Abstract
Two-photon microscopy of cellular autofluorescence intensity and lifetime (optical metabolic imaging, or OMI) is a promising tool for preclinical drug development. OMI, which exploits the endogenous fluorescence from the metabolic coenzymes NAD(P)H and FAD, is sensitive to changes in cell metabolism produced by drug treatment. Previous studies have shown that drug response, genetic expression, cell-cell communication, and cell signaling in 3D culture match those of the original in vivo tumor, but not those of 2D culture. The goal of this study is to use OMI to quantify dynamic cell-level metabolic differences in drug response in 2D cell lines vs. 3D organoids generated from xenograft tumors of the same cell origin. BT474 cells and Herceptin-resistant BT474 (HR6) cells were tested. Cells were treated with vehicle control, Herceptin, XL147 (PI3K inhibitor), and the combination. The OMI index was used to quantify response, and is a linear combination of the redox ratio (intensity of NAD(P)H divided by FAD), mean NADH lifetime, and mean FAD lifetime. The results confirm that the OMI index resolves significant differences (p<0.05) in drug response for 2D vs. 3D cultures, specifically for BT474 cells 24 hours after Herceptin treatment, for HR6 cells 24 and 72 hours after combination treatment, and for HR6 cells 72 hours after XL147 treatment. Cell-level analysis of the OMI index also reveals differences in the number of cell sub-populations in 2D vs. 3D culture at 24, 48, and 72 hours post-treatment in control and treated groups. Finally, significant increases (p<0.05) in the mean lifetime of NADH and FAD were measured in 2D vs. 3D for both cell lines at 72 hours post-treatment in control and all treatment groups. These whole-population differences in the mean NADH and FAD lifetimes are supported by differences in the number of cell sub-populations in 2D vs. 3D. Overall, these studies confirm that OMI is sensitive to differences in drug response in 2D vs. 3D, and provides further information on dynamic changes in the relative abundance of metabolic cell sub-populations that contribute to this difference.
OCIS codes: (180.2520) Fluorescence microscopy, (170.1530) Cell analysis, (170.1610) Clinical applications
1. Introduction
Many promising new drugs fail in clinical trials, highlighting a need for more robust pre-clinical screens that eliminate these drugs before costly clinical trials [1]. Currently, 2D cell monolayers are the predominant platform for assessing preclinical drug efficacy in vitro [2,3]. However, increasing evidence suggests that 2D cell monolayers may be insufficient to accurately recapitulate in vivo human drug response [3, 4]. Therefore, 3D tumor culture may present a better option, particularly for tissues with high structural organization such as breast cancer.
Differences in the microenvironment of 2D and 3D cultures, including unique molecular localization patterns and ECM composition, trigger different signal transduction cascades and result in different cell behaviors [5]. These differing behaviors in 2D vs. 3D cultures include apoptotic sensitivity, kinase phosphorylation, and regulation of the activity of receptors involved in neoplastic transformation, such as estrogen receptor (ER) and human epidermal growth factor receptor 2 (HER2) [4]. Additionally, 3D cultures better represent the oxygen, nutrient, lactate, and proliferation gradients of the in vivo microenvironment than 2D cultures [6–9]. It is also advantageous to include other, non-malignant cells representative of the heterogeneous cell populations in in vivo tumors, such as stromal cells, as they are known regulators of proliferation, quiescence, and drug sensitivity in nearby malignant cells [10].
Current drug development platforms often rely on markers for apoptosis and necrosis to gauge drug efficacy in cancer cells [2, 3]. Alternatively, cell metabolism is capable of dynamically resolving subtle changes induced by drug treatments that correlate to successful in vivo response [11]. NAD(P)H and FAD are autofluorescent metabolic co-enzymes. The ratio of the fluorescence intensities of NAD(P)H to FAD is the “optical redox ratio,” which is sensitive to malignancy, tumor aggressiveness, and drug response [12–18]. In fact, changes in the redox ratio induced by anti-cancer drugs predict in vivo tumor response long before changes in tumor volume can be measured [15]. The fluorescence lifetime, or the amount of time a fluorophore remains excited prior to decaying back to the ground state, provides complementary information to the redox ratio [16, 19]. The fluorescence lifetime is sensitive to changes in cell microenvironment, such as pH, protein binding, and proximity to fluorescence quenchers such as oxygen [20–22]. Fluorescence lifetime imaging microscopy (FLIM) is an optical imaging technique capable of resolving these changes on a single-cell basis [23]. Furthermore, multiphoton FLIM is advantageous for deeper imaging in highly-scattering samples such as 3D tumor models, due to superior optical sectioning and lower sample phototoxicity compared to single photon excitation [19]. Optical metabolic imaging (OMI) is two-photon imaging of the autofluorescence intensities and lifetimes of NAD(P)H and FAD.
OMI is currently under investigation for drug screening in 3D tumor cultures, including breast cancer. The differences between 2D and 3D breast cancer cultures have been well-characterized in previous studies using a variety of gold-standard methods, including RNA sequencing, Western blotting, and immunofluorescence assays [4, 5, 10]. This study aims to verify that OMI is sensitive to these known differences in drug response between 2D and 3D cultures, by conducting experiments in breast cancer cell monolayers in parallel to previous experiments conducted in 3D breast cancer tumor organoid cultures [15]. This cell-level imaging approach within intact samples also allowed for cell sub-populations to be compared in 2D vs. 3D cultures, thus enabling a comparison of cell-level heterogeneity in 2D vs. 3D cultures for control and treated groups. Additionally, absolute values of NAD(P)H and FAD mean fluorescence lifetimes were used as a direct point of comparison between the microenvironment of these molecules in 2D and 3D cultures. Previously published results showed that 3D organoid response accurately correlates to in vivo murine tumor growth, enabling the 2D monolayer results to be gauged against this standard as well [15]. To the best of our knowledge, this is the first study to use autofluorescence or FLIM to directly compare 2D and 3D cultures.
