, a hypercube of the leaf was obtained at a depth of 70 μm beneath the leaf surface. Figure 4(a) shows the image at a wavelength of 685 nm, with the use of Eq. (2). This image is equivalent to that acquired without pattern illumination in traditional confocal line-scanning method. In addition to the on-focus structures of the chloroplasts, strong background noises were contributed by the off-focus signals and the scattered on-focus signals and thus reduce the imaging contrast. On the other hand, Fig. 4(b) is the corresponding optical sectioning image reconstructed by applying Eq. (1). In comparison with Fig. 4(a), the background noises are apparently eliminated so as to increase the imaging contrast. Figures 4(c) and 4(d) show respectively the normalized intensity profiles along line A and line B indicated in both Figs. 4(a) and 4(b). It can be seen that the background signals are greatly reduced by applying structure illumination, giving it a better SNR. The SNR measured along line A in Fig. 4(a) is 2.37, while the SNR is improved to 18.36 in Fig. 4(b). In Fig. 4(d), the structure details within a single chloroplast (along line B) can be more easily distinguished in Fig. 4(b) than in Fig. 4(a) due to the notably enhanced imaging contrast. The contrast to distinguish the two peaks is 0.39 in Fig. 4(a), while improved to 0.86 in Fig. 4(b). Here the contrast is calculated by following the formula (Max-Min)/(Max + Min). Despite the improved SNR and contrast, signal fluctuations due to the subtraction operation in the reconstructed procedure can be observed in the intensity profiles from Fig. 4(b). With the same illuminating and detecting configurations of traditional confocal line-scanning method, this system should has the same penetrability in thick samples. However, as the depth increases, the aberration of the structured illumination pattern becomes more serious and the improvement of SNR and contrast decreases in consequence. According to the previous study [35], patterns’ contrast can be regained by increasing the pattern period and the improvement of SNR and contrast can be obtained in deeper layers.
Fig. 4.
Images of the Epipremnum aureum leaf at a wavelength of 685 nm, obtained at a depth of 70 μm beneath the leaf surface (a) without and (b) with applying optical sectioning. (c),(d) The intensity profiles along (c) line A and (d) line B indicated in (a) and (b).
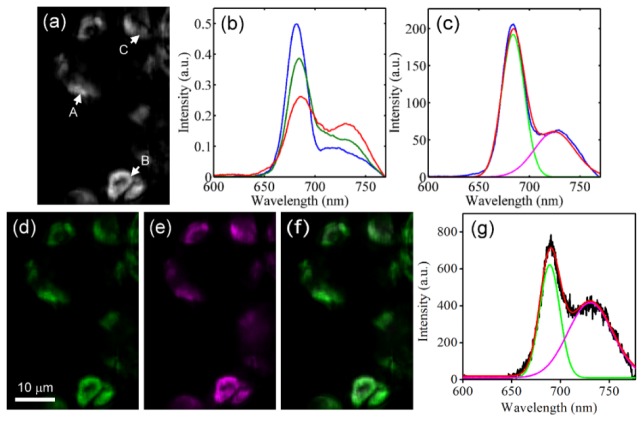
Chlorophyll fluorescence is known to consist of photosystem II (PSII) and photosystem I (PSI) fluorescence, with peaks at 685 nm and 730 nm, respectively. In the studies regarding photosynthesis, the intensity ratio between PSII and PSI can help to reveal the different states of the photosynthesis progresses [36]. In contrast to the spectroscopic methods recording only the spatially-averaged spectral information, the spatio-spectral information obtained by LHSI can further provides the spatially dependent photosynthesis states in cellular level. Figure 5(a) is the cropped image obtained from the hypercube used for Fig. 4, and the intensity of each pixel is calculated by integrating the intensity throughout whole spectral range. Figure 5(b) shows three exampled spectra acquired from points A, B and C indicated with arrows in Fig. 5(a). Each of them consists of PSII’s and PSI’s spectrum. The PSII-to-PSI ratio is considered as an important factor for photosynthesis, and the commonly-use method is to calculate the intensity ratio of 658-nm peak to 730-nm peak [36]. In the case that the intensity of 658-nm peak is much higher than that of 730-nm peak (blue line in Fig. 5(b)), the cross-talk from PSI to PSII can be ignored. However, it will become considerable when the intensity of two peaks are close to each other. To further reveal the contents of PSII and PSI without cross-talk problem, linear unmixing has to be applied and all the possible spectral bases contributing to the spectrum are required [37].
Fig. 5.
