Abstract
Carbonyl cyanide-p-trifluoro methoxyphenylhydrazone (FCCP) is a well-known mitochondrial uncoupling agent. We examined FCCP-induced fluorescence quenching of reduced nicotinamide adenine dinucleotide / nicotinamide adenine dinucleotide phosphate (NAD(P)H) in solution and in cultured HeLa cells in a wide range of FCCP concentrations from 50 to 1000µM. A non-invasive label-free method of hyperspectral imaging of cell autofluorescence combined with unsupervised unmixing was used to separately isolate the emissions of free and bound NAD(P)H from cell autofluorescence. Hyperspectral image analysis of FCCP-treated HeLa cells confirms that this agent selectively quenches fluorescence of free and bound NAD(P)H in a broad range of concentrations. This is confirmed by the measurements of average NAD/NADH and NADP/NADPH content in cells. FCCP quenching of free NAD(P)H in cells and in solution is found to be similar, but quenching of bound NAD(P)H in cells is attenuated compared to solution quenching possibly due to a contribution from the metabolic and/or antioxidant response in cells. Chemical quenching of NAD(P)H fluorescence by FCCP validates the results of unsupervised unmixing of cell autofluorescence.
OCIS codes: (110.4234) Multispectral and hyperspectral imaging, (170.2520) Fluorescence microscopy, (100.2960) Image analysis
1. Introduction
Cellular processes rely on energy produced during metabolism and a number of methodologies have been developed for its characterization. Nicotinamide Adenine Dinucleotide (NAD), a key coenzyme in metabolic pathways is frequently used as a metabolic fingerprint [1–3]. This soluble compound is found in its free and bound form in the cytosol and mitochondria, respectively [4, 5]. Its reduced form, NADH is a fluorophore which absorbs light in the 335-350 nm range, with the emission peak around 440-470 nm. Its fluorescence characteristics is virtually identical with that of nicotinamide adenine dinucleotide phosphate, (NADPH) [6]. These two compounds are jointly referred to as NAD(P)H. The protein-bound form of NAD(P)H shows small but detectable spectral differences compared to free NAD(P)H [7]. Non-invasive monitoring of NAD(P)H fluorescence to study metabolic processes has been first introduced by Chance [8, 9]. More recently, fluorescence lifetime imaging (FLIM) of endogenous cell fluorophores including NAD(P)H has been used to characterise the metabolic activity of cells and it has been related to free and bound NAD(P)H concentrations in cells [10, 11]. The advantage of FLIM is the independence of lifetime signatures on fluorescence intensity, but it requires more specialised instrumentation compared with fluorescence intensity-based methods. In contrast, our method uses low constant light irradiance and it does not require expensive systems.
Cells and tissues contain multiple endogenous fluorophores including NAD(P)H, retinoids, flavins, and other compounds which are involved in diverse cell functions and activities [12]. Identification of specific individual fluorophores in such a complex environment is not straightforward. A critical test is the ability to extinguish (quench) a specific fluorophore, ideally without affecting other fluorophores, for example by adding chemically-specific fluorescence quenchers. This approach cannot only help to confirm the prior existence of the quenched fluorophore, but may also provide other useful information, such as macromolecular conformation [13, 14]. Chemical quenching of NAD(P)H fluorescence can be achieved by its oxidation, taking advantage of the fact that only NAD(P)H and not NAD(P)+ is fluorescent [15]. This can be realised by standard oxidative agents, including sodium borohydrate, oxygen or FCCP [16]. However, in addition to their chemical activity as oxidising agents, these fluorescence quenchers exhibit various complex activities in cells. In particular, FCCP is known to be a mitochondrial inhibitor and uncoupler due to its interaction with mitochondrial lipid membranes [17–19]. Live cells rapidly respond to FCCP with a hydrolysis of ATP to stabilize the mitochondrial membrane potential [20]. It should be noted that FCCP oxidizes both free and bound NAD(P)H [21].
Recently, we advanced the idea, first put forward by Britton Chance, that intracellular monitoring of pyridine nucleotide enables a continuous measurement of the oxidation state [8], by decomposing the spectral images of cellular autofluorescence into individual fluorophore components [22]. This method was applied in the present work to determine chemical quenching effects of FCCP on NAD(P)H in cultured HeLa cells in comparison with FCCP quenching in solution. To this aim, we quantified free and bound NAD(P)H content in cell before and after exposure to FCCP in a broader range of FCCP concentrations (50-1000 µM) than previously used. This quantification was non-invasively carried out by hyperspectral fluorescence microscopy followed by spectral unmixing. NAD(P)H quenching in cells was independently confirmed by using NADH and NADPH detection kits. This result validates unsupervised unmixing of hyperspectral microscopy images as a way to determine fluorophore abundance.
2. Materials and methods
2.1 Cells preparation
HeLa cells from ATTC (CCL-2) were subcultured and maintained in the complete culture medium (Dulbecco’s modified Eagle’s medium (DMEM-high glucose, Sigma Aldrich, D5796) containing 10% fetal bovine serum (FbS; Gibco, Catalog No: 16000-044), penicillin/streptomycin (P/S; 100U/ml; Gibco, Catalog No: 15240-062). Cells were incubated at 37Co 5% CO2 incubator. Passaging of cells was performed once the confluency reached 80%. Cells were washed with phosphate buffered saline (PBS) and trypsinised with TrypLE (GIBCO, Australia, Catalog No: 12563-029). Following incubation with trypsin for 5 minutes at 37 Co, a complete medium was added to trypsinised cells. The cell suspension was centrifuged at 500 g for 5 minutes. After removing the supernatant, the cell pellet was resuspended in the complete medium. Cell viability testing was performed using Trypan blue 0.4% (Sigma Aldrich, Australia, Catalog No: T8154).