1. Materials and methods
BT474 (ER+/HER2 + ) cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Invitrogen) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin:streptomycin. Herceptin-resistant HR6 cells were derived from BT474 xenografts that were treated with Herceptin (trastuzumab), a HER2 antagonist, until in vivo resistance was achieved [24]. These cells were grown in media prepared as described above, further supplemented with 10 μg/mL Herceptin [25]. The organoids used for comparison in previous experiments were generated using the protocol described in [15]. Briefly, xenograft and primary tumors were mechanically dissociated and suspended in a PMEC and Matrigel solution for culture and imaging [15]. Cells were plated for imaging experiments on 35mm glass bottom dishes (BD Biosciences) at a concentration of 1 x 105 cells/mL. Twenty-four hours after plating, each dish was treated with one of the following: fresh DMEM media (control), Herceptin-supplemented media (H, 25 μg/mL dose), XL147-supplemented media (X, 10 nM dose), or combination treatment (H + X). Imaging experiments were conducted 24, 48, and 72-hour time points post-treatment. These time points were chosen based on previous studies [15] that showed agreement between drug response in organoids at 24, 48, and 72 hours post-treatment, and long-term (30 day) drug response in matched in vivo mouse models. Three representative fields of view were imaged and analyzed for each dish, and experiments were repeated three times for each cell line, providing nine total images for each cell line. 80-550 cells were imaged and analyzed per treatment group, with an average of n = 250 cells. 60-300 cells were imaged and analyzed per treatment group in the previous organoid studies [15].
Cell monolayers were imaged using a custom-built two-photon fluorescence lifetime microscope [16]. Excitation was provided by a Titanium:Sapphire tunable laser (Chameleon Ultra II, Coherent, Inc.) operated at wavelengths of 760 nm and 890 nm for NAD(P)H and FAD excitation, respectively [19]. Fluorescence emission was collected with a photomultiplier tube (PMT, Hamamatsu) using 440/80 nm and 550/100 nm bandpass emission filters for NAD(P)H and FAD, respectively. Laser power was monitored over the course of imaging experiments and maintained at 3.4-3.6 mW at the sample. The focused fluences incident on the sample for NAD(P)H and FAD excitation were 901 kW/cm2 and 657 kW/cm2, respectively. The photon count rates emitted over the course of an acquisition were also monitored to rule out the incidence of photobleaching, or the destruction of fluorophores. Count rates were observed to remain above 1 x 105 photons/s over the course of the entire 60-second acquisition period. A 4.8 μs pixel dwell time was used, and 256x256 pixel images were recorded for each field of view. The cells continued to divide post-imaging in control dishes, confirming no adverse effects from imaging, consistent with previous studies [26].
Time-correlated single photon counting electronics (TCSPC, Becker & Hickl) were used to record fluorescence lifetime data, which was subsequently analyzed with SPCImage. An instrument response function (IRF) was measured using the second-harmonic generated signal from a urea crystal, then deconvolved from the acquired sample decays. The time-resolved data was fit to a two-component exponential decay as described in previously published work, to model the distinct short and long fluorescence lifetimes of free and protein-bound NAD(P)H and FAD [13, 14, 16]. The fit model is I(t) = α1exp-t/τ1 + α2exp-t/τ2 + C, where I(t) is the time-resolved fluorescence intensity following the excitation pulse from the laser, α1 and α2 denote the relative contributions of the short and long lifetime components to the overall decay, and τ1 and τ2 are the short and long lifetime values. C accounts for constant background signal emanating from non-cellular components. This fit parameter was found to be consistent across pixels within individual images and across different images, suggesting minimal effects of background noise on lifetime and intensity data. The mean lifetime, τm, is calculated as the weighted sum of the short and long lifetimes, α1τ1 + α2τ2. The goodness of fit of this model was evaluated on a per-pixel basis for each NAD(P)H and FAD image and exported as a χ2 value. These values typically ranged from 1 to 2, with an average value of 1.6, indicating good fitting parameters. A fluorescent bead (Polysciences) with a monoexponential decay was imaged on each day of experiments as a reference standard to validate proper system functioning. The mean fluorescence lifetime of the beads (τ = 2100 +/− 5 ps) over sixteen total days of imaging was found to be in good agreement with published values [15, 19].
CellProfiler, a MATLAB-based open source image analysis software [27], was used for single-cell autofluorescence intensity and lifetime analysis as described previously [28]. Recorded images were first thresholded with an Otsu Global filter, then segmented to exclude nuclei from analysis. The redox ratio was calculated on a per-pixel basis for the cytoplasm as the NAD(P)H fluorescence intensity of that pixel divided by the FAD fluorescence intensity of that pixel. Cytoplasmic NAD(P)H and FAD lifetimes and the redox ratio were used to calculate the “OMI index.” The OMI index is a linear combination of the mean-normalized redox ratio, NAD(P)H τm, and FAD τm, with the respective coefficients (1, 1, −1) chosen to maximize the effect of treatment-induced changes, as described previously [15]. The mean OMI index was calculated for each treatment group at each time point for each cell line and used as the primary experimental endpoint. Significant changes in mean OMI index were assessed with respect to the daily control group using a Wilcoxon rank-sum test with α = 0.05. Additionally, absolute NAD(P)H and FAD τm were quantified for each group, compared directly to corresponding values measured in organoids [15], and analyzed for statistical significance using the same procedure. All bar graphs show mean values, with error bars representing standard error.
Population density curves were constructed to visualize sub-populations in OMI index, NAD(P)H τm, and FAD τm for each treatment group on a cellular-level. All data collected across each day of imaging for each group were fit to one-, two-, and three-component Gaussian curves. The Akaike Information Criteria (AIC) were used to assess relative goodness of fit for each model, and the model with the lowest AIC value was chosen as the most representative. The number of reported sub-populations corresponds to the number of Gaussian components in this chosen model. Measures were taken to verify that all analytical procedures performed on monolayer data were identical to those used to analyze the previously published organoid data [15].
2. Results
BT474 monolayers
The mean OMI index in 2D monolayers of each treatment group at each time point was computed and compared to that of the control at the same time point. The OMI index decreases in cells that are responding to a drug, and the OMI index increases or does not change in cells that are not responding to a drug. In BT474 2D monolayers, there was a significant decrease in OMI index in all treatment groups with respect to control at all time points (Fig. 1(a-c)), except there was a significant increase in OMI index in the Herceptin-treated group at 24 hours (Fig. 1(a)). Trends in the optical redox ratio for each treatment group at each time point were identical to those observed in OMI index for this cell line (see Appendix, Fig. 5(a-c)).
