Abstract
Reliable automated identification and susceptibility testing of clinically relevant bacteria is an essential routine for microbiology laboratories, thus improving patient care. Examples of automated identification systems include the Phoenix (Becton Dickinson) and the VITEK 2 (bioMérieux). However, more and more frequently, microbiologists must isolate “difficult” strains that automated systems often fail to identify. An alternative approach could be the genetic identification of isolates; this is based on 16S rRNA gene sequencing and analysis. The aim of the present study was to evaluate the possible use of MicroSeq 500 (Applera) for sequencing the 16S rRNA gene to identify isolates whose identification is unobtainable by conventional systems. We analyzed 83 “difficult” clinical isolates: 25 gram-positive and 58 gram-negative strains that were contemporaneously identified by both systems—VITEK 2 and Phoenix—while genetic identification was performed by using the MicroSeq 500 system. The results showed that phenotypic identifications by VITEK 2 and Phoenix were remarkably similar: 74% for gram-negative strains (43 of 58) and 80% for gram-positive strains were concordant by both systems and also concordant with genetic characterization. The exceptions were the 15 gram-negative and 9 gram-positive isolates whose phenotypic identifications were contrasting or inconclusive. For these, the use of MicroSeq 500 was fundamental to achieving species identification. In clinical microbiology the use of MicroSeq 500, particularly for strains with ambiguous biochemical profiles (including slow-growing strains), identifies strains more easily than do conventional systems. Moreover, MicroSeq 500 is easy to use and cost-effective, making it applicable also in the clinical laboratory.
Since the time when microbial identification (ID) was performed by using tube tests, much progress has been made. Initially, to assist microbiologists, miniaturized ID systems appeared, followed by innovative automatic ID systems such as VITEK 2 (bioMérieux, Marcy l'Étoile, France) and Phoenix (Becton Dickinson Microbiology Systems, Cockeysville, Md.) (1, 9). These are new, fully automated systems that provide accurate and reproducible IDs, as well as antimicrobial susceptibility tests. The systems possess either sophisticated software to identify microorganism phenotypes or “advanced expert systems” able to elaborate and validate the antimicrobial susceptibilities of the isolates (1, 2, 6, 7, 9, 10). In spite of the undoubtedly innovative results obtained with the widespread use of these automated systems, they do have some drawbacks, particularly when microbiologists need to identify microorganisms exhibiting biochemical features that do not fit into any known patterns of genus and species. These unusual isolates are quite common, especially when we consider that more and more strains isolated from patients that have undergone long-term antimicrobial therapy (such as hematological patients and those in intensive care units) can lose their typical biochemical characteristics and become extremely difficult to cultivate (19, 21, 23). DNA sequencing of the 16S rRNA gene and the consequent comparison of the gene sequences of bacterial species is a good method for identifying bacteria at the species level. An excellent example of these molecular methods is MicroSeq 500 (Perkin-Elmer Applied Biosystems Division [now Applera, Foster City, Calif.]) 16S rRNA sequencing (4, 12, 15, 19, 22). We sought to evaluate the possible use of MicroSeq 500 for identifying some “difficult” strains that conventional automated systems have failed to characterize either by furnishing an inconclusive ID or by exhibiting an unlikely (implausible) profile.
(These findings were presented in part at the 14th European Congress of Clinical Microbiology Infectious Diseases in Prague, Czech Republic, in 2004).
MATERIALS AND METHODS
Bacterial isolates.
Of 83 selected clinical isolates, 25 were gram positive and 58 were gram negative (including both fermenting and nonfermenting strains). All of the strains came from clinical samples collected from patients in intensive care or in hematology and were processed in the microbiological laboratories of the Policlinic of Tor Vergata. Of the 83 isolates, 37 strains (44.5%) were isolated from blood, 30 (36%) were isolated from lower respiratory samples (such as bronchial alveolar lavage and bronchial aspirate and sputum), 5 (6%) were isolated from pus, and 11 (13%) were isolated from venous catheters. The isolates were subcultured twice on Trypticase soy agar (Oxoid, Milan, Italy) supplemented with 5% sheep blood to ensure viability and purity.
ID methodologies.