(a) The image of the leaf with intensity integrated throughout the whole spectrum range; (b) three normalized spectra at points A, B and C indicated with arrows in (a). (c) Gaussian fitting was applied to a recorded spectrum (blue) to find the spectral bases of PSII (green) and PSI (magenta). Red line shows the fitting result. (d)-(f) The images of (d) PSII and (e) PSI contents (normalized to within 0 and 1) obtained with linear unmixing and (f) the combination of PSII and PSI without normalization. (g) The linear unmixing result of a spectrum (black line). PSII and PSI components are represented by green and magenta and red line is the fitting result.
To find out the spectral bases of PSII and PSI, Gaussian fitting was employed to a recorded chlorophyll fluorescence spectrum, as shown in Fig. 5(c) [38]. These bases were then adopted to perform the linear unmixing. Figures 5(d) and 5(e) show the distribution of the PSII and PSI contents obtained from the analysis, with PSII and PSI contents represented in green and magenta. Since the contents of PSI are generally lower than those of PSII, to show the PSI image with reasonable brightness and contrast, the contents shown in Figs. 5(d) and 5(e) were both normalized to within 0 and 1. Figure 5(f) is the combination of PSII and PSI contents without normalization. It can be seen that the chloroplasts mostly appear greenish, which agrees with the fact that the contents of PSII are generally higher. Figure 5(g) shows the linear unmixing result of an exampled spectrum. The intensity ratio of 658-nm peak to 730-nm peak is 1.81, while the ratio of PSII content to PSI content based on the linear unmixing result is only 1.51. This difference gives the evidence that applying linear unmixing can help to remove the impacts of cross-talk, especially for the case that the intensity of PSII and PSI peaks are close to each other.
4.2 Sectioned mouse skin with multiple staining
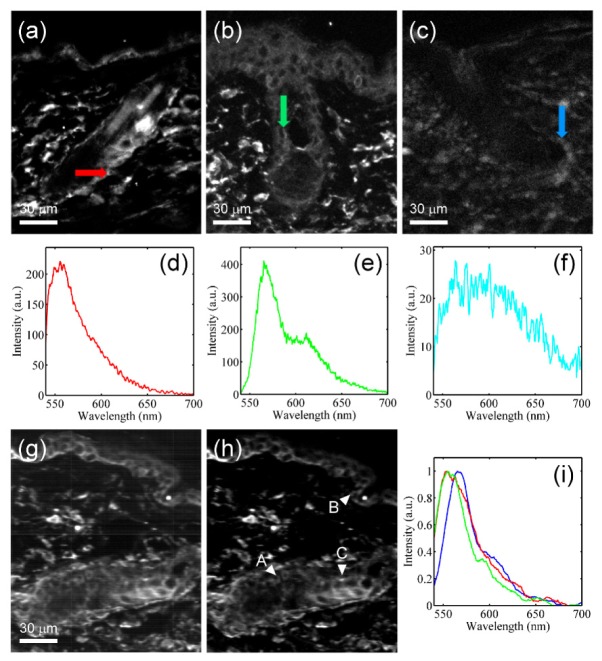
Multiple staining is a usual way in biomedical researches to reveal the distribution of two or more kinds of molecules or the interaction between them. If limited to single excitation, the available fluorophores could then suffer serious cross-talk issues. In the study of how hair follicle stem cells are related to hair growth, two fluorophores, Alexa 532 and Alexa 555, were chosen to target the epithelium cells and the follicle stem cells respectively, based on the green excitation. To acquire all the possible spectral bases, two samples singly stained with Alexa 532 and Alexa 555 and one sample without staining were prepared. Figures 6(a)-6(c) show the images of these three samples at a wavelength of 568 nm, respectively. The scanning range in the x-direction was 400 μm and the exposure time was set as 500 ms due to lower staining efficiency. The pattern period of the excitation line was set to 5.8 μm. In Fig. 6(a), the Alexa 532 fluorescence signals mostly come from the basal layer and parts of epithelium cells surrounding the hair follicle, while in Fig. 6(b), the Alexa 555 fluorescence signals give the cell morphology of the epidermis and the hair follicle bulge. Figures 6(d)-6(f) show the spectrum bases of Alexa 532, Alexa 555 and autofluorescence obtained from three samples at which indicated with arrows in Figs. 6(a)-6(c). Compared with two kinds of extrinsic fluorescence, intrinsic autofluorescence appear much weaker in intensity. Therefore, the contribution of autofluorescence to the spectrum bases of Alexa 532 and Alexa 555 can be ignored. These three spectra are found to have very similar peak wavelengths and overlap one-another seriously so that it is impossible to separate them apart just by using filters.