Fig. 1.
Optical metabolic imaging of BT474 2D monolayers. The OMI index was calculated at 24 (a), 48 (b), and 72 (c) hours post-drug treatment. Gold- and gray-colored bars represent changes versus control that are in agreement and disagreement, respectively, with prior 3D organoid studies [15]. Blue bars delineate the control group for comparison. Population density curves were also generated at 24 (d), 48 (e), and 72 (f) hour time points to resolve OMI index distributions across a population.
Population density curves were also generated for these 2D monolayers to visualize the presence of sub-populations that are not apparent when computing the average OMI index (Fig. 1(d-f)). These curves reveal subgroups of cells that respond to treatment differently, or fail to respond at all. A single population is representative of uniform treatment response, whereas resistant sub-populations may appear as secondary populations with a distinct mean OMI index. At 24 hours, the response is apparent for the XL147 and combination treatment group (Fig. 1(d)). At 48 hours, the combination treatment shows a sub-population with a higher OMI index (Fig. 1(e)). At 72 hours, the high OMI index sub-population in the combination treatment group is more prominent (Fig. 1(f)). The XL147-treated group also developed a small, higher-OMI index population at 72 hours (Fig. 1(f)) that was not resolvable at earlier time points.
It is difficult to compare redox ratio, and therefore OMI index, values across different experimental conditions due to the need to normalize values to a daily control. However, the fluorescence lifetime is independent of intensity, and thus comparable across experimental days. Therefore, absolute NAD(P)H and FAD mean fluorescence lifetime values were examined in each treatment group in these 2D monolayer experiments to enable comparisons to prior 3D organoid experiments in which the absolute fluorescence lifetime values for these fluorophores were also calculated [15]. All comparisons of absolute fluorescence lifetime values were made for the 72-hour post-treatment time point. For both NAD(P)H and FAD, τm values in BT474 2D monolayers were significantly greater than those in corresponding 3D organoids across all control and treatment groups (Fig. 2(a, b)). The greatest differences existed for the XL147 and combination treatment groups for both fluorophores. Population density curves for parallel 2D monolayer and 3D organoid groups (Fig. 2(c-j)) support the mean population trends observed in Fig. 2(a, b). For both fluorophores, the organoid τm values were generally best modeled as a single population with low standard deviation, although the control and Herceptin groups did show small sub-populations for FAD τm. In contrast, the monolayer population density curves had broader peaks with larger standard deviations, and more often showed the presence of multiple populations. Similar trends in both average values and population density were observed in the short (τ1) and long (τ2) lifetime values for both NAD(P)H (see Appendix, Fig. 6(a-j)) and FAD (Fig. 7(a-j)). However, the trends in NAD(P)H and FAD short lifetime components (α1) were less consistent, and treatment-induced changes were lesser in magnitude than those in 3D organoid τ1 and τ2 values (Fig. 8(a, b)). These comparisons were also made at the 72-hour time point.
Fig. 2.
Comparison of fluorescence lifetime values in BT474 3D organoids and 2D monolayers. Average values for NAD(P)H (a) and FAD (b) mean fluorescence lifetime (τm) in 2D monolayer and 3D organoid cultures at 72 hours post-treatment reveal significantly greater τm values in 2D monolayers with respect to corresponding 3D organoid groups (p<0.05). Population density curves for NAD(P)H (c-f) and FAD (g-j) τm further distinguish 2D monolayer and 3D organoid models. The organoid data is a subset of data previously published in [15].
HR6 monolayers
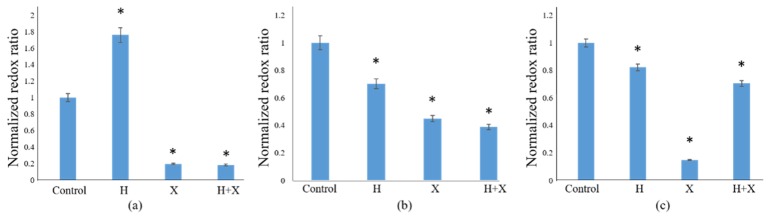
The response of Herceptin-resistant HR6 2D monolayers to the same panel of drug treatments was also examined. In general, the OMI index changes in HR6 monolayers were less uniform than those in BT474 monolayers (Figs. 1, 3). At the 24-hour time point, the only treatment group to differ significantly from control was the XL147-treated group (Fig. 3(a)). At 48 hours post-treatment, both the XL147 and combination treatment caused a significant decrease in OMI index, with that of the combination treatment being larger in magnitude (Fig. 3(b)). However, no significant changes in OMI index were resolvable at 72 hours post-treatment (Fig. 3(c)). The lack of change in OMI index in Herceptin-treated HR6 2D monolayers with respect to the control group at all points is an important validation in these Herceptin-resistant cells (Fig. 3(a-c)). The redox ratio results at the 24- and 48-hour time points show the same trends as the OMI index results for each treatment group (see Appendix, Fig. 9(a, b)). However, at the 72-hour time point, the redox ratio was significantly greater than that of the control group in all of the drug-treated groups (Fig. 9(c)).
Fig. 3.
Optical metabolic imaging of HR6 monolayers. The OMI index was calculated at 24 (a), 48 (b), and 72 (c) hours post-drug treatment. Gold- and gray-colored bars represent changes versus control that are in agreement and disagreement, respectively, with prior 3D organoid studies [15]. Blue bars delineate the control group for comparison. Population density curves were also generated at 24 (d), 48 (e), and 72 (f) hour time points to resolve OMI index distributions across a population.
Population density curves generated to visualize heterogeneity within the HR6 2D monolayers revealed mostly uniform curves representative of only a single population, particularly after the 24-hour time point (Fig. 3(d)). The similarity between all the curves is most evident at 72-hours post-treatment (Fig. 3(f)). The only treatment group to show a pronounced sub-population at any time point was the XL147 group at 24 hours (Fig. 3(d)), but these higher and lower OMI index groups later merged into a single population with an average OMI index closer to that of the other treatment groups and control (Fig. 3(e, f)). Monolayer experiments failed to show the presence of any sub-populations at either the 48 hour or 72 hour time point (Fig. 3(e, f)). The population density curves at 72 hours (Fig. 3(f)) support the lack of significant change among average OMI index values at this time point (Fig. 3(c)), most notably in the combination-treated group.