ID of the isolates was contemporaneously performed by using the VITEK 2 and Phoenix systems according to the manufacturers' instructions. Particularly, for the Phoenix system a suspension corresponding to a McFarland scale of 0.5 (accepted range, 0.5 to 0.6), adjusted by using a crystal nephelometer, was prepared in ID broth (Becton Dickinson) and poured, within 30 min, into the panel, which was then loaded into the instrument within 30 min. The Phoenix system gives an ID when a species or group of species is identified with >90% confidence. The confidence value is a measure of the likelihood that the issued ID is correct. The average time required to reach an ID ranges from 3 to 4 h. For the VITEK 2 system, a bacterial suspension was adjusted to a McFarland standard of 0.6 (range, 0.55 to 0.65) in 3 ml of 0.45% sodium chloride solution by using a densitometer (bioMérieux). This suspension was used to inoculate ID cards, IDGPC and IDGNB, respectively, to identify gram-positive and gram-negative isolates. Suspensions and cards were placed into a “smart tray,” which was then inserted into the VITEK 2 reader incubator module, where the cards are filled and sealed. Cards are automatically read every 15 min. In the VITEK 2 system, the confidence value is expressed as seven different categories of results: excellent ID, very good ID, good ID, or acceptable ID (each of these four categories shows only one ID result); low discrimination (more than one ID result is given, whereupon the software suggests performing additional tests such as oxidase, hemolysis, pigmentation, indole, and motility tests in order to obtain a correct ID); inconclusive ID; and unidentified. The time required for VITEK 2 to arrive at a final ID result is 3.5 h for gram-negative strains and 2.5 h for gram-positive strains.
All of the strains were maintained on Trypticase soy agar, supplemented with 5% sheep blood, until processed for 16S rRNA amplification.
Extraction of bacterial DNA.
A heavy suspension, corresponding to a McFarland scale of 1, of bacterial cells of each isolate was prepared in 1 ml of sterile distilled water. The suspension was centrifuged at 4,000 × g for 15 min. The pellet was suspended in 200 μl of PrepMan Ultra (Applera) and heat lysed at 100°C for 10 min, cooled at room temperature, and centrifuged at 12,000 × g for 3 min. Then, 2 μl of the supernatant of each bacterial extract was used for successive amplifications.
Amplification, sequencing, and analysis of the 16S rRNA by MicroSeq 500.
The MicroSeq 500 16S rRNA-based bacterial ID system (Applera) was designed for the genetic ID of bacteria. A 527-bp fragment of the 16S rRNA gene of the bacterial strains was amplified from the 5′ end of the gene in a reaction volume of 50 μl (25 μl of MicroSeq 500 PCR master mix containing 12.5 pmol of 005F and 531R primers, 23 μl of sterile distilled water, and 2 μl of the bacterial extract). Amplified products were purified by using a PCR purification kit (Qiagen, Valencia, Calif.), according to the manufacturer's recommendations, prior to sequencing. Forward- and reverse-sequencing reactions were performed for each amplified product. The sequencing reaction consisted of 13 μl of the MicroSeq 500 sequencing mix (containing 3.2 pmol of 005F and 531R primers), 4 μl of sterile distilled water, and 3 μl of purified amplified product. Sequencing reactions were purified with Centrispin columns (Princeton Separations, Princeton, N.J.) according to the manufacturer's instructions. All sequencing analysis were performed in an ABI Prism 310 genetic analyzer (Applera) (12, 22).
Sequence data analysis.
Sequencing data were analyzed by using MicroSeq 500 software (version 1.35). The analysis steps were (i) assembly at the forward and reverse sequences into a consensus sequence, (ii) editing of the consensus sequence to resolve discrepancies between the two strands by evaluation of the electropherograms, and (iii) comparison of the consensus sequence in the MicroSeq 500 database. The MicroSeq 500 database contains full-length 16S rRNA gene sequences for more than 1,400 different bacteria. In most cases, each species was represented by the type strains, thus ensuring that the database was created by using the “most typical” strain of the species. The MicroSeq libraries are tested in accordance with ISO 9001:2000 quality systems (11, 22). The database comparison, using the full alignment tool of the MicroSeq 500 software, generated a list of the closest matches with a distance score. This score indicated the percent difference between unknown sequences and the database sequence. To compare the original ID of an isolate to its MicroSeq 500 ID, MicroSeq 500 ID was considered to be the closest match in the MicroSeq 500 database no matter what the distance score was. The consensus sequences were also compared to universal databases in the NCBI data bank. No incongruence with MicroSeq 500 ID was found (22).