2.2 Free and bound NADH quenching in solution
To study the quenching effect of FCCP on bound-NADH (b-NADH) in solution, 50 µM β-NADH was prepared by binding the L-Malate Dehydrogenase (L-MDH, Sigma Aldrich #10127248001, from pig heart) protein. Both 50µM NADH (Sigma Aldrich # 10107735001) and 100 uM L-MDH was dissolved in 100 mM Mops (Sigma Aldrich # M1254) buffer (pH 7.0) to prepare 50µM β-NADH. Moreover, to prepare 50 µM β-NADPH, 50uM NADPH (Sigma Aldrich # 10107824001) and 100µM L-MDH was mixed in 100mM Mops buffer (pH 7.0). Meanwhile 50mM of FCCP (Sigma Aldrich # C2920-50MG) solution was prepared in dimethyl sulfoxide (DMSO, Sigma Aldrich # 8418-50ML). Appropriate amount of FCCP is mixed with the β-NADH solution to yield the following final concentration of FCCP: 0µM (control), 10 µM, 20µM, 30µM, 40µM, 50µM, 100µM, 150µM, 200µM, 250µM, 300µM, 500µM, 1mM, 2mM and 5mM. The same procedure was followed for the β-NADPH. The fluorescence emission was measured on a Cary Eclipse fluorescence spectrometer (Varian) by exciting the sample at 340 nm, while the emission was recorded in the range from 400 to 550 nm.
2.3 NAD(P)H quenching in plated cells
For quenching experiments on HeLa cells, a stock solution of 50 mM of FCCP (C2920-50MG, Sigma- Aldrich) was prepared in dimethyl sulfoxide (DMSO). All other concentrations (50-1000 µM) were prepared by diluting FCCP in Dulbecco phosphate buffer saline (DPBS) solution. The cells were cultured in a 96 well plate. The first three columns of the 96 well plate were cultured for 24 hours to achieve approximately 60% confluency. 150 µL of the FCCP solution with varying concentration from 50 µM to 1000 µM was added to each culture well and incubated for 60 minutes. After incubation of all the FCCP treated cells, each well was washed with PBS three times. The fluorescence quenching data were acquired on a plate reader Fluostar Galaxy (BMG Lab Technologies) at an available excitation wavelength of 360 ± 5 nm, (very close to the NADH excitation peak at 340 nm), and the emission was collected at 460 ± 5 nm.
2.4 Quantification of average NAD, NADH and NADPH in cells
NAD/NADH and NADP/NADPH were measured using NAD/NADH-Glo assay (catalog # G9071) and NADP/NADPH-Glo assay (catalog# G9081) supplied by Promega. The assays were performed following the protocol described in kit instruction. Standard curves were prepared for each set of assays. The cells (106 cells / ml) were seeded into 96 well culture plate, 50 microlitre in each well and incubated overnight. Controls and cells treated with FCCP in replicates were incubated with 50 microliters of NAD/ NADH and NADP/NADPH–Glo detection reagent in a white 96–well plate. After 30 minutes incubation, luminescence was measured with the SpectaraMax5 microplate luminometer at 1 second integration time per well. Each measure points represent average luminescence reaction measured in relative luminescence units (RLU).
2.5 Cell viability test
The viability test was performed by using Trypan Blue and Countess II Automated Cell Counter, AMQAX1000 (ThermoFisher Scientific). The samples were prepared by adding 10µl of cell suspension to 10µl of 0.4% trypan blue stain T10282 (ThermoFisher Scientific). The stained samples were mixed well and 10µl was loaded into the counting chamber slides. The viability values were obtained for control and FCCP treated samples. The cell viability is shown in Table 1, below.
Table 1. Trypan blue test and percentage viability after treatment with FCCP.
| FCCP Concentration (µM) | 0 | 50 | 100 | 150 | 200 | 300 | 500 | 1000 |
| % Viability | 80 ± 2.7 | 48 ± 2.2 | 44 ± 0.7 | 30 ± 1.9 | 28 ± 2.3 | 19 ± 0.8 | 19 ± 0.8 | 18 ± 0.1 |
2.6 Preparing cells for spectral imaging
To perform hyperspectral imaging, HeLa cells were cultured using the same protocol as described in Section 2.1. The cells were seeded into 35 mm plastic culture dishes with 18 mm well and # 1.5 cover slip bottoms (Cell E&G, USA, and # GDB0004-200). Each dish was seeded with 1 ml of cells (5000 cells/ cm2) and incubated at 37Co, 5% CO2 for 48 hours to reach 70% confluence. Prior to imaging, the media was removed and the cells were washed with PBS three times. The cells were then treated with (50, 100, 150, 200, 250, 300 and 1000) µM FCCP and incubated for 60 minutes at 37Co. The control cells were incubated in the same conditions without the FCCP treatment. The cells were washed three times with PBS prior to imaging, then 1 ml of Hanks Balanced Salt Solution (HBSS) was added into each dish. The imaging experiments were performed in duplicates for control and treated groups.