Fig. 6.
(a)-(c) The image of specimens singly stained with (a) Alexa 532, (b) Alexa 555 and (c) without staining, at a wavelength of 568 nm. (d)-(f) The spectrum bases of (d) Alexa 532, (e) Alexa 555 and (f) autofluorescence obtained from the positions indicated by arrows in (a)-(c). (g),(h) The image of a specimen with multiple staining, (g) with and (h) without optical sectioning. (i) Three spectra at positions A (blue), B (green) and C (red) indicated by arrowheads in (h).
Using a sectioned mouse skin stained with both Alexa 532 and Alexa 555 as specimen, the hypercube was acquired. The images at a wavelength of 568 nm without and with applying optical sectioning are shown in Figs. 6(g) and 6(h). The average SNR are measured as 1.02 and 4.58, respectively. Even in the ~5-μm specimen, applying optical sectioning can also provide a ~4.5-fold contrast enhancement. The normalized spectra of three positions A, B and C indicated with arrowheads in Fig. 6(h) can be seen in Fig. 6(i). Due to different contents contributed by Alexa 532, Alexa 555 and autofluorescence, three spectra differ slightly from one-another. The spectra at positions B and C have a shorter peak wavelength than at position A, while the tail of the spectrum at position C fits better with that at position A but not at position B. These properties may indicate the major composition at position A is Alexa 555, and at positions B and C is Alexa 532, while Alexa 555 also has contribution at position C.
To further analyze the constituents’ distribution, linear unmixing was applied. Based on the three bases from Figs. 6(d)-6(f), Figs. 7(a)-7(c) are the linear unmixing results corresponding to Fig. 6(h), referring to Alexa 532, Alexa 555 and autofluorescence, respectively. Figure 7(d) shows the combination of Figs. 7(a) to 7(c). Compared with Figs. 6(a) and 6(b), the distributions of Alexa 532 and Alexa 555 are found to accord with the results of samples with single staining. Which indicates the system’s spectral resolution is sufficient so that linear unmixing can work well to separate these seriously-overlapped spectrum bases. On the other hand, due to the relatively lower intensity of autofluorescence signals, the autofluorescence’s distribution (Fig. 7(c)) can hardly be noticed. Figures 7(e) and 7(f) shows the linear unmixing results of two exampled spectra at positions A and B indicated with arrowheads in Fig. 7(d). At position A, the content ratio of Alexa 532 to Alexa 555 is 1:2.14, while at position B, the content ratio is 13:1. The magenta lines in both figures show the spectra reconstructed based on the linear unmixing results. The root-mean-square deviations (RMSDs) of the reconstructed spectra at positions A and B are calculated as 0.68 and 0.16. Divided by the norm of each spectrum, the normalized RMSDs are then 0.081 and 0.082, providing a better comparison between spectra with different intensity scales.
Fig. 7.
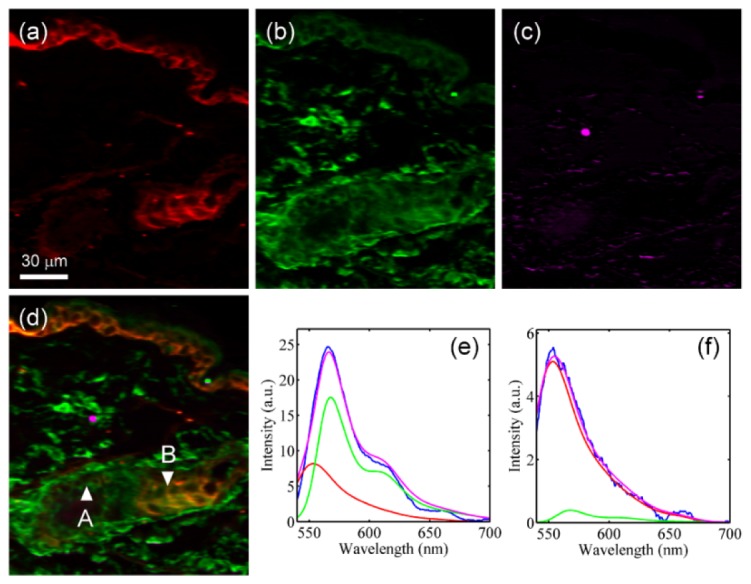
(a)-(c) The images of three constituents, (a) Alexa 532, (b) Alexa 555 and (c) autofluorescence, separated by applying linear unmixing. (d) The combination image of (a) to (c). (e),(f) The linear unmixing results of spectra (blue lines) at (e) position A and (f) position B indicated in (d). Red lines: Alexa 532; green lines: Alexa 555; magenta lines: reconstructed spectra.