RESULTS
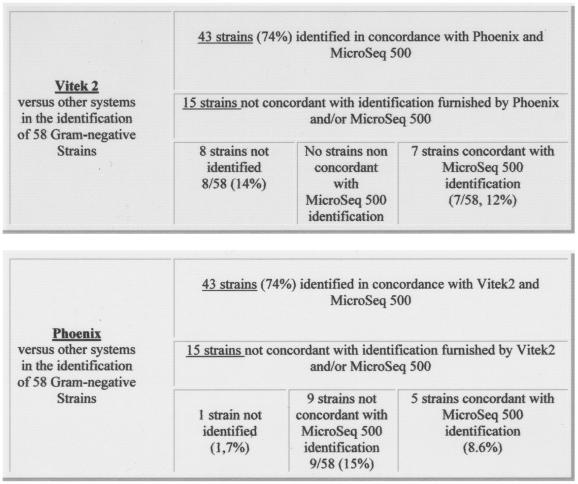
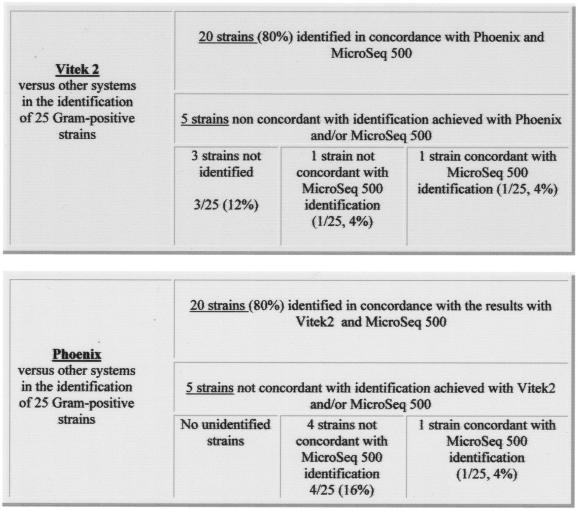
The results obtained for the 58 gram-negative isolates showed that both systems (VITEK 2 and Phoenix) achieved concordant IDs for 43 strains (43 of 58 [74%]). These IDs were also confirmed, at the species level, by using the MicroSeq 500, although for 15 gram-negative isolates the systems exhibited contradictory results when their data were compared to each other and to those obtained by the MicroSeq 500 (Fig. 1). In particular, in this group of 15 isolates, VITEK 2 showed the highest number of inconclusive IDs (“unidentified”): 8 unidentified strains (14% of the examined isolates). The IDs of seven isolates (of 58 [12%]), compared to MicroSeq 500, were confirmed to be correct. Therefore, VITEK 2 correctly identified 86% of the examined strains. The Phoenix system, in the group of 15 isolates, exhibited only one unidentified strain of 58 (1.7%). For 9 of 58 (15%) isolates the ID was not concordant either with VITEK 2 or with MicroSeq 500. Finally, 5 of 58 (8.6%) strains were confirmed to be correctly identified compared to MicroSeq 500. Therefore, the Phoenix system correctly identified 82.6% of the examined isolates. In Table 1 the respective IDs provided by the three systems are presented. In the gram-positive ID, VITEK 2 and Phoenix showed concordant results in 80% of the isolates (20 of 25). For the remaining five gram-positive isolates, the systems exhibited contradictory results. In particular, VITEK 2 was unable to identify three strains (i.e., 3 of 25 [12%]). Although for 2 isolates, one ID was correct and one was incorrect of 25 isolates tested (4%), a finding in disagreement with the IDs provided by the MicroSeq 500 (Fig. 2). Therefore, VITEK 2, for gram-positive isolates, furnished 84% conclusive IDs. In contrast, for the same group of gram-positive strains, the Phoenix system achieved an ID in all cases, but for four isolates (i.e., 4 of 25 [16%]) the ID was incorrect, being in disagreement with the IDs provided by the MicroSeq 500. The ID obtained by the Phoenix system was correct for only one strain of this group of five isolates (1 of 25 [4%]) (see Fig. 2). Hence, the Phoenix system also correctly identified 84% of the gram-positive isolates examined. The IDs provided by all of the systems for this group of five isolates are shown in Table 2.
FIG. 1.