2.7 Hyperspectral imaging
Single photon-excited autofluorescence of cells was measured by using an adapted wide-field fluorescence microscope (Olympus iX71) with a 40 × water U12 series objective with high transmission in the UV range. All images were captured by an Andor IXON camera (EMCCD, iXON 885 DU, Andor Technology Ltd., UK) operated below −65Co to minimise noise in the image data. Selected bands of excitation wavelengths produced by light emitting diodes (centred at 334 nm, 365 nm, 375 nm, 385 nm, 395 nm, 405 nm, 415 nm, 425 nm, 435 nm, 455 nm, 475 nm, and 495 nm, each about 10 nm wide) were used. The corresponding emission was collected with suitable optical filters and dichroic mirrors were used to separate excitation from emission. The 12 excitation sources and 3 (manually controlled) filter cubes form a total L = 18 specific optical channels [22] as detailed in Table 2. The samples were imaged in each of these spectral channels producing L images from each field of view. The total data collection time was about 2 minutes per hyperspectral image (all channels). The excitation sources were coupled by an optical fibre bundle with a 5 mm fused silica hexagonal homogenizer. The microscope system was calibrated with a mixture of 30 μM NADH and 5μM riboflavin whose spectrum spans across all our spectral channels. The excitation and emission spectra of the calibration fluid was measured in a Fluorolog Tau3 spectrometer (Jobin-Yvon-Horiba). The normalized calibrated spectrum thus obtained was used to correct the same spectrum measured with our hyperspectral system to be able to assign the unmixed fluorophores. Finally we used the image of the calibration fluid to flatten the uneven (approximately Gaussian) illumination of the field of view. In this work have obtained spectral images of eight groups of cells from 16 dishes, with duplicate dishes with each specific FCCP concentration. We have taken three images of each dish at three different locations and acquired a total of 48 images. All images have been corrected by subtracting the ‘background’ images of water. Moreover we employed a carefully constructed log-Gabour wavelet filter to remove the dominant Poisson noise, from the autofluorescence images [23]. After that, the cells were segmented to distinguish them from the dark background producing segmented images with cells only and background set to zero. The cells were segmented manually using the marque polygon selection using both the fluorescent image and DIC to determine the location of the cell membranes. This was necessary since the cells tended to grow in clusters and automatic methods produced unsatisfactory results. In this work, around 60 segmented cells were analysed per each FCCP concentration. Finally, spectral decorrelation of our L-dimensional image data set was carried out using the Principal Component Analysis.
Table 2. Spectral channel specifications for hyperspectral imaging.
| Ch.# | λ Exc (nm) | λ emit (nm) |
Dichoric mirror longpass (nm) | Power at objective, µW |
|---|---|---|---|---|
| 1 | 334 ± 5 | 447 ± 30 | 409 | 0.046 |
| 2 | 365 ± 5 | 447 ± 30 | 409 | 6.50 |
| 3 | 375 ± 5 | 447 ± 30 | 409 | 2.83 |
| 4 | 334 ± 5 | 587 ± 17 | 532 | 0.020 |
| 5 | 365 ± 5 | 587 ± 17 | 532 | 6.40 |
| 6 | 375 ± 5 | 587 ± 17 | 532 | 10.51 |
| 7 | 385 ± 5 | 587 ± 17 | 532 | 17.77 |
| 8 | 395 ± 5 | 587 ± 17 | 532 | 13.33 |
| 9 | 405 ± 5 | 587 ± 17 | 532 | 9.39 |
| 10 | 415 ± 5 | 587 ± 17 | 532 | 22.40 |
| 11 | 425 ± 5 | 587 ± 17 | 532 | 20.90 |
| 12 | 435 ± 5 | 587 ± 17 | 532 | 27.50 |
| 13 | 455 ± 5 | 587 ± 17 | 532 | 14.80 |
| 14 | 475 ± 5 | 587 ± 17 | 532 | 42.80 |
| 15 | 495 ± 5 | 587 ± 17 | 532 | 0.01 |
| 16 | 405 ± 5 | ≥700 | 635 | 9.50 |
| 17 | 455 ± 5 | ≥700 | 635 | 15.09 |
| 18 | 495 ± 5 | ≥700 | 635 | 9.97 |
2.8 Unsupervised unmixing method
The pixel spectra contain a mixture of fluorescent signatures from different native fluorescent metabolites, enzymes and co-factors in cells. An unmixing procedure is used to separate out their individual intensity contributions. In the presence of unavoidable image noise this procedure yields approximate spectra of these individual contributors called ‘endmembers’ which must then be verified to make sure they closely resemble the spectra of the actual reference fluorophores. The procedure produces relative intensity values of each of these endmembers which is proportional to their intensity fraction. The mathematics of unmixing is expressed in the language of vector spaces, where a fluorescence spectrum of any type, f, is represented by a vector , where are the excitation and emission wavelengths, at the centre of each channel. The formalism is based on the linear mixture model widely used in hyperspectral image analysis [24]. We denote the spectrum of the jth pixel as Ij, the abundance fraction of the kth endmember as Cjk, and the corresponding endmember spectrum as Ek. Here, where D is the total number of endmembers, and where J is the total number of pixels in the image, in our case over 106. Due to physical considerations, the abudance fractions must be non-negative (Eq. (1)) and they must add to unity [25,26] (Eq. (2)).