5. Conclusions
In this research, SIM optical sectioning has been combined into a LHSI system. With SIM, the off-focus signals and scattered on-focus signals along the slit’s long-side were successfully removed, and the axial imaging contrast was thus enhanced. In the unsectioned leaf, the imaging contrast was shown to be improved by ~8 times, while in the 5-μm sectioned mouse skin tissues, a 4.5-fold contrast enhancement can also be achieved. The system has lateral spatial resolution of 523.6 nm and 534.1 nm in x- and y-direction, respectively, and a spectral resolution of 1.15 nm. By applying linear unmixing to the chlorophyll fluorescence spectra, the spatial distribution of PSII and PSI contents can be obtained without cross-talk. With enhanced imaging contrast, the details within a single chloroplast can be revealed. Based on the 1.15-nm spectral resolution, the system shows its ability to separate heavily spectrally-overlapped fluorescence signals. Being able to accurately identify different constituents and remove the effects caused by autofluorescence can give great help to observations regarding multiple fluorescence staining.
Disclosures
The authors declare that there are no conflicts of interest related to this article.
Acknowledgments
This research was supported by the National Science Council under grants MOST 104-2221-E-008 −097 and MOST 105-2112-M-008 −020, and the National Central University’s Plan to Develop First-class Universities, Top-level Research Center Grant 102G903-2.
References and links
- 1.E. K. Hege, D. O’Connell, W. Johnson, S. Basty, and E. L. Dereniak, “Hyperspectral imaging for astronomy and space surviellance,” in Optical Science and Technology, SPIE's 48th Annual Meeting (SPIE, 2004), pp. 380–391. 10.1117/12.506426 [DOI] [Google Scholar]
- 2.O. Berné, A. Helens, P. Pilleri, and C. Joblin, “Non-negative matrix factorization pansharpening of hyperspectral data: An application to mid-infrared astronomy,” in 2010 2nd Workshop on Hyperspectral Image and Signal Processing: Evolution in Remote Sensing (IEEE, 2010), pp. 1–4. 10.1109/WHISPERS.2010.5594900 [DOI] [Google Scholar]
- 3.Wu D., Sun D.-W., “Advanced applications of hyperspectral imaging technology for food quality and safety analysis and assessment: A review—Part I: Fundamentals,” Innov. Food Sci. Emerg. Technol. 19, 1–14 (2013). 10.1016/j.ifset.2013.04.014 [DOI] [Google Scholar]
- 4.Wu D., Sun D.-W., “Advanced applications of hyperspectral imaging technology for food quality and safety analysis and assessment: a review—part II: applications,” Innov. Food Sci. Emerg. Technol. 19, 15–28 (2013). 10.1016/j.ifset.2013.04.016 [DOI] [Google Scholar]
- 5.Goetz A. F., “Three decades of hyperspectral remote sensing of the Earth: A personal view,” Remote Sens. Environ. 113, S5–S16 (2009). 10.1016/j.rse.2007.12.014 [DOI] [Google Scholar]
- 6.Goetz A. F., Vane G., Solomon J. E., Rock B. N., “Imaging spectrometry for Earth remote sensing,” Science 228(4704), 1147–1153 (1985). 10.1126/science.228.4704.1147 [DOI] [PubMed] [Google Scholar]
- 7.Liu Z., Yan J.-Q., Zhang D., Li Q.-L., “Automated tongue segmentation in hyperspectral images for medicine,” Appl. Opt. 46(34), 8328–8334 (2007). 10.1364/AO.46.008328 [DOI] [PubMed] [Google Scholar]
- 8.Liu H. Y., Li Q. L., Wang Y. T., Liu J. G., Xue Y. Q., “[Molecular hyperspectral imaging (MHSI) system and application in biochemical medicine],” Guangpuxue Yu Guangpu Fenxi 31(10), 2593–2597 (2011). [PubMed] [Google Scholar]
- 9.R. M. Levenson, P. J. Cronin, and N. R. Harvey, “Spectral imaging and biomedicine: new devices, new approaches,” in Applied Imagery Pattern Recognition Workshop, 2002. Proceedings. 31st (IEEE, 2002), pp. 105–111. 10.1109/AIPR.2002.1182262 [DOI] [Google Scholar]