Gram-negative ID: comparison of the results provided by using VITEK 2 versus Phoenix and MicroSeq 500 and Phoenix versus VITEK 2 and MicroSeq 500.
TABLE 1.
Fifteen gram-negative isolates with nonconcordant IDs among the systems used
| Isolate | IDa obtained by using:
|
||
|---|---|---|---|
| Phoenix | Vitek 2 | MicroSeq 500 | |
| 1 | Achromobacter xylosoxidans | ? | Achromobacter xylosoxidans |
| 2 | ? | Acinetobacter junii | Acinetobacter junii |
| 3 | Achromobacter xylosoxidans | ? | Achromobacter xylosoxidans |
| 4 | Shigella dysenteriae | Escherichia coli | Escherichia coli |
| 5 | Moraxella catarrhalis | Acinetobacter lwoffii | Acinetobacter lwoffii |
| 6 | Moraxella catarrhalis | ? | Moraxella catarrhalis |
| 7 | Pasteurella spp. | Escherichia coli | Escherichia coli |
| 8 | Acinetobacter spp. | ? | Acinetobacter junii |
| 9 | Moraxella catarrhalis | Acinetobacter lwoffii | Acinetobacter lwoffii |
| 10 | Moraxella catarrhalis | Acinetobacter lwoffii | Acinetobacter lwoffii |
| 11 | Stenotrophomonas maltophilia | ? | Stenotrophomonas maltophilia |
| 12 | Achromobacter xylosoxidans | ? | Sphingobacterium multivorum |
| 13 | Achromobacter xylosoxidans | ? | Burkholderia glathei |
| 14 | Moraxella catarrhalis | ? | Moraxella catarrhalis |
| 15 | Achromobacter xylosoxidans | Stenotrophomonas maltophilia | Stenotrophomonas maltophilia |
A question mark indicates a strain not identified by the automated system.
FIG. 2.
Gram-positive ID: comparison of the results furnished by using VITEK 2 versus Phoenix and MicroSeq 500 and Phoenix versus VITEK 2 and MicroSeq 500.
TABLE 2.
Five gram-positive isolates with nonconcordant IDs among the systems used
| Isolate | IDa obtained by using:
|
||
|---|---|---|---|
| Phoenix | Vitek 2 | MicroSeq 500 | |
| 1 | Corynebacterium diphtheriae | ? | Peptostreptococcus anaerobius |
| 2 | Streptococcus acidominimus | Streptococcus pneumoniae | Streptococcus oralis |
| 3 | Listeria grayi | ? | Listeria monocytogenes |
| 4 | Corynebacterium amycolatum | Staphylococcus aureus | Staphylococcus aureus |
| 5 | Streptococcus mitis | ? | Streptococcus mitis |
See Table 1, footnote a.
DISCUSSION
In this study we used a set of strains representative of the “difficult” isolates encountered by microbiologists when samples from the department of hematology or from intensive care units, in particular, must be evaluated. These isolates, obtained from patients undergoing treatment with several antibiotics, had lost their usual biochemical behavior, showing extremely slow growth on artificial media and exhibiting atypical biochemical profiles (13, 14, 17-23). These strains represent a challenge both for the automated system and for the microbiologist who needs to provide an answer to the clinician in the shortest time possible. Frequently, in fact, these isolates are responsible for sepsis or other complicated infections in immunosuppressed patients whose lives are at risk (3, 8, 14, 16, 20, 21, 23). The aim of the present study was to evaluate the ability of two well-known systems to detect “difficult” microorganisms and also to evaluate the use of these systems as a possible alternative to a genetic ID system (MicroSeq 500) (4, 12, 19, 22). We found that these systems provide concordant IDs both for gram-negative and for gram-positive isolates in the majority of cases (74 and 80%, respectively). Nevertheless, VITEK 2, which on the one hand presented the greater number of unfinalized IDs (range, 12 to 14%), is able, on the other hand, to furnish IDs in agreement with MicroSeq 500 in 4 to 12% of examined cases. The Phoenix system more easily achieves ID profiles, which in 15 to 16% cases are incorrect. Several authors, although they used predominantly standard reference strains (obtained from private or public strain collections) in their work, suggest resolving discrepancies in the ID provided by automated systems by using other commercial kits (both automated or manual), such as the API system (bioMérieux) (5, 7, 22). It is generally believed that the ID of particular strains (i.e., slow-growing or nonfermenting isolates) could be better achieved only by the helpful use of molecular methods, e.g., MicroSeq 500 (2, 11, 16, 22).