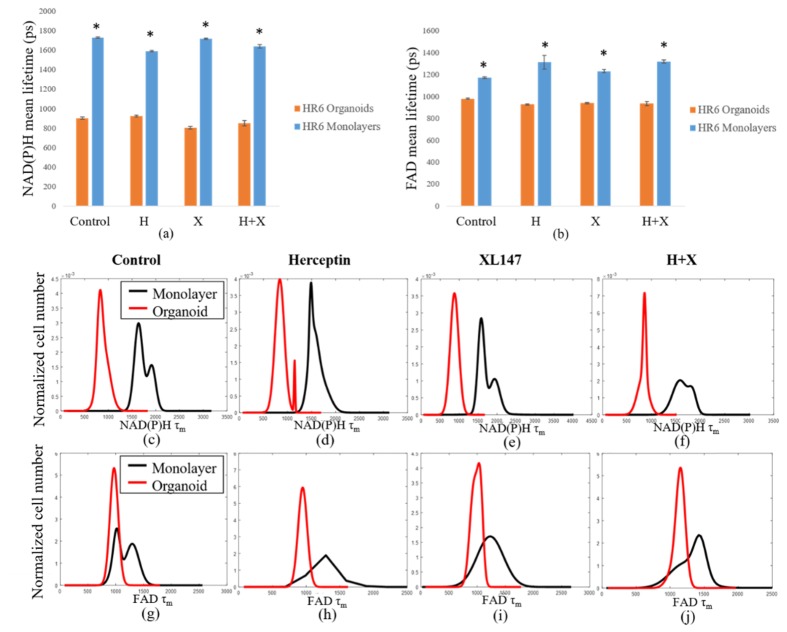
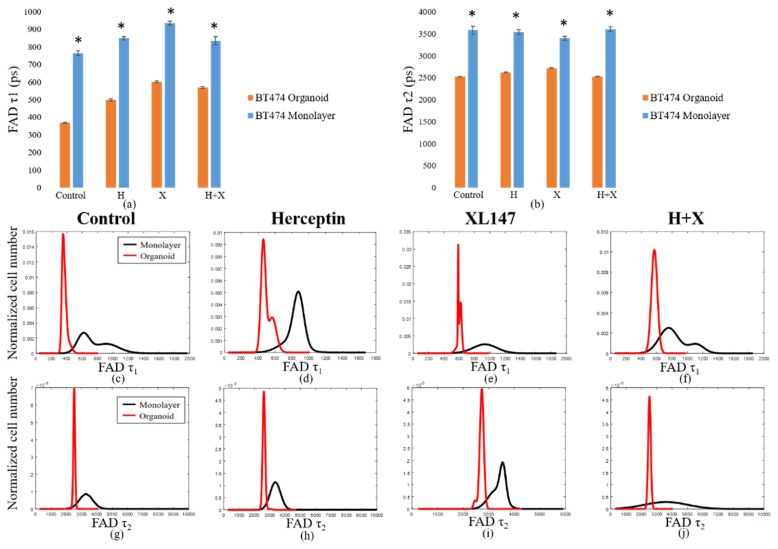
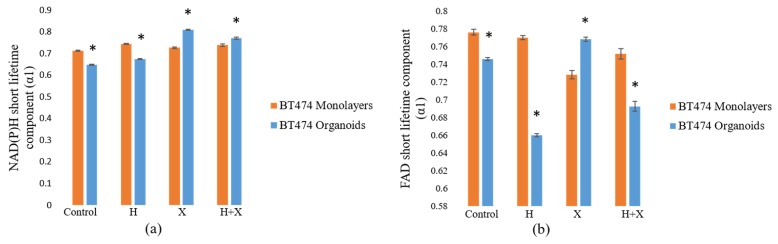
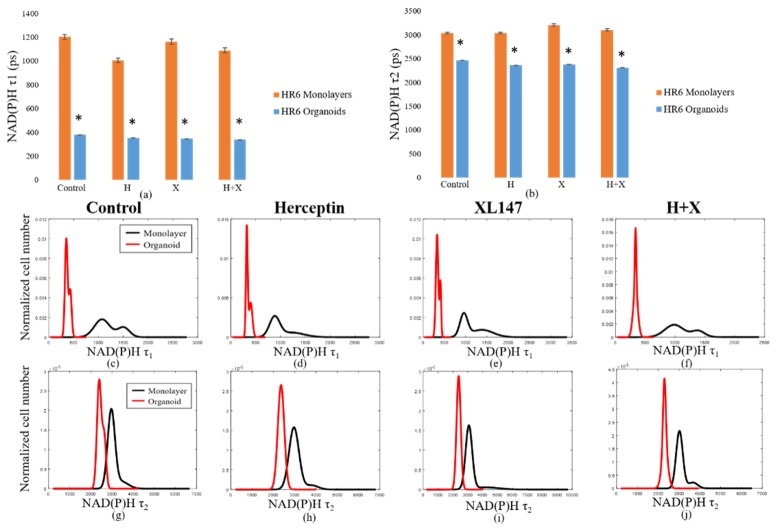
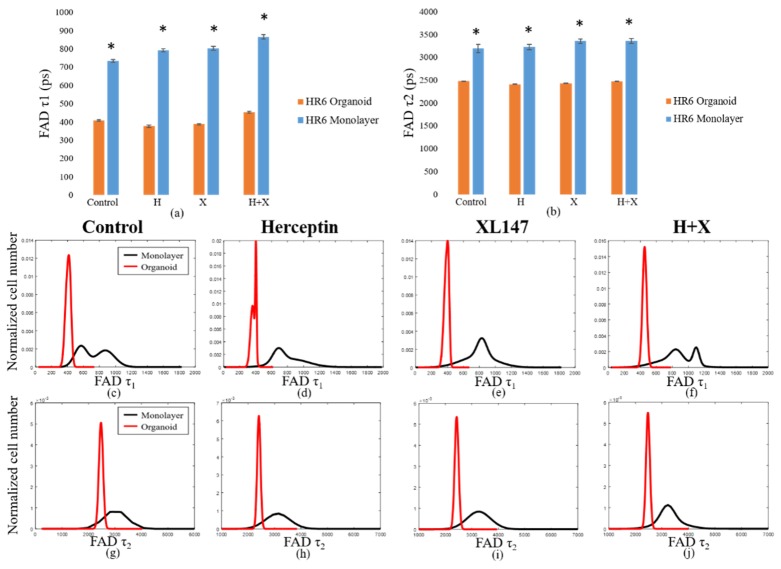
Like the BT474 results, the HR6 2D monolayers had significantly greater NAD(P)H and FAD τm than the 3D organoids for all treatment groups (Fig. 4(a-j)). These comparisons were also made at the 72-hour time point only. The differences in magnitude between 2D monolayer and 3D organoid NAD(P)H τm were similar across all treatment groups, ranging from 650 to 900 ps (Fig. 4(a)). The same was true for FAD τm comparisons, though the absolute differences between each pair were smaller, varying from 200 to 400 ps (Fig. 4(b)). For both NAD(P)H and FAD, the distributions of fluorescence lifetimes in 3D organoids were best modeled by a single, narrow population, with the exception of the Herceptin-treated organoids, which showed a small second peak (Fig. 4(c-j)). In 2D monolayer, the absolute values were better modeled by either a single, broader peak, or by two distinct sub-populations. As with the BT474 τm results, some of the HR6 2D monolayer populations showed a lower τm sub-population that overlapped well with the organoid distribution, but also had higher τm sub-populations (FAD τm in control, combination-treated groups, Fig. 4(g, j)). The average τ1 and τ2 values for both NAD(P)H (see Appendix, Fig. 10(a)) and FAD (Fig. 11(a)) were significantly higher for all treatment groups in 2D monolayers than in corresponding 3D organoid groups. Additionally, the distribution of τ1 in all 2D culture groups was consistently better modeled by two distinct populations (Fig. 10(c-f), Fig. 11(c-f)). However, the τ2 distribution was better modeled by a single population (Fig. 10(g-j), Fig. 11(g-j)). This trend held for both NAD(P)H and FAD at the 72-hour time point where these comparisons were made. The α1 values for NAD(P)H and FAD showed opposite trends in 2D and 3D models, with 3D organoids having a significantly greater NAD(P)H α1 (Fig. 12(a)), but 2D monolayers having a greater FAD α1 (Fig. 12(b)). In general, comparisons made across the HR6 2D and 3D models were like those made across the BT474 2D and 3D models, but differences between 2D and 3D for HR6 cells were more uniform and greater in magnitude, particularly for NAD(P)H τm.
Fig. 4.
Comparison of fluorescence lifetime values in HR6 3D organoids and 2D monolayers. Average values for NAD(P)H (a) and FAD (b) τm in 2D monolayers and 3D organoids at 72 hours post-treatment reveal significantly greater values in 2D monolayers with respect to corresponding 3D organoid groups (p<0.001). Population density curves for NAD(P)H (c-f) and FAD (g-j) τm further distinguish 2D monolayer and 3D organoid models. The organoid data is a subset of data previously published in [15].
3. Discussion
Previous studies have established that 2D and 3D breast tumor cultures exhibit differences in cell signaling, genetic expression, and drug response [4]. Our results confirm that autofluorescence imaging of NAD(P)H and FAD can non-invasively resolve these differences within intact samples. Our studies also provide new information on more subtle differences in metabolic cell subpopulations of varied response over a treatment time-course. The temporal evolution of these cell subpopulations also differs between 2D and 3D breast tumor cultures.