| (1) |
| (2) |
The fluorescence spectrum at each pixel and at each wavelength λ is assumed to be a linear combination of endmember spectra weighted by corresponding abundance fraction [27].
| (3) |
Moreover the spectrum of each pixel in each image is corrupted by the noise originating from the image sensor, illumination, and image processing electronics. etc., this noise is denoted by a random variable N. In this case the intensity of a mixed pixel at any wavelength λ should be described as in Eq. (4):
| (4) |
Equation (4) indicates that without the noise the observed spectral vectors form a simplex surrounded by a convex hull whose vertices correspond to the endmembers’ spectra [28]. These endmembers spectra are then identified with individual fluorophores present in the sample. The procedure also yields the abundance fractions of these fluorophores on a pixel level, and this makes it possible to calculate average cellular abundances of the fluorophores identified in the examined cells [22]. We emphasise that in the case of unmixing of autofluorescence, the abundance represents the product of fluorophore concentration by its quantum yield.
In this work, all pixels within the cells segmented areas undergo spectral unmixing producing an abundance value for each fluorophore for each pixel site. These pixel abundance values are averaged on a per cell basis producing a representative average abundance value for each cell.
3. Results and discussion
3.1 NAD(P)H quenching in solution
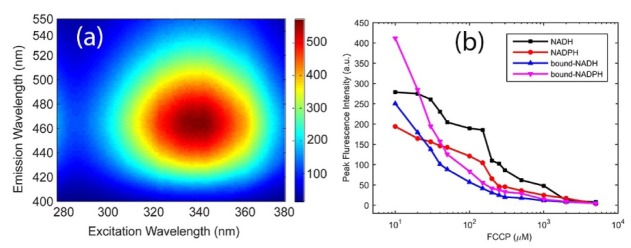
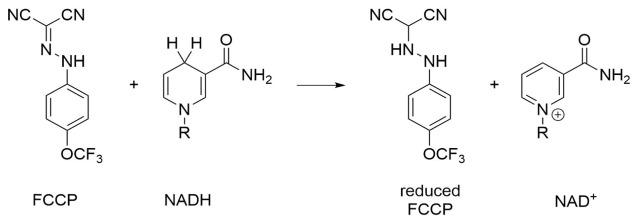
In order to characterise quenching of NAD(P)H by FCCP we first measured the excitation emission matrices (EEMs) of 50 uM NADH in an aqueous solution (Fig. 1(a)). To this aim, we excited the NADH in the range 280-380 nm and acquired its emission spectra between 400 and 550 nm. These ranges were selected to overlap with the free NADH absorption range from 300 to 380 nm and its fluorescence range from 420 to 480 nm described in the literature [29, 30]. The EEM for 50 µM NADH show that maximum of the excitation spectrum is at 340 ± 1 nm while the fluorescence spectrum peaks at 465 nm. We have therefore selected the excitation wavelength of 340 ± 1 nm and used varying concentrations of FCCP (10-5000 µM) to induce fluorescence quenching of NADH and NADPH. The fluorescence quenching results are presented in Fig. 1(b) where the fluorescence peak intensity is plotted versus FCCP concentration. Fluorescence quenching is clearly observed, consistent with the reaction shown in Fig. 2.
Fig. 1.
(a) Excitation-emission matrices (EEM) of the 50 µM NADH solution (b) Peak fluorescence intensity of free NADH and NADPH versus FCCP concentration(10 −5000 µM). Peak fluorescence intensity values for free NADH (black square), free NADPH (red circle), bound NADH (blue triangle) and bound NADPH (magenta triangle) are, respectively, 534, 352, 306 and 626. These data points could not be presented in Fig. 1(b) due to the log scale used in the plot.
Fig. 2.
Oxidation of NADH by FCCP [32].
3.2 Quenching of plated HeLa cells
Figure 3 shows the variation of fluorescence intensity of control HeLa cells and HeLa cells treated with (50, 100, 150, 200, 300, and 1000) μM FCCP excited at 360 ± 5 nm with emission collected at 460 ± 5 nm. The autofluorescence signal is the highest in the control HeLa cells. A marked decrease of peak fluorescence intensity appears after treatment with FCCP at 50 µM concentration and it generally continues across the whole range of FCCP concentrations. Similar quenching effects were previously reported in FCCP-treated cells but for a narrower range of concentrations, up up to 200 μM and for a shorter exposure time of 5 minutes [31]. FCCP also significantly interacts with cells: the literature indicates that at 100 nM it produces noticeable mitochondrial oxidation without affecting the mitochondrial membrane potential, while at 300 nM FCCP causes mitochondrial depolarisation [33].
Fig. 3.

Autofluorescence intensity (excitation at 360 ± 5 nm and emission at 460 ± 5 nm) of control (FCCP = 0) and FCCP -treated HeLa cells.
3.3 NAD+/NADH and NADP+/NADPH quantification
We quantified the intracellular level of NAD/NADH and NADP/NADPH in the HeLa cells with commercial assay kits. The kits use a luminescence –based technique.