The MicroSeq 500 system has been shown to be helpful in reliably identifying some strains that conventional systems fail to characterize. The relatively short time in which results can be obtained with MicroSeq 500 (DNA extraction, 1 h; amplification and sequencing, <10 h) and the low cost of each test ($13.79) make this system easily applicable in microbiology laboratories, particularly those that are expert in the use of molecular biologic procedures. The only disadvantage of MicroSeq 500, as Woo et al. and Cloud et al. reported, is the limitation of the 16S rRNA database; this has been addressed by the recent introduction of an expanded database (4, 11, 22). However, in the rare instances in which MicroSeq 500 does not correctly identify a strain, such strains can be identified by directly comparing their consensus sequences with the National Center for Biotechnology Information GenBank database (11).
Acknowledgments
We gratefully acknowledge Applera Italia and Becton Dickinson for providing the instrumentation and/or part of the reagents for this evaluation.
We thank Oriana Cicchetti, Alessandro Mauti, and Francesca Capalbo for technical assistance. We also thank Alison Inglis for helpful linguistic revision of the manuscript.
REFERENCES
- 1.Barenfanger, J., C. Drake, and G. Kacich. 1999. Clinical and financial benefits of rapid bacterial identification and antimicrobial susceptibility testing. J. Clin. Microb. 37:1415-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brisse, S., S. Stefani, J. Verhoef, A. V. Belkum, P. Vandamme, and W. Goessens. 2002. Comparative evaluation of the BD Phoenix and Vitek2 automated instruments for identification of isolates of the Burkholderia cepacia complex. J. Clin. Microb. 40:1743-1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clermont, D., C. Harmant, and C. Bizet. 2001. Identification of strains of Alcaligenes and Agrobacterium by a polyphasic approach. J. Clin. Microbiol. 39:3104-3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cloud, J. L., P. S. Conville, A. Croft, D. Harmsen, F. G. Witebsky, and K. C. Carroll. 2004. Evaluation of partial 16S ribosomal DNA sequencing for identification of Nocardia species by using the MicroSeq 500 system with an expanded database. J. Clin. Microbiol. 42:578-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donay, J. L., D. M. P. Fernandes, C. Prégermain, P. Bruel, A. Wargnier, I. Casin, F. X. Weill, P. H. Lagrange, and J. L. Herrmann. 2004. Evaluation of the automated Phoenix system for potential routine use in the clinical microbiology laboratory. J. Clin. Microbiol. 42:1542-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Endimiani, A., F. Luzzaro, A. Tamborini, G. Lombardi, V. Elia, R. Belloni, and A. Toniolo. 2002. Identification and antimicrobial susceptibility testing of clinical isolates of nonfermenting gram-negative bacteria by the PhoenixTM automated microbiology system. Microbiologica 25:323-329. [PubMed] [Google Scholar]
- 7.Fahr, A. M., U. Eigner, M. Armbrust, A. Caganic, G. Dettori, C. Chezzi, L. Bertoncini, M. Benecchi, and M. G. Menozzi. 2003. Two-center evaluation of the performance of the BD Phoenix Automated Microbiology System for identification and antimicrobial susceptibility testing of Enterococcus spp. and Staphylococcus spp. J. Clin. Microb. 41:1135-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandez Guerrero, M. L., J. M. Ramos, J. Marrero, M. Cuenca, R. Fernandez Roblas, and M. de Gorgolas. 2003. Bacteremic pneumococcal infections in immunocompromised patients without AIDS: the impact of beta-lactam resistance on mortality. Int. J. Infect. Dis. 7:46-52. [DOI] [PubMed] [Google Scholar]
- 9.Ferraro, M. J., and J. H. Jorgensen. 2003. Susceptibility testing instrumentation and computerized expert system for data analysis and interpretation, p. 208-233. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed. ASM Press, Washington, D.C.