In prior BT474 3D organoid experiments, each drug and combination treatment resulted in a significant decrease in OMI index at all three time points [15]. These decreases matched the outcomes seen in tumor growth curves, which showed a significant decrease in tumor volume over a 30-day period in all treatment groups. Overall, the trends in mean OMI index values in BT474 2D monolayers matched those seen in BT474 3D organoids fairly closely, particularly at 48 hours post-treatment (Fig. 1(a-c)). The similarity in average OMI index values at this time point is further supported by a comparison of the number of sub-populations in the 2D and 3D cultures (Table 1). The only inconsistency at 48 hours is the emergent second population in the combination treatment group in 2D monolayers (Table 1). However, the numbers of sub-populations differed greatly between 2D monolayers and 3D organoids for this cell line at 24 and 72 hours. As the multiple sub-populations resolved in BT474 3D organoids gave way to a more uniform response over time [15], the opposite trend occurred in BT474 2D monolayers. The number of sub-populations generally increased over time in BT474 2D monolayers. Interestingly, although the Herceptin-treated 3D organoids maintained two populations throughout the time course, the Herceptin-treated 2D monolayers were the only treatment group in the 2D experiments to merge into a single population at 72 hours.
Table 1. Comparison of number of sub-populations in 2D monolayer and 3D organoid BT474 cultures. Differences between 2D monolayers and 3D organoids are highlighted in bold.
| Number of subpopulations | |||
|---|---|---|---|
| Monolayers | Organoids | ||
| 24 hours | Control | 1 | 1 |
| Herceptin | 1 | 2 | |
| XL147 | 1 | 2 | |
| H + X | 1 | 2 | |
| 48 hours | Control | 1 | 1 |
| Herceptin | 2 | 2 | |
| XL147 | 1 | 1 | |
| H + X | 2 | 1 | |
| 72 hours | Control | 2 | 1 |
| Herceptin | 1 | 2 | |
| XL147 | 2 | 1 | |
| H + X | 2 | 1 | |
In general, OMI index changes in HR6 2D monolayers differed more from those of HR6 3D organoids (Fig. 3(a-c)) than the BT474 2D monolayers did from BT474 3D organoids (Fig. 1(a-c)). As in the BT474 2D monolayers, the average OMI index of the HR6 2D monolayers is most like that of the 3D organoids at the 48-hour time point (Fig. 3(b)). However, there are differences in numbers of sub-populations between 2D monolayers and 3D organoids at all time points (Table 2). In parallel 3D organoid experiments, two sub-populations were present in XL147-treated organoids at 48 hours, which merged to a single population at 72 hours (Table 2). The previous 3D organoid experiments also showed two populations at 72 hours after combination treatment (Table 2). Monolayer experiments failed to show the presence of any sub-populations at either the 48 hour or 72 hour time point (Fig. 3(e, f) and Table 2).