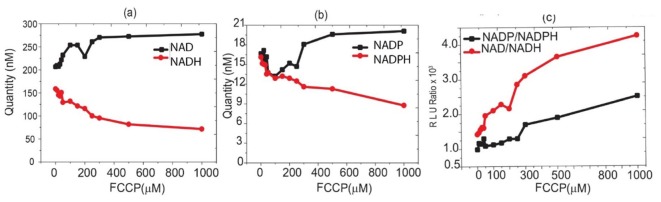
The results are shown in Fig. 4(a)-4(c). Figure 4(a) shows the variation of total NAD and NADH in HeLa cells with FCCP concentration. According to our kit results, HeLa cells without any treatment have the NADH and NAD levels of 158 nM and 206 nM /106 cells respectively. The value of NAD of 206 nM/106 cells is comparable with the value reported for rat liver cells of 1μM per gram of wet weight (approximately 106 cells) [34]. The levels of NADH and NADPH generally decrease with the increase in FCCP concentration. The NAD/NADH ratio (Fig. 4(c)) confirms that FCCP oxidises NADH but an active cell response at lower FCCP concentrations is also apparent.
Fig. 4.
(a) Variation of NAD and NADH levels as a function of FCCP concentration, (b) Variation of NADP and NADPH as a function of FCCP concentration (c) variation of the ratios NAD/NADH and NADP/NADPH as a function of FCCP concentration.
3.4 Hyperspectral imaging and unmixing results
Hyperspectral imaging and spectral unmixing makes it possible to spectrally resolve different cell-native fluorophores . Using this approach we have been able to differentiate free NAD(P)H from protein-bound forms of NAD(P)H whose spectrum is blue-shifted by 20 nm [7]. We were not able to differentiate between NADH and NADPH, thus the NAD(P)H components in this work (free or bound) represents both NADH and NADPH fluorescence.
The unmixing involves two major procedures, the endmember extraction, which identifies the presence of endmembers in the measured image, and a linear spectral mixture analysis, the inversion step applied to Eq. (4), which estimates the relative contribution of each endmember to the total intensity of each pixel [35, 36]. Linear unmixing is then used to produce the intensity contributions using a fast, fully constrained non negative unmixing algorithm [23]. Since the intensity contribution of each fluorophore is obtained for each pixel in the image, the unmixing procedure returns the spatial distribution of each endmember abundance in that image.
The principal component analysis of the pixel data indicates that the data variance could be largely explained based on four main component spectra as shown by the presence of four significant PCA eigenvalues (explaining, respectively 50%, 32%, 10% and 6% of data variability). These four components (endmembers) can be clearly identified in Fig. 5(a), where the pixel data were projected onto a PCA subspace, and the four components occupy the vertex positions. Figure 5(a) also indicates the location of the reference spectra of free NAD(P)H, FAD, bound NAD(P)H (whose spectrum is 20 nm blue shifted with respect to free NAD(P)H), and a fourth fluorophore, retinol or a mixture of retinoids (separately measured using a fluorimeter). Using the endmembers thus obtained and a linear mixture model we are able to explain the pixel spectra with greater than 92% accuracy.
Fig. 5.
(a) Data simplex obtained in unsupervised unmixing of hyperspectral images of HeLa cells exposed to FCCP concentration (50-1000 µM). All images are analysed together. Gray points represent the image pixels, red squares represent the reference spectra, the location of vertices indicates the endmember spectra. The lines mark the sides of the simplex, the axes represent the top 2 PCA components; (b) The results of mean cellular abundance of free NAD(P)H show that FCCP quenches its fluorescence.(c) Mean cellular abundance of bound NADH also shows FCCP quenching. Data for all cells are represented by box plots [37, 38].
Now we discuss the fluorophore content of untreated and treated HeLa cells with the same FCCP (50-1000) µM concentrations as used previously. These results are presented in Fig. 5(b)-5(c). The average abundance value of free NAD(P)H (Fig. 5(b)) is much lower compared to bound NAD(P)H (Fig. 5(c)), partly because free NADH has lower fluorescence quantum yield (0.02) as compared to bound NADH (0.1) [39, 40]. This is consistent with earlier reports that more than 80% of total NADH fluorescence is due to bound NADH [41, 42]. Figure 5(b) shows the variations of mean cellular fluorescence intensity of free NAD(P)H versus FCCP concentration. The fluorescence intensity of free NAD(P)H is highest prior to FCCP treatment, and it decreases with increasing concentration of FCCP by a factor of 5 for 1000 µM FCCP. A similar trend in fluorescence quenching was observed in the experiment with NADH in water solution shown in Fig. 1(b). Qualitatively, these two experiments show different quenching rates because (a) FCCP taken up by the HeLa cells may not match the FCCP concentrations obtained in the solution; (b) the cells metabolism is affected by FCCP via the decouping of mitochondria and (c) the cells may actively respond to oxidative insults. Figure 5(c) shows the mean intensity of bound NAD(P)H autofluorescence versus FCCP concentration. Similarly to free NAD(P)H, the control HeLa cells have the highest abundance of bound NADH. However, when exposed to FCCP bound NAD(P)H behaves differently compared to free NAD(P)H and the effects of FCCP are more attenuated. The abundance of free NAD(P)H is reduced by over 50% when exposed to 50µM FCCP while bound NAD(P)H is reduced by 12.5% only. The maximum quenching of free NAD(P)H achieved here was 80% but it was only 37.5% for bound NAD(P)H.