- 10.Guo N., Zeng L., Wu Q., “A method based on multispectral imaging technique for white blood cell segmentation,” Comput. Biol. Med. 37(1), 70–76 (2007). 10.1016/j.compbiomed.2005.10.003 [DOI] [PubMed] [Google Scholar]
- 11.Vo-Dinh T., Cullum B., Kasili P., “Development of a multi-spectral imaging system for medical applications,” J. Phys. D Appl. Phys. 36(14), 1663–1668 (2003). 10.1088/0022-3727/36/14/302 [DOI] [Google Scholar]
- 12.Hsieh Y.-F., Mang O.-Y., Chiou J.-C., Lin Y.-J., Tsai M.-H., Bau D.-T., Chiu C.-F., Teseng G.-C., Chang N.-W., Kao W.-C., Wu S.-D., “The new hyperspectral microscopic system for cancer diagnosis,” in SPIE BiOS (SPIE, 2011), pp. 79020J–79020J–5. [Google Scholar]
- 13.Kong S. G., Du Z., Martin M., Vo-Dinh T., “Hyperspectral fluorescence image analysis for use in medical diagnostics,” in Advanced Biomedical and Clinical Diagnostic Systems III (SPIE, 2005), pp. 21–28. [Google Scholar]
- 14.Lu G., Fei B., “Medical hyperspectral imaging: a review,” J. Biomed. Opt. 19(1), 010901 (2014). 10.1117/1.JBO.19.1.010901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sutherland V. L., Timlin J. A., Nieman L. T., Guzowski J. F., Chawla M. K., Worley P. F., Roysam B., McNaughton B. L., Sinclair M. B., Barnes C. A., “Advanced imaging of multiple mRNAs in brain tissue using a custom hyperspectral imager and multivariate curve resolution,” J. Neurosci. Methods 160(1), 144–148 (2007). 10.1016/j.jneumeth.2006.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vasefi F., Kaminska B., Brackstone M., Carson J. J., “Hyperspectral angular domain imaging for ex-vivo breast tumor detection,” in SPIE BiOS (SPIE, 2013), pp. 85870S–85870S–8. [Google Scholar]
- 17.Zuzak K. J., Francis R. P., Wehner E. F., Smith J., Litorja M., Allen D. W., Tracy C., Cadeddu J., Livingston E., “DLP hyperspectral imaging for surgical and clinical utility,” in SPIE MOEMS-MEMS: Micro-and Nanofabrication (SPIE, 2009), pp. 721006–721006–9. [Google Scholar]
- 18.Zavattini G., Vecchi S., Mitchell G., Weisser U., Leahy R. M., Pichler B. J., Smith D. J., Cherry S. R., “A hyperspectral fluorescence system for 3D in vivo optical imaging,” Phys. Med. Biol. 51(8), 2029–2043 (2006). 10.1088/0031-9155/51/8/005 [DOI] [PubMed] [Google Scholar]
- 19.Gowen A. A., Feng Y., Gaston E., Valdramidis V., “Recent applications of hyperspectral imaging in microbiology,” Talanta 137, 43–54 (2015). 10.1016/j.talanta.2015.01.012 [DOI] [PubMed] [Google Scholar]
- 20.Gao L., Kester R. T., Hagen N., Tkaczyk T. S., “Snapshot Image Mapping Spectrometer (IMS) with high sampling density for hyperspectral microscopy,” Opt. Express 18(14), 14330–14344 (2010). 10.1364/OE.18.014330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim H.-T., Murukeshan V. M., “A four-dimensional snapshot hyperspectral video-endoscope for bio-imaging applications,” Sci. Rep. 6(1), 24044 (2016). 10.1038/srep24044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sinclair M. B., Haaland D. M., Timlin J. A., Jones H. D., “Hyperspectral confocal microscope,” Appl. Opt. 45(24), 6283–6291 (2006). 10.1364/AO.45.006283 [DOI] [PubMed] [Google Scholar]
- 23.Wang P., Ebeling C. G., Gerton J., Menon R., “Hyper-spectral imaging in scanning-confocal-fluorescence microscopy using a novel broadband diffractive optic,” Opt. Commun. 324, 73–80 (2014). 10.1016/j.optcom.2014.03.044 [DOI] [Google Scholar]
- 24.Im K.-B., Kang M.-S., Kim J., Bestvater F., Seghiri Z., Wachsmuth M., Grailhe R., “Two-photon spectral imaging with high temporal and spectral resolution,” Opt. Express 18(26), 26905–26914 (2010). 10.1364/OE.18.026905 [DOI] [PubMed] [Google Scholar]