Table 2. Comparison of number of sub-populations in 2D monolayer and 3D organoid HR6 cultures. Differences between 2D monolayers and 3D organoids are highlighted in bold.
| Number of subpopulations | |||
|---|---|---|---|
| Monolayers | Organoids | ||
| 24 hours | Control | 1 | 1 |
| Herceptin | 1 | 1 | |
| XL147 | 2 | 2 | |
| H + X | 1 | 2 | |
| 48 hours | Control | 1 | 1 |
| Herceptin | 1 | 1 | |
| XL147 | 1 | 2 | |
| H + X | 1 | 1 | |
| 72 hours | Control | 1 | 1 |
| Herceptin | 1 | 1 | |
| XL147 | 1 | 1 | |
| H + X | 1 | 2 | |
The most meaningful discrepancy between 3D organoid and 2D monolayer results was the outcome of combination-treated HR6 2D monolayers at 72 hours. The Herceptin-resistant HR6 3D organoids did not maintain a response to XL147 treatment alone at 72 hours, but did respond to combination treatment of XL147 and Herceptin [15]. XL147 inhibits the phosphatidylinositol-3-kinase pathway, which acts downstream of HER2 [29]. Therefore, the combination treatment of XL147 and Herceptin is under clinical consideration as a potentially viable treatment for HER2-positive patients [24]. However, the lack of response at 72 hours in 2D monolayers treated with this combination of drugs (Fig. 3(b)) is inconsistent with 3D organoid and in vivo results [15], demonstrating that the predictive value of the 2D model is inferior to that of the 3D organoid model in this context.
Resolving the presence of cell sub-populations through population density modeling helped to explain some of the discrepancies seen in OMI index trends between 2D and 3D cultures (Tables 1 & 2). In general, 3D organoids showed a decrease in the number of sub-populations over time in groups that had been treated successfully. This trend shows an intuitive disappearance of responsive sub-populations with continued treatment. For both cell lines, there were few discernable trends in the number of sub-populations over time in 2D monolayer cultures, despite the overall agreement in average OMI index between BT474 2D monolayers and 3D organoids. This observation further suggests that the 2D monolayer model may be inferior to the 3D organoid model, particularly in applications studying trends in drug response and resistance on a single-cellular level over time.
Large differences were observed in the absolute mean NAD(P)H and FAD fluorescence lifetime values between 2D monolayers and 3D organoids for both cell lines (Figs. 2, 4). These values provide a direct, quantitative comparison of 2D and 3D microenvironments. Previous experiments conducted in head and neck squamous cell carcinoma (HNSCC) compared 3D primary tumor organoids to in vivo tumor xenografts of the same cell origin (FaDu) [29]. The NAD(P)H and FAD τm were found to be significantly greater in 3D organoids with respect to in vivo xenografts [30]. The difference in τm between HNSCC organoids and in vivo xenografts were on the order of approximately 200 ps and 500 ps for NAD(P)H and FAD, respectively. Similarly, in the current study, NAD(P)H and FAD τm were greater in 2D monolayers than in 3D organoids (Figs. 2, 4). Differences in τm between 2D and 3D ranged from 200 to 500 ps for NAD(P)H τm in the BT474 line and for FAD τm in both lines. Larger differences were measured for NAD(P)H τm in the HR6 line, which potentially correlate to the greater differences in OMI index and sub-population number between the 2D and 3D cultures for this line. Given that fluorescence lifetime is sensitive to the oxygen and pH gradients that exist to a much lesser degree in 2D monolayer cultures than in 3D organoids, it is not surprising that these values differ greatly between 2D and 3D models. It is important to note that the media used to culture the monolayers and organoids was not identical; the media used for 3D organoid culture was supplemented with epidermal growth factor (EGF), hydrocortisone, insulin, and matrigel [15]. However, we nonetheless believe that inherent differences in cell signaling were more responsible for these lifetime differences than growth conditions alone.
The good agreement between OMI index values in BT474 2D monolayers, 3D organoid cultures, and in vivo xenografts, supports the continued use of cell monolayers for studying endpoints in some cell lines on some platforms. As 2D cultures continue to dominate drug development platforms, it is important to understand when their usage is appropriate, as they offer many practical advantages for high-throughput applications over 3D organoid cultures. Nonetheless, the findings of this study support the existing evidence that 3D analogues offer a superior method for analyzing drug efficacy and resistance within in vitro populations over time. In particular, single-cell based population modeling and comparisons of absolute fluorescence lifetime values helped to reveal differences between 2D and 3D cultures that global average values could not. Fluorescence lifetime and redox ratio measurements can resolve subtle differences in microenvironment between drug-treated 2D and 3D BT474 and HR6 cultures that may not be evident by examining overall structure or gene expression alone. The findings of these experiments conducted in breast cancer cell monolayers and their comparative analysis to parallel organoid experiments validate the use of optical metabolic imaging as a sensitive indicator of drug response. These results show that OMI can resolve differences in drug response between 2D and 3D cultures, and provide further evidence that OMI could be a powerful tool for drug screening on a single-cell level, particularly when applied to 3D breast cancer models.
Acknowledgments
We acknowledge Tiffany Heaster for helpful discussions and review of the manuscript.
Appendix: Additional figures
Fig. 5.
Redox ratios of BT474 2D monolayers. The normalized redox ratio was calculated at 24 (a), 48 (b), and 72 (c) hours post-drug treatment. The redox ratio significantly decreased (p<0.05) with treatment in all groups excluding the Herceptin group at the 24-hour time point. These trends are in good agreement with corresponding OMI index results (Fig. 1(a-c)).
Fig. 6.
Comparison of short (τ1) and long (τ2) NAD(P)H fluorescence lifetime values in BT474 3D organoids and 2D monolayers. Average values for NAD(P)H τ1 (a) and τ2 (b) in 2D monolayers and 3D organoids at 72 hours post-treatment reveal significantly greater values in 2D monolayers with respect to corresponding 3D organoid groups (p<0.05). Population density curves for NAD(P)H τ1 (c-f) and τ2 (g-j) further distinguish 2D monolayer and 3D organoid models. The organoid data is a subset of data previously published in [15].