We now comment on the observed fluorophore trends by using well-established fluorescent signatures of metabolic pathways and their changes upon alteration of mitochondrial machinery. Cells exhibit dynamic changes in their NAD(P)H in response to conditions affecting cell metabolism [7]. Their levels reflect the varying balance of glycolysis and oxidative phosphorylation, depending on environmental signals (availability of oxygen, and/or lactate/pyruvate, the presence of inhibitors or uncouplers, etc) and genetic makeup of cells (Warburg effect) [43, 44]. We emphasise that in this work, we interfere with the activity of the mitochondria by using FCCP at such high levels that they, eventually, become inactive leading to cell death (see cell viability data in Table 1), due to increased production of reactive oxygen species (ROS) and other effects [45, 46]. A purely chemical effect of NADH/NADPH oxidation, which decreases NADH/NADPH fluorescence, as quantified in Fig. 1, underpins all the observable changes, and it is the only effect observed in cells that are no longer alive. Our results (Figs. 3-5) are affected by the balance between these processes.
4. Conclusions
We applied non-invasive hyperspectral imaging of cell autofluorescence to the study of fluorescence quenching by FCCP in HeLa cells. The FCCP quenching of total NAD(P)H in cells was first confirmed by a biochemical assay and the effect of FCCP on mitochondrial membrane potential confirmed that cells have taken up FCCP. After unmixing of the hyperspectral autofluorescence images of control and FCCP-treated cells, we observed that both free and bound NAD(P)H are quenched by FCCP within the 10-1000 µM concentration range. The effect of quenching was more pronounced for free NAD(P)H than for bound NAD(P)H. In summary, our data show that unmixing of the hyperspectral autofluorescence images yields correct identification of free and bound NAD(P)H in cells. Thus, hyperspectral imaging used in this work in combination with fluorescence spectroscopy and chemical quenching provides a new methodology to investigate cell metabolism.
Funding
This work was partially supported by the Australian Research Council (CE140100003), International Macquarie University Research Excellence Scholarship (IMQRES) – No. 2014030, National Natural Science Foundation of China (Grant 21575045), and by CCNU (CCNU15A02015).
References and links
- 1.Andersson H., Baechi T., Hoechl M., Richter C., “Autofluorescence of living cells,” J. Microsc. 191(1), 1–7 (1998). 10.1046/j.1365-2818.1998.00347.x [DOI] [PubMed] [Google Scholar]
- 2.Fjeld C. C., Birdsong W. T., Goodman R. H., “Differential binding of NAD+ and NADH allows the transcriptional corepressor carboxyl-terminal binding protein to serve as a metabolic sensor,” Proc. Natl. Acad. Sci. U.S.A. 100(16), 9202–9207 (2003). 10.1073/pnas.1633591100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stringari C., Edwards R. A., Pate K. T., Waterman M. L., Donovan P. J., Gratton E., “Metabolic trajectory of cellular differentiation in small intestine by Phasor Fluorescence Lifetime Microscopy of NADH,” Sci. Rep. 2, 568 (2012). 10.1038/srep00568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Estabrook R. W., “Fluorometric measurement of reduced pyridine nucleotide in cellular and subcellular particles,” Anal. Biochem. 4(3), 231–245 (1962). 10.1016/0003-2697(62)90006-4 [DOI] [PubMed] [Google Scholar]
- 5.Paul R. J., Schneckenburger H., “Oxygen concentration and the oxidation-reduction state of yeast: determination of free/bound NADH and flavins by time-resolved spectroscopy,” Naturwissenschaften 83(1), 32–35 (1996). 10.1007/BF01139308 [DOI] [PubMed] [Google Scholar]
- 6.Jérôme D. R., Maureen F., Johan W., Perpèteb A., Julien P., Denis J., “Towards the understanding of the absorption spectra of NAD(P)H/NAD(P)+ as a common indicator of dehydrogenase enzymatic activity,” Chem. Phys. Lett. 450(1–3), 119–122 (2007). [Google Scholar]
- 7.Piersma S. R., Visser A. J. W. G., de Vries S., Duine J. A., “Optical spectroscopy of nicotinoprotein alcohol dehydrogenase from Amycolatopsis methanolica: a comparison with horse liver alcohol dehydrogenase and UDP-galactose epimerase,” Biochemistry 37(9), 3068–3077 (1998). 10.1021/bi972115u [DOI] [PubMed] [Google Scholar]
- 8.Chance B., Cohen P., Jobsis F., Schoener B., “Intracellular oxidation-reduction states in vivo,” Science 137(3529), 499–508 (1962). 10.1126/science.137.3529.499 [DOI] [PubMed] [Google Scholar]
- 9.Chance B., Lieberman M., “Intrinsic fluorescence emission from the cornea at low temperatures: evidence of mitochondrial signals and their differing redox states in epithelial and endothelial sides,” Exp. Eye Res. 26(1), 111–117 (1978). 10.1016/0014-4835(78)90159-8 [DOI] [PubMed] [Google Scholar]