- 25.Nakamura R., Izumi Y., Kajiyama S., Kobayashi A., Kanematsu Y., “Line-scanning microscopy for time-gated and spectrally resolved fluorescence imaging,” J. Biol. Phys. 34(1-2), 51–62 (2008). 10.1007/s10867-008-9113-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Z., Ma S., Ji Y., Liu L., Guo J., He Y., “Parallel scan hyperspectral fluorescence imaging system and biomedical application for microarrays,” J. Phys. Conf. Ser. 277, 012023 (2011). 10.1088/1742-6596/277/1/012023 [DOI] [Google Scholar]
- 27.Kumazaki S., Hasegawa M., Ghoneim M., Shimizu Y., Okamoto K., Nishiyama M., Oh-Oka H., Terazima M., “A line-scanning semi-confocal multi-photon fluorescence microscope with a simultaneous broadband spectral acquisition and its application to the study of the thylakoid membrane of a cyanobacterium Anabaena PCC7120,” J. Microsc. 228(2), 240–254 (2007). 10.1111/j.1365-2818.2007.01835.x [DOI] [PubMed] [Google Scholar]
- 28.Chen S.-Y., Lu C.-S., Yeh C.-H., “Non-de-scanned parallel recording two-photon hyperspectral microscopy with high spectral and spatial resolution,” Biomed. Opt. Express 5(2), 338–347 (2014). 10.1364/BOE.5.000338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang D., Chen Y., Wang Y., Liu J. T. C., “Comparison of line-scanned and point-scanned dual-axis confocal microscope performance,” Opt. Lett. 38(24), 5280–5283 (2013). 10.1364/OL.38.005280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neil M. A., Juškaitis R., Wilson T., “Method of obtaining optical sectioning by using structured light in a conventional microscope,” Opt. Lett. 22(24), 1905–1907 (1997). 10.1364/OL.22.001905 [DOI] [PubMed] [Google Scholar]
- 31.Agard D. A., “Optical sectioning microscopy: cellular architecture in three dimensions,” Annu. Rev. Biophys. Bioeng. 13(1), 191–219 (1984). 10.1146/annurev.bb.13.060184.001203 [DOI] [PubMed] [Google Scholar]
- 32.Conchello J.-A., Lichtman J. W., “Optical sectioning microscopy,” Nat. Methods 2(12), 920–931 (2005). 10.1038/nmeth815 [DOI] [PubMed] [Google Scholar]
- 33.Keahey P., Ramalingam P., Schmeler K., Richards-Kortum R. R., “Differential structured illumination microendoscopy for in vivo imaging of molecular contrast agents,” Proc. Natl. Acad. Sci. U.S.A. 113(39), 10769–10773 (2016). 10.1073/pnas.1613497113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dan D., Lei M., Yao B., Wang W., Winterhalder M., Zumbusch A., Qi Y., Xia L., Yan S., Yang Y., Gao P., Ye T., Zhao W., “DMD-based LED-illumination super-resolution and optical sectioning microscopy,” Sci. Rep. 3(1), 1116 (2013). 10.1038/srep01116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen S.-Y., Hsu Y. J., Yeh C.-H., Chen S.-W., Chung C.-H., “Pico-projector-based optical sectioning microscopy for 3D chlorophyll fluorescence imaging of mesophyll cells,” J. Opt. 17(3), 035301 (2015). 10.1088/2040-8978/17/3/035301 [DOI] [Google Scholar]
- 36.Baker N. R., “Chlorophyll fluorescence: a probe of photosynthesis in vivo,” Annu. Rev. Plant Biol. 59(1), 89–113 (2008). 10.1146/annurev.arplant.59.032607.092759 [DOI] [PubMed] [Google Scholar]
- 37.Schlachter S., Schwedler S., Esposito A., Kaminski Schierle G. S., Moggridge G. D., Kaminski C. F., “A method to unmix multiple fluorophores in microscopy images with minimal a priori information,” Opt. Express 17(25), 22747–22760 (2009). 10.1364/OE.17.022747 [DOI] [PubMed] [Google Scholar]
- 38.Subhash N., Mohanan C. N., “Curve-fit analysis of chlorophyll fluorescence spectra: Application to nutrient stress detection in sunflower,” Remote Sens. Environ. 30(60), 347–356 (1997). 10.1016/S0034-4257(96)00217-9 [DOI] [Google Scholar]