Fig. 7.
Comparison of short (τ1) and long (τ2) FAD fluorescence lifetime values in BT474 3D organoids and 2D monolayers. Average values for FAD τ1 (a) and τ2 (b) in 2D monolayers and 3D organoids at 72 hours post-treatment reveal significantly greater values in 2D monolayers with respect to corresponding 3D organoid groups (p<0.05). Population density curves for FAD τ1 (c-f) and τ2 (g-j) further distinguish 2D monolayer and 3D organoid models. The organoid data is a subset of data previously published in [15].
Fig. 8.
Comparison of the NAD(P)H and FAD fractional short lifetime components (α1) in BT474 2D monolayers and 3D organoids. The NAD(P)H short lifetime component (a) changed less with treatment in 2D monolayers than in 3D organoids. The changes in the FAD short lifetime component (b) were also lesser in magnitude and showed similarly inconsistent trends with respect to corresponding 3D organoid α1 values. The organoid data is a subset of data previously published in [15].
Fig. 9.
Redox ratios of HR6 2D monolayers. The normalized redox ratio was calculated with respect to control at 24 (a), 48 (b), and 72 (c) hours post-drug treatment. Trends in redox ratio in this cell line agree with trends observed in OMI index at the 24-hour (Fig. 3(a)) and 48-hour (Fig. 3(b)) time points. However, the redox ratio increased significantly (p<0.05) in each of the treatment groups after 72 hours, whereas no significant changes were measured in OMI index at this time point (Fig. 3(c)).
Fig. 10.
Comparison of short (τ1) and long (τ2) NAD(P)H fluorescence lifetime values in HR6 3D organoids and 2D monolayers. Average values for NAD(P)H τ1 (a) and τ2 (b) in 2D monolayers and 3D organoids at 72 hours post-treatment reveal significantly greater values in 2D monolayers with respect to corresponding 3D organoid groups (p<0.05). Population density curves for NAD(P)H τ1 (c-f) and τ2 (g-j) further distinguish 2D monolayer and 3D organoid models. The organoid data is a subset of data previously published in [15].
Fig. 11.
Comparison of short (τ1) and long (τ2) FAD fluorescence lifetime values in HR6 3D organoids and 2D monolayers. Average values for NAD(P)H τ1 (a) and τ2 (b) in 2D monolayers and 3D organoids at 72 hours post-treatment reveal significantly greater values in 2D monolayers with respect to corresponding 3D organoid groups (p<0.05). Population density curves for FAD τ1 (c-f) and τ2 (g-j) further distinguish 2D monolayer and 3D organoid models. The organoid data is a subset of data previously published in [15].
Fig. 12.
Comparison of the NAD(P)H and FAD fractional short lifetime components (α1) in HR6 2D monolayers and 3D organoids. The NAD(P)H short lifetime component (a) was significantly lesser in 2D monolayers than in 3D organoids. FAD α1 values (b) in 2D monolayers were significantly greater than those in corresponding 3D organoids. The organoid data is a subset of data previously published in [15].
Funding
NSF Graduate Research Fellowship (DGE-0909667), NIH/NCI (R01 CA185747, R01 CA205101), DoD BCRP (BC121998), NSF (CBET-1554027), and Mary Kay Foundation (067-14).
References and links
- 1.Paul S. M., Mytelka D. S., Dunwiddie C. T., Persinger C. C., Munos B. H., Lindborg S. R., Schacht A. L., “How to improve R&D productivity: the pharmaceutical industry’s grand challenge,” Nat. Rev. Drug Discov. 9(3), 203–214 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Suggitt M., Bibby M. C., “50 years of preclinical anticancer drug screening: empirical to target-driven approaches,” Clin. Cancer Res. 11(3), 971–981 (2005). [PubMed] [Google Scholar]
- 3.Bhadriraju K., Chen C. S., “Engineering cellular microenvironments to improve cell-based drug testing,” Drug Discov. Today 7(11), 612–620 (2002). 10.1016/S1359-6446(02)02273-0 [DOI] [PubMed] [Google Scholar]
- 4.Kenny P. A., Lee G. Y., Myers C. A., Neve R. M., Semeiks J. R., Spellman P. T., Lorenz K., Lee E. H., Barcellos-Hoff M. H., Petersen O. W., Gray J. W., Bissell M. J., “The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression,” Mol. Oncol. 1(1), 84–96 (2007). 10.1016/j.molonc.2007.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bissell M. J., Radisky D. C., Rizki A., Weaver V. M., Petersen O. W., “The organizing principle: microenvironmental influences in the normal and malignant breast,” Differentiation 70(9-10), 537–546 (2002). 10.1046/j.1432-0436.2002.700907.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavnar S. P., Xiao A., Gibbons A. E., Rickelmann A. D., Neely T., Luker K. E., Takayama S., Luker G. D., “Imaging sensitivity of quiescent cancer cells to metabolic perturbations in bone marrow spheroids,” Tomography 2(2), 146–157 (2016). 10.18383/j.tom.2016.00157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirschhaeuser F., Menne H., Dittfeld C., West J., Mueller-Klieser W., Kunz-Schughart L. A., “Multicellular tumor spheroids: an underestimated tool is catching up again,” J. Biotechnol. 148(1), 3–15 (2010). 10.1016/j.jbiotec.2010.01.012 [DOI] [PubMed] [Google Scholar]
- 8.Longati P., Jia X., Eimer J., Wagman A., Witt M. R., Rehnmark S., Verbeke C., Toftgård R., Löhr M., Heuchel R. L., “3D pancreatic carcinoma spheroids induce a matrix-rich, chemoresistant phenotype offering a better model for drug testing,” BMC Cancer 13(1), 95 (2013). 10.1186/1471-2407-13-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pampaloni F., Reynaud E. G., Stelzer E. H. K., “The third dimension bridges the gap between cell culture and live tissue,” Nat. Rev. Mol. Cell Biol. 8(10), 839–845 (2007). 10.1038/nrm2236 [DOI] [PubMed] [Google Scholar]