- 10.Stringari C., Wang H., Geyfman M., Crosignani V., Kumar V., Takahashi J. S., Andersen B., Gratton E., “In vivo single-cell detection of metabolic oscillations in stem cells,” Cell Reports 10(1), 1–7 (2015). 10.1016/j.celrep.2014.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vergen J., Hecht C., Zholudeva L. V., Marquardt M. M., Hallworth R., Nichols M. G., “Metabolic imaging using two-photon excited NADH intensity and fluorescence lifetime imaging,” Microsc. Microanal. 18(4), 761–770 (2012). 10.1017/S1431927612000529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skala M. C., Riching K. M., Bird D. K., Gendron-Fitzpatrick A., Eickhoff J., Eliceiri K. W., Keely P. J., Ramanujam N., “In vivo multiphoton fluorescence lifetime imaging of protein-bound and free nicotinamide adenine dinucleotide in normal and precancerous epithelia,” J. Biomed. Opt. 12(2), 024014 (2007). 10.1117/1.2717503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammen P. K., Allali-Hassani A., Hallenga K., Hurley T. D., Weiner H., “Multiple conformations of NAD and NADH when bound to human cytosolic and mitochondrial aldehyde dehydrogenase,” Biochemistry 41(22), 7156–7168 (2002). 10.1021/bi012197t [DOI] [PubMed] [Google Scholar]
- 14.Lehrer S. S., “The selective quenching of tryptophan fluorescence in proteins by iodide ion: lysozyme in the presence and absence of substrate,” Biochem. Biophys. Res. Commun. 29(5), 767–772 (1967). 10.1016/0006-291X(67)90284-7 [DOI] [PubMed] [Google Scholar]
- 15.Held P., “Determination of NADH concentrations with the Synergy™ 2 multi-detection microplate reader using fluorescence or absorbance” (Biotech, 2007). [Google Scholar]
- 16.Lakowicz J. R., Principles of Fluorescence Spectroscopy (Springer Science & Business Media, 2007). [Google Scholar]
- 17.Buckler K. J., Vaughan-Jones R. D., “Effects of mitochondrial uncouplers on intracellular calcium, pH and membrane potential in rat carotid body type I cells,” J. Physiol. 513(3), 819–833 (1998). 10.1111/j.1469-7793.1998.819ba.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griffiths E. J., Lin H., Suleiman M. S., “NADH fluorescence in isolated guinea-pig and rat cardiomyocytes exposed to low or high stimulation rates and effect of metabolic inhibition with cyanide,” Biochem. Pharmacol. 56(2), 173–179 (1998). 10.1016/S0006-2952(98)00016-1 [DOI] [PubMed] [Google Scholar]
- 19.Kessler R. J., Vande Zande H., Tyson C. A., Blondin G. A., Fairfield J., Glasser P., Green D. E., “Uncouplers and the molecular mechanism of uncoupling in mitochondria,” Proc. Natl. Acad. Sci. U.S.A. 74(6), 2241–2245 (1977). 10.1073/pnas.74.6.2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nicholls D. G., Budd S. L., “Mitochondria and neuronal survival,” Physiol. Rev. 80(1), 315–360 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Monteiro J. P., Martins A. F., Lúcio M., Reis S., Geraldes C. F., Oliveira P. J., Jurado A. S., “Interaction of carbonylcyanide p-trifluoromethoxyphenylhydrazone (FCCP) with lipid membrane systems: a biophysical approach with relevance to mitochondrial uncoupling,” J. Bioenerg. Biomembr. 43(3), 287–298 (2011). 10.1007/s10863-011-9359-2 [DOI] [PubMed] [Google Scholar]
- 22.Gosnell M. E., Anwer A. G., Cassano J. C., Sue C. M., Goldys E. M., “Functional hyperspectral imaging captures subtle details of cell metabolism in olfactory neurosphere cells, disease-specific models of neurodegenerative disorders,” Biochim. Biophys. Acta 1863(1), 56–63 (2016). 10.1016/j.bbamcr.2015.09.030 [DOI] [PubMed] [Google Scholar]
- 23.Gosnell M. E., “Unlocking the potential of spectral imaging for the characterisation of cell and stem cell populations / Martin E. Gosnell,” P. Macquarie University. Department of Physics &Astronomy, (Sydney, Australia: Macquarie University, 2014). [Google Scholar]
- 24.Harsanyi J. C., Chang C.-I., “Hyperspectral image classification and dimensionality reduction: an orthogonal subspace projection approach,” IEEE Trans. Geosci. Remot. Sens. 32(4), 779–785 (1994). 10.1109/36.298007 [DOI] [Google Scholar]
- 25.Manolakis D., Siracusa C., Shaw G., “Hyperspectral subpixel target detection using the linear mixing model,” IEEE Trans. Geosc Remot. Sens 39(7), 1392–1409 (2001). 10.1109/36.934072 [DOI] [Google Scholar]
- 26.Nascimento J. M., Bioucas-Dias J. M., “Hyperspectral unmixing based on mixtures of Dirichlet components,” IEEE Trans. Geosc Remot. Sens. 50(3), 863–878 (2012). 10.1109/TGRS.2011.2163941 [DOI] [Google Scholar]