- 10.Ligozzi, M., C. Bernini, M. G. Borona, M. de Fatima, J. Zulliani, and R. Fontana. 2002. Evaluation of the Vitek 2 system for identification and antimicrobial susceptibility testing of medically relevant gram-positive cocci. J. Clin. Microb. 40:1681-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mellmann, A., J. L. Cloud, S. Andrees, K. Blackwood, K. C. Carrol, A. Kabani, A. Roth, and D. Harmsen. 2003. Evaluation of Ridom, MicroSeq, and Genbank services in the molecular identification of Nocardia species. Int. J. Med. Microbiol. 293:359-370. [DOI] [PubMed] [Google Scholar]
- 12.Patel, J. B., D. G. B. Leonard, X. Pan, J. M. Musser, R. E. Berman, and I. Nachamkin. 2000. Sequence-based identification of Mycobacterium species using the MicroSeq 500 16S rDNA bacterial identification. J. Clin. Microbiol. 38:246-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pugliese, A., B. Pacris, P. E. Schoch, and B. A. Cunha. 1993. Oligella urethralis urosepsis. Clin. Infect. Dis. 17:1070-1071. [DOI] [PubMed] [Google Scholar]
- 14.Qian, Q, Y. W. Tang, C. P. Kolbert, C. A. Torgerson, J. G. Hughes, E. A. Vetter, W. S. Harmsen, S. O. Montgomery, F. R. Cockerill III, and D. H. Persing. 2001. Direct identification of bacteria from positive blood cultures by amplification and sequencing of the 16S rRNA gene: evaluation of BACTEC 9240 instrument true-positive and false-positive results. J. Clin. Microbiol. 39:3578-3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rantakokko-Jalava, K., S. Nikkari, J. Jalava, E. Eerola, M. Skurnik, O. Meurman, O. Ruuskanen, A. Alanen, E. Kotilainen, P. Toivanen, and P. Kotilainen. 2000. Direct amplification of rRNA genes in diagnosis of bacterial infections. J. Clin. Microbiol. 38:32-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosset, R. 2001. Microbial growth and cold. Study of the particular case of Listeria monocytogenes. Bull. Acad. Natl. Med. 185:287-298. [PubMed] [Google Scholar]
- 17.Shittu, A., J. Lin, D. Morrison, and D. Kolawole. 2004. Isolation and molecular characterization of multiresistant Staphylococcus sciuri and Staphylococcus haemolyticus associated with skin and soft-tissue infections. J. Med. Microbiol. 53:51-55. [DOI] [PubMed] [Google Scholar]
- 18.Spanu, T., M. Sanguinetti, D. Ciccaglione, T. D'Inzeo, L. Romano, F. Leone, and G. Fadda. 2003. Use of the Vitek 2 system for rapid identification of clinical isolates of staphylococci from bloodstream infection. J. Clin. Microbiol. 41:4259-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang, Y. W., N. M. Ellis, M. K. Hopkins, D. H. Smith, D. E. Dodge, and D. H. Persing. 1998. Comparison of phenotypic and genotypic technique for identification of unusual aerobic pathogenic gram-negative bacilli. J. Clin. Microb. 36:3674-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang, Y. W., A. Von Graevenitz, M. G. Waddington, M. K. Hopkins, D. H. Smith, H. Li, C. P. Kolbert, S. O. Montgomery, and D. H. Persing. 2000. Identification of coryneform bacterial isolates by ribosomal DNA sequence analysis. J. Clin. Microbiol. 38:1676-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woo, P. C. Y., E. Y. L. Cheung, K. W. Leung, and K. Y. Yuen. 2001. Identification by 16S ribosomal RNA gene sequencing of an Enterobacteriaceae species with ambiguous biochemical profile from renal transplant recipient. Diagn. Microbiol. Infect. Dis. 29:85-93. [DOI] [PubMed] [Google Scholar]
- 22.Woo, P. C. Y., H. L. Ng Kenneth, S. K. P. Lau, K. Yip, A. M. Y. Fung, K. L. Leung, D. M. W. Tam, T. Que, and K. Yuen. 2003. Usefulness of the MicroSeq 500 16S ribosomal DNA-based bacterial identification of clinically significant bacterial isolates with ambiguous biochemical profiles. J. Clin. Microbiol. 41:1463-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woo, P. C. Y., P. K. L. Leung, K. W. Leung, and K. Y. Yuen. 2000. Identification by 16S ribosomal RNA gene sequencing of an Enterobacteriaceae species from a bone marrow transplant recipient. Mol. Pathol. 53:211-215. [DOI] [PMC free article] [PubMed] [Google Scholar]