- 10.Bissell M. J., Rizki A., Mian I. S., “Tissue architecture: the ultimate regulator of breast epithelial function,” Curr. Opin. Cell Biol. 15(6), 753–762 (2003). 10.1016/j.ceb.2003.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galluzzi L., Kepp O., Vander Heiden M. G., Kroemer G., “Metabolic targets for cancer therapy,” Nat. Rev. Drug Discov. 12(11), 829–846 (2013). 10.1038/nrd4145 [DOI] [PubMed] [Google Scholar]
- 12.Chance B., Schoener B., Oshino R., Itshak F., Nakase Y., “Oxidation-reduction ratio studies of mitochondria in freeze-trapped samples. NADH and flavoprotein fluorescence signals,” J. Biol. Chem. 254(11), 4764–4771 (1979). [PubMed] [Google Scholar]
- 13.Shah A. T., Demory Beckler M., Walsh A. J., Jones W. P., Pohlmann P. R., Skala M. C., “Optical metabolic imaging of treatment response in human head and neck squamous cell carcinoma,” PLoS One 9(3), e90746 (2014). 10.1371/journal.pone.0090746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walsh A. J., Castellanos J. A., Nagathihalli N. S., Merchant N. B., Skala M. C., “Optical imaging of drug-induced metabolism changes in murine and human pancreatic cancer organoids reveals heterogeneous drug response,” Pancreas 45(6), 863–869 (2016). 10.1097/MPA.0000000000000543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walsh A. J., Cook R. S., Sanders M. E., Aurisicchio L., Ciliberto G., Arteaga C. L., Skala M. C., “Quantitative optical imaging of primary tumor organoid metabolism predicts drug response in breast cancer,” Cancer Res. 74(18), 5184–5194 (2014). 10.1158/0008-5472.CAN-14-0663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walsh A. J., Cook R. S., Manning H. C., Hicks D. J., Lafontant A., Arteaga C. L., Skala M. C., “Optical metabolic imaging identifies glycolytic levels, subtypes, and early-treatment response in breast cancer,” Cancer Res. 73(20), 6164–6174 (2013). 10.1158/0008-5472.CAN-13-0527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alhallak K., Rebello L. G., Muldoon T. J., Quinn K. P., Rajaram N., “Optical redox ratio identifies metastatic potential-dependent changes in breast cancer cell metabolism,” Biomed. Opt. Express 7(11), 4364–4374 (2016). 10.1364/BOE.7.004364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shah A. T., Diggins K. E., Walsh A. J., Irish J. M., Skala M. C., “In vivo autofluorescence imaging of tumor heterogeneity in response to treatment,” Neoplasia 17(12), 862–870 (2015). 10.1016/j.neo.2015.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skala M. C., Riching K. M., Gendron-Fitzpatrick A., Eickhoff J., Eliceiri K. W., White J. G., Ramanujam N., “In vivo multiphoton microscopy of NADH and FAD redox states, fluorescence lifetimes, and cellular morphology in precancerous epithelia,” Proc. Natl. Acad. Sci. U.S.A. 104(49), 19494–19499 (2007). 10.1073/pnas.0708425104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lakowicz J. R., Szmacinski H., “Fluorescence lifetime-based sensing of pH, Ca2 +, KS and glucose,” Sens. Actuators 11(1-3), 133–143 (1993). 10.1016/0925-4005(93)85248-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blacker T. S., Mann Z. F., Gale J. E., Ziegler M., Bain A. J., Szabadkai G., Duchen M. R., “Separating NADH and NADPH fluorescence in live cells and tissues using FLIM,” Nat. Commun. 5, 3936 (2014). 10.1038/ncomms4936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lakowicz J. R., Principles of Fluorescence Spectroscopy (Springer, 2006). [Google Scholar]
- 23.Walsh A. J., Skala M. C., “Optical metabolic imaging quantifies heterogeneous cell populations,” Biomed. Opt. Express 6(2), 559–573 (2015). 10.1364/BOE.6.000559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chakrabarty A., Bhola N. E., Sutton C., Ghosh R., Kuba M. G., Dave B., Chang J. C., Arteaga C. L., “Trastuzumab-resistant cells rely on a HER2-PI3K-FoxO-survivin axis and are sensitive to PI3K inhibitors,” Cancer Res. 73(3), 1190–1200 (2013). 10.1158/0008-5472.CAN-12-2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vogel C. L., Cobleigh M. A., Tripathy D., Gutheil J. C., Harris L. N., Fehrenbacher L., Slamon D. J., Murphy M., Novotny W. F., Burchmore M., Shak S., Stewart S. J., Press M., “Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer,” J. Clin. Oncol. 20(3), 719–726 (2002). 10.1200/JCO.20.3.719 [DOI] [PubMed] [Google Scholar]
- 26.Squirrell J. M., Wokosin D. L., White J. G., Bavister B. D., “Long-term two-photon fluorescence imaging of mammalian embryos without compromising viability,” Nat. Biotechnol. 17(8), 763–767 (1999). 10.1038/11698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.CellProfiler, Broad Institute (2016). Available at: www.cellprofiler.org. (Accessed: 2nd September 2016)
- 28.Walsh A. J., Skala M. C., “An automated image processing routine for segmentation of cell cytoplasms in high-resolution autofluorescence images,” in Proc. SPIE 8948 (eds. Periasamy A., So P. T. C., König K.) 8948, (2014). [Google Scholar]
- 29.Junttila T. T., Akita R. W., Parsons K., Fields C., Lewis Phillips G. D., Friedman L. S., Sampath D., Sliwkowski M. X., “Ligand-independent HER2/HER3/PI3K complex is disrupted by trastuzumab and is effectively inhibited by the PI3K inhibitor GDC-0941,” Cancer Cell 15(5), 429–440 (2009). 10.1016/j.ccr.2009.03.020 [DOI] [PubMed] [Google Scholar]
- 30.Shah A. T., Heaster T. M., Skala M. C., “Metabolic imaging of head and neck cancer organoids,” PLoS One 12(1), e0170415 (2017). 10.1371/journal.pone.0170415 [DOI] [PMC free article] [PubMed] [Google Scholar]