- 27.Keshava N., Mustard J. F., “Spectral unmixing,” Sig. Proc. Mag., IEEE 19(1), 44–57 (2002). 10.1109/79.974727 [DOI] [Google Scholar]
- 28.Nascimento J. M., Bioucas-Dias J. M., “Hyperspectral signal subspace estimation,” in Proceedings of IEEE Conference on Geoscience and Remote Sensing Symposium, (IEEE; ), pp. 3225–3228 (2007). [Google Scholar]
- 29.Mayevsky A., Rogatsky G. G., “Mitochondrial function in vivo evaluated by NADH fluorescence: from animal models to human studies,” Am. J. Physiol. Cell Physiol. 292(2), C615–C640 (2006). 10.1152/ajpcell.00249.2006 [DOI] [PubMed] [Google Scholar]
- 30.Cordeiro P. G., Kirschner R. E., Hu Q.-Y., Chiao J. J., Savage H., Alfano R. R., Hoffman L. A., Hidalgo D. A., “Ultraviolet excitation fluorescence spectroscopy: a noninvasive method for the measurement of redox changes in ischemic myocutaneous flaps,” Plast. Reconstr. Surg. 96(3), 673–680 (1995). 10.1097/00006534-199509000-00021 [DOI] [PubMed] [Google Scholar]
- 31.Eng J., Lynch R. M., Balaban R. S., “Nicotinamide adenine dinucleotide fluorescence spectroscopy and imaging of isolated cardiac myocytes,” Biophys. J. 55(4), 621–630 (1989). 10.1016/S0006-3495(89)82859-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galkin A. S., Grivennikova V. G., Vinogradov A. D., “→H+/2e- stoichiometry in NADH-quinone reductase reactions catalyzed by bovine heart submitochondrial particles,” FEBS Lett. 451(2), 157–161 (1999). 10.1016/S0014-5793(99)00575-X [DOI] [PubMed] [Google Scholar]
- 33.Brennan J. P., Berry R. G., Baghai M., Duchen M. R., Shattock M. J., “FCCP is cardioprotective at concentrations that cause mitochondrial oxidation without detectable depolarisation,” Cardiovasc. Res. 72(2), 322–330 (2006). 10.1016/j.cardiores.2006.08.006 [DOI] [PubMed] [Google Scholar]
- 34.Reiss P. D., Zuurendonk P. F., Veech R. L., “Measurement of tissue purine, pyrimidine, and other nucleotides by radial compression high-performance liquid chromatography,” Anal. Biochem. 140(1), 162–171 (1984). 10.1016/0003-2697(84)90148-9 [DOI] [PubMed] [Google Scholar]
- 35.Keshava N., Kerekes J. P., Manolakis D. G., Shaw G. A., “Algorithm taxonomy for hyperspectral unmixing,” in Proceeding of AeroSense 2000 (International Society for Optics and Photonics; ), pp. 42–63(2000) 10.1117/12.410362 [DOI] [Google Scholar]
- 36.J. M. Bioucas-Dias and A. Plaza, “Hyperspectral unmixing: Geometrical, statistical, and sparse regression-based approaches,” in Proceedings of Remote Sensing (International Society for Optics and Photonics), pp. 78300A–78315(2010) [Google Scholar]
- 37.Frigge M., Hoaglin D. C., Iglewicz B., “Some Implementations of the Boxplot,” Am. Stat. 43(1), 50–54 (1989). [Google Scholar]
- 38.Benjamini Y., “Opening the Box of a Boxplot,” Stat 42(4), 257–262 (1988). [Google Scholar]
- 39.Lindqvist L., Czochralska B., Grigorov I., “Determination of the mechanism of photoionization of NADH in aqeous solution on laser excitation at 355 nm,” Chem. Phys. Lett. 119(6), 494–498 (1985). 10.1016/0009-2614(85)85375-6 [DOI] [Google Scholar]
- 40.Lakowicz J. R., Principles of Fluorescence Spectroscopy, 3rd ed. (Springer, 2006) pp. 63–67. [Google Scholar]
- 41.Blinova K., Carroll S., Bose S., Smirnov A. V., Harvey J. J., Knutson J. R., Balaban R. S., “Distribution of mitochondrial NADH fluorescence lifetimes: steady-state kinetics of matrix NADH interactions,” Biochemistry 44(7), 2585–2594 (2005). 10.1021/bi0485124 [DOI] [PubMed] [Google Scholar]
- 42.Schweitzer D., Schenke S., Hammer M., Schweitzer F., Jentsch S., Birckner E., Becker W., Bergmann A., “Towards metabolic mapping of the human retina,” Microsc. Res. Tech. 70(5), 410–419 (2007). 10.1002/jemt.20427 [DOI] [PubMed] [Google Scholar]
- 43.Zeng W., Liu P., Pan W., Singh S. R., Wei Y., “Hypoxia and hypoxia inducible factors in tumor metabolism,” Cancer Lett. 356(2 2 Pt A), 263–267 (2015). 10.1016/j.canlet.2014.01.032 [DOI] [PubMed] [Google Scholar]
- 44.Vander Heiden M. G., Cantley L. C., Thompson C. B., “Understanding the Warburg effect: the metabolic requirements of cell proliferation,” Science 324(5930), 1029–1033 (2009). 10.1126/science.1160809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Datta R., Alfonso-García A., Cinco R., Gratton E., “Fluorescence lifetime imaging of endogenous biomarker of oxidative stress,” Sci. Rep. 5, 9848 (2015), doi:. 10.1038/srep09848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li L. Z., Xu H. N., Ranji M., Nioka S., Chance B., “Mitochondrial redox imaging for cancer diagnostic and therapeutic studies,” J. Innov. Opt. Health Sci. 2(4), 325–341 (2009). 10.1142/S1793545809000735 [DOI] [PMC free article] [PubMed] [Google Scholar]